Abstract
Objective
Severe infection can lead to organ dysfunction and sepsis. Identifying subphenotypes of infected patients is essential for personalized management. It is unknown how different time series clustering algorithms compare in identifying these subphenotypes.
Materials and Methods
Patients with suspected infection admitted between 2014 and 2019 to 4 hospitals in Emory healthcare were included, split into separate training and validation cohorts. Dynamic time warping (DTW) was applied to vital signs from the first 8 h of hospitalization, and hierarchical clustering (DTW-HC) and partition around medoids (DTW-PAM) were used to cluster patients into subphenotypes. DTW-HC, DTW-PAM, and a previously published group-based trajectory model (GBTM) were evaluated for agreement in subphenotype clusters, trajectory patterns, and subphenotype associations with clinical outcomes and treatment responses.
Results
There were 12 473 patients in training and 8256 patients in validation cohorts. DTW-HC, DTW-PAM, and GBTM models resulted in 4 consistent vitals trajectory patterns with significant agreement in clustering (71–80% agreement, P < .001): group A was hyperthermic, tachycardic, tachypneic, and hypotensive. Group B was hyperthermic, tachycardic, tachypneic, and hypertensive. Groups C and D had lower temperatures, heart rates, and respiratory rates, with group C normotensive and group D hypotensive. Group A had higher odds ratio of 30-day inpatient mortality (P < .01) and group D had significant mortality benefit from balanced crystalloids compared to saline (P < .01) in all 3 models.
Discussion
DTW- and GBTM-based clustering algorithms applied to vital signs in infected patients identified consistent subphenotypes with distinct clinical outcomes and treatment responses.
Conclusion
Time series clustering with distinct computational approaches demonstrate similar performance and significant agreement in the resulting subphenotypes.
Keywords: infection, sepsis, vital signs, subphenotypes, phenotypes
INTRODUCTION
Severe infection can lead to organ dysfunction and sepsis, which is associated with high morbidity, mortality, and costs.1 Decades of clinical trials have failed to identify one-size-fits-all treatments that improve mortality for patients with severe infection and sepsis, which is likely due to the significant heterogeneity in this population.2,3 This has resulted in research focused on identifying subphenotypes (ie, subgroups) that may benefit from targeted treatments, leading to a precision medicine approach to treating patients with infection.4
In addition to being heterogeneous, the physiological responses to infection are dynamic and rapidly evolve over minutes to hours.5,6 There has been recent focus on subphenotyping patients using time series clustering algorithms to capture this dynamic heterogeneity. Most work has applied time series clustering to univariate longitudinal data such as temperature measurements, ventilator parameters, vasopressor requirements, and severity of illness scores to identify subphenotypes within critically ill patients.7–11 Our recent work used group-based trajectory modeling (GBTM) to identify sepsis subphenotypes using multivariate longitudinal vital signs (ie, temperature, heart rate, respiratory rate, and blood pressure) from the first 8 h of hospitalization.12 The vitals trajectory subphenotypes had distinct clinical characteristics and outcomes such as ICU admission and mortality. Importantly, the vitals trajectory subphenotypes demonstrate heterogeneity of treatment responses to normal saline versus balanced crystalloids, representing the first clinically feasible phenotyping method demonstrating heterogeneous responses to intravenous fluids.
Although GBTM is a popular classical approach to time series clustering, it has some limitations that may restrict the discovery of subphenotypes.13,14 First, GBTM assumes a polynomial shape underlying trajectories (eg, linear vs quadratic) instead of allowing for an unrestricted trajectory form. Second, GBTM aligns patients without temporal warping (eg, vital signs at hour 1 of a patient are aligned with vital signs at hour 1 of another patient), and thus may fail to recognize similar but temporally shifted sequences. Alternative time series clustering algorithms using dynamic time warping (DTW) may overcome these limitations. DTW is an algorithm that computes the distance between temporal sequences by warping the sequences to an optimal alignment.15 Clustering algorithms are then applied to the DTW distances to identify trajectory subphenotypes. This approach has the advantage of discovering trajectories with nonpolynomial shapes and identifying temporally distorted but similar trajectories. DTW-based clustering has been used to identify organ failure trajectories and biomarker trajectories in COVID-19 and sepsis patients.11,16,17 Despite the theoretical differences between GBTM and DTW-based models, it is unknown whether these distinct algorithms would identify different or similar subphenotypes of patients with infection using longitudinal vital signs.
OBJECTIVE
The objectives of this study were: (1) to develop and validate vital sign trajectory models using DTW-based clustering algorithms, (2) to compare the subphenotype clustering agreement between the DTW-based models and a previously published GBTM model,12 (3) to compare clustering performance between models using model fit metrics (ie, mean squared error and Davies–Bouldin index), and (4) to compare clustering performance between models using clinical validity metrics (ie, associations of subphenotypes with clinical outcomes and treatment responses).
METHODS
Study cohort
The study cohort included adult patients presenting to the Emergency Department with suspected infection on admission between January 2014 and December 2019 to 4 hospitals in the Emory Healthcare system. Suspected infection was defined as a combination of antibiotic administration within 6 h of presentation and body fluid culture collection. The following exclusion criteria were applied: (1) patients who died or were discharged within the first 8 h, (2) patients transferred to other hospitals during the admission, and (3) patients with less than 3 complete sets of vital signs.12 The study cohort was partitioned into training and validation cohorts by admission year: (1) training—admissions between 2014 and 2017 and (2) validation—admissions between 2018 and 2019. The temporally separate validation cohort was designed to simulate prospective implementation of the subphenotype model to evaluate stability over time with potential changes in patient population, hospital practices, and outcomes.
