Abstract
With the advent of volumetric EM techniques, large connectomic datasets are being created, providing neuroscience researchers with knowledge about the full connectivity of neural circuits under study. This allows for numerical simulation of detailed, biophysical models of each neuron participating in the circuit. However, these models typically include a large number of parameters, and insight into which of these are essential for circuit function is not readily obtained. Here, we review two mathematical strategies for gaining insight into connectomics data: linear dynamical systems analysis and matrix reordering techniques. Such analytical treatment can allow us to make predictions about time constants of information processing and functional subunits in large networks.
SIGNIFICANCE STATEMENT This viewpoint provides a concise overview on how to extract important insights from Connectomics data by mathematical methods. First, it explains how new dynamics and new time constants can evolve, simply through connectivity between neurons. These new time-constants can be far longer than the intrinsic membrane time-constants of the individual neurons. Second, it summarizes how structural motifs in the circuit can be discovered. Specifically, there are tools to decide whether or not a circuit is strictly feed-forward or whether feed-back connections exist. Only by reordering connectivity matrices can such motifs be made visible.
Keywords: connectomics, network dynamics, neural circuits
Introduction
The quest for understanding the function and dysfunction of organs, including the brain, has been classically subdivided into two disciplines, anatomy and physiology, which examine, respectively, structural and functional properties from separate but complementary perspectives. Of course, this historical dichotomy was never absolute as probably best evidenced by the neurobiological pioneer Santiago Ramon y Cajal (1911), who always studied anatomic structures in the light of current functional insights. Modern developments of functional imaging techniques (Kim and Schnitzer, 2022) even make such disciplinary and methodological distinctions virtually impossible. Nevertheless, our limited success in understanding brain function has often been excused by technological shortcomings that only allow studying reduced systems, preventing us from seeing the whole anatomic and physiological complexity at the same time. With the advent of connectomics (Denk and Horstmann, 2004; Lichtman and Denk, 2011; Abbott et al., 2020), these limitations are being overcome, and the anatomic connections of large brain regions and complete functional subcircuits (White et al., 1986; Briggman et al., 2011; Helmstaedter et al., 2013; Takemura et al., 2017; Shinomiya et al., 2019) have been revealed. At the same time, we witness simultaneous optical (Ghosh et al., 2011; Aimon et al., 2019; Kim and Schnitzer, 2022) and electrical (Buzsaki et al., 2015; Jun et al., 2017; Steinmetz et al., 2019; Allen et al., 2019; Gardner et al., 2022) recordings of many hundreds of neurons. Yet these spectacularly valuable datasets have not readily uncovered the remaining mysteries of neural function, but instead clearly revealed the next boundaries we need to cross: They are theoretical, data analytical, and mathematical.
Although we are far from well equipped to master all challenges imposed on us by new connectomic data and large-scale single-unit recordings, we do not need to start from scratch. Computational neuroscientists have explored tools to study activity in interacting neural networks over many decades (Herz et al., 2006) and have established a set of standard mathematical tools by which such data can be viewed (Dayan and Abbott, 2005). Since the methods encompass large-scale simulation techniques (Markram et al., 2015; Lazar et al., 2021), mean-field approaches (Nakagawa et al., 2013; Gerstner, 2000), and mathematical and graph-theoretical (Sporns et al., 2000; Rajan and Abbott, 2006) techniques, no single review is able to treat of all of those with due respect.
In this review, we will lay our main focus on dynamical systems approaches for linear networks, since they combine a number of advantages that make them attractive as a standard tool set to systems neuroscientists. First, they can be clearly related to underlying anatomic and physiological elements (see below). Second, they have been demonstrated to successfully explain physiological phenomena (Seung et al., 2000a,b; Usher and McClelland, 2001; Goldman et al., 2002; White et al., 2004). Third, the simplifications made in these models are obvious, and numerical simulations of a nonsimplified (i.e., nonlinear) system can be readily compared with the theoretical predictions. And finally, there exist established mathematical tools from a standard repertoire that is taught to physicists, computer scientists, engineers, chemists, and to an increasing extent to biologists as well.
Model design
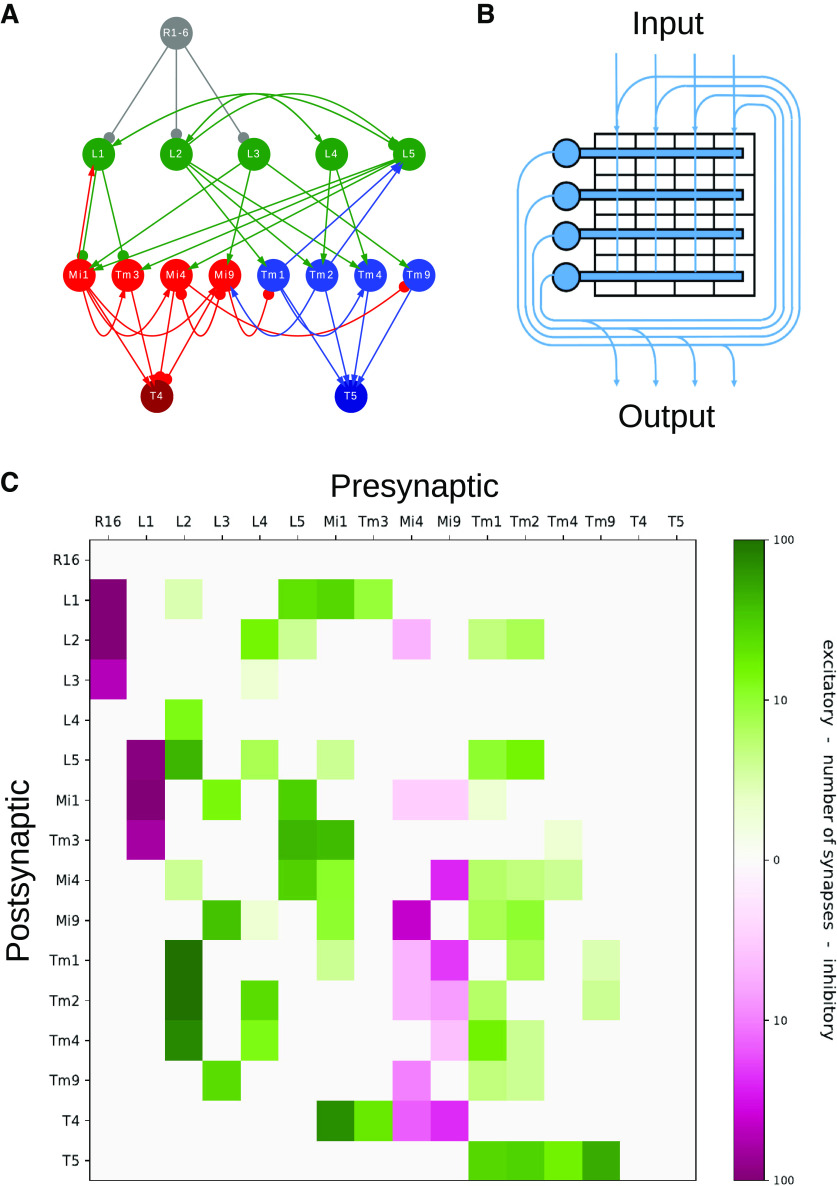
For reasons of better illustration, we illustrate mathematical modeling for the small, but well-studied neural circuit of fly motion processing (Fig. 1A). At its core, it is composed of 15 genetically, anatomically, and physiologically characterized neurons that are all located within each of the 750 columns of the Drosophila optic lobe. The output neurons of the circuit, T4 and T5, are the primary, motion-sensitive neurons of the fly, which compute the direction of ON- (T4) and OFF- (T5) edges along one of the four cardinal directions (Maisak et al., 2013; for review, see Borst et al., 2020a). We also know about the type and response characteristics of the retinal input (R) that drives the circuit. All connections between the various circuit elements have been thoroughly quantified by volumetric EM studies (Shinomiya et al., 2019) and are summarized in a 16 × 16 matrix (Fig. 1B). The rows of this matrix collect all inputs to a specific cell, the columns all output from a specific cell. Since the retinal input cells do not receive input from the rest of the circuit, the first row is empty. The retinal cells are the only ones that receive external visual input, and thus are the only ones that receive a contribution from the input vector (Fig. 1C).
Figure 1.
Network architecture, connectivity matrix, and general layout of a recurrent network. A, Circuit diagram of the fly motion vision system. Retinal input (gray, only one is shown) arrives at laminar neurons (green), which project to medulla (red) and lobula (blue) neurons. From there, activity reaches the T4 (dark red) and T5 (dark blue) cells. In this system, the action of the neurotransmitters is known: arrows indicate excitation; discs indicate inhibition. B, The number of synapses between neuron types is summarized in a connectivity matrix. C, If one interprets the number of synapses between each pair of cells as the total synaptic strength of this connection, the matrix in B defines a recurrent neural network in which inputs from all presynaptic cells are collected at “dendrites” (horizontal bars). The retinal input I(t) provides additional external drive to each neuron. A, B, Reproduced from Borst et al. (2020b), with permission.
