Abstract
Objective
We propose a system, quEHRy, to retrieve precise, interpretable answers to natural language questions from structured data in electronic health records (EHRs).
Materials and Methods
We develop/synthesize the main components of quEHRy: concept normalization (MetaMap), time frame classification (new), semantic parsing (existing), visualization with question understanding (new), and query module for FHIR mapping/processing (new). We evaluate quEHRy on 2 clinical question answering (QA) datasets. We evaluate each component separately as well as holistically to gain deeper insights. We also conduct a thorough error analysis for a crucial subcomponent, medical concept normalization.
Results
Using gold concepts, the precision of quEHRy is 98.33% and 90.91% for the 2 datasets, while the overall accuracy was 97.41% and 87.75%. Precision was 94.03% and 87.79% even after employing an automated medical concept extraction system (MetaMap). Most incorrectly predicted medical concepts were broader in nature than gold-annotated concepts (representative of the ones present in EHRs), eg, Diabetes versus Diabetes Mellitus, Non-Insulin-Dependent.
Discussion
The primary performance barrier to deployment of the system is due to errors in medical concept extraction (a component not studied in this article), which affects the downstream generation of correct logical structures. This indicates the need to build QA-specific clinical concept normalizers that understand EHR context to extract the “relevant” medical concepts from questions.
Conclusion
We present an end-to-end QA system that allows information access from EHRs using natural language and returns an exact, verifiable answer. Our proposed system is high-precision and interpretable, checking off the requirements for clinical use.
Keywords: question answering, electronic health records, natural language processing, artificial intelligence, machine learning, FHIR
BACKGROUND AND SIGNIFICANCE
A tremendous amount of useful patient information in electronic health records (EHRs) is frequently accessed by clinicians to provide care. However, issues associated with EHRs (related to their usability1 and navigation2) hinder accessing information from these systems.3 Efforts to tackle these issues rely on visualization4 (eg, showing information as charts) or information retrieval5 (IR) (surfacing information through keyword-based searches). Though such methods improve information access, for a given information need (eg, lab value, procedure status), they still present users excess information (eg, a table full of lab values, a long list of procedures) over what is needed (a single lab value or procedure). In other words, these methods are unable to grasp the exact information needs of users. To this end, question answering (QA) provides a natural way to identify information needs and return an exact, verifiable answer.6
Information in EHRs is present in 2 primary formats: structured (eg, lab values) and unstructured (eg, clinical notes), where each type requires different tools for querying. Unstructured notes certainly contain a wealth of patient information, and they are highly amenable to many state-of-the-art QA methods.7–9 Importantly, the methods for unstructured data are not amenable to structured data, which is also far less studied.
Structured information is generally stored in databases with sophisticated schemas. Keyword-based searches are unable to take full advantage of such schemas as they simply match text.10 However, the power underlying the complexity of such schemas can be harnessed using query mechanisms understood by such databases (eg, SQL, FHIR).11 However, most clinicians are not well-versed with such query languages and it would be burdensome to train them to interact using such queries. Instead, a more intuitive approach is to let clinicians pose information needs using natural language to a system capable of retrieving answers to those exact needs from EHRs.12,13
EHR-based IR is well-studied,5,10,14–16 where a user is presented with a list of results, which may not even contain an answer.17 On the contrary, our QA system aims to return a single, exact, verifiable answer (focusing on precision). This corresponds to the QA task known as factoid QA, which is amenable to the types of structured data found in EHRs. Notably, in order to have a single, exact answer, factoid-type questions are relatively unambiguous and have comparatively simple syntactic structures (which aligns well with our methodology). Moreover, traditional EHR IR systems are largely tailored for unstructured EHR data18 and fail to take advantage of the full set of capabilities offered by structured databases (present in EHRs). There are efforts to use semantics with IR to include structured EHR data in the searches.5,10,16 However, these methods still depend on text-based searches, and thus are unable to grasp exact information needs. Furthermore, such simple text searching techniques are unable to provide any feedback on how user queries are interpreted. Our system, on the contrary, is able to show an exact representation of predicted information needs for the input question.
At a high level, most biomedical QA systems are focused on either biomedical literature data,19,20 consumer health data,21–23 or EHR data.7,8 Most biomedical literature QA systems focus on identifying medical evidence for clinicians or researchers, while consumer health QA systems focus on providing easy-to-understand medical information to nonexperts. Surveys of biomedical QA systems24,25 indicate that EHR QA methods have received relatively little attention, and no system to our knowledge has presented an end-to-end QA system that could be integrated directly into an EHR. Most prior work in EHR QA for structured data is aimed at creating datasets (manually26–29 or automatically7,30–32), or focuses on individual components and not an end-to-end EHR QA solution. Most studies deal with the semantic parsing component (mapping questions to logical forms [LFs]) using rule-based,26,33,34 traditional machine learning,35 and advanced deep learning30,36 techniques. Still, identifying a machine-understandable LF is not practically useful unless it is mapped to an EHR-understandable query language to retrieve actual answers. One of the main contributions of this work includes designing and developing a query module that is capable of handling a wide variety of logical constructs and FHIR resources.
Other essential parts of end-to-end EHR QA for structured data include concept normalization and time frame classification. Concept normalization is a well-studied task in the clinical domain,37–42 however it is largely explored independent of a target application (such as QA in our case). To our knowledge, no work has focused on this task for clinical questions. Similarly, temporal information extraction is a widely explored topic,43,44 yet no previous work has pursued this task for EHR QA.
To our knowledge, no previous work has looked at the problems and challenges underlying EHR QA as an end-to-end process that starts from a question and ends at an exact answer. It is critical to tackle EHR QA in a practical, end-to-end, manner as ultimately that is how these systems will be useful in practice. This is accomplished by converting the question to a corresponding machine-understandable form that then is mapped to FHIR45 (Fast Healthcare Interoperability Resources) queries (an EHR query language). Additionally, our system is interpretable by displaying understood information needs in the form of an easily and rapidly understandable diagram. Such interpretability is of paramount importance in the clinical domain as it enables clinicians to understand and rely on such advanced query mechanisms.46–48 Moreover, quEHRy is a high-precision system that, to the best of its ability, refrains from giving an incorrect answer. Like the important principle of “non-maleficence” in medical ethics to “do no harm,” quEHRy strives to “give no incorrect answer.” Specifically, rather than providing likely incorrect answers to the clinician, it favors responses such as “No answer available” or “Unable to understand the question,” thereby further increasing trust in the system.49
OBJECTIVE
Our objectives of this article are to:
Build an end-to-end EHR QA pipeline taking in a natural language question and returning an exact answer with high precision along with an interpretable visualization highlighting underlying information needs.
Convert LFs to FHIR queries for fetching structured information from EHRs.
Integrate a third-party concept normalizer, a new time frame classifier, and an existing semantic parser for end-to-end LF generation.
Evaluate the QA components in isolation and end-to-end on 2 clinical datasets for a comprehensive performance assessment.
While the data28,29 and the semantic parsing component35 have been published separately, all other parts of the QA system, including the contributions listed above, are novel. Further, we make our datasets and source code publicly available.
DATA
We use 2 clinical QA datasets in this study.28,29 Each consists of questions answerable using EHR data along with their corresponding LFs. An LF represents the meaning of a natural language utterance unambiguously (thus, machine-understandable). In our datasets, these representations are based on λ-calculus, consisting of logical predicates and their arguments. There are 2 broad categories of logical predicates, concept and nonconcept predicates, which differ in terms of function. Concept predicates (eg, has_concept()) retrieve information from the EHR while nonconcept predicates (eg, latest()) manipulate this information. There may be many or few predicates in a LF depending upon the complexity or information requirement of a question. Concept predicates assume 3 types of arguments: concept variable (an instantiation of the concept type), concept code (from standard ontology such as Unified Medical Language System [UMLS]), and implicit time frame (temporal restrictions on the events such as visit for events that happened during the current encounter). For other types of predicates, arguments such as location references (eg, BICU) and measurements (eg, 38C) are used to supply the required information for retrieving an exact answer. For instance, a natural language question “What is the result of the CT scan?” is semantically represented as LF “latest(λx.has_concept(x, 0040405, visit))” where latest() and has_concept() are logical predicates with x (a variable of type concept), 0040405 (UMLS concept unique identifier [CUI] for X-Ray Computed Tomography which is matched with the concept codes found within the FHIR resources) and visit (implicit time frame signifying the current hospital visit) as arguments. Table 1 provides more examples, while further details can be found in prior work.27,29
Table 1.