Measurement of vital signs
Vital signs (ie, oral temperature, heart rate, respiratory rate, systolic, and diastolic blood pressure) from the first 8 h of hospitalization were used in the analysis. Nonphysiological vital signs were excluded.18 The vital signs in the training cohort were standardized to the mean and standard deviation of the training cohort, and the vital signs in the validation cohort were standardized to the mean and standard deviation of the validation cohort. Standardization of variables ensures that all variables are on comparable scales, which can minimize the impact of one variable on the DTW distance metric (eg, temperature does not influence the distance more than respiratory rate). Additionally, standardization can also reduce the impact of outliers, so that the DTW calculation is more robust to extreme values. Vital signs were binned into 8 1-h blocks of time; the mean measurement of each vital sign was used if multiple measurements were available in the 1-h time block. Missing vital sign values were imputed using last observation carried forward (LOCF). If there remained missing data after imputation with LOCF (eg, vital sign at the first hour was missing), next observation carried backward (NOCB) was used. Carry forward imputation uses the conservative assumption that the most likely value of the missing data is the same as the closest-in-time measured value of that vital sign (eg, the most likely temperature measurement is the temperature measurement that is temporally closest to the missing value). Further, carry forward imputation has been used in similar studies of time series clustering using DTW in critically ill patients.11,16
Algorithm development and model selection
The original vitals trajectory subphenotypes were developed and validated using GBTM, a finite mixture model that is used to identify clusters following similar trajectories of variables over time.13,14 The algorithm computes the underlying coefficients for the polynomial functions describing the trajectories of the vital signs over time for each of the groups. As reported, a 4-group model fit the training and validation data best. In this study, 2 additional time series clustering algorithms were developed and compared to the original GBTM model: (1) dynamic time warping with hierarchical clustering (DTW-HC) and (2) dynamic time warping with partitioning around medoids (DTW-PAM). DTW is an algorithm that computes the distance between 2 multivariate temporal sequences by warping the sequences to an optimal alignment. The DTW algorithm computes a distance matrix between pairs of patients and this distance matrix is combined with an additional clustering algorithm (ie, HC and PAM) to identify clusters.
To select the optimal number of clusters and to capture stable clustering assignments from DTW-HC and DTW-PAM, we tested 2 through 6-group models using consensus clustering.19 In consensus clustering, the algorithm (eg, DTW-HC, DTW-PAM) is run 100-times with varying subsamples of patients (subsampled at 80% for each run). The cumulative results for each clustering solution across the 100 runs result in a consensus matrix capturing the distance between patients (ie, the proportion of runs a pair of patients was clustered together). A cumulative density plot of the consensus results was used to calculate the area under the cumulative density function (CDF) curve. A delta area plot was used to evaluate the relative change in the area under the CDF curve with each additional cluster number, and the optimal cluster number was the number of clusters at which there was the highest delta change in area under the CDF curve. Once the optimal number of clusters were selected for DTW-HC and DTW-PAM using the above criteria, a hierarchical clustering algorithm with complete linkage was applied to the consensus matrix to obtain the final cluster assignments for both models. The code used to generate the DTW-HC and DTW-PAM models presented in this manuscript is available on GitHub at https://github.com/siva-bhavani122/Sepsis_Project
Model agreement
Model agreement between the 3 distinct clustering algorithms (GBTM, DTW-HC, and DTW-PAM) was calculated to evaluate whether all algorithms converged on the same underlying physiological trajectories of patients with infection. Adjusted Rand Index (ARI) was used to evaluate the similarity in clustering results between the 3 models.20 An ARI of 0 indicates that the models resulted in nonsimilar clustering and any similarities in matches are due to chance. An ARI of 1 indicates that the models resulted in perfectly matched clusters. Significance of the ARI between models (ie, testing nondifference from 0) was calculated using a published permutational procedure.21 A heatmap was used to visualize the agreement between subphenotypes across models.
Internal clustering metrics of model performance
Since there is no ground truth to determine the “best” model, we used both internal and external metrics of model performance. The internal metrics (ie, model fit metrics) used were mean squared error and Davies–Bouldin index.22,23 A patient’s mean squared error from their assigned subphenotype was calculated as the sum of the squared differences between predicted vital signs (ie, trajectory centroid values) and observed vital signs over the 8-h period. Pairwise testing was performed (ie, patients’ mean squared error from 2 models were compared) across the 3 combinations of models (GBTM and DTW-HC; GBTM and DTW-PAM; DTW-PAM and DTW-HC).
The Davies–Bouldin index is a ratio of within cluster and between cluster separation, with lower values reflecting better performance (ie, patients within clusters are separated by smaller distances while patients in different clusters are separated by larger distances). The Davies–Bouldin index was measured at each time point and visualized over the 8-h period for the 3 models.
External clinical metrics of model performance
Using chi-squared testing, the DTW-HC subphenotypes and the DTW-PAM subphenotypes were evaluated for association of subphenotypes with outcomes (30-day inpatient mortality, ICU admission, vasopressor and inotrope requirement, renal replacement therapy, mechanical ventilation). All outcomes were dichotomous variables. Vasopressor use was defined as the use of norepinephrine, vasopressin, epinephrine, dopamine, or phenylephrine, without a threshold for total daily dose. Inotrope use was defined as the use of milrinone or dobutamine, without a total daily dose threshold. Renal replacement therapy was defined as requiring one or more sessions of hemodialysis.