The dynamics of the membrane potentials Vi(t) relative to the resting potential of each neuron can then be computed by the differential equation as follows:
| (1) |
Here, we assume passive membrane properties only (i.e., without the participation of any voltage-activated ion channels). If we compare Equation 1 to the one of a first order low-pass filter as follows:
| (2) |
where x(t) is the input and y(t) is the output signal of the filter, we realize that the input to each neuron consists of two parts, that is, the external, visual input Ii(t) and the internal one, which is the sum of all presynaptic elements weighted by their respective number of synapses, as defined by the connectivity matrix M. Furthermore, the activity of a presynaptic neuron is passed through a transfer function f, that captures the translation from the presynaptic voltage Vj(t) to synaptic transmission. For many graded response neurons in the fly visual system, the function f is well approximated by a rectilinear function (Groschner et al., 2022); that is, it prohibits transmission below a certain threshold (approximately resting potential) and is linear above this threshold. In spiking neural networks, f is considered to be the input-output function that translates membrane depolarization into firing rate. The latter simplification is a good approximation if synaptic integration acts on a slower time scale than membrane dynamics, that is, if τ is large compared to the time scale of fluctuations in the entries of I (Dayan and Abbott, 2005). In any case, Equation 1 can be solved numerically by replacing the differential with and, choosing a small enough time step Δt and starting from a given value of Vi(t = 0), by iterating through time.
Before going into further mathematical detail, we should consider the general purpose of such connectomics-based network modeling. While in an ideal experimental world the connectivity matrix M would be fully and accurately determined, allowing for quantitative predictions of the system response to any time-dependent stimulus I(t), real connectomic data mostly provide only the number of connections or may only include part of the circuit elements. Even if the respective neurotransmitters are known, the real connection strength in terms of the postsynaptic potential amplitude, and particularly the time course, is mostly a matter of guessing, although synapse sizes are becoming increasingly available from a number of EM datasets, which allows inference about connection strength for different cell types (Motta et al., 2019). Furthermore, synaptic plasticity involving, for example, changes in presynaptic release dynamics or neuromodulation, as well as long-range synaptic inputs from nonimaged brain regions, is entirely out of scope. Thus, the matrix M is never fully experimentally constrained, which is exactly the reason why dynamical systems modeling is so crucial: It allows us to set further constraints on the matrix elements of M (the synaptic connection strengths) as well as on the parameters determining single-cell dynamics, by comparing between model outcomes and measurements. There are two general strategies for making this comparison: biophysical dynamical systems approaches and regression-based approaches from machine learning. In Linear models, we will treat biophysical approaches; in Matrix exegesis, we will provide a brief introduction to nondynamical graph-theoretical approaches; and regression-based approaches are briefly referred to in the Discussion.
Linear models
Linear approximations of dynamical systems allow for in-depth mathematical treatment. Moreover, unbiased information processing requires a linear regime (e.g., Atick, 1992), since only in such regimes are changes in the input always translated into proportional changes in the output; there is no information transfer if the membrane voltage Vj is below threshold or so large that it consistently evokes maximal response. As a note of caution, we would like to stress that linearization is a coarse approximation, which, of course, introduces inaccuracies and errors. One cannot expect quantitative agreement between a linear model and the true biological circuit dynamics. However, in addition to the advantage of analytical tractability we will exploit next, linearization allows for a reduction of parameters, combining multiple physiological properties into one effective weight, which greatly increases the feasibility of parameter fitting. Matrix entries of M thus not only reflect synaptic efficacy, but also the slope of the f-I curve of the input neuron, which depends on multiple morphological parameters as well as cellular conductances and their current modulatory states. Linear models thus always reflect an approximation to the current regime of operation of the circuit. Conversely, the real biophysical circuit may generally correspond to multiple linear models (for threshold linear neurons, see Curto et al., 2019), depending on its mode of operation. Linear models can hence make qualitative predictions about which regimes are possible in a circuit and, together with the connectivity matrix, constrain the contribution of individual neurons for each of those regimes.
Formally, the linear counterpart of the dynamical system from Equation 1 is obtained by setting f(V) = V. Since not all neurons possess the same membrane time constant τ, one may straightforwardly generalize the dynamical equation to multiple time constants by replacing τ in Equation 1 by a diagonal matrix T with the different time constants on the diagonal. This then leads to a system of linear differential equations in matrix form as follows:
| (3) |
that can be solved numerically and allows for a direct comparison with the nonlinear system. In addition, it opens the route to analytical treatment. For example, the steady state of the system for a constant input I can be computed by setting to zero and solving the resulting system of linear equations as follows:
| (4) |
for V. Here, the symbol stands for the identity matrix, which carries 1s along its diagonal and is zero elsewhere.
Introducing the dynamical matrix , Equation 3 simplifies to the following:
| (5) |
which has the standard form of a linear differential equation with a matrix A taking the role of a growth rate.
Analytical solution and eigen decomposition
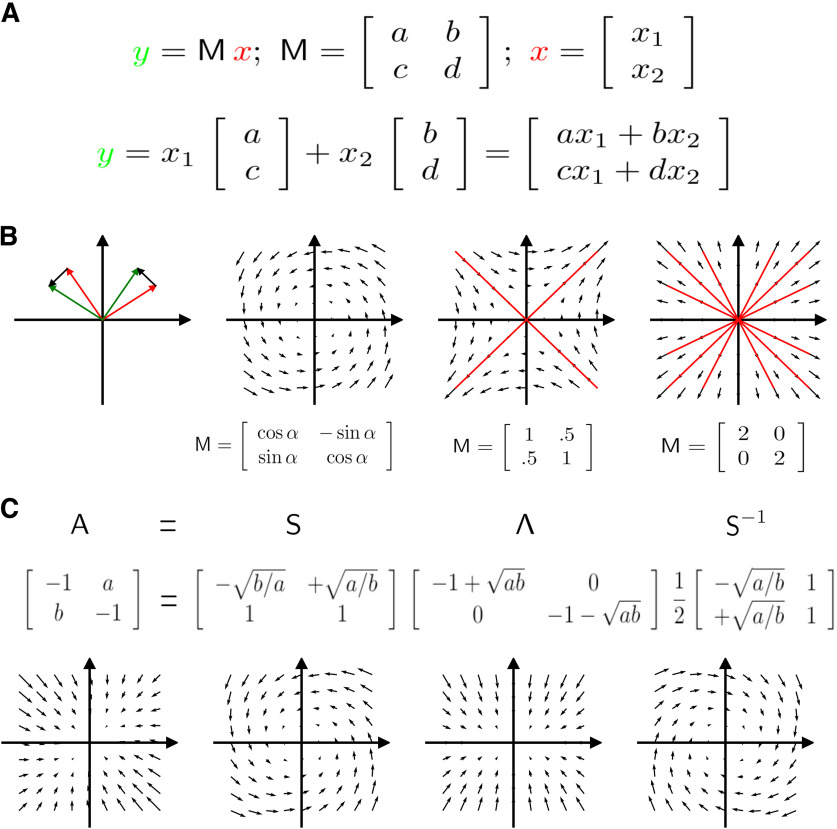
The benefit of linearity in dynamical system analysis is that closed-form solutions can be found. For stationary systems (i.e., all synaptic weights are constant), these solutions are solely determined by the eigenvalues and eigenvectors of the matrix A. For an explanation of eigenvectors and eigenvalues, see Figure 2. Let us first consider the situation when there is no input, that is, I(t) = 0. Then, the solution of the differential Equation 3 equals the following:
| (6) |
with the matrix containing all eigenvectors of A, and the diagonal matrix all its eigenvalues. Since
| (7) |
Equation 6 reflects the exponential decay of the voltage from its current state V(0) to the resting potential (V = 0) in the absence of further input. For unconnected neurons (M = 0 → A = –T–1), each of the neurons decays to rest with its intrinsic membrane time constant. If the connectivity matrix M is not zero, we still predict exponential decay to rest, as long as all eigenvalues λ have negative real parts. In this case, however, the decay time constants depend on both the intrinsic membrane time constants and the connectivity of the neuron within the circuit. In cases where the eigenvalues are complex, , the model predicts oscillatory solutions, since . If the real part of only one of the λs is positive, the model predicts exponential growth and the voltages may settle at a new steady state that is outside the realm of the linear part of the transfer function f. Despite the fact that having no input is a relatively uninteresting situation physiologically, it nevertheless provides us already with fundamental insight into the connectivity matrix. In particular, we realize that the eigenvalues of A must all have negative real parts if the circuit should express a stable zero state in the case of no input.
Figure 2.