Examples and descriptive statistics
| (a) Examples from the datasets used in this study. Q: question; LF: logical form. | |
|---|---|
| Corpus | Example questions with logical forms |
| FHIRDATA | Q: Does the patient have any history of Otitis media? |
| LF: delta(λx.has_concept(x, C0029882, history)) | |
| Q: When did she last visit the clinic? | |
| LF: time(latest(λx.has_concept(x, C0422299, pmh))) | |
| Q: What was the dose for amoxicillin? | |
| LF: dose(latest(λx.has_concept(x, C1298551, pmh))) | |
| Q: When did the Alzheimer’s disease start? | |
| LF: time(earliest(λx.has_concept(x, C0002395, pmh))) | |
| Q: What was the highest hemoglobin A1c value in the past 4 years? | |
| LF: max(lambda(λx.has_concept(x, C0366781, history) ^ time_within(x, ‘in the past 4 years’))) | |
| Q: Has his hemoglobin A1c ever been less than 6? | |
| LF: delta(λx.has_concept(x, C0366781, history) ^ less_than(x, ‘6’)) | |
| ICUDATA | Q: Do they have any chest drain? |
| LF: delta(λx.has_concept(x, C0008034, visit)) | |
| Q: Do we know any positive microscopy for this patient? | |
| LF: delta(positive(λx.has_concept(x, C0026018, visit))) | |
| Q: What is his mental status? | |
| LF: latest(λx.has_concept(x, C0278060, status)) | |
| Q: What was the pre-op echocardiogram result? | |
| LF: latest(λx.has_concept(x, C0013516, visit) and time_within(x, preoperative)) | |
| Q: How many blood products have been administered in the past 25 hours? | |
| LF: sum(λx.has_concept(x, C0456388, visit) and time_within(x, ‘past 25 hours’)) | |
| Q: Did the patient temperature exceed 38C in last 48 hours? | |
| LF: delta(λx.has_concept(x, C0005903, visit) and greater_than(x, ‘38 C’) and time_within(x, ‘last 48 hours’)) | |
| (b) Descriptive statistics of the datasets. #: count. | ||
|---|---|---|
| Metric | Corpus |
|
| FHIRDATA | ICUDATA | |
| # of queries | 966 | 400 |
| # of unique predicates | 20 | 30 |
| Mean # of predicates per query | 3.59 | 3.33 |
| # of unique words | 749 | 467 |
| Mean # of words per query | 7.79 | 6.04 |
One of the datasets was constructed by a physician and an informaticist using an annotation tool based on FHIR.28 We refer to this dataset as FHIRDATA. The annotators were asked to create questions that could be answered using the data from the FHIR server that they were able to review. They were also asked to indicate the answers to these constructed questions via the annotation tool. The CUI for the concepts in the question are automatically populated based on the selected answer from FHIR. Notably, this can result in a more granular concept than is identifiable from the question alone. This is a representative dataset for the task as it originates from a FHIR server itself. The questions in the other dataset, which we refer to as ICUDATA, were collected from shadowing clinicians in an intensive care unit (ICU) setting26 and further annotated with their corresponding LFs.29 This dataset captures the realistic information needs of the task. Some examples and the descriptive statistics for both datasets are presented in Table 1. We used a local FHIR instance with synthetic data50 for FHIRDATA. For ICUDATA, we manually inserted answers to the questions into this same server. More details about the datasets are included in Supplementary Section S1.
MATERIALS AND METHODS
System overview
An overall view of our system is presented in Figure 1. Again, the design philosophy is a high-precision tool that “knows what it does not know” as best as possible to foster user trust in the answers. This means that questions with misspellings, out-of-vocabulary terminology, or unseen structures will intentionally fail (since QA is interactive, the user still can reformulate the question). This is accomplished primarily by a carefully maintained lexicon mapping nonmedical terms to logical functions.35 Additionally, the user is presented with a visual interpretation of the LF to validate the question was understood correctly.
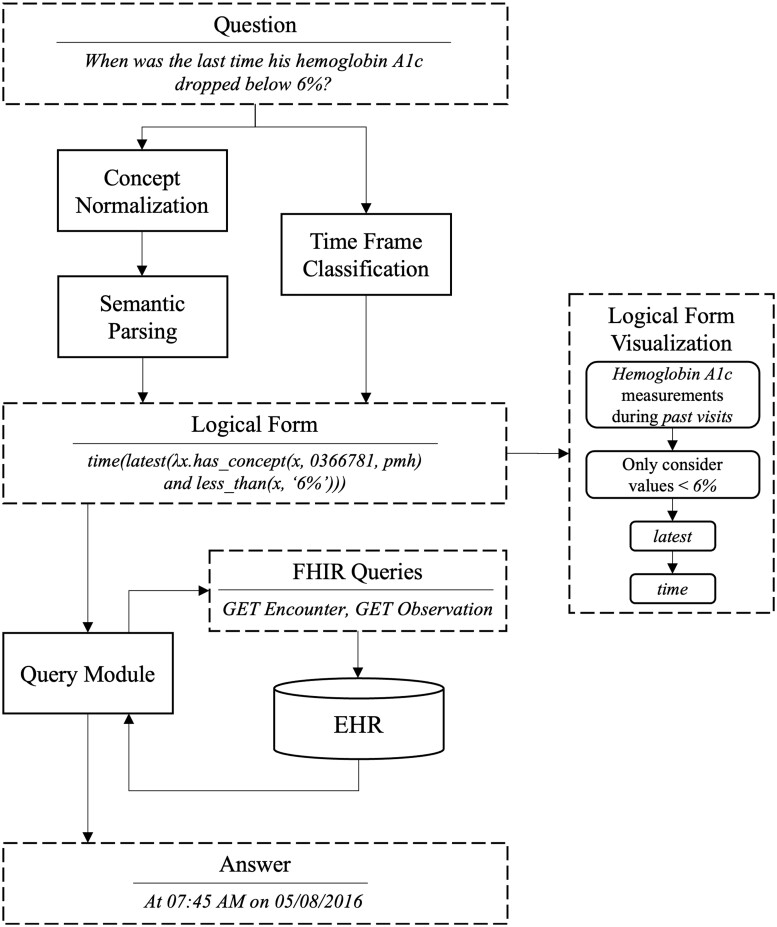
Figure 1.
System overview.
If used as a black box, our proposed system will return an exact answer and its justification for a given natural language question (see Figure 3). If a question is unanswerable using the EHR data or the system is unable to understand it, no answer is returned (favoring precision over recall). Instead, an explanation regarding this inability is returned to help the user understand what went wrong (eg, what words were not understood) instead of returning a wrong or meaningless answer. A question is passed through the 3 main components of the system—concept normalization, semantic parsing, and time frame classification—all of which play an important role in constructing a LF. The predicted LF is further passed to a query mapping module that translates it to FHIR queries and fetches required information from the EHR in the form of FHIR resources. The returned resources are further processed by this module to build the answer in a human-comprehensible format. The system also returns a graphical representation of the predicted LF that serves as a justification for the answer. This visualization helps the user verify whether the system properly understood their query.
