Abstract
Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of the ςB transcription factor. We investigated Obg's cellular associations by differential centrifugation of crude B. subtilis extracts, using an anti-Obg antibody as a probe to monitor Obg during the fractionation, and by fluorescent microscopy of a B. subtilis strain in which Obg was fused to green fluorescent protein. The results indicated that Obg is part of a large cytoplasmic complex. In subsequent analyses, Obg coeluted with ribosomal subunits during gel filtration of B. subtilis lysates on Sephacryl S-400 and specifically bound to ribosomal protein L13 in an affinity blot assay. Probing the gel filtration fractions with antibodies specific for ςB and its coexpressed regulators (Rsb proteins) revealed coincident elution of the upstream components of the ςB stress activation pathway (RsbR, -S, and -T) with Obg and the ribosomal subunits. The data implicate ribosome function as a possible mediator of the activity of Obg and the stress induction of ςB.
ςB is a Bacillus subtilis transcription factor that controls the bacterium's general stress regulon, a collection of at least 22 operons whose products confer multiple stress resistances on the organism (11, 24, 34, 37). Induction of the regulon occurs by the activation of ςB itself (15, 38). ςB is present but inactive in unstressed B. subtilis, due to an association with RsbW, an inhibitory anti-ςB protein (4, 5, 8). ςB is released from RsbW when another protein, RsbV, binds to RsbW in its place (8). The availability of RsbV thus determines the activity state of ςB (38). During growth, RsbV is not available to activate ςB due to an RsbW-dependent phosphorylation (2, 8, 38). When cultures are exposed to either physical stress (e.g., heat shock, acid shock, osmotic shock, or ethanol treatment) or a drop in energy charge (entry into stationary phase), stress- or stationary phase-specific phosphatases reactivate RsbV to drive the release of ςB (15, 33, 35, 38, 39, 42). The physical stress pathway of ςB activation is controlled by the products of five genes (rsbR, -S, -T, -U, and -X) which are cotranscribed with rsbV, rsbW, and the ςB structural gene (sigB) as an eight gene operon (1, 6, 10, 14, 15, 35, 41, 42). The operon is constitutively transcribed from a promoter (PA) recognized by B. subtilis' housekeeping ς factor (ςA), with an internal ςB-dependent promoter (PB) enhancing the expression of the rsbV, rsbW, sigB, and rsbX genes during periods of ςB activity (i.e., PA rsbR rsbS rsbT rsbU PB rsbV rsbW sigB rsbX) (4, 6, 15, 41). A model for stress activation of ςB is depicted in Fig. 1. RsbT is the key stress activator for ςB induction (42). In unstressed B. subtilis, RsbT is complexed to RsbS and is inactive. Following exposure of B. subtilis to environmental stress, RsbT becomes empowered to inactivate RsbS by phosphorylation and then activate RsbU, the stress pathway's RsbV-P phosphatase (42). RsbR is thought to mediate the RsbT-RsbS interaction, but its exact role in this process is not clear (1, 10). The stress induction of ςB is limited by RsbX, which can dephosphorylate RsbS-P and reestablish the RsbS-dependent inhibition of RsbT (29, 36, 42).
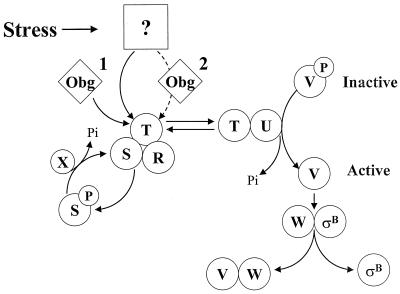
FIG. 1.
Model for stress activation of ςB. ςB is held inactive in unstressed B. subtilis as a complex with an anti-ςB protein, RsbW (W). ςB is freed from RsbW when a release factor, RsbV (V), binds to RsbW. In unstressed B. subtilis, RsbV is inactive due to an RsbW-catalyzed phosphorylation (V-P). Environmental stress activates an RsbV-P phosphatase, RsbU (U), which reactivates RsbV. RsbT (T) is the RsbU activator. RsbT is normally bound to a negative regulator, RsbS (S), which inhibits its activity. RsbR (R) also binds to RsbS and -T and is believed to facilitate their interactions. Upon exposure to stress, RsbT phosphorylates and inactivates RsbS and then activates the RsbU phosphatase. Obg, an essential GTP binding protein, is required for stress to trigger the activation of RsbT. It is unknown whether an Obg-dependent process is a cofactor for stress to activate RsbT (step 1) or whether stress communicates directly to RsbT through Obg (step 2). RsbS-P is dephosphorylated and reactivated by a phosphatase, RsbX (X), that is encoded by one of the genes downstream of the sigB operon's ςB-dependent promoter. RsbX levels become elevated when ςB is active, which may facilitate a return of RsbT to an inactive complex with RsbS. The model is based on references 1, 3, 5, 6, 8, 15, 27, 35, 38, and 42).