Brief introduction to linear algebra. A, Matrix-vector-multiplication. The result y (green) of multiplying a matrix M with a vector x (red) is a linear combination of the columns of the matrix M. B, The difference vectors y – x (black) provide a graphical interpretation of the action of a matrix, which is illustrated for three example matrices: a rotational matrix, and an isotropic diagonal matrix. Vectors (red) for which the result y does not change directions compared with x are called eigenvectors. A 2 × 2 matrix has at most 2 eigenvectors. The constant λ, which determines the multiplicative scaling of the output y, is called eigenvalue. Eigenvalues are calculated by solving the equation , which leads to . This requires that . Solving this so-called “characteristic polynome” yields the eigenvalues λ. The corresponding eigenvectors are obtained by inserting them into and solving for x. Contractions/expansions have eigenvalues smaller/larger than 1, and correspond to arrows pointing inwards/outwards, respectively. C, Most square matrices A can be expressed by an eigenvector decomposition (Golub and Wilkinson, 1976). The basis transformation S–1 rotates the vector x into a coordinate system in which the eigenvectors of the transformed matrix are parallel to the coordinate axes. The matrix S rotates the output back to the original coordinate system. For further reading, see Strang (2009) and Christodoulou and Vogels (2022) or the video series linear algebra from 3blue1brown.
In the more relevant and interesting case, the network is driven by a non-zero input I(t). In the example of the fly visual system, this would correspond to the situation when the fly is visually stimulated and the photoreceptors thus become active. Then, a closed solution can also be found using the previously introduced elements as follows:
| (8) |
This equation reflects a convolution of the input T−1I(t) with the impulse response matrix . In the above equations, the reader may have noted a similarity to simple linear filters like the first order low-pass filter described by the differential equation in Formula 2: for zero input, the filter decays to zero from whatever value it has been before, for non-zero input, the output is obtained by convolving the input with the impulse response of the filter. The difference from a 1-dimensional filter (one input signal x(t), one output signal y(t)) is that we now deal with a multidimensional filter, where input and output signals are described as vectors as a function of time. The filter properties then depend on both the connectivity and the intrinsic time constants of the elements, and the calculation requires a projection of the input signal into the so-called “eigenspace” of the connectivity matrix and back again.
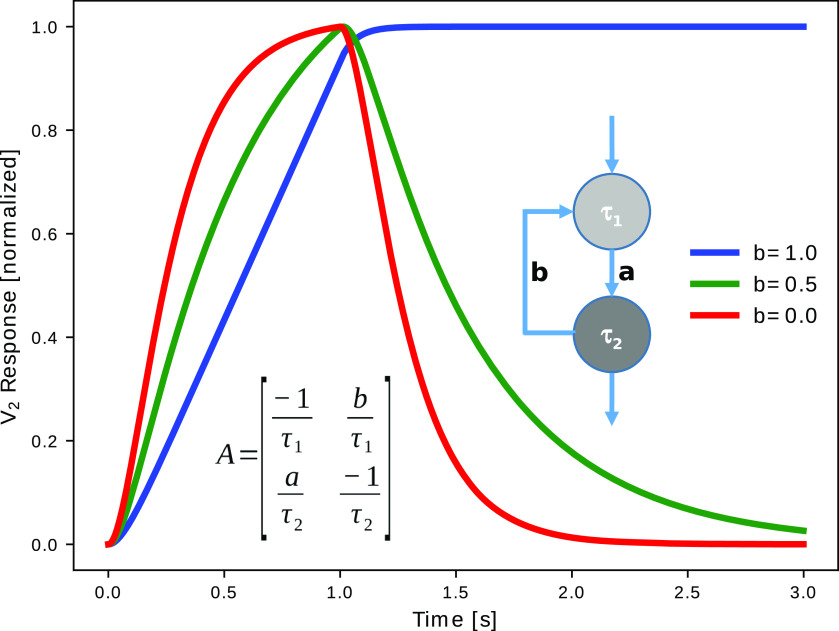
The solution of Equation 8 is exemplified in Figure 3A for a simple neural circuit that consists of two neurons connected in sequence with a feedback synapse of strength b. The circuit is stimulated via a 1-s-long depolarizing pulse to neuron 1. The voltage propagates from neuron 1 to neuron 2 as indicated by the delay. In the phase plane (V 1, V 2) (Fig. 3B), the initial stimulus drives the voltage to some state in the top right quadrant. After the stimulus is shut off, the trajectory follows the vector representation of the matrix and converges back to the origin along the direction of one of the eigenvectors (red). Both eigenvectors (red and blue) point toward the steady-state rendering the circuit stable.
Figure 3.
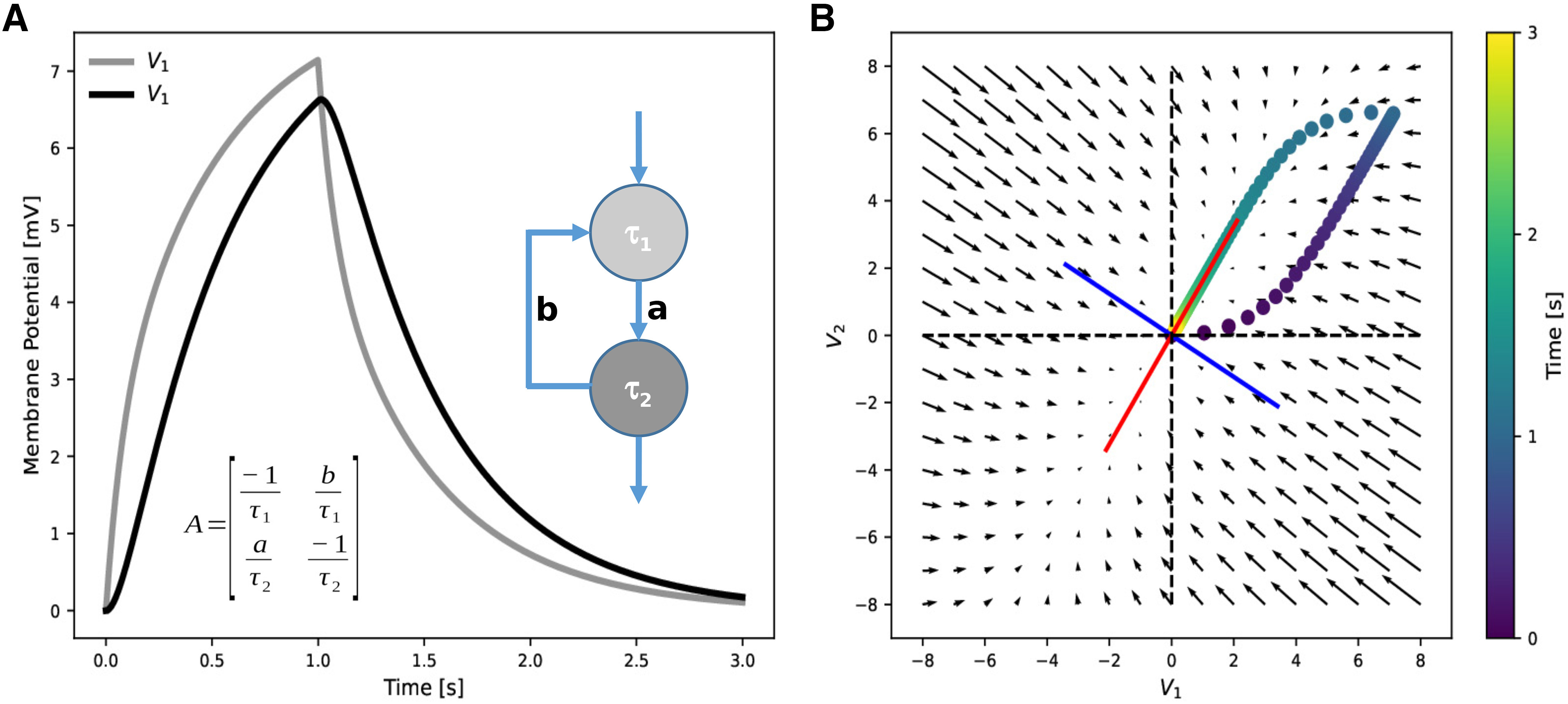
Time course of voltage signals and phase space of a 2-neuron circuit. A, Analytical solutions of the membrane potentials of two neurons (light and dark gray) in a feedback circuit on a 1 s constant input to neuron 1. The dynamical matrix A includes both neuronal (τ1, τ2) and connectivity (a,b) parameters. Solutions were obtained for a = 1, b = 0.5, τ1 = 0.1 s, τ2 = 0.2 s. B, Graphical representation of the matrix from A (arrows) and its eigenvectors (red, blue). The trajectory V2 versus V1 from A (color dots) follows the vectors representing the matrix after the stimulus is switched off (1 s) and converges to the direction of the red eigenvector.
