Abstract
Disruption in neurogenesis and neuronal migration can influence the assembly of cortical circuits, affecting the excitatory-inhibitory balance and resulting in neurodevelopmental and neuropsychiatric disorders. Using ventral cerebral organoids and dorsoventral cerebral assembloids with mutations in the extracellular matrix gene LGALS3BP, we show that extracellular vesicles released into the extracellular environment regulate the molecular differentiation of neurons, resulting in alterations in migratory dynamics. To investigate how extracellular vesicles affect neuronal specification and migration dynamics, we collected extracellular vesicles from ventral cerebral organoids carrying a mutation in LGALS3BP, previously identified in individuals with cortical malformations and neuropsychiatric disorders. These results revealed differences in protein composition and changes in dorsoventral patterning. Proteins associated with cell fate decision, neuronal migration, and extracellular matrix composition were altered in mutant extracellular vesicles. Moreover, we show that treatment with extracellular vesicles changes the transcriptomic profile in neural progenitor cells. Our results indicate that neuronal molecular differentiation can be influenced by extracellular vesicles.
The extracellular matrix protein LGALS3BP and extracellular vesicles regulate interneuron specification.
INTRODUCTION
Neurogenesis and neuronal migration are essential processes for mammalian brain development, especially for the correct assembly of neuronal circuits. Cortical circuitry function relies on an adequate excitatory/inhibitory balance, coordinated by glutamatergic and GABAergic neuronal activities, such that the regulation of migration of these respective cell types during development is critical (1, 2).
The mammalian neocortex is mainly populated by glutamatergic excitatory neurons, while GABAergic inhibitory neurons [here, “interneurons” (INs)] are present in smaller proportions (3). In rodents, approximately 15 to 20% of neocortical neurons are INs (4), while primates, including humans, have higher proportions (circa 24%) (5–7).
In humans, excitatory neurons are generated from apical and basal radial glia cells (aRGs and bRGs, respectively) as well as intermediate progenitors (IPs) located in the germinal zones: the ventricular (VZ), subventricular (SVZ), and outer subventricular zone (OSVZ). Using the basal process of aRGs and bRGs as a scaffold, they migrate radially into the cortical layers. This process relies on the integrity of aRGs, and defects in radial migration can lead to the disorganization of the cortical layers or ectopically located neurons, which are hallmarks of cortical malformations (CMs) (8–10).
While excitatory neurons migrate and remain in the dorsal forebrain, most cortical INs migrate over long distances from their birthplace to their final positions in the neocortex. Specifically, INs are born in the ganglionic eminences (GEs) in the ventral forebrain from the proliferative zone of the medial GE (MGE) and the caudal GE (CGE) (3, 11). As they become postmitotic, INs migrate tangentially into the neocortex. INs are highly polarized, and during their migration, they extend branches from the leading process. The stabilization of one of the branches leads to cytoskeleton remodeling resulting in nucleus displacement in a process called nucleokinesis. This cyclic movement is characteristic of IN migration and, in particular, of their saltatory migratory behavior (3, 12, 13).
The extracellular space, composed of the extracellular matrix (ECM), soluble molecules, and extracellular vesicles (EVs), plays a crucial role in brain development. Specifically, the ECM, a complex and dynamic cellular microenvironment, regulates progenitor proliferation, differentiation, morphogenesis, and neuronal migration (14). Moreover, IN migration is guided by extracellular attractive and repulsive cues, such as extrinsic signals, including molecules and proteins secreted into the extracellular space or by cell-cell contact. Eph and ephrins, as well as Robo, Slit, and netrins, are attractive and repulsive cues that guide cell migration (3, 15–17).
In this study, we analyze the role of the extracellular environment in IN specification and migration, focusing on the ECM component LGALS3BP. Within the ECM, LGALS3BP interacts with integrins, fibronectins, galectins, laminins, and tetraspanins (18, 19).
We have previously shown that LGALS3BP is enriched in human neuronal progenitor cells (NPCs), and its product is secreted via EVs (20). A de novo variation in exon 5 of LGALS3BP has been diagnosed in a patient presenting CM, developmental delay, autism, dysarthria, ataxia, and focal seizures, suggesting a crucial role during brain development. Moreover, we recently showed that LGALS3BP is critical for human corticogenesis, shaping the extracellular environment and regulating NPC delamination and neuronal migration through its extrinsic function (20). Together, these previous findings highlight LGALS3BP as an interesting candidate gene for the investigation of non–cell-autonomous regulation in brain development, as well as in both physiological and pathological conditions.