The means by which diverse stresses communicate with the components of the ςB induction pathway is unknown. Based on reconstitution studies with Escherichia coli, the Rsb proteins appear to be inadequate, in themselves, to sense stress (e.g., by changes in conformation and/or stability) and activate ςB (28). In E. coli, where stress induction processes are better characterized, protein denaturation and chaperone activation play key roles in communicating environmental stress to stress-responsive transcription factors (reviewed in reference 44). Although similar processes appear to control some stress-induced processes in B. subtilis, i.e., the repressor-mediated regulation of B. subtilis chaperone gene (groEL and dnaK) expression (21, 22), we and others have found no obvious correlation between chaperone activity and ςB induction (22, 28). These results argue that the mechanism that communicates environmental stress to the ςB induction pathway is likely to employ novel Bacillus-specific factors.
By using the yeast Gal4 dihybrid system to identify B. subtilis proteins that could interact with Rsb proteins, a GTP binding protein, Obg, was discovered to be an Rsb interactor and a necessary factor for stress activation of RsbT (27). Members of the Obg subfamily of GTP binding proteins have been found in a number of bacteria (described in reference 23), where they are speculated to monitor the state of intracellular GTP levels and serve as a switch to promote growth when associated with GTP but not when bound to GDP (18, 19, 23). Obg's explicit function is unknown, but it is essential for both B. subtilis growth and sporulation (17, 30, 32, 40).
Given the Obg requirement for stress activation of ςB, we sought to learn more of Obg's properties with the expectation that such data could provide clues as to how stress triggers ςB induction. In the present study we describe the fractionation of crude B. subtilis extracts and the discovery that Obg cofractionates with the bacterium's ribosomes, binding specifically to ribosome protein L13. A similar fractionation analysis of ςB and its Rsb regulators revealed that approximately half of the extracts' RsbR and RsbS, as well as most of the detectable RsbT, elute in the Obg-ribosome fractions. These data present the possibility that ribosome-mediated processes are involved in both the function of Obg and the generation of the signal for stress activation of ςB.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. The BSA and BSJ strains are derivatives of PY22. Bacteria were grown in Luria-Bertani medium (LB) (25) or Difco sporulating medium at 37°C with shaking. Transformation of competent B. subtilis was performed as described by Yasbin et al. (43).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Construction or source |
|---|---|---|
| Bacillus strains | ||
| PY22 | trpC2 | Laboratory strain |
| SMY | trpC | Laboratory strain |
| BSA46 | trpC2 SPβ ctc::lacZ | 3 |
| BSJ37 | trpC2 PSPAC::obg::mgfp | pJM46→PY22 |
| S28Egm | PSPAC::sigE55::mgfp | p28Egm→SMY |
| S28gm | PSPAC::mgfp | p28gm→SMY |
| Plasmids | ||
| pT7-5 | bla PT7 | J. Baseman, University of Texas Health Science Center at San Antonio |
| pDG28 | bla erm PSPAC lacI | P. Stragier, Institut de Biologie Physico-Chimie, Paris, France |
| pF1 | PdacF::sigE55::gfp | 13 |
| pMUTGFP2 | gfp F64L S65T (mgfp) | P. Fawcett, University of Georgia, Athens |
| pJM32 | bla PT7::obg[his]6 | This study |
| pJM46 | bla erm PSPAC::obg::mgfp lacI | This study |
| pJM55 | bla PT7::rplM bla | This study |
| p28gm | bla erm PSPAC::mgfp lacI | This study |
| p28Egm | bla erm PSPAC::sigE55::mgfp lacI | This study |
| pSI1-sigE | cat PSPAC::sigE lacI | 12 |
Construction of green fluorescent protein (GFP) fusions.
mgfp was PCR amplified from plasmid pMUTGFP2 by using the oligonucleotide primers 5′GFPXba (TGGTACCTCTAGAAAAA) and 3′GFPSphI (GGCTGCAGGCATGCTACGAATGC). The resulting 700-bp fragment was cloned into pDG28 downstream of PSPAC by using the 5′ XbaI and 3′ SphI sites inserted during the amplification (p28gm). obg was PCR amplified from PY22 chromosomal DNA by using Obg5′HIII (TGATTGAAGCTTGGGTTGGAC) and Obg3′XbaGFP (CAACTTGATCTAGATCAATAAATTC) primers. The resulting 1.2-kb piece contained 40 bp upstream of obg, including the ribosomal binding site but with the Obg termination codon eliminated. The fragment was cloned into the HindIII and XbaI sites of p28gm. This created an in-frame translational fusion of Obg::mGFP downstream of PSPAC (pJM46). The fusion was verified by DNA sequencing. p28Egm was formed by PCR amplification of sigE55::mgfp from pF-1 using 5′sigEDIII (TCGGGCAAGCTTGTCAAACA) and 3′GFPSphI. The piece was cloned into the HindIII and SphI sites of pDG28 by using sites introduced in the amplification. mgfp was removed from p28Egm by using XbaI and SphI and inserted into pDG28 by using the same restriction sites to create p28gm (PSPAC::mgfp).
Construction of the L13-expressing plasmid vector.
rplM was PCR amplified from PY22 chromosomal DNA by using the oligonucleotide primers rplM5′Eco (GTGTTGTGAATTCGAACGTAATCG) and rplM3′Bam (ACACGGGATCCAGAGCTTTTACG), to yield a 575-bp piece that included the ribosomal binding site of rplM. The fragment was cloned into pT7-5 by using EcoRI and BamHI sites that were introduced during the PCR amplification. This placed rplM downstream of the vector's inducible promoter as plasmid pJM55.
Preparation of Obg-[His]6 antigen and antibody production.