In logistic regression, the primary outcome of 30-day inpatient mortality was evaluated for association with subphenotypes when adjusting for age, sex, race, and comorbidities (congestive heart failure, chronic pulmonary disease, diabetes mellitus, hypertension, chronic kidney disease, liver disease, and metastatic cancer).
Model performance in validation cohort
Each of the vitals trajectory subphenotypes in both DTW-HC and DTW-PAM models can be represented as a set of 8 centroids for each of the 5 vital signs (ie, the mean value of that vital sign for that subphenotype at every hour from hour 0 to hour 7). Patients in the validation cohort were assigned to the vitals trajectory subphenotype that resulted in the lowest mean squared error from the centroids as done in prior work.7,12 The associations between subphenotypes and outcomes were evaluated in the validation cohort as described above for the training cohort.
Model performance in randomized controlled trial data
The Isotonic Solutions and Major Adverse Renal Events Trial (SMART) was a randomized controlled trial (RCT) comparing balanced crystalloids versus normal saline in critically ill patients.24 In this secondary analysis of the SMART trial, the DTW-HC and DTW-PAM models were applied to the first 8 h of hospitalization vital signs from sepsis patients in the study, by assigning patients to the subphenotype that resulted in the lowest mean squared error.12 The primary outcome was 30-day inpatient mortality for each subphenotype compared between the balanced crystalloid and normal saline treatment arms using a logistic regression model accounting for baseline covariates as prespecified in prior work.25 Heterogeneity of treatment effect (HTE) was calculated using the ANOVA likelihood ratio test between a full logistic regression model predicting mortality including interaction terms between the subphenotype and treatment assignment compared to a nested model without the interaction terms.
RESULTS
There were 20 729 patients with suspected infection in the study cohort: 12 473 patients in the training cohort and 8256 patients in the validation cohort (Supplementary Figure S1). The training cohort was a median of 62 years (IQR 48–75 years), with 51% males, 38% Black patients, 55% White patients, and 7% other race, and with a 2.1% mortality rate. The validation cohort was a median of 62 years (IQR 47–74 years), with 53% males, 39% Black patients, 53% White patients, and 8% other race, and with a 2.2% mortality rate.
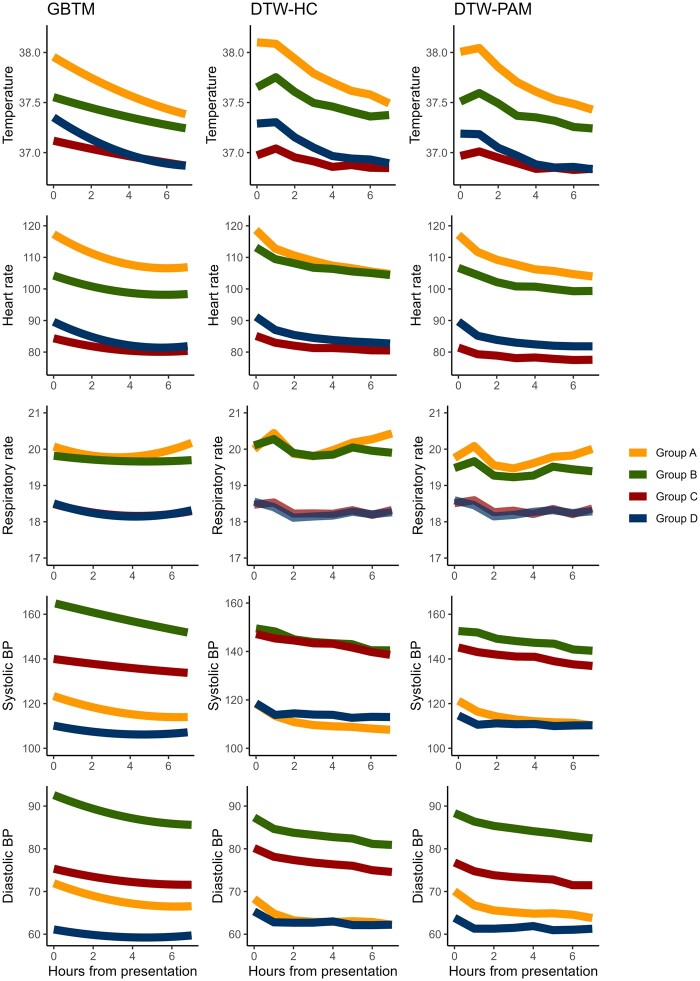
The 4-group model had the highest delta change in area under the CDF curve for both DTW-HC and DTW-PAM algorithms (Supplementary Figures S2 and S3). The 4 subphenotypes identified using DTW-HC and DTW-PAM qualitatively matched the previously developed GBTM subphenotypes: group A was hyperthermic, tachycardic, and tachypneic, and were relatively hypotensive. Group B was also hyperthermic, tachycardic and tachypneic, but not as pronounced as group A, and was hypertensive. Group C and group D had lower temperatures, heart rates, and respiratory rates, with group C being normotensive and group D being the most hypotensive subphenotype (Figure 1).
Figure 1.