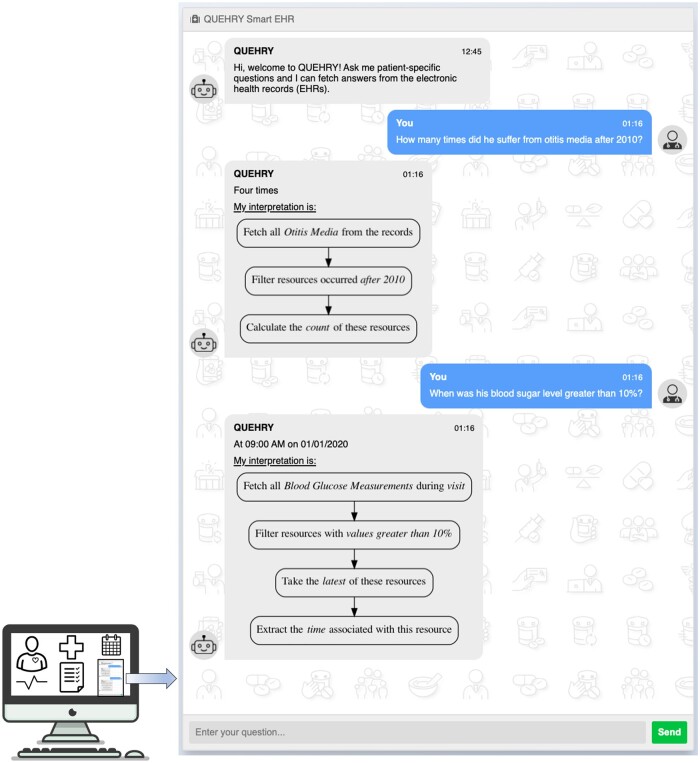
Figure 3.
quEHRy interface where a user inputs their questions and the system responds with answers from the EHR.
Concept normalization
This component is responsible for extracting different types of concepts from a question. This separates the task of semantic parsing from concept normalization and assists the semantic parser by providing specific arguments to fill in a predicted logical tree (a LF with placeholders for the arguments).35 Specifically, we extract patient references (eg, he, she), temporal references (eg, today), location references (eg, BICU), measurements (eg, 30C), and other hospital events (eg, admission, preoperative) using a simple rule-based system. The medical concepts (eg, diabetes) in questions are extracted with MetaMap,51 a tool to extract and map biomedical concepts to UMLS CUIs.52 We experiment using gold concepts directly, picking the top-ranked/longest concept from MetaMap, filtering candidates using EHR concepts before passing to the next step (semantic parsing), and picking the longest concept from EHR-filtered concepts. In the case where all EHR-filtered concepts are passed, the concept prediction from candidates is postponed until after the semantic parsing is completed.
Semantic parsing
We take a hybrid approach to convert questions into their corresponding logical trees using a combination of rule-based and machine learning techniques.35 Candidate logical trees are generated using a dependency tree-based lexicon and generation/filtering rules, then a support vector machine (SVM) classifier chooses a single tree. To generate the logical trees, a lexicon maps natural language phrases to λ-calculus predicates (eg, “when” → time(), “today” → time_within()). For details, see Supplementary Section S2 or Roberts and Patra.35 We explored other neural models for the task, however, interestingly, they perform worse than the employed lexicon-based approach.53
Implicit time frame detection
The semantic parser results in a logical tree that needs to be filled with a time frame before it can be executed against a FHIR server. Thus, we built a separate SVM to predict the implicit time frame for a given question using simple n-gram features.
Query module
EHRs interfaced using a FHIR server expose a set of RESTful (REpresentational State Transfer style) APIs (Application Programming Interfaces). We refer to these API calls as FHIR queries. These queries return data in the form of FHIR resources (eg, Encounter, Observation). The query module is responsible for mapping the predicted LF from earlier steps to its corresponding FHIR queries and executing them against a FHIR server to access EHR data (see flowchart in Figure 2). This module also processes the responses from the FHIR server and returns an answer.
Figure 2.
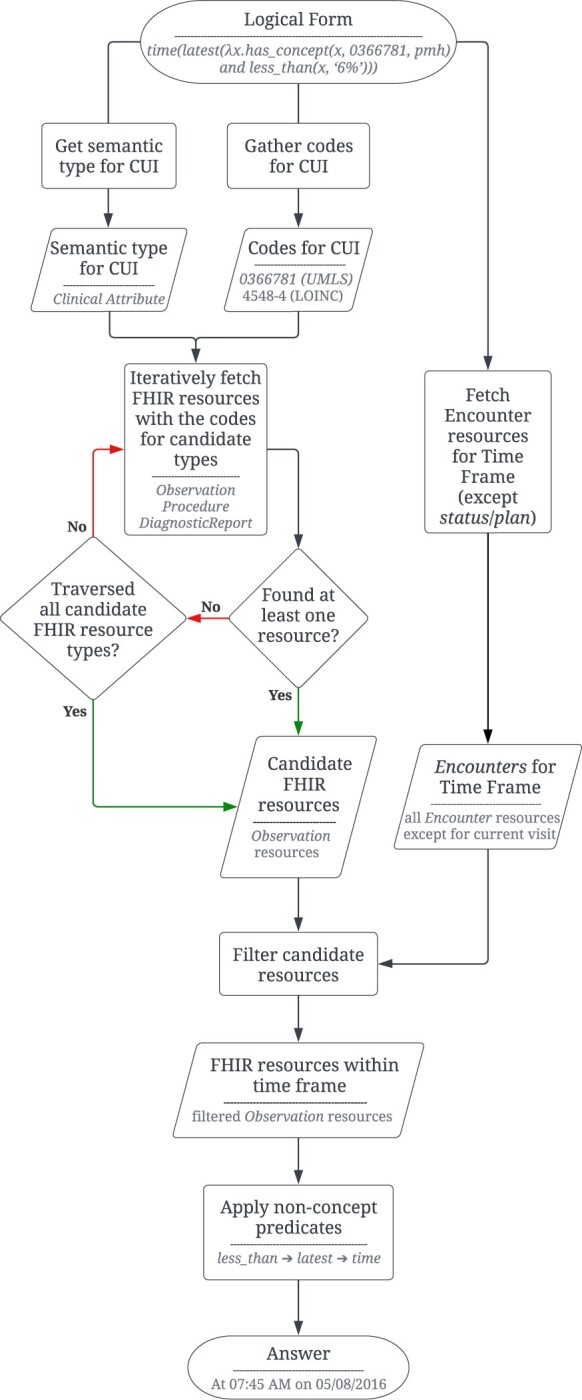
Flowchart showing the steps in the query module component of quEHRy. The example question used in the flowchart is “When was the last time his hemoglobin A1c dropped below 6%?”.
Given the complex information needs of a clinical question, it is often not possible that a single FHIR query can extract all the required information. Thus, a series of FHIR queries is constructed for a given LF based on the different logical predicates and their parameters. First, the UMLS semantic type of the medical concepts passed to the has_concept() predicate is used to determine the different types of FHIR resources to be extracted. For example, for semantic type “Laboratory or Test Result,” Observation resources are fetched. Due to the inherent ambiguity in storing information using the FHIR standard,28 the semantic types are oftentimes mapped to multiple FHIR resources. For example, for the semantic type “Finding,” both Observation and Condition resources are fetched. A complete list of such mappings is provided in Supplementary Table S1. Second, the CUI is mapped to synonymous medical codes from different vocabularies (eg, LOINC, SNOMED CT) using UMLS. This is necessary as different types of information are stored using different vocabularies.
Further, the implicit time frame is used to restrict the temporal search space for fetching the FHIR resources. This is achieved using the Encounter resources (used to record patient encounters) and the status or time of resources. For example, for the implicit time frame visit, we restrict the search to FHIR resources associated with the current patient Encounter resource. The descriptions for the other time frames are available in Supplementary Table S2.