Obg was amplified using the oligonucleotides Obg5′Bam (GGCAGAATGGATCCGAGGACG) and Obg3′6His (ATTGGATCCTTAATGATGATGATGATGATGATCAATAAATTCAAATTCAA) to generate a 1.4-kb piece containing the ribosomal binding site and the complete obg sequence plus a stretch of six histidine codons at its 3′ end. The fragment was cloned, using BamHI and PstI sites that had been introduced in the amplification, into pT7-5 (pJM32). The construction was verified by DNA sequencing. Obg-[His]6 was overexpressed in E. coli BL21(DE3)(pLysS) as follows. The recombinant strain was grown in LB to an optical density at 540 nm (OD540) of 0.7. IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and the culture was incubated for an additional 5 h. The cells were harvested over ice chips, and the protein was purified as described previously (4), using an Ni-nitrilotriacetic acid resin and a denaturing buffer. The antigen was extensively dialyzed into phosphate-buffered saline and used to immunize BALB/c mice. The antibodies were produced as previously described (4), using the non-immunoglobulin-secreting NS1 BALB/c myeloma cell line to produce hybridomas. The resulting antibodies detected a single Obg-size protein band in crude B. subtilis and E. coli extracts prepared from strains expressing obg (data not shown).
Renaturation of Obg-[His]6.
After elution from Ni-nitrilotriacetic acid resin, the fractions containing [His]6 protein were pooled and renatured as described by Burgess (7). The fractions were dialyzed against buffer A (50 mM Tris-HCl [pH 7.9], 5% glycerol, 0.1 mM EDTA, 0.1 mM dithiothreitol [DTT], 50 mM NaCl) with 0.4% Sarkosyl for 2 h at 4°C. The sample was then diluted 10-fold with buffer A without Sarkosyl in 2-fold increments every 10 to 15 min with stirring at 4°C. The diluted sample was then dialyzed into buffer A without Sarkosyl at 4°C overnight. The dialyzed Obg solution was spun at 8,000 rpm for 30 min to remove aggregated protein. The supernatant was loaded onto a DEAE-Sepharose column, which was washed with buffer (150 mM Tris [pH 7.5], 1 mM EDTA) and then eluted using a linear gradient of KCl (0 to 0.5 M) in column buffer. Fractions containing Obg were pooled, dialyzed into storage buffer (50 mM Tris [pH 7.9], 50% glycerol, 0.1 mM EDTA, 0.1 mM DTT, 50 mM NaCl), and held at −20°C until needed.
Obg affinity blotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and incubated for 30 min at room temperature in BLOTTO without NaCl (10 mM Tris [pH 7.6], 50 μM EDTA, 1.5 mM MgCl2, 2.5% milk). Obg-[His]6 was added to a concentration of 2.5 μg of protein/ml, and the incubation was continued for 2 h. The blots were then rinsed extensively with double-distilled water (ddH2O) and incubated with anti-Obg polyclonal immunoglobulin. Bound antibody was detected using an alkaline phosphatase-conjugated goat immunoglobulin against mouse immunoglobulin G (American Qualex).
Gel filtration chromatography.
One liter of exponentially growing B. subtilis (OD540 = 0.5) was harvested into an equal volume of ice chips, concentrated by centrifugation 400-fold in low-salt buffer (10 mM Tris [pH 8.0], 50 μM EDTA, 1.5 mM MgCl2, 1 mM DTT) supplemented with 0.03% phenylmethylsulfonyl fluoride, and disrupted by passage through a French pressure cell. The cell extract was centrifuged for 10 min at 8,000 rpm to remove cell debris. A 2.5-ml portion of the supernatant was loaded onto a 250-ml Sephacryl S-400 column (Sigma) and eluted at 4°C using low-salt buffer. Fractions of 2.5 ml were collected. Aliquots of the fractions were ethanol precipitated and analyzed by Western blot techniques.
Fluorescence microscopy.
Fluorescent images were obtained as described previously (13). Cells from a colony that had formed overnight at 30°C on an LB agar plate supplemented with IPTG (100 mM) were suspended in 1 μl of water on a microscope slide and compressed tightly with a coverslip. Cells were viewed with a Zeiss Axiophoto epifluorescence microscope. Images were captured with an AlphaImager 2000 (Alpha Innotech Corp., San Leandro, Calif.) and processed with Adobe Photoshop version 4.0.
Protein sequencing.
The proteins of interest were separated by SDS-PAGE and transferred to a 0.22-μm-pore-size polyvinylidene difluoride membrane (Micron Separations, Inc.) by electrophoresis for 90 min in transfer buffer (25 mM Tris, 192 mM glycine, 15% methanol), as described by the company. After transfer, the membrane was rinsed with ddH2O, and stained with 0.2% Ponceau-S in 1% acetic acid for 5 min. After destaining in ddH2O, the protein bands of interest were excised and microsequenced by the Protein Core Facility at the University of Texas Health Science Center at San Antonio, using Edman degradation. Sequences obtained were compared to the B. subtilis Genome Database (http://www.pasteur.fr/Bio/SubtiList) for identification of the proteins.
Sedimentation analyses.