Trajectories of vital signs in the first 8 h of admission compared between models. All 3 models converged on 4 visually similar vitals trajectory subphenotypes: group A was hyperthermic, tachycardic, tachypneic, and hypotensive. Group B was also hyperthermic, tachycardic, and tachypneic, but not as pronounced as group A, and were hypertensive. Groups C and D had lower temperatures, heart rates, and respiratory rates, with group C having normal blood pressure and group D being the most hypotensive subphenotype.
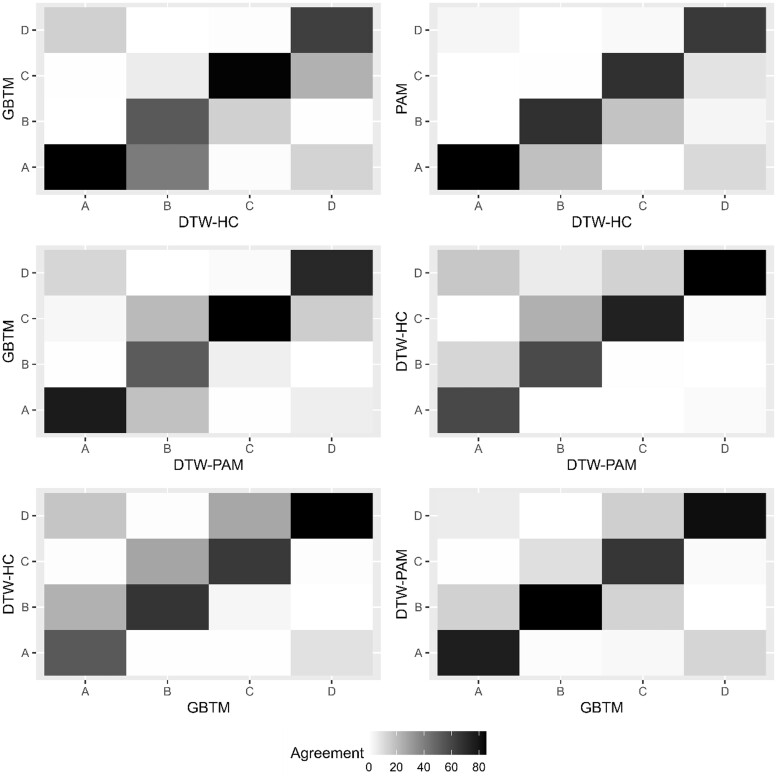
The distributions for subphenotype membership in DTW-HC were group A (N = 2282, 18%), group B (N = 2093, 17%), group C (N = 3273, 26%), and group D (N = 4825, 39%). For DTW-PAM: group A (N = 3297, 26%), group B (N = 2468, 20%), group C (N = 3011, 24%), and group D (N = 3697, 30%). For the previously published GBTM algorithm: group A (N = 3483, 28%), group B (N = 1578, 13%), group C (N = 4044, 32%), and group D (N = 3368, 27%). The ARI was significant between all 3 models (P < .001), suggesting substantial interalgorithm agreement in classification. Between DTW-HC and DTW-PAM, 80% of patients were classified into the same subphenotype, with an ARI 0.68. Between DTW-HC and GBTM, there was 71% agreement and ARI 0.41, and between DTW-PAM and GBTM, there was 79% agreement and ARI 0.55 (Figure 2).
Figure 2.
Heatmap of agreement in subphenotype classification between models. The heatmap presents the percentage of cross-classification of groups A through D compared between the 3 models. The percentage represents the percent of patients in a subphenotype of the model labeled on the x-axis that were classified in the subphenotype of the model labeled on the y-axis, with the diagonal representing agreement in classification (eg, group A in one model classified as group A in the other model). Darker shades represent higher percentages.
Both DTW-HC and DTW-PAM had significantly lower mean squared error compared to GBTM (P = .04 and P < .001, respectively). DTW-PAM also had significantly lower mean squared error compared to DTW-HC (P < .001) (Supplementary Figure S4). Davies–Bouldin index was lowest for GBTM at all time points, signifying better performance (Supplementary Figure S5). Thus, DTW methods had a better fit (ie, lower mean squared error) to individual trajectories, while GBTM had better intergroup separation and intragroup cohesion (ie, lower Davies–Bouldin index).