Eigenvalues as inverse time constants
The analytical solution in Equation 8 not only allows us to make qualitative statements about the global system behavior, but in some simple cases, it also allows us to derive formulas for the eigenvalues (Fig. 4) and thereby discuss the dependence of global circuit behavior on the connectivity parameters. In the model circuit from Figure 3, we vary the strength b of the feedback synapse while keeping a = 1 and find that an increased feedback prolongs the decay constant of the pulse response. In networks without feedback, the eigenvalues are always the inverse cellular time constants. Feedback generally generates new time constants and thereby allows for signal processing at time scales that go beyond the ones of the isolated elements. This is reflected by the fact that the eigenvalues now depend not only on the cellular time constants, but also on the connectivity parameters. The time scale of activity decay in the network may substantially exceed the single neuron time scales, extending over seconds to minutes and thereby provide a substrate of working memory (Seung, 1996, 2003). In the extreme case of b = 1, that is, if the feedback connection is as strong as the feedforward connection, one eigenvalue becomes 0; and, thus, the time constant goes to infinity (Fig. 4, blue curve). This case corresponds to a perfect integration of the input signal, as argued for the oculomotor system (Seung et al., 2000a; Goldman et al., 2002).
Figure 4.
Feedback prolongs time constants. Responses of the same circuit as in Figure 3 to a 1 s constant input to neuron 1 for varying feedback strength b. The eigenvalues define the new effective time constants of the feedback circuit and depend on the connectivity parameters a and b. Without feedback (b = 0), the time constants are identical to the cellular time constants and the circuit becomes a second order low-pass filter. For a = b = 1, the time constant reaches , which means perfect integration (blue). For b > 1 (a = 1), λ2 becomes negative, indicating unstable self-excitation leading to unbounded voltage growth (as long as the linear regimen is valid).
Eigenvalues determine the input gain of network modes
Eigenvalues of A not only determine the time constants by which activity patterns rise and decay, they also define an “input resistance” (i.e., gain constant) of the pattern, equivalently to the relation between membrane time constant and input resistance of nerve membranes. For a constant input current (vector) I, the steady state solution from Equation 4 can be expressed in eigenmodes as follows:
| (9) |
with denoting the rows of . The overlap thereby measures how much of the n-th eigenmode is present in the input pattern. For stable modes , the contribution of the n-th mode to the steady state output pattern V is thus weighted by the positive gain factor , which can be considered to be the input resistance of this eigenmode. If is large (time constant small), the gain factor is small and corresponds to a “leaky” pattern with little contribution to the output pattern. If is small (time constant large), the gain factor is large and corresponds to an integrator pattern with large contribution to the output pattern.
Eigenvalues of the connectivity matrix thus allow us to translate intuitions from single-cell electrophysiology to the realm of activity patterns in networks.
Matrix exegesis
We have argued that feedback has profound effects on circuit computation, even for linear models. It, therefore, would be helpful to readily identify feedback from a circuit diagram (connectome) without simulations or analytical assessment of network dynamics. How would a connectivity matrix of a pure feedforward circuit differ from the one of a circuit that has feedback? In a pure feedforward network, the first neuron does not receive input from any of the following neurons; that is, the top row contains 0's only (Fig. 5, top right). The second neuron receives input from the first one only, so in row 2, there is only one entry in column 1. The third neuron receives input at most from neuron one and two, so maximum entries in row 3 are in columns 1 and 2. Thus, in general, in a pure feedforward circuit, the output of any neuron n has no effect on its own activity and only influences neurons m > n. This is reflected in its connectivity matrix M that has only entries below the diagonal. Such a matrix is said to be “lower triangular.” Unfortunately, however, neurons are numbered in an arbitrary way, so the connectivity matrix is scrambled. In order to see whether it is purely feedforward or not, the circuit elements need to be reordered. For systems without feedback, there always exists a reordering such that the matrix M has lower-triangular form (Fig. 5, top row).
Figure 5.
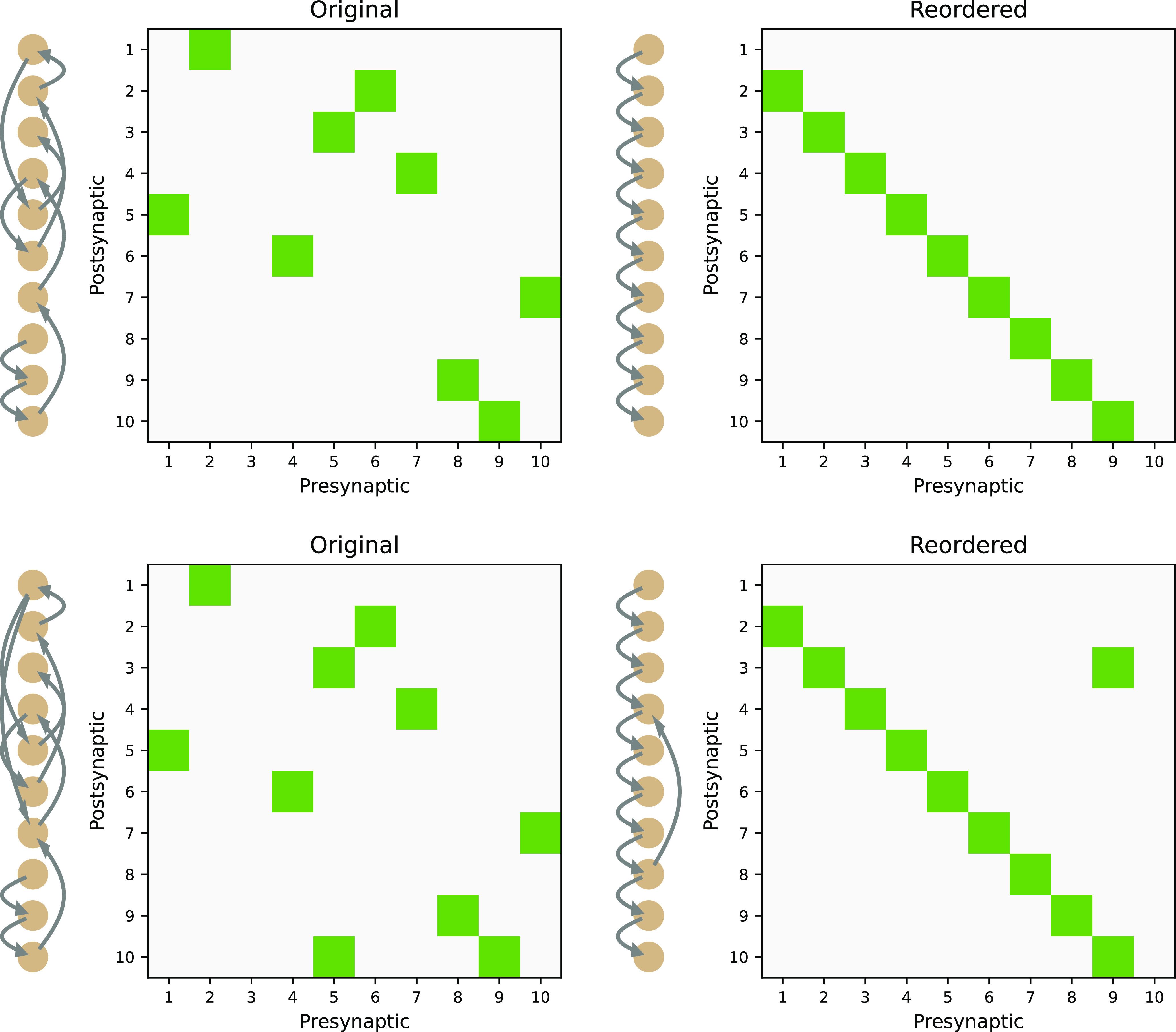
Matrix reordering. The numbering of neurons in a circuit is arbitrary. Random ordering (left column) does not readily reveal feedforward and feedback structure in the connectivity matrix. It can, however, be revealed by appropriate reordering (right column).
True feedback connections are immune to such reordering of the connectivity matrix (Fig. 5, bottom row) and cannot be brought into lower-triangular form. A simple criterion to check whether a connectome is feedforward and the connectivity matrix M can be reordered into lower-triangular form is to test for nilpotence of M: Whenever there exists a finite integer power a, such that Ma = 0, the connectome is strictly feedforward. Physiologically, the condition Ma = 0 reflects the fact that any activity in a pure feedforward network inevitably dies out after a finite amount of time steps. Recurrent loops, however, can keep the network activity indefinitely, if the synapses are strong enough.
Evidently, connectomes are never strictly feedforward; however, approximate feedforward organization has been reported for subcircuits (e.g., Borst et al., 2020a), and even for some cell classes in the whole Caenorhabditis elegans connectome (Durbin, 1987). Approximate feedforwardness also seems reasonable given that, on a large scale, an animal needs to select its motor output depending on sensory input. Finding feedforward, or “almost” feedforward motifs in the connectivity matrix, hence, can reveal important insight into the local flow of information.
While for small circuits with few neurons (e.g., Fig. 5), reordering into lower-triangular form could be done by visual inspection and trial-and-error, such practices are intractable for larger connectomes: here, numerical tools are clearly required. A classical method from linear algebra that has been successfully applied to neural circuit connectivity (Goldman, 2009) is the Schur decomposition. This method uses the fact that, for any diagonalizable matrix A, there exists a unitary basis transformation matrix U (where ), such that
| (10) |
is a lower-triangular matrix. Typical software packages for linear algebra include algorithms for Schur decomposition. However, the standard mathematical literature (and thus many numerical packages) considers R as being an upper-triangular matrix, which can be trivially turned into a lower-triangular matrix by transposition.