B. subtilis extracts, prepared in 10 mM Tris (pH 8.0)–50 μM EDTA–10 mM MgCl2–1 mM DTT–0.03% phenylmethylsulfonyl fluoride by disruption through a French pressure cell, were centrifuged twice for 10 min at 10,000 rpm in a Sorvall SS-34 rotor to remove cell debris. The supernatant was then diluted into either the same buffer, buffer containing 0.5 M KCl, or buffer containing 0.5% Triton X-100 and centrifuged for 2 h at 40,000 rpm and 4°C in an SW 50.1 rotor. Equivalent portions of pellet and supernatant fractions were analyzed by Western blotting.
RESULTS
Sedimentation analysis of Obg in crude B. subtilis extracts.
Ultracentrifugation and immunoelectron microscopy studies argued that the Obg ortholog of Streptomyces coelicolor is a membrane-associated protein (23). We therefore analyzed crude B. subtilis extracts by differential centrifugation to ask whether B. subtilis Obg might be similarly membrane bound. In order to monitor Obg's distribution in the crude cell extracts, we prepared anti-Obg antibodies for use as probes. As controls in this experiment, we used two gene products, pro-ςE and RsbX, for which we had specific antibody probes and which should display distinct sedimentation properties. pro-ςE, the inactive precursor to a sporulation-specific transcription factor, is tethered to the inner surface of the B. subtilis cytoplasmic membrane (13), the proposed cellular location for S. coelicolor Obg (23). RsbX, a ςB regulatory protein which does not associate with any cellular component, should serve as a marker for free cytoplasmic elements (9).
Crude extracts of vegetatively growing B. subtilis, expressing pro-ςE from an inducible promoter, were prepared by disruption in a low-salt buffer and centrifuged under conditions (approximately 150,000 × g for 2 h) which would pellet membrane and large subcellular components. The pelleted and supernatant fractions were probed by Western blotting using antibodies specific for the Obg, pro-ςE, and RsbX proteins.
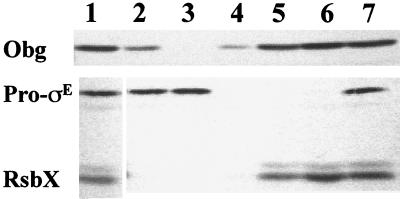
Virtually all of the pro-ςE, a portion of the Obg, and none of the RsbX were pelleted by our centrifugation conditions (Fig. 2, lane 2 [pellet] versus lane 5 [supernatant]). When the experiment was repeated using an extract that had been prepared in a high-salt buffer (0.5 M KCl), the sedimentation of pro-ςE was unaffected, but, as was seen in the Streptomyces Obg study (23), the Obg was no longer pelleted (Fig. 2, lanes 3 and 6). A similar centrifugation experiment done in the presence of Triton X-100 (0.5%) blocked the pelleting of pro-ςE but had only a partial effect on the sedimenting of Obg (Fig. 2, lane 4). The failure of Triton to solubilize Obg was unexpected. If Obg had been pelleted in the low-salt buffer due to an electrostatic (i.e., salt-sensitive) interaction with a membrane component, the addition of detergent should extract the component, as it did for pro-ςE, and allow Obg to remain in the supernatant. This result argues that the pelleting of Obg that we observed in these experiments is not necessarily an indication of membrane association. The B. subtilis Obg may be tethered to the cytoplasmic membrane, but if so, it must still be as part of a complex that is not dissociated by detergent and is sufficiently large to be pelleted by the centrifugation conditions that we employed.
FIG. 2.
Sedimentation analysis of B. subtilis extracts. PY22(pSI1-sigE), grown in LB plus 1 mM IPTG, was harvested late in exponential phase (OD540, 0.6). Extracts were prepared and subjected to ultracentrifugation (2 h, 40,000 rpm, SW50.1 rotor) as described in Materials and Methods. Equivalent amounts of unfractionated extract (lane 1), pellet (lanes 2 to 4), and supernatant (lanes 5 to 7) fractions were analyzed by Western blotting using antibodies specific for Obg, pro-ςE, and RsbX. Lanes 2 and 5, low-salt buffer; lanes 3 and 6, 0.5 M KCl; lanes 4 and 7, 0.5% Triton.
An Obg::GFP fusion protein is cytoplasmic.
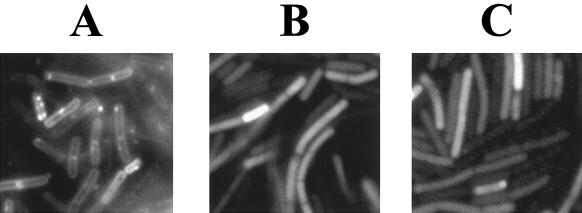
We had previously used a fusion of pro-ςE to GFP to visualize its membrane association in B. subtilis (13). This prompted us to construct a chimera of the full-length Obg protein to GFP to ask whether a similar fusion to Obg might also reveal Obg's placement on the B. subtilis membrane. B. subtilis cells expressing Obg::GFP as their sole source of Obg are viable (data not shown). Thus, the Obg::GFP is able to provide the essential Obg function and likely occupies the normal Obg site within the cell. B. subtilis expressing either pro-ςE::GFP, Obg::GFP, or GFP without additional B. subtilis sequences was examined by fluorescence microscopy (Fig. 3). The pro-ςE::GFP displayed the anticipated membrane location (Fig. 3A); however, the Obg::GFP did not (Fig. 3C), resembling instead the case for a B. subtilis strain that expressed free GFP as a cytoplasmic protein (Fig. 3B). This result, when taken with the detergent resistance of the fast-sedimenting Obg complex, indicates that B. subtilis Obg protein is more likely to be associated with a large cytoplasmic component rather than with the B. subtilis cell membrane.