Model performance in training and validation cohorts
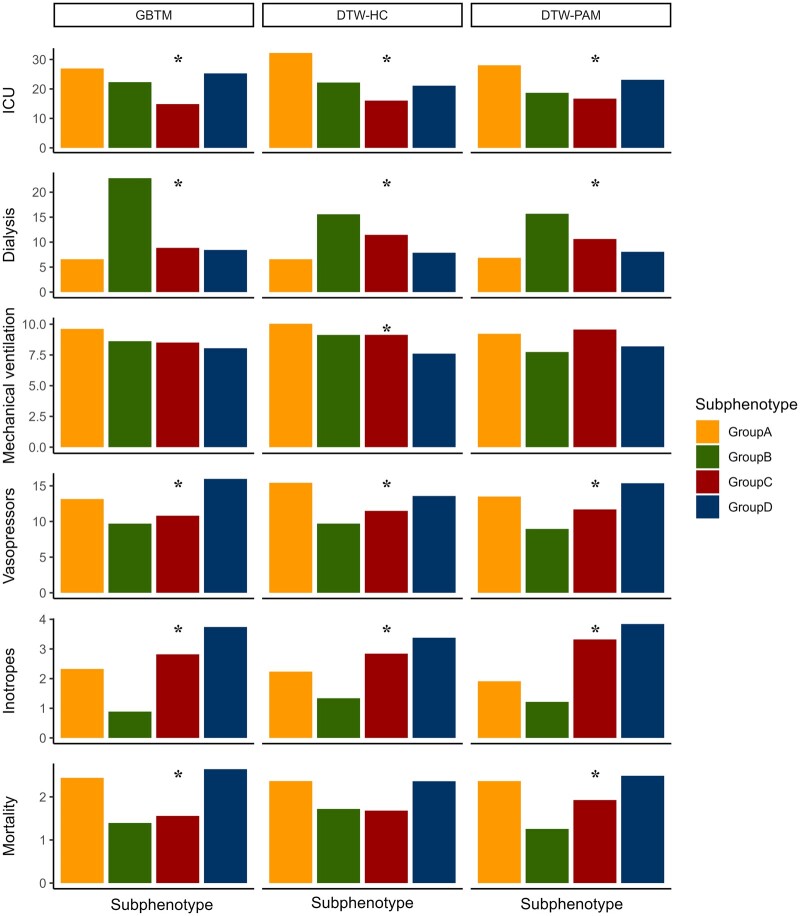
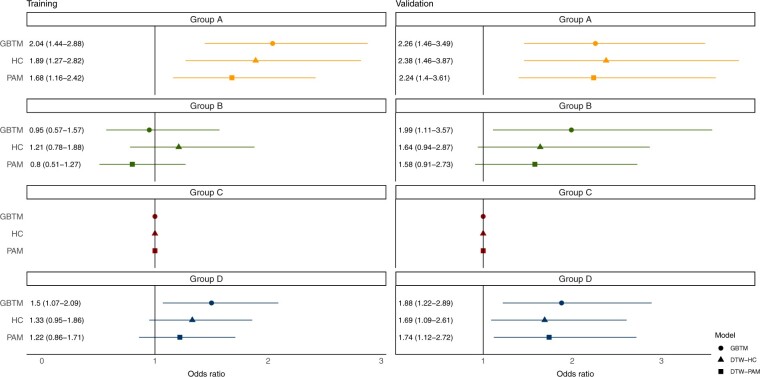
Clinical characteristics and outcomes were consistent across all 3 models: groups A and B were younger, while groups C and D were older (P < .001). Group A had the fewest baseline comorbidities, with the lowest prevalence of congestive heart failure, diabetes mellitus, hypertension, and chronic kidney disease (P < .001). Group B had the highest dialysis requirement (P < .001). groups A and D had higher rates of vasopressor use, ICU transfers, and 30-day mortality (P < .001, Supplementary Tables S1–S3 and Figure 3). Consistent with the GBTM results, on logistic regression, group A had higher odds ratio (OR) of 30-day inpatient mortality in both DTW-HC and DTW-PAM in the training cohort (DTW-HC—OR 1.89, 95% CI 1.27–2.82, P = .002; DTW-PAM—OR 1.68, 95% CI 1.16–2.42, P = .006) and validation cohort (DTW-HC—OR 2.38, 95% CI 1.46–3.87, P < .001; DTW-PAM—OR 2.24, 95% CI 1.40–3.61, P < .001). Group D had higher 30-day mortality only in the validation cohort (DTW-HC—OR 1.69, 95% CI 1.09–2.61, P = .02; DTW-PAM—OR 1.74, 95% CI 1.12–2.72, P = .01; Figure 4).
Figure 3.
Clinical outcomes in subphenotypes in the training cohort. Presented are the clinical outcomes (in percentage) for requiring ICU transfer, dialysis, mechanical ventilation, vasopressors, inotropes, and for 30-day mortality across the 3 models. Asterisk denotes that the outcome was significantly associated with subphenotype in that model. There was significant association between subphenotype and ICU transfer, dialysis, vasopressors, and inotropes in all 3 models. Groups A and D had high ICU transfers and vasopressor use. Group D had high inotrope use. Group B had high incidence of requiring dialysis. Groups A and D had higher 30-day mortality in all models, with significant associations in the GBTM and DTW-PAM models. DTW-PAM: dynamic time warping-partition around medoids; GBTM: group-based trajectory model.
Figure 4.
Odds ratio of 30-day mortality in the training and validation cohorts. Presented are the odds ratio (OR) of 30-day mortality in a logistic regression model adjusting for demographics and comorbidities, with group C as the reference subphenotype. The results are presented for the 3 models in the training and validation cohorts. Group A had higher 30-day mortality in all 3 models in the training and validation cohorts. Group D had higher 30-day mortality in the GBTM model in both training and validation cohorts, and only in the validation cohort in the DTW-HC and DTW-PAM models. DTW-HC: dynamic time warping-hierarchical clustering; DTW-PAM: dynamic time warping-partition around medoids; GBTM: group-based trajectory model.