After fetching a set of FHIR resources using the concept predicates, the functions derived from the nonconcept predicates are applied over this set to construct an answer. Different nonconcept predicates accept single or multiple resources as input and call for different sets of operations to be performed. For example, the latest() predicate returns the most recent resource from the set of resources (using time-related information from the FHIR resources). Another example of a nonconcept predicate is time() that extracts the timestamp from a given resource. Note that the structure of all the FHIR resources is different and thus the same type of information are often stored as different attributes. For example, time in the Observation resource is stored as effectiveDateTime or effectivePeriod (depending upon its type) while for Condition it is stored as onsetDateTime and/or abatementDateTime (based on its resolution status). Other than the aforementioned example of nonconcept predicates, there are more that operate on different (and oftentimes more than one) parts of the resources such as location() that operate on bodySite in Condition and Procedure. Our source code includes all such attribute mappings. An end-to-end example is included in Supplementary Section S5.
Interface
To emphasize our vision for quEHRy and emulate its use in the real world, we also implemented a chat-like interface (Figure 3). This is part of a pilot Graphical User Interface.
Evaluation
We evaluate the 4 primary components (concept normalization, semantic parsing, implicit time frame detection, and query module) and end-to-end performance to assess each component’s impact on the overall pipeline.
For concept normalization, we calculate accuracy by exact boundary matching, marking the question as correct when all the concepts are predicted accurately. For medical concepts, the predicted CUI is further matched with the ground truth. For time frame detection, we similarly mark the question as correct when all time frames are predicted correctly.
For semantic parsing, we calculate coverage (recall) and accuracy for the intermediate logical trees and the final LF (including CUIs and time frames). We perform leave-one-out cross-validation for all the steps to maximize data use.
The answers produced by the query module are compared against the ground truth answers to calculate accuracy. Since quEHRy returns a single answer, ranking-based metrics (eg, MRR, NDGC) are not appropriate. We, moreover, calculate precision of the overall system by determining false positives, ie, when the system returns an incorrect answer instead of refraining from giving an answer. We also calculate the accuracy of fetching the correct FHIR resources. Further, we calculate its coverage, ie, the proportion of LFs that can be successfully handled by the query module.
RESULTS
The results of the system components are shown in Tables 2 and 3. Using gold concepts, the precision (the most important measure according to our QA user model philosophy) in providing an exact answer is 98.33% and 90.91% for FHIRDATA and ICUDATA, respectively. Interestingly, the precision remains at excellent levels (94.03% and 87.79%) even after employing MetaMap to extract the concepts. The recall of MetaMap for concept boundaries (83.85% and 93.0%) is better than CUIs (41.51% and 90.25%). For FHIRDATA, concept prediction accuracy drops significantly when exact CUIs are matched (40.06%) as opposed to concept boundaries (63.66%), both of which are low-to-moderate. Note: while not shown here, we additionally experimented with cTAKES54 instead of MetaMap (as it generally has higher performance in many clinical settings), but cTAKES only generated the correct concepts for 32% of the questions in ICUDATA (as opposed to the 93% recall for MetaMap).
Table 2.
Coverage and prediction results of different components of the QA pipeline on FHIRDATA
| Component | Using gold concept |
Top MetaMap score |
Longest concept |
Filter using EHR concepts |
Longest after filter using EHR concepts |
|---|---|---|---|---|---|
| Result: % (#) [total = 966] | |||||
| MetaMap generated concepts include gold Boundary (recall) | – | 83.85% (810) | |||
| MetaMap generated concepts include gold CUI (recall) | – | 41.51% (401) | |||
| Predicted concepts match gold Boundary (accuracy) | – | 54.04% (522) | 49.59% (479) | 49.38% (477) | 63.66% (615) |
| Predicted concepts match gold CUI (accuracy) | – | 23.50% (227) | 20.50% (198) | 39.13% (378) | 40.06% (387) |
| Generated logical trees include gold (recall) | 100.00% (966) | 83.64% (808) | 78.88% (762) | 99.17% (958) | 87.06% (841) |
| Predicted logical tree matches gold (accuracy) | 97.41% (941) | 81.57% (788) | 39.03% (377) | 96.27% (930) | 84.89% (820) |
| Predicted time frame matches gold (accuracy) | 88.30% (853) | ||||
| Generated logical forms include gold (recall) | 100.00% (966) | 45.45% (439) | 47.10% (455) | 42.34% (409) | 54.45% (526) |
| Predicted logical form matches gold (accuracy) | 86.23% (833) | 19.25% (186) | 9.21% (89) | 33.54% (324) | 34.16% (330) |
| Predicted FHIR response matches gold (accuracy) | 97.41% (941) | 22.77% (220) | 10.14% (98) | 38.51% (372) | 39.13% (378) |
| Predicted answer matches gold (accuracy) | 97.41% (941) | 22.98% (222) | 10.14% (98) | 38.61% (373) | 39.34% (380) |
| Predicted answer matches gold (precision) [# correct responses/all responses] | 98.33% [941/957] | 94.42% [220/233] | 50.52% [98/194] | 94.67% [373/394] | 94.03% [378/402] |
Table 3.
Coverage and prediction results of different components of the QA pipeline on ICUDATA
| Component | Using gold concept |
Top MetaMap score |
Longest concept |
Filter using EHR concepts |
Longest after filter using EHR concepts |
|---|---|---|---|---|---|
| Result: % (#) [total = 400] | |||||
| MetaMap generated concepts include gold Boundary (recall) | – | 93.00% (372) | |||
| MetaMap generated concepts include gold CUI (recall) | – | 90.25% (361) | |||
| Predicted concepts match gold Boundary (accuracy) | – | 66.50% (266) | 60.75% (243) | 67.00% (268) | 86.50% (346) |
| Predicted concepts match gold CUI (accuracy) | – | 65.75% (263) | 58.00% (232) | 76.50% (306) | 90.25% (361) |
| Generated logical trees include gold (recall) | 100.00% (400) | 89.00% (356) | 87.75% (351) | 98.50% (394) | 97.00% (388) |
| Predicted logical tree matches gold (accuracy) | 87.25% (349) | 79.00% (316) | 78.75% (315) | 87.00% (348) | 86.00% (344) |
| Predicted time frame matches gold (accuracy) | 85.00% (340) | ||||
| Generated logical forms include gold (recall) | 100.00% (400) | 85.00% (340) | 83.75% (335) | 92.50% (370) | 92.50% (370) |
| Predicted logical form matches gold (accuracy) | 74.75% (299) | 48.75% (195) | 43.75% (175) | 56.50% (226) | 67.50% (270) |
| Predicted FHIR response matches gold (accuracy) | 87.75% (351) | 55.50% (222) | 50.00% (200) | 63.50% (254) | 73.25% (293) |
| Predicted answer matches gold (accuracy) | 87.75% (351) | 57.25% (229) | 52.00% (208) | 66.75% (267) | 75.75% (303) |
| Predicted answer matches gold (precision) [# correct responses/all responses] | 90.91% [350/385] | 88.76% [229/258] | 91.19% [207/227] | 81.85% [266/325] | 87.79% [302/344] |
The errors from concept normalization step seep into subsequent layers of quEHRy, especially the query module. Logical tree recall (100% using gold concepts, 87.06% and 97.0% otherwise) highlights the power of the employed lexicon whereas their prediction performance (84.89% and 86.0%), specifically the difference of prediction accuracy from recall, indicates the efficacy of the semantic parsing. Time frame detection is separate from concept normalization and semantic parsing, and thus has the same accuracy (88.3% and 85.0%) across the different variations. LF coverage (100.0% using gold concepts, 54.45% and 92.5% otherwise) is essentially the amalgamation of MetaMap and logical tree recall. The query module depends on the underlying CUIs in a LF to generate correct FHIR responses and answers, thus a performance drop in CUI prediction (gold → 40.06% and gold → 90.25%) affects the module’s accuracy (97.41% → 39.34% and 87.75% → 75.75%).