Here, using human cerebral organoids and assembloids as a model system, we show that IN specification is influenced by secreted factors released into the extracellular environment. Our findings suggest that the extracellular protein LGALS3BP can regulate the patterning of ventral progenitors and migration of INs. Moreover, we propose that the non–cell-autonomous function of LGALS3BP is mediated by EVs.
RESULTS
LGALS3BP E370K-mutant ventral organoids show alterations in cell identity
To investigate whether the specification of INs might be influenced by secreted factors, we chose LGALS3BP as a model to study this phenomenon because of its known secretion and function during neurodevelopment (20). LGALS3BP is expressed in GE human progenitor cells and IN precursors from cerebral organoids (21), and in fetal brain tissue, it is enriched in MGE progenitors (22). It is detected in all cortical IN subtypes, with a higher expression in LGE and CGE INs (5). On the contrary, its expression in the developing mouse brain is undetectable (23).
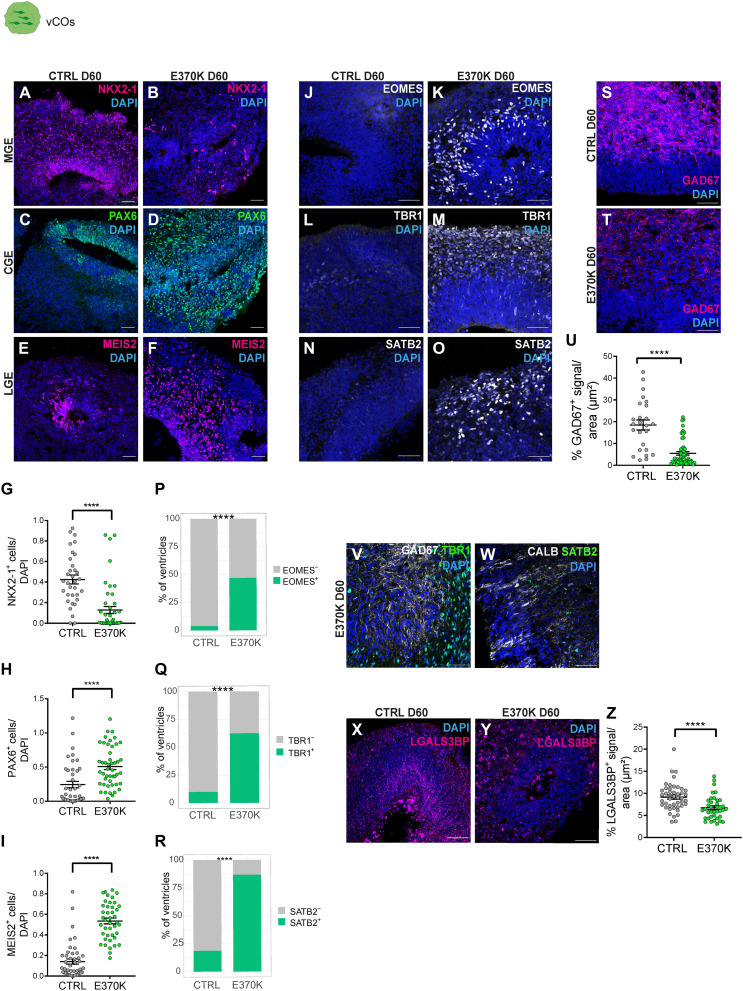
To this end, we generated ventral cortical organoids (vCOs) (24), which serve as a model to investigate cell fate and differentiation of ventral progenitors into INs. The ventral proliferative zones of MGE and CGE generate different subpopulations of INs (3), while lateral GE (LGE) mostly gives rise to medium spiny neurons (fig. S1A) (25). We used an induced pluripotent stem cell (iPSC) line carrying the LGALS3BP variant in heterozygosity (E370K) previously described (fig. S1B) (20). We generated vCOs from an isogenic control (CTRL) and the E370K iPSC line and characterized both at different stages of development. In particular, we analyzed the expression of typical markers of MGE (NKX2-1), CGE (PAX6), and LGE (MEIS2) (7, 24, 25) in vCOs at day 30 (D30), D45, and D60 of culture (Fig. 1, A to F, and fig. S1, C to N). At D30, E370K-vCOs were highly enriched in progenitors and did not show differences in the number of NKX2-1+ cells and PAX6+ cells (fig. S1, G and H). The LGE marker MEIS2 was not detected in D30 vCOs. We observed a significant decrease in NKX2-1+ cells and a significant increase in PAX6+ and MEIS2+ cells in D45 and D60 E370K-vCOs (Fig. 1, G to I, and fig. S1, O to Q).