Model performance in RCT cohort
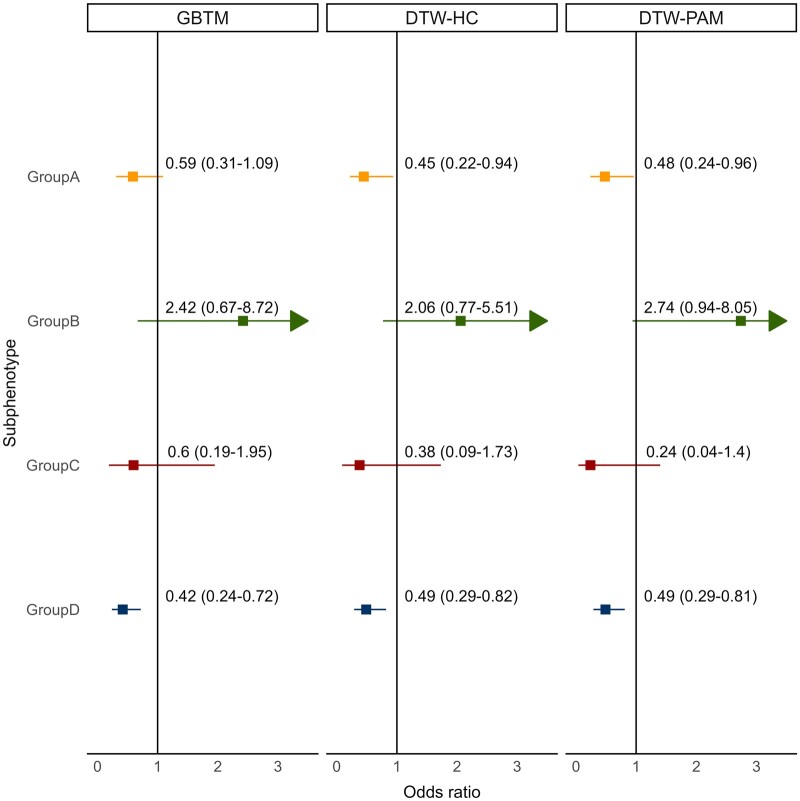
In the SMART secondary analysis, there was significant HTE of balanced crystalloids versus saline in all 3 algorithms (P = .03 for GBTM, P = .04 for DTW-HC, and P = .02 for DTW-PAM). Group D had lower OR of mortality with balanced crystalloids compared to saline in all 3 models (GBTM—OR 0.42, 95% CI 0.24–0.72, P = .002; DTW-HC—OR 0.49, 95% CI 0.29–0.82, P = .006; DTW-PAM—OR 0.49, 95% CI 0.29–0.81, P = .005; Figure 5 and Supplementary Figure S6). Group B trended towards higher OR of mortality with balanced crystalloids in all 3 models (GBTM—OR 2.42, 95% CI 0.67–8.72, P = .2; DTW-HC—OR 2.06, 95% CI 0.77–5.51, P = .2; DTW-PAM—OR 2.74, 95% CI 0.94–8.05, P = .07).
Figure 5.
Odds ratio of 30-day mortality with balanced crystalloids compared to saline. Presented are the odds ratio (OR) of 30-day mortality in patients in each subphenotype treated with balanced crystalloids compared to saline across the 3 models (GBTM, DTW-HC, and DTW-PAM). All 3 models showed significant heterogeneity of treatment effect (P < .05), and group D had a significantly lower OR of mortality with balanced crystalloids compared to saline in all 3 models (GBTM—OR 0.42, 95% CI 0.24–0.72, P = .002; DTW-HC—OR 0.49, 95% CI 0.29–0.82, P = .006; DTW-PAM—OR 0.49, 95% CI 0.29–0.81, P = .005). Since the entire confidence interval for group B could not be presented in the figure, the arrow signifies that the confidence interval extends beyond the presented axis. DTW-HC: dynamic time warping-hierarchical clustering; DTW-PAM: dynamic time warping-partition around medoids; GBTM: group-based trajectory model.
DISCUSSION
In this multicenter study comparing 3 multivariate time series clustering algorithms applied to vital signs from patients with infection, we found substantial consistency in the resulting trajectory subphenotypes. First, the optimal number of clusters were 4 subphenotypes for all 3 models. Second, there was significant agreement in subphenotype classification between models. Third, all models found subphenotypes with similar trajectory shapes. Fourth, all models found subphenotypes with similar distribution of clinical outcomes. Fifth, all 3 models demonstrated HTE to intravenous fluids and identified a consistent subphenotype with a significant mortality benefit from balanced crystalloids. This consistency is clinically significant and provides evidence that the vitals trajectory subphenotypes have a physiological basis independent of the computational approach.
Since there is no definitive method of confirming the “best” subphenotyping algorithm, a multifaceted approach was used to evaluate the algorithms for: (1) agreement in subphenotype classification and consistency of underlying trajectory patterns, (2) comparison of internal model fit metrics, and (3) comparison of external clinical metrics of model performance using associations with clinical outcomes and treatment responses. The most important finding was that all 3 algorithms found consistent subphenotypes with 71–80% agreement in classification using longitudinal vital signs. The 4 subphenotypes were: group A—higher temperature, heart rate, and respiratory rate, and lower blood pressure; group B—higher temperature, heart rate, and respiratory rate (although not as pronounced as group A), and high blood pressure. Group C—relatively lower temperature, heart rate and respiratory rate, and normal blood pressure. Group D—relatively lower temperature, heart rate, respiratory rate, and the lowest blood pressure. The trajectory shapes for these subphenotypes were consistent in both the training and validation cohorts in all 3 models.