FIG. 3.
Fluorescence microscopy of B. subtilis expressing Obg::Gfp. B. subtilis strains expressing pro-ςE::GFP (PSPAC::sigE55::mgfp) (A), gfp (PSPAC::mgfp) (B), or Obg::GFP (PSPAC::obg::mgfp) (C) were visualized by fluorescence microscopy as described in Materials and Methods.
Obg cofractionates with ribosomes.
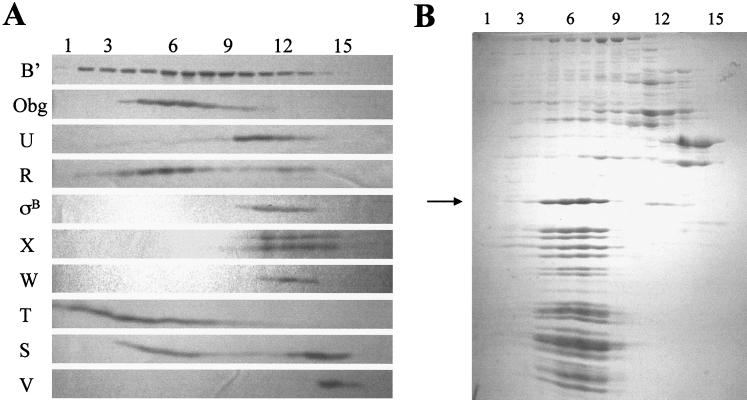
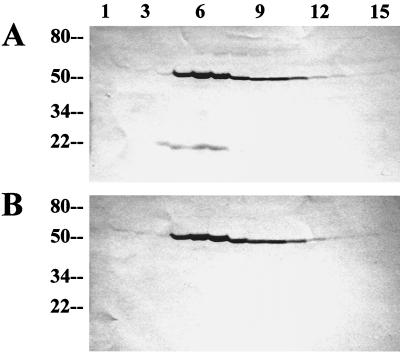
In an attempt to identify the putative cytoplasmic component with which Obg associates in crude B. subtilis extracts, we fractionated a portion of such an extract by gel filtration in a low-salt buffer on Sephacryl S-400 (fractionation range, 8 × 103 to 20 × 103 kDa). Western blot analyses of the fractions obtained revealed that the peak of Obg protein was exiting the column slightly before the peak of RNA polymerase (approximately 5 × 105 Da) (Fig. 4A, row β′ versus row Obg). When the protein components of these fractions were visualized by Coomassie blue staining, abundant low-molecular-weight proteins (<30 kDa) were found to peak coincident with Obg (Fig. 4B). The sizes of complexes eluting at this position of the gradient are difficult to determine with accuracy, due to the lack of adequate molecular mass standards in this size range; however, the Obg peak occurs around the fractions at which a blue dextran marker (average mass, 2 × 106 Da) exited the column. The large size of the complex with which Obg is associated (approximately 2 × 106 Da) and the abundance of low-molecular-mass proteins in these same fractions suggest that the Obg-associated complex could include the extract's ribosomal subunits. To test whether the coeluting proteins were ribosomal subunits, we extracted the largest of the abundant proteins (approximately 30 kDa), whose elution profile coincided with that of Obg (Fig. 4B) and subjected it to amino-terminal sequencing. The partial sequence of this protein (AIKKYKPT) is a seven-of-eight match with the known sequence (MAIKKYKPS) of the 30.2-kDa rplB gene product, the 50S ribosomal protein, L2 (Bacillus subtilis Genome Database [http://www.pasteur.fr/Bio/SubtiList]). Thus, Obg appears to be eluting from the gel filtration column in the same fractions as the extract's ribosomal subunits.
FIG. 4.
Gel filtration chromatography fractions of crude B. subtilis extracts. BSA46 was harvested in exponential phase (OD540 = 0.5). Extracts were prepared as described in Materials and Methods and loaded onto a Sephacryl S-400 column. Samples (0.5 ml) of the included fractions were ethanol precipitated and analyzed. Numbers at the top indicate fraction numbers, with 1 being the earliest-eluting fraction. (A) Western blot of the fractions probed with antibodies specific for the proteins indicated on the left. (B) Coomassie blue-stained gel of the same fractions. The arrow indicates the largest abundant protein, which was subjected to amino-terminal sequencing (see text).
Obg binds to ribosomal protein L13.
The observation that Obg exits the Sephacryl column coincident with the ribosomal subunits implies the Obg either is associated with ribosomal proteins or is complexed to a similarly sized entity. In an attempt to identify the components to which Obg was joined in the Sephacryl fractions, we modified our Western blot protocol for use as an affinity blot assay. The gel filtration fractions, illustrated in Fig. 4B, were separated by SDS-PAGE. Proteins from duplicate gels were transferred to nitrocellulose membranes which were then blocked, as described in Materials and Methods, with BLOTTO lacking added NaCl. Before probing with anti-Obg antibody, one of the nitrocellulose membranes was incubated with purified Obg-[His]6. We thought it possible that Obg might bind to whatever protein was responsible for its unusual position of elution from the gel filtration column, even though that protein was now immobilized on nitrocellulose. After several low-salt washes, both of the nitrocellulose membranes were probed with anti-Obg antibody. Aside from the band at the position of Obg itself, the Western blot revealed a band at the position of a protein of approximately 20 kDa on the nitrocellulose membrane that had been preincubated with Obg (Fig. 5). The abundance of this band peaked in concert with that of the Obg protein in the elution profile from the gel filtration column, a characteristic expected of the protein interaction responsible for Obg's presence in these fractions.