While Schur decomposition identifies feedforward motifs in networks, it has the downside that it only does so in a transformed neuron space, that is, it comes along with a transformation from activities of single neurons to activity of neuronal populations , for which the system is feedforward. Each entry in P denotes a population activity variable, which arises from a linear combination of the original neuronal activities V. The feedforward matrix R can thus be considered as connecting neural ensembles (or population patterns) in sequence. Generally, the matrix A will nevertheless be recurrent; that is, the neurons within an ensemble will be recursively coupled. If and only if the transformation matrix U is a permutation matrix (a square binary matrix that has exactly one entry of 1 in each row and each column and 0's elsewhere), the matrix R is a reordered version of A and, hence, truly feedforward with no single neuron's activity having an effect on itself. Thus, while the Schur decomposition provides a robust solution of the reordering problem at the level of population patterns, it is insufficient to do so at the single-cell level.
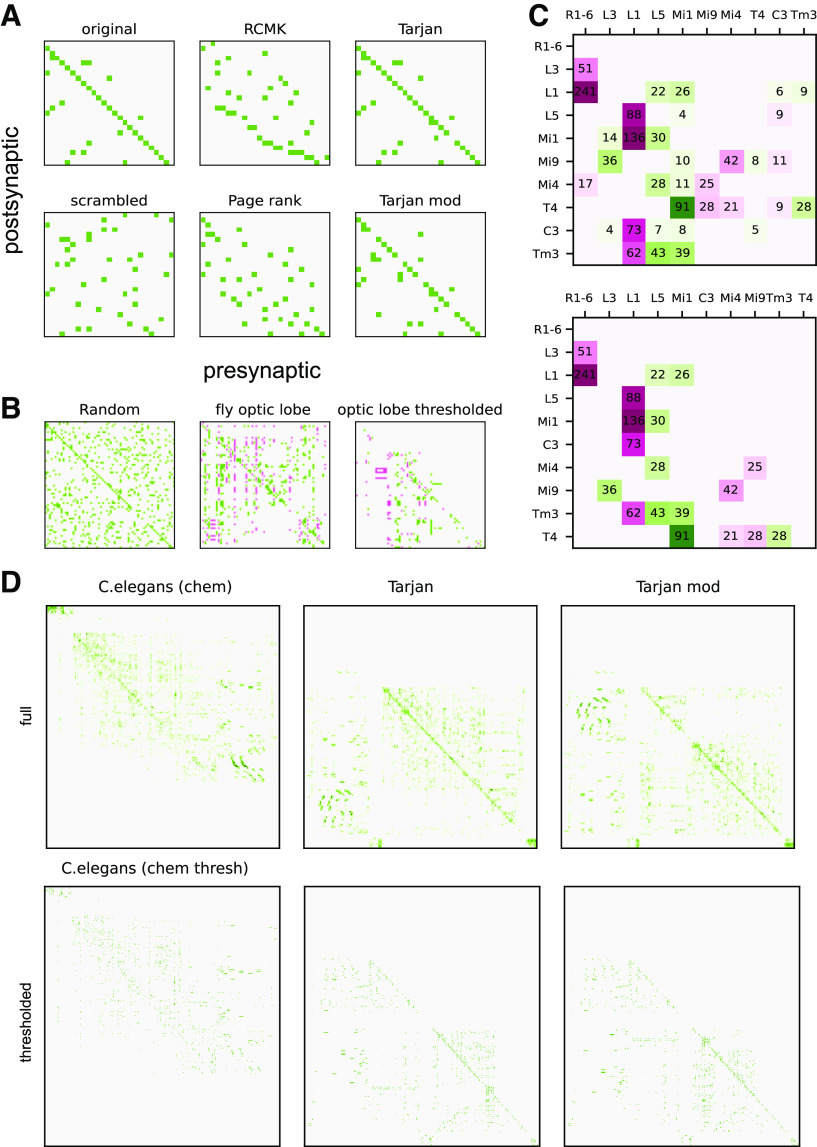
A more direct interpretation in terms of single neurons, however, can be achieved by classical matrix reordering techniques (Robinson, 1951; for review, see Liiv, 2010; Behrisch et al., 2016), which determine index permutations such that the shape of the matrix optimizes some objective. In general, both row and column indices can be reordered independently, but for connectomic analyses only algorithms that use the same permutation for columns and rows (presynaptic and postsynaptic neurons) are meaningful. A standard objective used to find optimal permutations is bandwidth minimization (Leung et al., 1984), that is, keeping all matrix elements in as few bands as possible close to the diagonal. Such minimal bandwidth matrices readily allow translation into circuit schemes. Classical solutions are based on the Cuthill-McKee algorithm (Cuthill and McKee, 1969), improved versions of which (George, 1971; Gibbs et al., 1976) are available in standard numerics packages as Scipy (Virtanen et al., 2020) (Fig. 6A, top middle).
Figure 6.
Matrix reordering methods. A, Left, Example of a matrix implementing a single feedforward sequence, which is distorted by a few random feedback and out-of-sequence feedforward components (top). Random index shuffling yields a scrambled version of the original matrix (bottom). Middle, Outcome of matrix reordering algorithms applied to the scrambled matrix based on bandwidth minimization (top, reverse Cuthill McKee) and propagating ranks (bottom). Right, Outcome of matrix reordering using the python package Tarjan (top), and a custom-modified version minimizing the upper triangular elements in loops (bottom). B, Application of the modified Tarjan procedure to a 66 × 66 random matrix (sparseness 0.15), the full connectome of the fly optic lobe (red represents hyperpolarizing connections), and a thresholded version of the optic lobe connectome only considering connections with >10 synapses. C, Modified Tarjan reordering of the fly subcircuit for ON-motion detection (top) and a reduced version with a threshold at 20 synapses (bottom). Saturation of colors (green represents excitatory; red represents inhibitory) illustrates synapse numbers (black). D, Top, Original connectome derived from chemical synapses of the entire worm hermaphrodite C. elegans (454 node) (Cook et al., 2019), reordered with the original (middle) and modified (right) Tarjan algorithm. Bottom, Same as in top, but only taking into account connections with 4 and more synapses (independent of neurotransmitter).
Although the problem is solved in principle, in a neuroscience context (Carmel et al., 2004; Seung, 2009; Varshney et al., 2011), the algorithms are numerically costly (Monien and Sudborough, 1985) and not very well feasible for large connectomic data. Therefore, several new approaches are actively being developed, including heuristics (Wang et al., 2014), kernel-based methods (Bollen et al., 2018), and deep neural networks (Watanabe and Suzuki, 2021), usually for independent column and row index permutations.
“Feedforwardness,” however, not only requires small bandwidth of the connectivity matrix, but also a serial arrangement of connections. The latter point is illustrated by the page-rank algorithm (Page et al., 1999) (Fig. 6A, bottom middle). This algorithm is used in Internet searches to quantify the importance of websites, with the most important one having the last index. While page rank graphs are computationally highly efficient and also tend to reduce upper triangle entries, they are not good in sorting feedforward dependencies in the less important (earlier) nodes.
A family of algorithms that both detect feedforwardness and reduce upper-triangle entries are flow-graph approaches. Developed in the 1970s for analyzing the command flow of computer programs to prove theorems of program termination, these approaches are based on a “depth-first search” (considered within the class of “topological sorting” algorithms) that reduces graphs into loops (Tarjan, 1972, 1974) (Fig. 6A, right). A flow graph-based algorithm even finds feedforward structures in random graphs (Fig. 6B, left) and also identifies sequential structure in the full connectome of the fly optic lobe (66 nodes) (Shinomiya et al., 2019) (Fig. 6B, middle). The detected order, however, still provides no immediate recipe for functional interpretation.
A simple strategy for extracting physiological insight from matrix reordering is to set a threshold on the synapse numbers. Only considering connections above that threshold effectively disregards weaker routes of information flow. Even a moderately low threshold of 10 synapses gives rise to a mostly feedforward matrix for the full connectome of the fly optic lobe (Fig. 6B, right), which allows identification of circuit modules that can be studied independently of the rest of the network. This idea is illustrated by the ON-motion subcircuit (Takemura et al., 2017), which comprises those cells that provide input to T4 cells. Figure 6C depicts two versions of this subcircuit: one including all connections (top) and one including only connections with >20 synapses (bottom). The reduced matrix identifies a subnetwork of recurrently connected Mi4 and Mi9 cells that drive no other neurons than T4 cells and receives input from L3 and L5 only. The thresholded matrix also identifies one large feedback loop via L1. Application of the Tarjan-derived reordering to the whole worm connectome (Cook et al., 2019) (Fig. 6D) yields consistent conclusions, in that thresholded connectivity is largely feedforward (originally observed in Durbin, 1987). Functional analysis using these identified submodules of course can post hoc be checked with simulations or eigenvalue analysis of the full connectome.