Internal metrics of model performance were compared using mean squared error and Davies–Bouldin index, without a consistent “winner”. Both DTW algorithms had lower mean squared error compared to GBTM, with DTW-PAM having the lowest mean squared error. However, GBTM had the lowest Davies–Bouldin index, suggesting reduced within cluster separation and increased between cluster separation. DTW offers the advantage of identifying unrestricted trajectory shapes, while GBTM is restricted to the specified polynomial function.13,15 This potential advantage likely resulted in lower mean squared error for the DTW models. However, this advantage may be offset by the computational cost of DTW, especially if there is no additional benefit in clinical metrics of model performance. DTW operates on quadratic time and requires the pairwise comparison of all patients in the training cohort—resulting in the building of a 12 473 by 12 473 distance matrix for this study’s training cohort. With larger datasets, this type of model building may not be feasible.26
The subphenotypes had similar clinical outcomes and treatment responses in all 3 models, with groups A and D having the highest mortality, and the highest vasopressor and ICU requirement. Group B had the highest dialysis requirement. Group C had the lowest ICU requirement. These outcomes were similarly distributed in the validation cohorts in all 3 models. Additionally, all models discovered the same subphenotype (group D) in RCT data with significant benefit from balanced crystalloids. Intravenous fluids are one of the most common interventions in sepsis, but after multiple RCTs enrolling over 35 000 patients, there is still uncertainty in what type of intravenous fluids (balanced crystalloids vs saline) should be given to which patients.27 In our study, all 3 models found that group D had a number needed to treat with balanced crystalloids of 6–8 patients for a reduction in 30-day mortality. This is a significant clinical finding, and suggests that regardless of computational approach, there may be an underlying physiological pattern that can inform clinical practice and portends a significant mortality benefit from balanced crystalloids.
Despite differences in internal model fit metrics, the consistency in trajectory shapes, clinical outcomes, and responses to treatments suggest similar clinical performance in the 3 algorithms. For high frequency oscillating data such as continuous vitals monitoring, DTW’s unrestricted shape may be advantageous in identifying distinct latent subphenotypes that GBTM would not be able to identify. Additionally, for data over a longer observation window, DTW’s nonlinear temporal warping and matching may identify patterns that would otherwise be missed with GBTM. For sparse clinical measurements over a short observation window such as the data used in our study, we recommend GBTM given its parsimony and relative computational simplicity. The appropriate time series clustering algorithm for different critical care settings requires further research.
The study had several limitations. First, the 3 algorithms evaluated are in no way comprehensive, and represent a small portion of the growing number of multivariate time series clustering algorithms available; some of these algorithms may identify consistent subphenotypes while others may discover distinct latent subphenotypes. Second, all hospitals in the study were within a single health system. Third, missing vitals data were present since the data were collected as clinically indicated, which may have introduced inaccuracies in subphenotype membership assignment.
This study is the first of its kind to compare multiple multivariate time series clustering algorithms in identifying subphenotypes of patients with infection. The findings of this study provide important insights for investigating dynamic subphenotypes in critically ill patients. The results suggest that time series clustering can be used to discover dynamic subphenotypes of infected patients with distinct clinical characteristics, outcomes, and responses to treatments. The most clinically important finding is the identification of a consistent subphenotype with a significant mortality benefit from balanced crystalloids compared to saline.
In conclusion, this study provides an evaluation framework for comparing time series clustering algorithms. A holistic evaluation should include comparison of intermodel agreement in clustering, internal metrics of clustering fit, and external metrics of association with relevant clinical outcomes. Additionally, selecting the optimal model should take into consideration implementation factors such as model parsimony, computational expense, and feasibility for real-time patient subphenotyping.
Supplementary Material
Contributor Information
Sivasubramanium V Bhavani, Department of Medicine, Emory University, Atlanta, Georgia, USA; Emory Critical Care Center, Atlanta, Georgia, USA.
Li Xiong, Department of Computer Science, Emory University, Atlanta, Georgia, USA.
Abish Pius, Department of Computational & Systems Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Matthew Semler, Department of Medicine, Vanderbilt University, Nashville, Tennessee, USA.
Edward T Qian, Department of Medicine, Vanderbilt University, Nashville, Tennessee, USA.
Philip A Verhoef, Department of Medicine, University of Hawaii John A. Burns School of Medicine, Honolulu, Hawaii, USA; Hawaii Permanente Medical Group, Honolulu, Hawaii, USA.
Chad Robichaux, Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
Craig M Coopersmith, Emory Critical Care Center, Atlanta, Georgia, USA; Department of Surgery, Emory University, Atlanta, Georgia, USA.
Matthew M Churpek, Department of Medicine, University of Wisconsin, Madison, Wisconsin, USA; Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, Wisconsin, USA.
FUNDING
This work was supported by NIH/NIGMS (GM144867), NIGMS (GM145330), Department of Defense (W81XWH-21-1-0009), NHLBI (R01HL157262), NIDDK (DK126933), NIGMS (GM072808, GM104323, GM148217), NHLBI (T32HL087738), and NCATS (UL1TR002378).
AUTHOR CONTRIBUTIONS
Study concept and design: SVB, MMC, CMC; acquisition of data: SVB, CR, EQ, MS; analysis and interpretation of data: all authors; first drafting of the manuscript: SVB; critical revision of the manuscript for important intellectual content: all authors; statistical analysis: SVB, AP; administrative, technical, and material support: SVB, CR, MS; study supervision: MMC, CMC; data access and responsibility: All authors take responsibility for the integrity of the data and the accuracy of the data analysis.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTERESTS STATEMENT
MMC is a named inventor on a patent for a risk stratification algorithm for hospitalized patients (US patent # 11 410 777). The other authors have no competing interests to declare.