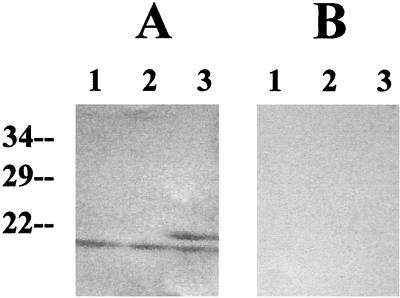
FIG. 5.
Obg affinity blot of gradient fractions. The gel filtration gradient fractions from Fig. 4 were separated by SDS-PAGE and transferred to nitrocellulose. The nitrocellulose membranes with transferred proteins were processed as described in Materials and Methods. The membrane depicted in panel A was incubated with Obg-[His]6 before the polyclonal antibody to Obg was added, while that depicted in panel B was not. Numbers on the left correspond to protein molecular masses in kilodaltons; numbers at the top represent the gradient fraction present in each lane.
The protein band, which electrophoresed to a position equivalent to that of the band that bound Obg in the affinity blot experiment, was excised from a companion gel and sequenced. The amino-terminal sequence of the putative Obg binding protein was determined to be MRTTPMAASTI. This matches the amino-terminal sequence of the B. subtilis rplM gene product (MRTTPMANASTI) (Bacillus subtilis Genome Database [http://www.pasteur.fr/Bio/SubtiList]). rplM encodes the 50S ribosome subunit L13. In order to verify that the protein to which Obg bound was L13 and not an undetected contaminant at a similar position in the original gel, the rplM gene was amplified from the B. subtilis chromosome, cloned into an E. coli vector (pT7-5), and expressed in E. coli. Extracts prepared from E. coli cultures that either expressed or did not express the B. subtilis rplM gene were examined using the Obg affinity assay. A band corresponding in size to L13 was detected on the nitrocellulose blot of the culture extract expressing rplM if the nitrocellulose had been incubated with Obg prior to antibody probing (Fig. 6A, lane 3) and was absent from the blots of extracts that did not express rplM (Fig. 6A, lanes 1 and 2). The band was also not detected in any blots that had not been incubated with Obg protein (Fig. 6B). In addition to the putative RplM band, all of the E. coli extracts contained an Obg binding protein with a slightly higher mobility in SDS-PAGE than the B. subtilis L13. We assume that the protein is the E. coli ribosome protein L13. E. coli L13 is approximately the same size as its B. subtilis homolog and has a very similar structure (74% similar or identical amino acids over its entire length).
FIG. 6.
Obg affinity blot to L13 overexpressed in E. coli. BL21 containing pJM55 (PT7::rplM) was grown until the OD540 reached 0.5. IPTG (1 mM) was added to a portion of the cells, and the cells were incubated for 2 h longer. Cells were pelleted, resuspended in SDS-PAGE loading buffer, and boiled. Each lane contains the equivalent of 200 μl of crude cell extract. Lanes: 1, BL21; 2, BL21/pJM55, uninduced; 3, BL21/pJM55, induced. (A) Incubation with Obg-[His]6 before addition of polyclonal antibody to Obg; (B) no incubation with Obg-[His]6 (see Materials and Methods). Numbers to the left represent protein molecular masses in kilodaltons.
The finding that Obg can bind to the 50S ribosome subunit protein L13 in the affinity blot assay, when taken with the coincident elution of Obg with the ribosomal subunits during gel filtration, argues that Obg's high-molecular-weight association in B. subtilis extracts reflects binding to B. subtilis ribosomes, presumably to the 50S subunit by an interaction that involves at least L13.
Elution of RsbR, -S, and -T with Obg and ribosomal subunits.
Obg was identified as a protein that could interact with several of the ςB regulators and was necessary for environmental stress to trigger the RsbT-dependent activation of a ςB (27). In an earlier study, in which possible interactions among the ςB regulators were analyzed by Sephacryl S-200 chromatography, a significant proportion of the crude extract's RsbR and -S was found in the fractions excluded from the gel matrix (i.e., in aggregates of greater than 250 kDa) (9). These two observations prompted us to analyze the S-400 gel filtration fractions for ςB and the Rsb proteins to determine whether any of the ςB regulators would coelute with Obg in the ribosome fractions.