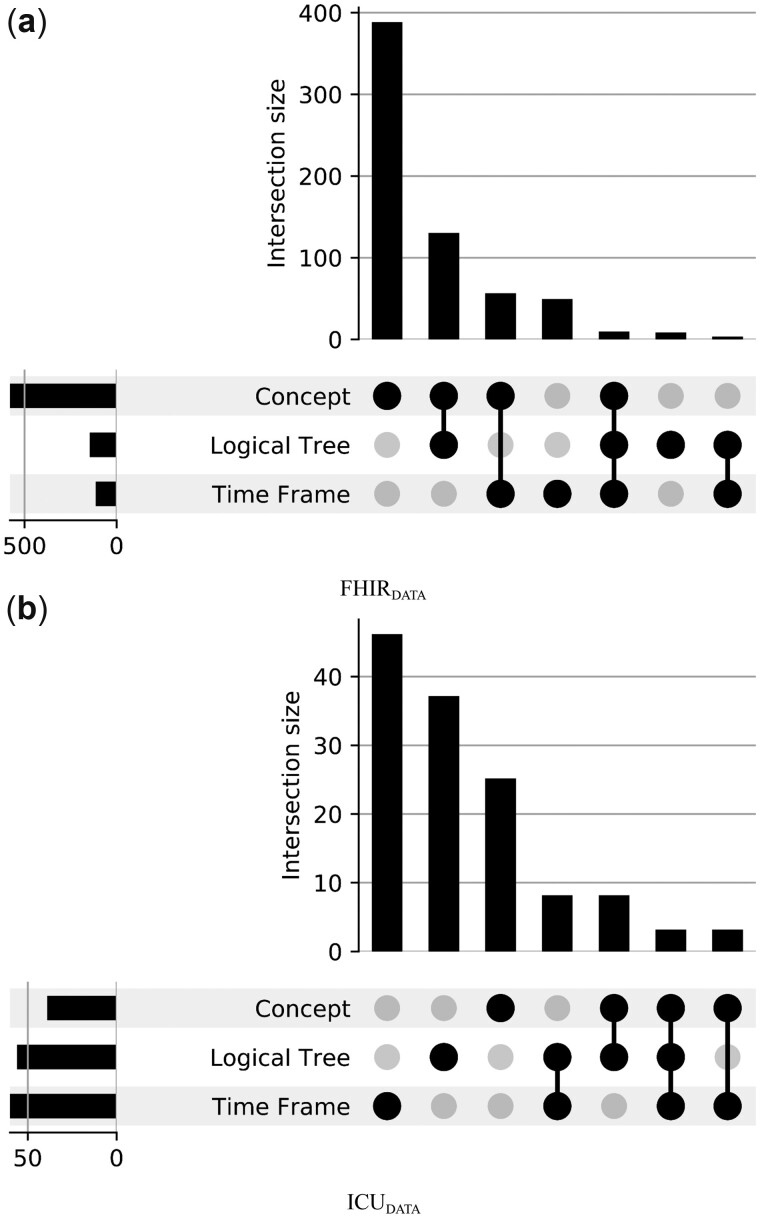
A correctly predicted LF plays the most important role in the whole pipeline. To understand the impact of errors made during the prediction of various LF components, we use UpSet plots (Figure 4). This chart helps identify which components contribute to the performance (or error) of overall LF prediction.
Figure 4.
LF prediction errors for the different components of the system pipeline.
We also conduct an error analysis of the incorrectly predicted concepts (Table 4). For FHIRDATA, a majority of the incorrectly predicted concepts are broader than the gold concepts, eg, for the same concept boundary “diabetes” the gold concept was “Diabetes Mellitus, Non-Insulin-Dependent—C0011860” while the predicted concept was “Diabetes—C0011847.” See Supplementary Table S5 for the complete output of MetaMap. We experimented with simple methods using UMLS to automatically expand these broad concepts to include narrower concepts in the FHIR queries, but this approach greatly damaged precision for only a moderate gain in recall. We thus leave this specific problem to future work.
Table 4.
Error analysis of incorrectly predicted CUIs by the best performing variant that selects the longest concept after filtering using EHR concepts
| (a) FHIRDATA | |||
|---|---|---|---|
| Boundary match | Semantic type match | Example | Count |
| ✓ | ✗ | Ques: What was the dose for amoxicillin? | 212 |
| Gold: Amoxicillin 200 MG Oral Tablet—C1298551 [Clinical Drug] | |||
| Pred: Amoxicillin—C0002645 [Antibiotic] | |||
| ✓ | ✓ | Ques: When was the onset of her diabetes? | 44 |
| Gold: Diabetes Mellitus, Non-Insulin-Dependent—C0011860 [Disease or Syndrome] | |||
| Pred: Diabetes—C0011847 [Disease or Syndrome] | |||
| ✗ | ✗ | Ques: How many times has he followed up on his asthma? | 311 |
| Gold: Asthma follow-up—C1273970 [Health Care Activity] | |||
| Pred: Asthma—C0004096 [Disease or Syndrome] | |||
| ✗ | ✓ | Ques: Was he ever admitted to an ER? | 12 |
| Gold: Emergency room admission—C0583237 [Health Care Activity] | |||
| Pred: Admitted to—C4482331 [Health Care Activity] | |||
| (b) ICUDATA | |||
|---|---|---|---|
| Boundary match | Semantic type match | Example | Count |
| ✓ | ✗ | Ques: When was the drainageapplied? | 8 |
| Gold: Drainage procedure—C0013103 [Therapeutic or Preventive Procedure] | |||
| Pred: Body Fluid Discharge—C0012621 [Body Substance] | |||
| ✓ | ✓ | Ques: How old is the line? | 4 |
| Gold: Intravenous Catheters—C0745442 [Medical Device] | |||
| Pred: Intravascular line—C0700221 [Medical Device] | |||
| ✗ | ✗ | Ques: Does she have a diaphragmatic tear? | 25 |
| Gold: Rupture of diaphragm—C0238088 [Injury or Poisoning] | |||
| Pred: Respiratory Diaphragm—C0011980 [Body Part, Organ, or Organ Component] | |||
| ✗ | ✓ | – | 0 |
Note: Gold concept boundaries are bolded while the predicted boundaries are underlined.
Ques: question; Gold/Pred: gold/predicted concept in the form of Display Name—CUI [Semantic Type].
For ICUDATA, most predicted concepts are neither too broad nor too narrow than the gold concepts. Since ICUDATA was not built off of an EHR server, the gold-annotated concepts are not always specific, in fact, the concepts were manually normalized by searching UMLS and thus likely to be subjective. For example, for the question “Was she hypertensive?,” the gold concept boundary is hypertensive with CUI C0520539 (Hypertensive episode, semantic type disease) while a predicted concept boundary was hypertensive with CUI C0857121 (Hypertensive [finding], semantic type finding). Here, the concept boundary is predicted correctly but the predicted CUI is different. Any performance drop because of such mismatches is an artifact of the subjective nature of manual concept normalization annotations. For example, if the annotator instead annotated the same information of hypertension as a finding (instead of disease), this prediction would have been correct. This highlights the vulnerability of such systems to the different ways in which the same medical information can be represented. Thus, we annotated an additional set of medical concepts for each question in ICUDATA to compensate for such annotation bias. Specifically, MetaMap was used to generate top 5 candidate concept CUIs for each concept boundary in the dataset and an annotator marked which among these are valid. We found that the CUI coverage improved significantly after this relaxation, where we marked a predicted concept as correct if it belonged to the set of gold concepts for a question (as opposed to matching it with just a gold concept). We take these additional concepts into account during our evaluation for ICUDATA.