DATA AVAILABILITY
The patient data used in this study cannot be made publicly available due to privacy and ethical concerns. However, the data can be made available to researchers upon reasonable request and subject to a Data Use Agreement (DUA) that restricts the use of the data for research purposes only and prohibits any attempt to identify individual patients.
REFERENCES
- 1. Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: for the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315 (8): 762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med 2014; 20 (4): 195–203. [DOI] [PubMed] [Google Scholar]
- 3. Scicluna BP, Baillie JK.. The search for efficacious new therapies in sepsis needs to embrace heterogeneity. Am J Respir Crit Care Med 2019; 199 (8): 936–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shah FA, Meyer NJ, Angus DC, et al. A research agenda for precision medicine in sepsis and acute respiratory distress syndrome: an official American Thoracic Society Research Statement. Am J Respir Crit Care Med 2021; 204 (8): 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cazalis MA, Lepape A, Venet F, et al. Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive Care Med Exp 2014; 2 (1): 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maslove DM, Wong HR.. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol Med 2014; 20 (4): 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhavani SV, Carey KA, Gilbert ER, Afshar M, Verhoef PA, Churpek MM.. Identifying novel sepsis subphenotypes using temperature trajectories. Am J Respir Crit Care Med 2019; 200 (3): 327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhavani SV, Wolfe KS, Hrusch CL, et al. Temperature trajectory subphenotypes correlate with immune responses in patients with sepsis. Crit Care Med 2020; 48 (11): 1645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bos LDJ, Sjoding M, Sinha P, et al. ; PRoVENT-COVID collaborative group. Longitudinal respiratory subphenotypes in patients with COVID-19-related acute respiratory distress syndrome: results from three observational cohorts. Lancet Respir Med 2021; 9 (12): 1377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perizes EN, Chong G, Sanchez-Pinto LN.. Derivation and validation of vasoactive inotrope score trajectory groups in critically ill children with shock. Pediatr Crit Care Med 2022; 23 (12): 1017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Z, Mao C, Su C, et al. Sepsis subphenotyping based on organ dysfunction trajectory. Crit Care 2022; 26 (1): 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhavani SV, Semler M, Qian ET, et al. Development and validation of novel sepsis subphenotypes using trajectories of vital signs. Intensive Care Med 2022; 48 (11): 1582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagin DS, Jones BL, Passos VL, Tremblay RE.. Group-based multi-trajectory modeling. Stat Methods Med Res 2018; 27 (7): 2015–23. [DOI] [PubMed] [Google Scholar]
- 14. Nagin DS, Odgers CL.. Group-based trajectory modeling in clinical research. Annu Rev Clin Psychol 2010; 6 (1): 109–38. [DOI] [PubMed] [Google Scholar]
- 15. Berndt DJ, Clifford J, eds.. Using Dynamic Time Warping to Find Patterns in Time Series. KDD Workshop; 1994: 359–70.
- 16. Su C, Xu Z, Hoffman K, et al. Identifying organ dysfunction trajectory-based subphenotypes in critically ill patients with COVID-19. Sci Rep 2021; 11 (1): 15872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burke H, Freeman A, O’Regan P, et al. Biomarker identification using dynamic time warping analysis: a longitudinal cohort study of patients with COVID-19 in a UK tertiary hospital. BMJ Open 2022; 12 (2): e050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Churpek MM, Zadravecz FJ, Winslow C, Howell MD, Edelson DP.. Incidence and prognostic value of the systemic inflammatory response syndrome and organ dysfunctions in ward patients. Am J Respir Crit Care Med 2015; 192 (8): 958–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilkerson MD, Hayes DN.. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010; 26 (12): 1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santos JM, Embrechts M.. On the Use of the Adjusted Rand Index as a Metric for Evaluating Supervised Classification. Berlin, Heidelberg: Springer; 2009: 175–184. [Google Scholar]
- 21. Qannari EM, Courcoux P, Faye P.. Significance test of the adjusted Rand index. Application to the free sorting task. Food Qual Prefer 2014; 32: 93–7. [Google Scholar]
- 22. Aghabozorgi S, Seyed Shirkhorshidi A, Ying Wah T.. Time-series clustering—a decade review. Inform Syst 2015; 53: 16–38. [Google Scholar]
- 23. Davies DL, Bouldin DW.. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 1979; PAMI-1 (2): 224–7. [PubMed] [Google Scholar]
- 24. Semler MW, Self WH, Wanderer JP, et al. Balanced crystalloids versus saline in critically ill adults. N Engl J Med 2018; 378 (9): 829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown RM, Wang L, Coston TD, et al. Balanced crystalloids versus saline in sepsis. A secondary analysis of the SMART clinical trial. Am J Respir Crit Care Med 2019; 200 (12): 1487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silva DF, Batista G. Speeding up all-pairwise dynamic time warping matrix calculation. In: Proceedings of the 2016 SIAM International Conference on Data Mining (SDM); Miami, FL; 2016: 837–45.
- 27. Hammond NE, Zampieri FG, Tanna GLD, et al. Balanced crystalloids versus saline in critically ill adults: a systematic review with meta-analysis. NEJM Evid 2022; 1 (2): EVIDoa2100010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The patient data used in this study cannot be made publicly available due to privacy and ethical concerns. However, the data can be made available to researchers upon reasonable request and subject to a Data Use Agreement (DUA) that restricts the use of the data for research purposes only and prohibits any attempt to identify individual patients.