Proteins from the fractions illustrated in Fig. 4B were probed with monoclonal antibodies against ςB and the seven Rsb regulatory proteins. As can be seen in Fig. 4A, most of the RsbR and approximately half of the RsbS proteins eluted in the fractions containing Obg and the ribosomal subunits. All of the RsbT eluted as an apparent component of high-molecular-weight complexes. Although RsbT was included in the ribosomal protein fractions, it did not peak with the ribosome proteins, as did RsbR and -S. A significant portion of the RsbT appeared to elute as even higher-molecular-weight complexes. It is not evident from the stained gel (Fig. 4B) what these complexes might be. Although the data raise the possibility that RsbT could be part of a very large unknown complex, the abundance of the ribosome proteins and the similarity in size of several of them to RsbT suggest a possible alternative explanation for the displacement of the RsbT peak from the ribosome fractions: that the abundance of the ribosome proteins in these fractions interferes with the transfer and detection of RsbT in the Western blot analysis. Such a situation had initially impaired our ability to reproducibly detect RsbS in these fractions (data not shown), until we employed higher-resolution (10 to 17.5%) gradient gels in the analysis. It is possible that a similar circumstance is restricting our ability to properly gauge the abundance of RsbT in the ribosome fractions; however, repeated alterations in the conditions of electrophoresis have not yet altered RsbT's apparent elution profile (data not shown). Unlike RsbR, -S, and -T, ςB and the remaining Rsb proteins exited the column as the smaller complexes and unbound proteins that we had described previously (9).
In order to verify that the profiles of the Rsb proteins, ribosomal proteins, and Obg reflect their coincident elution as components of complexes of very similar size and not a peculiarity of the filtration characteristics of the S-400 column, we repeated the experiment with a column matrix (Sephacryl S-500) with a greater inclusion limit (40 to 20,000 kDa). When the fractions filtered through this matrix were analyzed, the coelution profiles seen with the S-400 column persisted, although the resolution of the column was not as great (i.e., the proteins were concentrated among fewer fractions) (data not shown).
DISCUSSION
The mechanism by which environmental stress is sensed by B. subtilis and communicated to the regulators of the ςB transcription factor is unknown. The activities of the principal chaperone proteins (DnaK and GroEL), which play important roles in stress activation in other systems, are dispensable for ςB induction (22, 28). Additionally, the known components of the regulatory cascade that activates ςB are inadequate, in themselves, to detect stress and activate ςB (28). Obg, an essential GTP binding protein of unknown function, is the only B. subtilis gene product, aside from the regulators encoded within the sigB operon, that has been shown to be needed for stress to trigger ςB activation (27).
We have now presented evidence that Obg and the most upstream members (RsbR, RsbS, and possibly RsbT) of the ςB stress pathway regulators cofractionate with ribosomes in crude B. subtilis extracts and that Obg can specifically bind to ribosomal protein L13 in an affinity blot assay. This raises the intriguing possibility that ribosome-mediated processes are involved both in Obg function and in stress-dependent signaling to the Rsb proteins. The finding that Obg associates with ribosomal subunits is not startling. Bacterial GTP binding proteins have long been associated with translational processes (14, 20). In addition, an E. coli GTP binding protein (Era), which is similar to Obg in being a small monomeric GTPase with an unknown essential function, has recently been found to bind to ribosomes (26). Assuming that the interaction between Obg and ribosomes, which we observe in crude extracts, has biological significance, it is unclear whether the interaction reflects Obg communicating to the ribosome or receiving a regulatory input from that structure. Members of the Obg subfamily of GTP binding proteins are thought to monitor the state of GTP levels in bacteria and serve as a switch to promote growth when bound to GTP but not when associated with GDP (18, 19, 23). In vitro, purified Obg proteins from B. subtilis (Obg) and Caulobacter crescentus (CgtA) have high GDP-GTP dissociation-exchange rates (Kd, ∼1.5 s−1) but relatively low GTP hydrolysis rates (half-life, ∼23 min for CgtA) (18, 40). This finding suggests that the nucleotide state of CgtA or Obg is controlled by the intracellular GDP-GTP pool rather than by these proteins' GTPase activities (18). Thus, members of the Obg group would respond to changes in the GTP level and communicate these changes to cell processes. In the case of B. subtilis, this communication could be to the ribosome to influence continued growth and, either directly or indirectly, to both the sporulation and ςB induction pathways.
Although this may be the simplest model, it is possible that the GTPase activity of Obg, like those of other members of the Ras family GTP binding proteins, is stimulated by undefined GTPase-activating proteins (20). It is plausible that a ribosome component could have GTPase-activating activity and stimulate the GTPase of Obg under particular conditions of altered translation. In such a model, the signal would not be sent to the ribosome by Obg but rather would be sent from the ribosome via Obg to other cellular functions. The sorting out of these possibilities awaits detailed biochemical analyses.
Although the directionality of putative signaling between Obg and the ribosome is at present arbitrary, the likely directionality of any potential ribosome-Rsb interaction is less so. The finding that Obg, as well as all three of the most upstream Rsb components of the ςB stress activation pathway, is found in ribosome-containing fractions suggests that some stress-induced perturbation in translation (i.e., changes in translation initiation, ribosome-associated chaperone activity, or nascent protein misfolding, etc.) could be a component of whatever signal is being sensed by the Rsb proteins to activate ςB. Ribosomes have, for example, been implicated as sensors of heat and cold shock in E. coli (31). Perhaps they are more general stress sensors in B. subtilis. In such a model a ribosome-mediated process becomes the unknown stress target represented in Fig. 1. Whether the putative signal is conveyed directly to the Rsb proteins by the ribosomes, with Obg as a required secondary input (Fig. 1, step 1), or through Obg to the Rsb proteins (Fig. 1, step 2) remains to be determined. Studies to test the biological relevance of Rsb and Obg protein binding to ribosomes, as well as to determine possible targets for RsbR, -S, and -T within the ribosome, are under way.