Of course, thresholding also changes the optimal sequence order of the neurons, but we would argue that stronger connections should also be assigned a larger weight in matrix reordering.
The practical feasibility of matrix reordering techniques strongly depends on how the algorithm scales with matrix size and thus the required computing resources. In the case of the original Tarjan algorithm (Tarjan, 1972, 1974) computing time was shown to efficiently scale with the number S of synapses as (Fig. 7). For comparison, we also used a brute force approach, in which the number of truly recurrent connections is identified in two steps: In the first step, the cost function to be minimized calculates the sum of the lengths (i.e., index differences) of all connections in the upper triangle of the connectivity matrix; in the second step, the cost function only calculates the number of entries in the upper triangle, regardless of the length of the connection. In each of the two steps, permutations are tested in a systematic way, ordered by the lengths of the exchanged sequences. Naturally, this causes the algorithm's computing time to scale much worse with the size of the matrix than the graph theoretical approaches, but it reaches the best results regarding a minimization of the upper triangular entries (Fig. 7, bottom). The graph theoretical approaches excel in finding a single connected feedforward structure (first lower subdiagonal).
Figure 7.
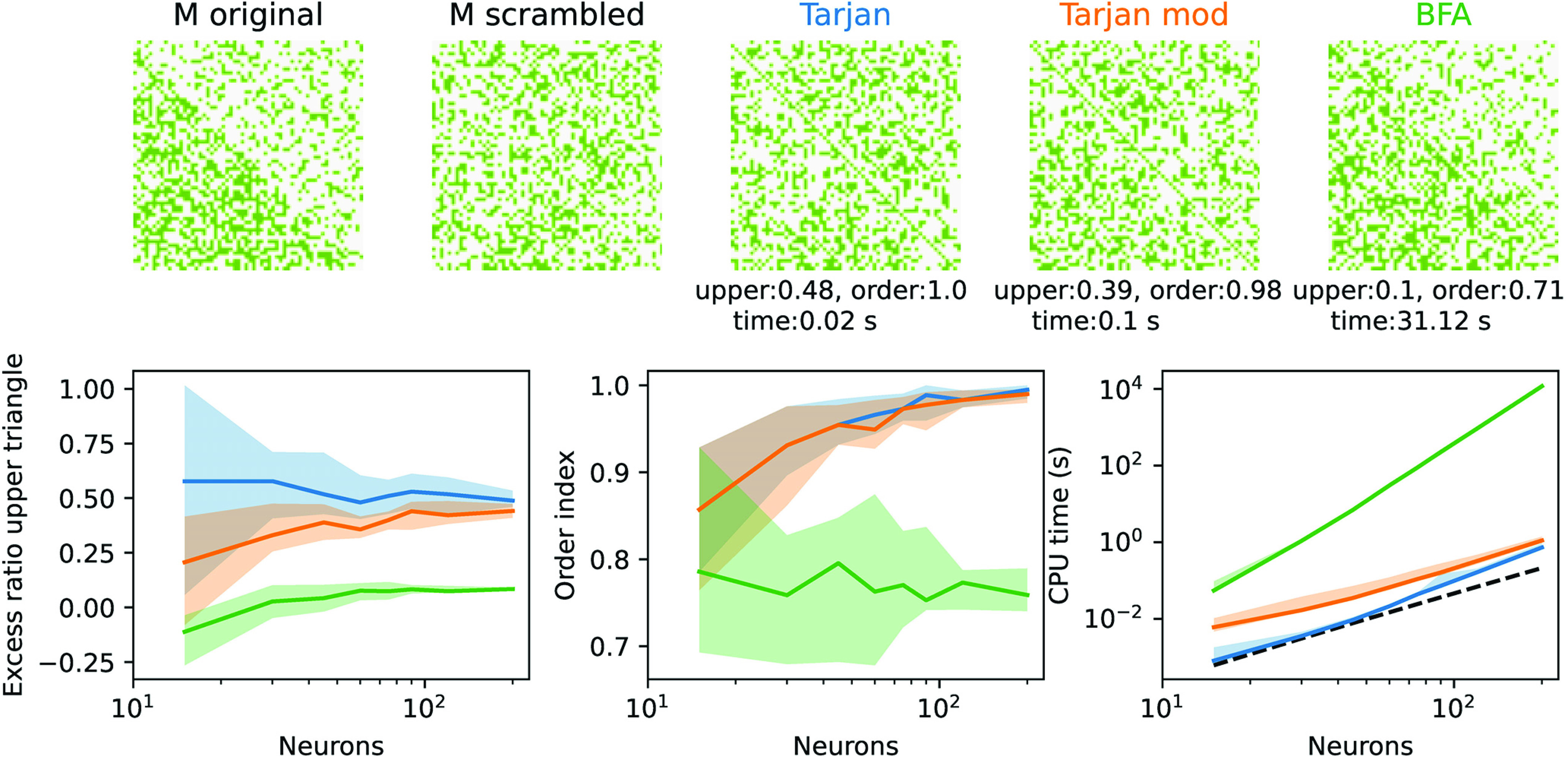
Scaling of reordering algorithms. Top, Example of a random binary 60 × 60 matrix M with half of the edges filled in the lower and a quarter of edges filled in the top triangle (and no self-connections). After random index permutation (scrambled), we applied three reordering algorithms, the two graph-based methods from Figure 6A, and a brute-force approach (BFA) minimizing the entries in the top triangle as explained in the text. The performance of the reordering is quantified by three measures. The excess ratio in the top triangle (top) measures the increase of entries in the top triangle relative to the original matrix. The Order index measures the fraction of filled entries in the first subdiagonal in the bottom triangle and reveals the extent to which the algorithm has identified a closed path from a first to a last node. Finally, the CPU time used on a standard PC indicates the applicability of the algorithm to large connectomes. Bottom, Three performance measures as a function of node number (colors as on top). Solid lines indicate the median and the area of the 90th percentile obtained from 15 repetitions. Dashed black line on the right indicates the theoretical scaling law (multiplied by an arbitrary constant for better comparison).
Apart from CPU time, quantitative comparisons of algorithms require definition of quality measures for their outcome. In Figure 7, we choose two measures to compare different algorithms: the first measure (“recurrency”) is defined by the increase of entries in the upper triangle of the matrix ordered by the respective algorithm relative to the original matrix, the second measure (“feedforwardness”) is defined as the (normalized) number of matrix entries in the first subdiagonal. Optimizing for one does not necessarily optimize the other (Behrisch et al., 2020). Large feedforwardness, hence, may reflect a graph of many feedforwardly ordered loops, whereas low recurrency reduces feedback but does not necessarily identify the optimal order of information flow.
Discussion and Outlook
The connectivity matrix resulting from volumetric EM analysis contains much information about the functional properties of a neural circuit. Yet, these are not obvious at first sight. Linear algebra provides a lot of tools useful in two ways: (1) they allow for detecting functional substructures of a circuit (e.g., by separating feedforward from feedback connections); and (2) they allow for predicting time constants of particular elements within the circuit that arise from network architecture, and not from intrinsic membrane properties.
Open problems and success stories
Although linear models provide important tools to dissect specific network regimens, they are unarguably limited as they cannot explain transitions between these regimens as, for example, in multistability, or they ignore nonlinearities of spike generation (e.g., Lehnert et al., 2014) and synaptic transmission (e.g., Wienecke et al., 2018; Mishra et al., 2023). Some progress has been made in the development of analytical tools for threshold linear networks by dividing up the state space into multiple subspaces with linear dynamics (Curto et al., 2019; Santander et al., 2021). To our knowledge, however, these have not been applied to connectomics data. Tschopp et al. (2018) used a connectome-constrained (rectified linear) network model of the fly visual system to optimize the synaptic weights to specific computer vision tasks, nicely illustrating how EM connectivity data can be used as a seed to facilitate parameter estimation (for review, see Litwin-Kumar and Turaga, 2019). In a similar vein, Klinger et al. (2021) compared the synaptic connections resulting from simple network models to connectomic data of the neocortical column, using approximate Bayesian estimation to judge which of the models best explains the observed connectivity. Qualitative insight into the dynamical effects of connectivity parameters is less straightforward when directly optimizing parameters of nonlinear models. A promising approach is deep density estimators (Goncalves et al., 2020; Bittner et al., 2021), which predict distributions for parameters of mechanistic models when trained with activity data.
Large-scale connectomics data do not necessarily rely on single-neuron models but can use population models with smooth nonlinear activation functions. For example, dynamical mean field models (Deco et al., 2008; Deco and Jirsa, 2012) incorporate nonlinearities f that are microscopically derived from averaging activity of pools of spiking-neuron models (Brunel and Hakim, 1999), and the objective of fitting is to replicate the functional connectivity matrix (activity covariance) obtained from whole-brain fMRI. Thereby, constraining M as much as possible by functional connectivity turned out to be essential to achieve good fits (Cabral et al., 2017), indicating the general importance of the underlying large-scale network structure. By design, however, dynamical mean fields are not applicable to microscopic single-cell–based models, let alone for circuits consisting of graded response neurons, such as in the fly visual pathway.