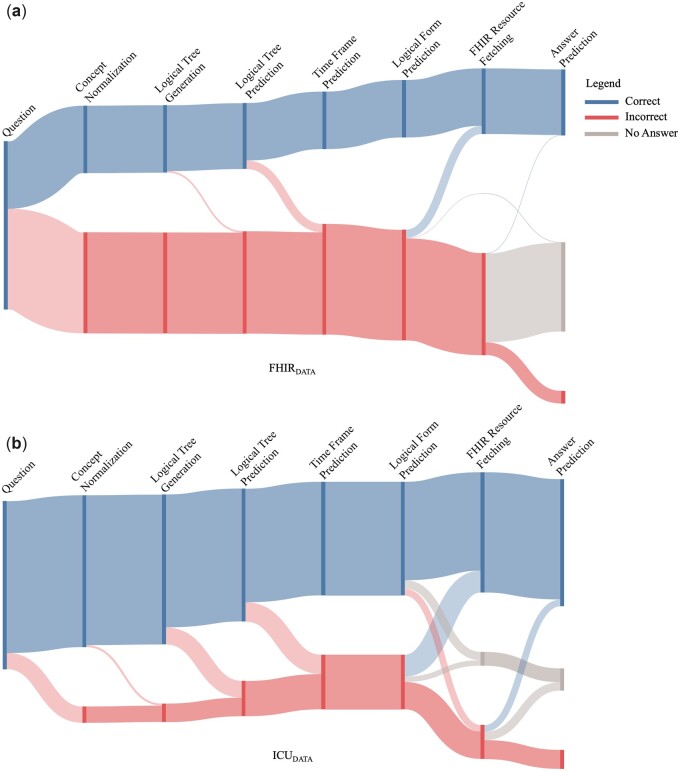
Finally, the flow of errors through quEHRy is shown in Figure 5. Note that for FHIRDATA, the system gracefully handles a majority of the questions in the “Incorrect” flow by responding with “No Answer.” For ICUDATA, almost half of the “Incorrect” flow questions move toward “Correct” (eg, a correct answer is returned for an incorrect yet plausible time frame) or “No Answer.”
Figure 5.
The flow of errors through quEHRy for both of the datasets. The length of the bars corresponding to the different components represents a cumulative measure of correct predictions or generations (eg, at any correct bar in the graph, the proportional number of questions had correct responses for all the previous steps).
DISCUSSION
We designed an end-to-end QA system that takes a natural language question as input and precisely returns an exact answer from structured EHR data along with a visualization depicting the system’s interpretation of the question. To achieve this, we developed an EHR QA-tailored time frame classifier and also a query module that enables us to query the EHRs using the predicted LFs—by converting them to FHIR queries for fetching information from EHRs.
Identification of medical concepts in the questions contributed to the majority of errors in the system’s performance. We annotated additional CUIs for ICUDATA, however, the medical concept normalization errors in FHIRDATA surface a deeper issue with using application-agnostic tools. The issue reflects how the information stored in EHRs are “specific” while the concepts extracted from questions are “broad.” While one may argue that the questions do not contain sufficient information in order to map their underlying concepts to the specific concepts actually present in EHRs, it is usually not the case with naturally asked questions (that tend to be broad). Thus, there is instead a need to build a QA-specific concept normalizer that understands the context (with respect to the variety of concepts used in an EHR) while predicting a concept. As this is not the sole focus of this work and is a well-studied research area,55 we leave this interesting problem to future research.
The coverage (and thereby prediction) of logical trees relies heavily on the extracted concept boundaries. For the best performing variant for automatically selecting a concept (filter using EHR concepts and choose longest), among the questions where the gold logical tree was not generated, most were with an incorrect concept boundary prediction (124 out of 125 for FHIRDATA, 8 out of 12 for ICUDATA). For example, for “What is the dose of metoprolol?”, the gold boundary is “metoprolol” but the predicted concept boundary is “dose of metoprolol.” Thus, while the gold logical tree for this question is dose(latest(λx.has_concept(x))) (corresponding to the concept-substituted question “What is the dose of [concept]?”), the semantic parser incorrectly predicted latest(λx.has_concept(x)), corresponding to the concept-substituted question “What is the [concept]?”.
QA work involving structured data in the general domain falls broadly into 2 categories based on the underlying data structure, namely, graph56,57 and table.58 EHRs, on the other hand, are rarely in the form of standardized graph or table structures. However, several studies explored methods with EHR data formatted as graphs31,59 or tables,30,60,61 perhaps inspired by the public availability of EHR data in these formats. Differently, in this work, we tackle the problem of QA from EHRs using a data model in use in real-life EHRs, ie, the FHIR standard. Ours is also the first work to explore an end-to-end QA solution from FHIR servers.
There are several limitations to this work. The size of the evaluated datasets is relatively small (with 966 and 400 questions) and a larger dataset may better estimate the system’s performance. The proposed technique relies on a lexicon, which needs to be hand-crafted to maintain the quality of the overall system. This intentionally limits the ability of such systems to generalize to the kinds of questions that were not covered by a lexicon. However, a lexicon provides a robust mechanism that enables a QA system to “know what it does not know” (improves both reliability and interpretability). Additionally, the philosophical choice of returning a single, exact answer or nothing at all is admittedly an assumption, which will require user testing to validate. Finally, since our focus here is structured data and factoid questions, the system is not designed for more open-ended questions that are more likely to be answerable using unstructured EHR data.
Future work can explore the techniques to automatically harvest lexicon from existing datasets,62,63 however, this will first require building a sizable EHR QA dataset. Further, though our query module is exhaustive enough to capture most logical predicates, its capability depends on the variety of logical predicates that are successfully handled or, in other words, present in our datasets. To expand the variety of questions, one may need to extend the lexicon and/or the query module (to define mappings for new logical predicates). That said, expanding the current lexicon and query module is simple and merely requires adding a few mappings from phrases to logical predicates and from logical predicates to FHIR resources, respectively. More generally, future work will also explore the usability of this system when deployed in a clinical setting, as well as the integration of QA approaches for unstructured notes to complement this study’s focus on structured information.
CONCLUSION
We constructed an end-to-end QA system, quEHRy, to allow users to query EHRs using natural language questions. It consists of multiple components, yet it is a high-precision and interpretable system that fits clinical use-cases. To further improve the performance and coverage of the proposed system, a future direction of research is to focus on building QA-specific concept normalization systems.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dina Demner-Fushman, Braja Gopal Patra, Hua Xu, and Peter Killoran for their feedback.
Contributor Information
Sarvesh Soni, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, Texas, USA.
Surabhi Datta, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, Texas, USA.
Kirk Roberts, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, Texas, USA.
FUNDING
This work was supported by the U.S. National Library of Medicine, National Institutes of Health (NIH), under awards R00LM012104, R21EB029575, and R01LM011934; the Bridges Family Doctoral Fellowship Award from UTHealth Houston; and the UTHealth Innovation for Cancer Prevention Research Training Program Predoctoral Fellowship, under award CPRIT RP210042.
AUTHOR CONTRIBUTIONS
SS and KR built the end-to-end pipeline, conducted the experiments, and drafted the initial manuscript. SD built the initial version of the query module. KR conceived the idea, edited the manuscript, supervised the project, and secured funding. All the authors reviewed and approved of the final manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The datasets and code underlying this article are available in GitHub at https://github.com/krobertslab/olympia-quehry.