ACKNOWLEDGMENT
This work was supported by NIH grant GM-48220.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 3.Benson A K, Haldenwang W G. Characterization of a regulatory network that controls ςB expression in Bacillus subtilis. J Bacteriol. 1992;174:749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess R R. Purification of overproduced E. coli RNA polymerase ς factors by solubilizing inclusion bodies and refolding in sarkosyl. Methods Enzymol. 1996;273:145–149. doi: 10.1016/s0076-6879(96)73014-8. [DOI] [PubMed] [Google Scholar]
- 8.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-ς factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour A, Voelker U, Voelker A, Haldenwang W G. Relative levels and fractionation properties of Bacillus subtilis ςB and its regulators during balanced growth and stress. J Bacteriol. 1996;178:3701–3709. doi: 10.1128/jb.178.13.3701-9sigma.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaidenko T A, Yang X, Lee Y M, Price C W. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the stress signaling pathway of Bacillus subtilis. J Mol Biol. 1999;288:29–39. doi: 10.1006/jmbi.1999.2665. [DOI] [PubMed] [Google Scholar]
- 11.Hecker M, Schumann W, Voelker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 12.Jonas R M, Peters III H K, Haldenwang W G. Phenotypes of Bacillus subtilis mutants altered in the precursor-specific region of ςE. J Bacteriol. 1990;172:4178–4186. doi: 10.1128/jb.172.8.4178-4186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju J, Luo T, Haldenwang W G. Bacillus subtilis pro-ςE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J Bacteriol. 1997;179:4888–4893. doi: 10.1128/jb.179.15.4888-4893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaziro Y, Itoh H, Kazasa T, Nakafuku M, Satoh T. Structure and function of signal-transducing GTP-binding proteins. Annu Rev Biochem. 1991;60:349–400. doi: 10.1146/annurev.bi.60.070191.002025. [DOI] [PubMed] [Google Scholar]
- 17.Kok J, Trach K A, Hoch J A. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J Bacteriol. 1994;176:7155–7160. doi: 10.1128/jb.176.23.7155-7160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin B, Covalle K L, Maddock J R. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J Bacteriol. 1999;181:5825–5832. doi: 10.1128/jb.181.18.5825-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddock J, Bhatt A, Koch M, Skidmore J. Identification of an essential Caulobacter cresentus gene encoding a member of the Obg family of GTP-binding proteins. J Bacteriol. 1997;179:6426–6431. doi: 10.1128/jb.179.20.6426-6431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.March P E. Membrane-associated GTPases in bacteria. Mol Microbiol. 1992;6:1253–1257. doi: 10.1111/j.1365-2958.1992.tb00845.x. [DOI] [PubMed] [Google Scholar]
- 21.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mogk A, Voelker A, Engelmann S, Hecker M, Schumann W, Voelker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto S, Ochi K. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol Microbiol. 1998;30:107–119. doi: 10.1046/j.1365-2958.1998.01042.x. [DOI] [PubMed] [Google Scholar]
- 24.Petersohn A, Bernhardt J, Gerth U, Hoper D, Koburger T, Voelker U, Hecker M. Identification of ςB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J Bacteriol. 1999;181:5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sayed A, Matsuyama S, Inouye M. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem Biophys Res Commun. 1999;264:51–54. doi: 10.1006/bbrc.1999.1471. [DOI] [PubMed] [Google Scholar]
- 27.Scott J M, Haldenwang W G. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor ςB. J Bacteriol. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott J M, Smirnova N, Haldenwang W G. A Bacillus-specific factor is needed to trigger the stress-activated phosphatase/kinase cascade of ςB induction. Biochem Biophys Res Commun. 1999;257:106–110. doi: 10.1006/bbrc.1999.0418. [DOI] [PubMed] [Google Scholar]
- 29.Smirnova N, Scott J, Voelker U, Haldenwang W G. Isolation and characterization of Bacillus subtilis sigB operon mutations that suppress the loss of the negative regulators RsbX. J Bacteriol. 1998;180:3671–3680. doi: 10.1128/jb.180.14.3671-3680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trach K, Hoch J A. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J Bacteriol. 1989;171:1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Bogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidwans S J, Ireton K, Grossman A D. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3308–3311. doi: 10.1128/jb.177.11.3308-3311.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PA5 domain is required to convey signals of energy stress to the ςB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 34.Voelker U, Engelmann S, Maul B, Riethdorf S, Voelker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 35.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelker U, Luo T, Smirnova N, Haldenwang W G. Stress activation of Bacillus subtilis ςB can occur in the absence of the ςB negative regulator RsbX. J Bacteriol. 1997;179:1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voelker U, Maul B, Hecker M. Expression of the ςB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol. 1999;181:3942–3948. doi: 10.1128/jb.181.13.3942-3948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voelker U, Voelker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsh K, Trach K A, Folger C, Hoch J A. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J Bacteriol. 1994;176:7161–7168. doi: 10.1128/jb.176.23.7161-7168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 43.Yasbin R E, Wilson G A, Young F E. Transformation and transfection of lysogenic strains of Bacillus subtilis 168. J Bacteriol. 1973;113:540–548. doi: 10.1128/jb.113.2.540-548.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]