Generalized linear models (GLMs), which have been successfully applied to small- and medium-scale circuits (Paninski, 2004; Pillow et al., 2008; Pho et al., 2018), could be considered as a compromise between brute-force nonlinear fitting and qualitative interpretability of connectivity parameters. In GLMs, the voltage variable V(t) arises from the input I in very much the same way as in the example from Equation 8. Therefore, qualitative analysis of eigenvalues of A can be compared with the best-fit solution obtained from mapping V(t) into a mean firing rate by a so-called inverse link function f(V). The inverse link function models the spiking nonlinearity and is determined by the firing statistics of the model. For Poisson noise, f is exponential and generally interpreted as the hazard function of the Poisson process. For graded neurons, however, there is so far no general agreement on the choice of noise model and inverse link function. Also, the application of GLMs to large connectomes is limited by numerical optimization in high-dimensional nonlinear parameter spaces.
In conclusion, although which single approach to connectome-based nonlinear modeling (if any) will become the gold standard remains to be determined, techniques from linear algebra can be advantageously applied to a simplified linear circuit derived from a well-constrained connectivity matrix. In combination with numerical simulations of a biophysically more realistic circuit (e.g., taking into account voltage-gated ion channels and further nonlinear characteristics of neural transmission), this approach can reveal deep insight into the particular function of a circuit and the microscopic biological mechanisms on which it is based.
Footnotes
We thank Lukas Groschner and Moritz Helmstaedter for carefully reading the manuscript.
The authors declare no competing financial interests.
References
- Abbott LF, et al. (2020) The mind of a mouse. Cell 182:1372–1376. 10.1016/j.cell.2020.08.010 [DOI] [PubMed] [Google Scholar]
- Aimon S, Katsuki T, Jia T, Grosenick L, Broxton M, Deisseroth K, Sejnowski TJ, Greenspan RJ (2019) Fast near-whole-brain imaging in adult Drosophila during responses to stimuli and behavior. PLoS Biol 17:e2006732. 10.1371/journal.pbio.2006732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WE, Chen MZ, Pichamoorthy N, Tien RH, Pachitariu M, Luo L, Deisseroth K (2019) Thirst regulates motivated behavior through modulation of brainwide neural population dynamics. Science 364:eaav3932. 10.1126/science.aav3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atick JJ (1992) Could information theory provide an ecological theory of sensory processing? Network 3:213–251. 10.1088/0954-898X_3_2_009 [DOI] [PubMed] [Google Scholar]
- Behrisch M, Bach B, Riche HN, Schreck T, Fekete JD (2016) Matrix reordering methods for table and network visualization. Comput Graph Forum 35:693–716. 10.1111/cgf.12935 [DOI] [Google Scholar]
- Behrisch M, Schreck T, Pfister H (2020) GUIRO: user-guided matrix reordering. IEEE Trans Vis Comput Graph 26:184–194. [DOI] [PubMed] [Google Scholar]
- Bittner SR, Palmigiano A, Piet AT, Duan CA, Brody CD, Miller KD, Cunningham J (2021) Interrogating theoretical models of neural computation with emergent property inference. Elife 10:e56265. 10.7554/eLife.56265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen T, Leurquin G, Nijssen S (2018) Convomap: Using convolution to order Boolean data. In: Advances in intelligent data analysis XVII (Duivesteijn W, Siebes A, Ukkonen A, eds), pp 62–74. Cham: Springer International. [Google Scholar]
- Borst A, Haag J, Mauss AS (2020a) How fly neurons compute the direction of visual motion. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 206:109–124. 10.1007/s00359-019-01375-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A, Drews M, Meier M (2020b) The neural network behind the eyes of a fly. Curr Opin Physiol 16:33–42. 10.1016/j.cophys.2020.05.004 [DOI] [Google Scholar]
- Briggman KL, Helmstaedter M, Denk W (2011) Wiring specificity in the direction-selectivity circuit of the retina. Nature 471:183–188. 10.1038/nature09818 [DOI] [PubMed] [Google Scholar]
- Brunel N, Hakim V (1999) Fast global oscillations in networks of integrate-and-fire neurons with low firing rates. Neural Comput 11:1621–1671. 10.1162/089976699300016179 [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Stark E, Berényi A, Khodagholy D, Kipke DR, Yoon E, Wise KD (2015) Tools for probing local circuits: high-density silicon probes combined with optogenetics. Neuron 86:92–105. 10.1016/j.neuron.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral J, Kringelbach ML, Deco G (2017) Functional connectivity dynamically evolves on multiple time-scales over a static structural connectome: models and mechanisms. Neuroimage 160:84–96. 10.1016/j.neuroimage.2017.03.045 [DOI] [PubMed] [Google Scholar]
- Carmel L, Harel D, Koren Y (2004) Combining hierarchy and energy for drawing directed graphs. IEEE Trans Visual Comput Graphics 10:46–57. 10.1109/TVCG.2004.1260757 [DOI] [PubMed] [Google Scholar]
- Christodoulou G, Vogels T (2022) The eigenvalue value (in neuroscience). OSF Preprints 10.31219/osf.io/evqhy. [DOI] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KC, Tang LT, Bayer EA, Duerr JS, Bülow HE, Hobert O, Hall DH, Emmons SW (2019) Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571:63–71. 10.1038/s41586-019-1352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto C, Geneson J, Morrison K (2019) Fixed points of competitive threshold-linear networks. Neural Comput 31:94–155. 10.1162/neco_a_01151 [DOI] [PubMed] [Google Scholar]
- Cuthill E, McKee J (1969) Reducing the bandwidth of sparse symmetric matrices. In: Proceedings of the 1969 24th National Conference, ACM '69, pp 157–172. New York: Association for Computing Machinery. 10.1145/800195.805928 [DOI] [Google Scholar]
- Dayan P, Abbott LF (2005) Theoretical neuroscience: computational and mathematical modeling of neural systems. Cambridge, MA: Massachusetts Institute of Technology. [Google Scholar]
- Deco G, Jirsa VK (2012) Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. J Neurosci 32:3366–3375. 10.1523/JNEUROSCI.2523-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, Robinson PA, Breakspear M, Friston K (2008) The dynamic brain: from spiking neurons to neural masses and cortical fields. PLoS Comput Biol 4:e1000092. 10.1371/journal.pcbi.1000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Horstmann H (2004) Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol 2:e329. 10.1371/journal.pbio.0020329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM (1987) Studies on the development and organisation of the nervous system of Caenorhabditis elegans. PhD dissertation, Cambridge, UK: University of Cambridge. [Google Scholar]
- Gardner RJ, Hermansen E, Pachitariu M, Burak Y, Baas NA, Dunn BA, Moser MB, Moser EI (2022) Toroidal topology of population activity in grid cells. Nature 602:123–128. 10.1038/s41586-021-04268-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JA (1971) Computer Implementation of the finite element method. PhD dissertation, Stanford, CA: Stanford University. [Google Scholar]
- Gerstner W (2000) Population dynamics of spiking neurons: fast transients, asynchronous states, and locking. Neural Comput 12:43–89. 10.1162/089976600300015899 [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ (2011) Miniaturized integration of a fluorescence microscope. Nat Methods 8:871–878. 10.1038/nmeth.1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs NE, Poole WG, Stockmeyer PK (1976) An algorithm for reducing the bandwidth and profile of a sparse matrix. SIAM J Numer Anal 13:236–250. 10.1137/0713023 [DOI] [Google Scholar]
- Goldman MS (2009) Memory without feedback in a neural network. Neuron 61:621–634. 10.1016/j.neuron.2008.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MS, Kaneko CR, Major G, Aksay E, Tank DW, Seung HS (2002) Linear regression of eye velocity on eye position and head velocity suggests a common oculomotor neural integrator. J Neurophysiol 88:659–665. 10.1152/jn.2002.88.2.659 [DOI] [PubMed] [Google Scholar]
- Golub GH, Wilkinson JH (1976) Ill-conditioned eigensystems and the computation of the Jordan canonical form. SIAM Rev 18:578–619. 10.1137/1018113 [DOI] [Google Scholar]
- Goncalves PJ, Lueckmann JM, Deistler M, Nonnenmacher M, Öcal K, Bassetto G, Chintaluri C, Podlaski WF, Haddad SA, Vogels TP, Greenberg DS, Macke JH (2020) Training deep neural density estimators to identify mechanistic models of neural dynamics. Elife 9:e56261. 10.7554/eLife.56261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschner LN, Malis JG, Zuidinga B, Borst A (2022) A biophysical account of multiplication by a single neuron. Nature 603:119–123. 10.1038/s41586-022-04428-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W (2013) Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500:168–174. 10.1038/nature12346 [DOI] [PubMed] [Google Scholar]