REFERENCES
- 1. Zhang J, Walji M (eds). Better EHR: Usability, Workflow and Cognitive Support in Electronic Health Records. Houston: The National Center for Cognitive Informatics & Decision Making in Healthcare, The University of Texas Health Science Center at Houston School of Biomedical Informatics; 2014. [Google Scholar]
- 2. Roman LC, Ancker JS, Johnson SB, et al. Navigation in the electronic health record: a review of the safety and usability literature. J Biomed Inform 2017; 67: 69–79. doi: 10.1016/J.JBI.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 3. Khairat S, Coleman C, Ottmar P, et al. Association of electronic health record use with physician fatigue and efficiency. JAMA Netw Open 2020; 3 (6): e207385. doi: 10.1001/jamanetworkopen.2020.7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shneiderman B, Plaisant C, Hesse BW.. Improving healthcare with interactive visualization. Computer 2013; 46 (5): 58–66. doi: 10.1109/MC.2013.38 [DOI] [Google Scholar]
- 5. Hanauer DA, Mei Q, Law J, et al. Supporting information retrieval from electronic health records: a report of University of Michigan’s nine-year experience in developing and using the Electronic Medical Record Search Engine (EMERSE). J Biomed Inform 2015; 55: 290–300. doi: 10.1016/j.jbi.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ely JW, Osheroff JA, Chambliss ML, et al. Answering physicians’ clinical questions: obstacles and potential solutions. J Am Med Inform Assoc 2005; 12 (2): 217–24. doi: 10.1197/jamia.M1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pampari A, Raghavan P, Liang J, et al. emrQA: a large corpus for question answering on electronic medical records. EMNLP. Brussels, Belgium: Association for Computational Linguistics; 2018: 2357–68. doi: 10.18653/v1/D18-1258. [DOI] [Google Scholar]
- 8. Soni S, Roberts K.. Evaluation of dataset selection for pre-training and fine-tuning transformer language models for clinical question answering. In: LREC. Marseille, France: European Language Resources Association; 2020: 5534–40. https://www.aclweb.org/anthology/2020.lrec-1.679. Accessed September 26, 2022. [Google Scholar]
- 9. Datta S, Roberts K.. Fine-grained spatial information extraction in radiology as two-turn question answering. Int J Med Inform 2021; 158: 104628. doi: 10.1016/j.ijmedinf.2021.104628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prager JM, Liang JJ, Devarakonda MV.. SemanticFind: locating what you want in a patient record, not just what you ask for. In: AMIA Joint Summits on Translational Science Proceedings. San Francisco, CA: American Medical Informatics Association; 2017: 249–58. http://www.ncbi.nlm.nih.gov/pubmed/28815139. Accessed September 26, 2022. [PMC free article] [PubMed] [Google Scholar]
- 11. Kamath A, Das R.. A survey on semantic parsing. In: Automated Knowledge Base Construction. Amherst, MA; 2019. https://openreview.net/forum?id=HylaEWcTT7. Accessed September 26, 2022. [Google Scholar]
- 12. Ely JW, Osheroff JA, Ebell MH, et al. Obstacles to answering doctors’ questions about patient care with evidence: qualitative study. BMJ 2002; 324 (7339): 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Del Fiol G, Workman TE, Gorman PN.. Clinical questions raised by clinicians at the point of care a systematic review. JAMA Intern Med 2014; 174 (5): 710–8. doi: 10.1001/jamainternmed.2014.368 [DOI] [PubMed] [Google Scholar]
- 14. Voorhees EM, Tong R.. Overview of the TREC 2011 medical records track. In: The Twentieth Text REtrieval Conference Proceedings. Online: National Institute of Standards and Technology; 2011. [Google Scholar]
- 15. Voorhees EM, Hersh W.. Overview of the TREC 2012 medical records track. In: The Twenty-First Text REtrieval Conference Proceedings. Online: National Institute of Standards and Technology; 2012. https://trec.nist.gov/pubs/trec21/papers/MED12OVERVIEW.pdf. Accessed September 26, 2022. [Google Scholar]
- 16. Hanauer DA, Wu DTY, Yang L, et al. Development and empirical user-centered evaluation of semantically-based query recommendation for an electronic health record search engine. J Biomed Inform 2017; 67: 1–10. doi: 10.1016/j.jbi.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee M, Cimino J, Zhu HR, et al. Beyond information retrieval—medical question answering. AMIA Annu Symp Proc 2006; 2006: 469–73. [PMC free article] [PubMed] [Google Scholar]
- 18. Chamberlin SR, Bedrick SD, Cohen AM, et al. Evaluation of patient-level retrieval from electronic health record data for a cohort discovery task. JAMIA Open 2020; 3 (3): 395–404. doi: 10.1093/jamiaopen/ooaa026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsatsaronis G, Balikas G, Malakasiotis P, et al. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinformatics 2015; 16: 138. doi: 10.1186/s12859-015-0564-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin Q, Dhingra B, Liu Z, et al. PubMedQA: a dataset for biomedical research question answering. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). Hong Kong, China: Association for Computational Linguistics; 2019: 2567–77. doi: 10.18653/v1/D19-1259 [DOI] [Google Scholar]
- 21. Liu F, Antieau LD, Yu H.. Toward automated consumer question answering: automatically separating consumer questions from professional questions in the healthcare domain. J Biomed Inform 2011; 44 (6): 1032–8. doi: 10.1016/j.jbi.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demner-Fushman D, Mrabet Y, Ben Abacha A.. Consumer health information and question answering: helping consumers find answers to their health-related information needs. J Am Med Inform Assoc 2020; 27 (2): 194–201. doi: 10.1093/jamia/ocz152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savery M, Abacha AB, Gayen S, et al. Question-driven summarization of answers to consumer health questions. Sci Data 2020; 7 (1): 322. doi: 10.1038/s41597-020-00667-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Athenikos SJ, Han H.. Biomedical question answering: a survey. Comput Methods Programs Biomed 2010; 99 (1): 1–24. doi: 10.1016/j.cmpb.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 25. Jin Q, Yuan Z, Xiong G, et al. Biomedical question answering: a survey of approaches and challenges. ACM Comput Surv 2023; 55 (2): 1–36. doi: 10.1145/3490238 [DOI] [Google Scholar]
- 26. Patrick J, Li M.. An ontology for clinical questions about the contents of patient notes. J Biomed Inform 2012; 45 (2): 292–306. doi: 10.1016/j.jbi.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 27. Roberts K, Demner-Fushman D.. Toward a natural language interface for EHR questions. In: AMIA Summits on Translational Science Proceedings. San Francisco, CA: American Medical Informatics Association; 2015: 157–61. http://www.ncbi.nlm.nih.gov/pubmed/26306260. Accessed September 26, 2022. [PMC free article] [PubMed] [Google Scholar]
- 28. Soni S, Gudala M, Wang DZ, et al. Using FHIR to construct a corpus of clinical questions annotated with logical forms and answers. In: AMIA Annual Symposium Proceedings. Washington, DC: American Medical Informatics Association; 2019: 1207–15. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7153115/. Accessed September 26, 2022. [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts K, Demner-Fushman D.. Annotating logical forms for EHR questions. In: LREC. Portorož, Slovenia: NIH Public Access; 2016: 3772–8. https://www.aclweb.org/anthology/L16-1598. Accessed September 26, 2022. [PMC free article] [PubMed] [Google Scholar]
- 30. Wang P, Shi T, Reddy CK.. Text-to-SQL generation for question answering on electronic medical records. In: Proceedings of the Web Conference. New York, NY: Association for Computing Machinery; 2020: 350–61. doi: 10.1145/3366423.3380120 [DOI] [Google Scholar]
- 31. Park J, Cho Y, Lee H, et al. Knowledge graph-based question answering with electronic health records. In: Proceedings of the 6th Machine Learning for Healthcare Conference. Virtual: PMLR; 2021: 36–53. https://proceedings.mlr.press/v149/park21a.html. Accessed September 26, 2022. [Google Scholar]