Fig. 1. LGALS3BP E370K-mutant ventral organoids show alterations in cell identity.
(A to F) Micrograph of D60 CTRL and E370K-vCOs sections immunostained for NKX2-1 (A) and (B), PAX6 (C) and (D), and marker MEIS2 (E) and (F). (G to I) Quantification of the number of NKX2-1+ cells (G), PAX6+ cells (H), and MEIS2+ cells (I) normalized by 4′,6-diamidino-2-phenylindole (DAPI) in D60 E370K-vCOs. Statistical significance was based on two-tailed Mann-Whitney U test; ****P < 0.0001. (J to O) Micrograph of D60 CTRL and E370K-vCOs sections immunostained for IP marker EOMES (J) and (K), deep layer cortical neuron TBR1 (L) and (N), and upper layer cortical neuron SATB2 (N) and (O). (P to R) Quantification of the percentage of ventricles with EOMES+ cells (P), TBR1+ cells (Q), and SATB2+ cells (R), showing the unexpected expression of EOMES in D60 E370K-vCOs. Statistical significance was based on the exact binomial test; ****P < 0.0001. (S and T) Micrograph of D60 CTRL and E370K-vCO sections immunostained for GAD67. (U) Quantification of the percentage of GAD67+ signal normalized per area (μm2) in E370K-vCOs. Statistical significance was based on two-tailed Mann-Whitney U test; ****P < 0.0001. (V and W) Micrographs of D60 E370K-vCOs sections immunostained GAD67 (gray) and TBR1 (green) (V) as well as for CALB1 (gray) and SATB2 (green) (W). (X and Y) Micrographs of D60 CTRL and E370K-vCOs sections immunostained for LGALS3BP. (Z) Quantification of the percentage of LGALS3BP+ signal normalized per area (μm2). Statistical significance was based on the two-tailed Mann-Whitney U test; ****P < 0.001. Each individual data point represents one GE unit (ventricle). Data are shown as means ± SEM. Scale bars, 50 μm.
These data suggest that E370K-vCOs generate less MGE and more CGE and LGE regions, resulting in a different proportion of IN precursors compared to CTRL. However, PAX6 is also a marker for dorsal cortical progenitors in the VZ and SVZ. To understand the regional identity of the PAX6+ cells found in D45 and D60 E370K-vCOs, we performed immunohistochemistry for EOMES, a dorsal marker for cortical IPs (Fig. 1, J and K, and fig. S1, R and S). Unexpectedly, E370K-vCOs express EOMES, specifically EOMES-labeled cells that were identified in 25% of the observed ventricles in D45 vCOs (fig. S1T) and in 50% of the observed ventricles in D60 vCOs (Fig. 1P). To investigate the regional identity of neurons, we also evaluated the expression of TBR1—a marker for deep-layer cortical neurons—and SATB2—a marker for upper-layer cortical neurons in D60 vCOs (Fig. 1, L to O), where neurons are more abundant. TBR1 was expressed in approximately 60% of the ventricles (Fig. 1Q) and SATB2 in approximately 90% of the ventricles (Fig. 1R), indicating that E370K-vCOs unexpectedly express dorsal cortical markers.
The presence of cortical IPs and neurons was combined with a significant decrease in GAD67+, CALB1+, and CALB2+ INs in D60 E370K-vCOs (Fig. 1, S to U, and fig. S1, U to Z). To confirm whether the neurons expressing TBR1 and SATB2 in E370K-vCOs also coexpressed interneuronal markers, we performed an analysis in D60 E370K-vCOs using TBR1 and GAD67, or SATB2 and CALB1 (Fig 1, V and W). Mutant cells did not coexpress dorsal and ventral neuronal markers. Moreover, mutant vCOs were characterized by a significantly decreased LGALS3BP expression compared to CTRL (Fig. 1, X to Z).