- Herz AV, Gollisch T, Machens CK, Jaeger D (2006) Modeling single-neuron dynamics and computations: a balance of detail and abstraction. Science 314:80–85. 10.1126/science.1127240 [DOI] [PubMed] [Google Scholar]
- Jun JJ, et al. (2017) Fully integrated silicon probes for high-density recording of neural activity. Nature 551:232–236. 10.1038/nature24636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Schnitzer MJ (2022) Fluorescence imaging of large-scale neural ensemble dynamics. Cell 185:9–41. 10.1016/j.cell.2021.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger E, Motta A, Marr C, Theis FJ, Helmstaedter M (2021) Cellular connectomes as arbiters of local circuit models in the cerebral cortex. Nat Commun 12:2785. 10.1038/s41467-021-22856-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar AA, Liu T, Turkcan MK, Zhou Y (2021) Brain in the connectomic and synaptomic era. Elife 10:e62362. 10.7554/eLife.62362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S, Ford MC, Alexandrova O, Hellmundt F, Felmy F, Grothe B, Leibold C (2014) Action potential generation in an anatomically constrained model of medial superior olive axons. J Neurosci 34:5370–5384. 10.1523/JNEUROSCI.4038-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JY, Vornberger O, Witthoff JD (1984) On some variants of the bandwidth minimization problem. SIAM J Comput 13:650–667. 10.1137/0213040 [DOI] [Google Scholar]
- Lichtman JW, Denk W (2011) The big and the small: challenges of imaging the brain's circuits. Science 334:618–623. 10.1126/science.1209168 [DOI] [PubMed] [Google Scholar]
- Liiv I (2010) Seriation and matrix reordering methods: an historical overview. Stat Anal Data Min 3:70–91. [Google Scholar]
- Litwin-Kumar A, Turaga SC (2019) Constraining computational models using electron microscopy wiring diagrams. Curr Opin Neurobiol 58:94–100. 10.1016/j.conb.2019.07.007 [DOI] [PubMed] [Google Scholar]
- Maisak MS, Haag J, Ammer G, Serbe E, Meier M, Leonhardt A, Schilling T, Bahl A, Rubin GM, Nern A, Dickson BJ, Reiff DF, Hopp E, Borst A (2013) A directional tuning map of Drosophila elementary motion detectors. Nature 500:212–216. 10.1038/nature12320 [DOI] [PubMed] [Google Scholar]
- Markram H, et al. (2015) Reconstruction and simulation of neocortical microcircuitry. Cell 163:456–492. 10.1016/j.cell.2015.09.029 [DOI] [PubMed] [Google Scholar]
- Mishra A, Serbe E, Borst A, Haag J (2023) Voltage to calcium transformation enhances direction selectivity in Drosophila T4 neurons. J Neurosci 43:2497–2514. 10.1523/JNEUROSCI.2297-22.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monien B, Sudborough IH (1985) Bandwidth constrained np-complete problems. Theor Comput Sci 41:141–167. 10.1016/0304-3975(85)90068-4 [DOI] [Google Scholar]
- Motta A, Berning M, Boergens KM, Staffler B, Beining M, Loomba S, Hennig P, Wissler H, Helmstaedter M (2019) Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science 366:eaay3134. 10.1126/science.aay3134 [DOI] [PubMed] [Google Scholar]
- Nakagawa TT, Jirsa VK, Spiegler A, McIntosh AR, Deco G (2013) Bottom up modeling of the connectome: linking structure and function in the resting brain and their changes in aging. Neuroimage 80:318–329. 10.1016/j.neuroimage.2013.04.055 [DOI] [PubMed] [Google Scholar]
- Page L, Brin S, Motwani R, Winograd T (1999) The pagerank citation ranking: bringing order to the web. Technical Report 1999-66, Stanford InfoLab. [Google Scholar]
- Paninski L (2004) Maximum likelihood estimation of cascade point-process neural encoding models. Network 15:243–262. 10.1088/0954-898X_15_4_002 [DOI] [PubMed] [Google Scholar]
- Pho GN, Goard MJ, Woodson J, Crawford B, Sur M (2018) Task-dependent representations of stimulus and choice in mouse parietal cortex. Nat Commun 9:2596. 10.1038/s41467-018-05012-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillow JW, Shlens J, Paninski L, Sher A, Litke AM, Chichilnisky EJ, Simoncelli EP (2008) Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature 454:995–999. 10.1038/nature07140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan K, Abbott LF (2006) Eigenvalue spectra of random matrices for neural networks. Phys Rev Lett 97:188104. 10.1103/PhysRevLett.97.188104 [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S (1911) Histologie du systéme nerveux de l'homme & des vertébrés, Vol 1. Paris: Maloine. https://www.biodiversitylibrary.org/bibliography/48637. [Google Scholar]
- Robinson WS (1951) A method for chronologically ordering archaeological deposits. Am Antiq 16:293–301. 10.2307/276978 [DOI] [Google Scholar]
- Santander DE, Ebli S, Patania A, Sanderson N, Burtscher F, Morrison K, Curto C (2021) Nerve theorems for fixed points of neural networks. arXiv 2102.11437. [Google Scholar]
- Seung HS (1996) How the brain keeps the eyes still. Proc Natl Acad Sci USA 93:13339–13344. 10.1073/pnas.93.23.13339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS (2003) Amplification, attenuation, and integration. Handb Brain Theory Neural Netw 2:94–97. [Google Scholar]
- Seung HS (2009) Reading the book of memory: sparse sampling versus dense mapping of connectomes. Neuron 62:17–29. 10.1016/j.neuron.2009.03.020 [DOI] [PubMed] [Google Scholar]
- Seung HS, Lee DD, Reis BY, Tank DW (2000a) Stability of the memory of eye position in a recurrent network of conductance-based model neurons. Neuron 26:259–271. 10.1016/S0896-6273(00)81155-1 [DOI] [PubMed] [Google Scholar]
- Seung HS, Lee DD, Reis BY, Tank DW (2000b) The autapse: a simple illustration of short-term analog memory storage by tuned synaptic feedback. J Comput Neurosci 9:171–185. 10.1023/A:1008971908649 [DOI] [PubMed] [Google Scholar]
- Shinomiya K, et al. (2019) Comparisons between the ON- and OFF-edge motion pathways in the Drosophila brain. Elife 8:e40025. 10.7554/eLife.40025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman GM (2000) Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb Cortex 10:127–141. 10.1093/cercor/10.2.127 [DOI] [PubMed] [Google Scholar]
- Steinmetz NA, Zatka-Haas P, Carandini M, Harris KD (2019) Distributed coding of choice, action and engagement across the mouse brain. Nature 576:266–273. 10.1038/s41586-019-1787-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang G (2009) Introduction to linear algebra, Ed 4. Wellesley, MA: Wellesley-Cambridge. [Google Scholar]
- Takemura SY, Nern A, Chklovskii DB, Scheffer LK, Rubin GM, Meinertzhagen IA (2017) The comprehensive connectome of a neural substrate for 'ON' motion detection in Drosophila. Elife 6:e24394. 10.7554/eLife.24394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarjan RE (1972) Depth-first search and linear graph algorithms. SIAM J Comput 1:146–160. 10.1137/0201010 [DOI] [Google Scholar]
- Tarjan RE (1974) Testing flow graph reducibility. J Comput Syst Sci 9:355–365. 10.1016/S0022-0000(74)80049-8 [DOI] [Google Scholar]
- Tschopp FD, Reiser MB, Turaga SC (2018) A connectome based hexagonal lattice convolutional network model of the drosophila visual system. arXiv 1806.04793. [Google Scholar]
- Usher M, McClelland JL (2001) The time course of perceptual choice: the leaky, competing accumulator model. Psychol Rev 108:550–592. 10.1037/0033-295X.108.3.550 [DOI] [PubMed] [Google Scholar]
- Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB (2011) Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol 7:e1001066. 10.1371/journal.pcbi.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P, et al. SciPy 1.0 Contributors (2020) SciPy 1.0: fundamental algorithms for scientific computing in python. Nat Methods 17:261–272. 10.1038/s41592-020-0772-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Xu C, Lisser A (2014) Bandwidth minimization problem. In: MOSIM 2014, 10ème Conférence Francophone de Modélisation, Optimisation et Simulation, Nancy, France: Colloque avec actes et comité de lecture. internationale. [Google Scholar]
- Watanabe C, Suzuki T (2021) Deep two-way matrix reordering for relational data analysis. arXiv 10.48550/ARXIV.2103.14203. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314:1–340. [DOI] [PubMed] [Google Scholar]
- White OL, Lee DD, Sompolinsky H (2004) Short-term memory in orthogonal neural networks. Phys Rev Lett 92:148102. 10.1103/PhysRevLett.92.148102 [DOI] [PubMed] [Google Scholar]
- Wienecke CF, Leong JC, Clandinin TR (2018) Linear summation underlies direction selectivity in Drosophila. Neuron 99:680–688.e4. 10.1016/j.neuron.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]