- 32. Raghavan P, Liang JJ, Mahajan D, et al. emrKBQA: A Clinical Knowledge-Base Question Answering Dataset. In: Proceedings of the 20th Workshop on Biomedical Language Processing. Online: Association for Computational Linguistics; 2021: 64–73. doi: 10.18653/v1/2021.bionlp-1.7 [DOI] [Google Scholar]
- 33. Schwertner MA, Rigo SJ, Araujo DA, et al. Fostering natural language question answering over knowledge bases in oncology EHR. In: IEEE Computer-Based Medical Systems. Cordoba, Spain: IEEE; 2019: 501–6. doi: 10.1109/CBMS.2019.00102. [DOI] [Google Scholar]
- 34. Ruan T, Huang Y, Liu X, et al. QAnalysis: a question-answer driven analytic tool on knowledge graphs for leveraging electronic medical records for clinical research. BMC Med Inform Decis Mak 2019; 19 (1): 82. doi: 10.1186/s12911-019-0798-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberts K, Patra BG.. A semantic parsing method for mapping clinical questions to logical forms. In: AMIA Annual Symposium Proceedings. Washington, DC: American Medical Informatics Association; 2017: 1478–87. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5977685/. Accessed September 26, 2022. [PMC free article] [PubMed] [Google Scholar]
- 36. Neuraz A, Rance B, Garcelon N, et al. The impact of specialized corpora for word embeddings in natural langage understanding. Stud Health Technol Inform 2020; 270: 432–6. doi: 10.3233/SHTI200197 [DOI] [PubMed] [Google Scholar]
- 37. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. doi: 10.1136/amiajnl-2011-000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013. In: Information Access Evaluation. Multilinguality, Multimodality, and Visualization: 4th International Conference of the CLEF Initiative. Valencia, Spain: Springer; 2013: 212–31. doi: 10.1007/978-3-642-40802-1_24 [DOI] [Google Scholar]
- 39. Kelly L, Goeuriot L, Suominen H, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2014. In: Information Access Evaluation. Multilinguality, Multimodality, and Visualization: 5th International Conference of the CLEF Initiative. Sheffield, UK: Springer; 2014: 172–91. doi: 10.1007/978-3-319-11382-1_17 [DOI] [Google Scholar]
- 40. Pradhan S, Elhadad N, Chapman W, et al. SemEval-2014 task 7: analysis of clinical text. In: Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014). Stroudsburg, PA: Association for Computational Linguistics; 2014: 54–62. doi: 10.3115/v1/S14-2007 [DOI] [Google Scholar]
- 41. Elhadad N, Pradhan S, Gorman S, et al. SemEval-2015 Task 14: analysis of clinical text. In: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015). Stroudsburg, PA: Association for Computational Linguistics; 2015: 303–10. doi: 10.18653/v1/S15-2051 [DOI] [Google Scholar]
- 42. Leaman R, Islamaj Dogan R, Lu Z.. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics 2013; 29 (22): 2909–17. doi: 10.1093/bioinformatics/btt474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. J Am Med Inform Assoc 2013; 20 (5): 806–13. doi: 10.1136/amiajnl-2013-001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bethard S, Savova G, Chen W-T, et al. SemEval-2016 task 12: clinical TempEval. In: Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016). Stroudsburg, PA: Association for Computational Linguistics; 2016: 1052–62. doi: 10.18653/v1/S16-1165 [DOI] [Google Scholar]
- 45. Health Level Seven International. Welcome to FHIR. https://www.hl7.org/fhir/. Accessed September 26, 2022. [Google Scholar]
- 46. Yadav P, Steinbach M, Kumar V, et al. Mining electronic health records (EHRs): a survey. ACM Comput Surv 2018; 50 (6): 1–40. doi: 10.1145/3127881 [DOI] [Google Scholar]
- 47. Tonekaboni S, Joshi S, McCradden MD, et al. What clinicians want: contextualizing explainable machine learning for clinical end use. In: Doshi-Velez F, Fackler J, Jung K, et al. , eds. Proceedings of the 4th Machine Learning for Healthcare Conference. Ann Arbor, Michigan: PMLR; 2019: 359–80. http://proceedings.mlr.press/v106/tonekaboni19a.html. Accessed September 26, 2022 [Google Scholar]
- 48. Rowe M. An introduction to machine learning for clinicians. Acad Med 2019; 94 (10): 1433–6. doi: 10.1097/ACM.0000000000002792 [DOI] [PubMed] [Google Scholar]
- 49. Bussone A, Stumpf S, O’Sullivan D.. The role of explanations on trust and reliance in clinical decision support systems. In: 2015 International Conference on Healthcare Informatics. Dallas, TX: IEEE; 2015: 160–9. doi: 10.1109/ICHI.2015.26 [DOI] [Google Scholar]
- 50. Walonoski J, Kramer M, Nichols J, et al. Synthea: an approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J Am Med Inform Assoc 2018; 25 (3): 230–8. doi: 10.1093/jamia/ocx079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 17 (3): 229–36. doi: 10.1136/jamia.2009.002733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lindberg DAB, Humphreys BL, McCray AT.. The unified medical language system. Methods Inf Med 1993; 32 (4): 281–91. doi: 10.1136/jamia.1998.0050001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soni S, Roberts K. Toward a neural semantic parsing system for EHR question answering. In: AMIA Annual Symposium Proceedings. Washington, DC: American Medical Informatics Association; 2022:. 1002–11. doi: 10.48550/arXiv.2211.04569 [DOI] [PMC free article] [PubMed]
- 54. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. doi: 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fu S, Chen D, He H, et al. Clinical concept extraction: a methodology review. J Biomed Inform 2020; 109: 103526. doi: 10.1016/j.jbi.2020.103526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Diefenbach D, Lopez V, Singh K, et al. Core techniques of question answering systems over knowledge bases: a survey. Knowl Inf Syst 2018; 55 (3): 529–69. doi: 10.1007/s10115-017-1100-y [DOI] [Google Scholar]
- 57. Lan Y, He G, Jiang J, et al. Complex knowledge base question answering: a survey. IEEE Trans Knowl Data Eng2022; 1–20. doi: 10.1109/TKDE.2022.3223858 [DOI]
- 58. Jin N, Siebert J, Li D, et al. A survey on table question answering: recent advances. In: Knowledge Graph and Semantic Computing: Knowledge Graph Empowers the Digital Economy. Singapore: Springer Nature; 2022: 174–86. doi: 10.1007/978-981-19-7596-7_14 [DOI]
- 59. Wang P, Shi T, Agarwal K, et al. Attention-based aspect reasoning for knowledge base question answering on clinical notes. In: Proceedings of the 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. Northbrook, IL: Association for Computing Machinery; 2022: 1–6. doi: 10.1145/3535508.3545518 [DOI] [Google Scholar]
- 60. Lee G, Hwang H, Bae S, et al. EHRSQL: A Practical Text-to-SQL Benchmark for Electronic Health Records. In: Advances in Neural Information Processing Systems. New Orleans, LA: Curran Associates, Inc; 2022: 15589–601. https://proceedings.neurips.cc/paper_files/paper/2022/file/643e347250cf9289e5a2a6c1ed5ee42e-Paper-Datasets_and_Benchmarks.pdf. Accessed September 26, 2022. [Google Scholar]
- 61. Pan Y, Wang C, Hu B, et al. A BERT-based generation model to transform medical texts to SQL queries for electronic medical records: model development and validation. JMIR Med Inform 2021; 9 (12): e32698. doi: 10.2196/32698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cai Q, Yates A.. Large-scale semantic parsing via schema matching and lexicon extension. In: Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Sofia, Bulgaria: Association for Computational Linguistics; 2013: 423–33. https://www.aclweb.org/anthology/P13-1042. Accessed September 26, 2022. [Google Scholar]
- 63. Berant J, Chou A, Frostig R, et al. Semantic parsing on freebase from question-answer pairs. In: Proceedings of the 2013 Conference on Empirical Methods in Natural Language Processing. Seattle, WA: Association for Computational Linguistics; 2013: 1533–44. https://aclanthology.org/D13-1160. Accessed September 26, 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and code underlying this article are available in GitHub at https://github.com/krobertslab/olympia-quehry.