As we have previously shown that LGALS3BP controls apical detachment of radial glial cells dorsally (20), we analyzed delamination in D60 CTRL and E370K-vCOs by performing immunohistochemistry for a pH3 antibody. We found no difference in the position of pH3+ cells in V-SVZ and MZ, suggesting no differences in the delamination process in ventral progenitor cells (fig. S1, AA to DD).
Additional mutations located in exon 5 of LGALS3BP have been associated with brain malformations (20). We, therefore, generated a genetically edited iPSC line lacking exon 5 (Y366Lfs) in homozygosity (fig. S1B) (20). Similarly, to E370K-vCOs, D60 Y366Lfs-vCOs showed reduced expression of LGALS3BP (fig. S2, A and B) and expressed EOMES, TBR1, and SATB2 (fig. S2, C to K), suggesting that exon 5 of LGALS3BP is essential for its function on the correct specification of INs. These results show that LGALS3BP plays a role in dorsoventral patterning, regulating cell differentiation in the ventral forebrain.
LGALS3BP variation causes alterations in cell fate and developmental trajectory
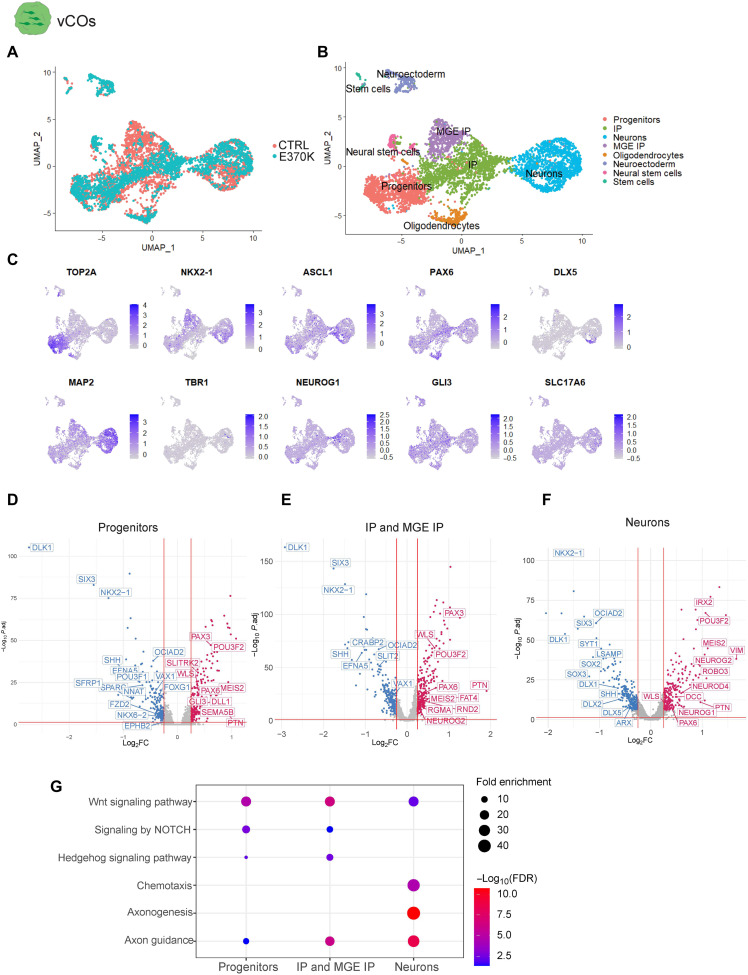
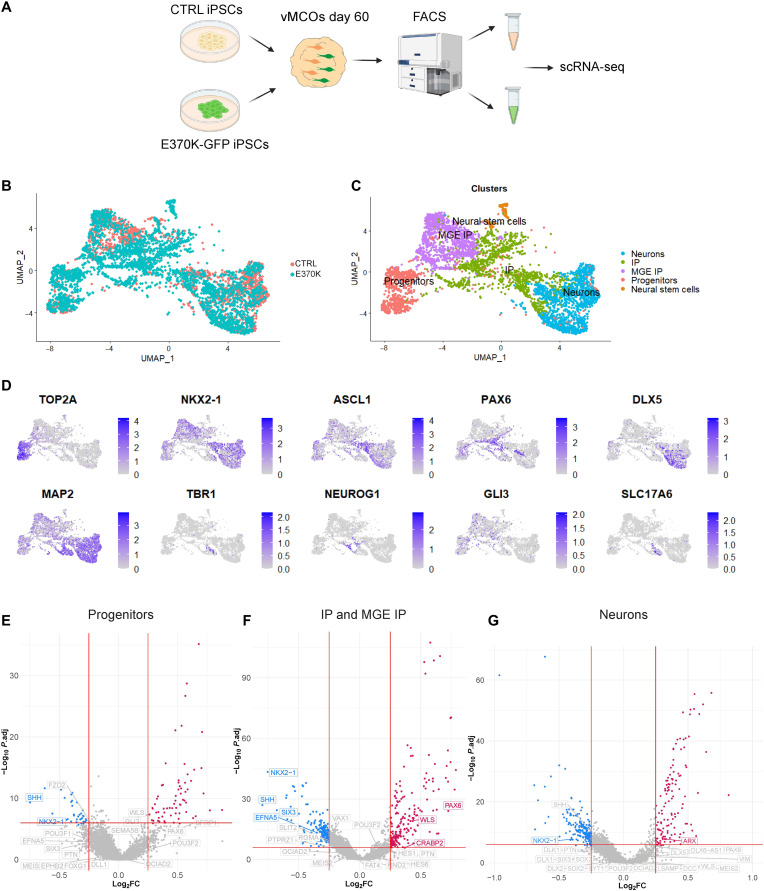
To dissect the transcriptional signatures of altered cells in E370K-vCOs, we performed single-cell RNA sequencing (scRNA-seq) analysis in D60 CTRL-vCOs and E370K-vCOs. The 5369 identified cells clustered into eight main groups (Fig. 2, A and B), including progenitors expressing TOP2A and IPs expressing NKX2-1, ASCL1, and PAX6, as well as neurons expressing MAP2 and DLX5 (Fig. 2C). We also identified cortical genes, such as TBR1, NEUROG1, GLI3, and SLC17A6 being expressed in E370K-vCOs (Fig. 2C and fig. S3A), confirming their dorsal identity also at the transcriptomic level.
Fig. 2. LGALS3BP variation causes alterations in cell fate and developmental trajectory.
(A) Uniform manifold approximation and projection (UMAP) visualization of scRNA-seq data of D60 and E370K-vCOs (n = 5369 cells from a pool of five organoids per condition). (B) UMAP visualization of scRNA-seq clusters of D60 CTRL and E370K-vCOs. Each color cluster represents a different cell population. (C) Feature plots depicting the expression of progenitor markers (TOP2A), IP markers (NKX2-1, ASCL1, and PAX6), and neuron markers (DLX5, MAP2, TBR1, NEUROG1, GLI3, and SLC17A6) in D60 vCOs. (D) Volcano plot showing differentially expressed genes in E370K progenitors, plotting the negative log10 adjusted P value (P.adj) of all genes against their log2 fold change (log2FC) (E370K- versus CTRL-vCOs). Significantly expressed genes (P.adj < 0.01) are labeled. Up-regulated genes are labeled in red, while down-regulated are labeled in blue. (E) Volcano plot showing differentially expressed genes in E370K IPs and MGE IPs, plotting the negative log10 P.adj of all genes against their log2 fold change (E370K- versus CTRL-vCOs). Significantly expressed genes (P.adj < 0.01) are labeled. Up-regulated genes are labeled in red, while down-regulated are labeled in blue. (F) Volcano plot showing differentially expressed genes in E370K neurons, plotting the negative log10 P.adj of all genes against their log2 fold change (E370K- versus CTRL-vCOs). Significantly expressed genes (P.adj < 0.01) are labeled. Up-regulated genes are labeled in red, while down-regulated are labeled in blue. (G) GO term analysis of dysregulated pathways in progenitors, in IPs and MGE IPs, and in neurons in E370K-vCOs. The size of the dots represents the number of genes enriched in the GO term, and the color of the dots represents the −log10 [false discovery rate (FDR)].
Then, to investigate whether the LGALS3BP variation could cause developmental trajectory alterations, ventral telencephalic cells were aligned on a developmental pseudo-differentiation axis showing the trajectory from progenitors to neurons (fig. S3, B and C). We could not detect major changes in the pseudo-differentiation trajectory in mutant vCOs (fig. S3C). Using Monocle 3 (https://cole-trapnell-lab.github.io/monocle3/) (26), we identified pseudo-time trajectories in both CTRL-vCOs and E370K-vCOs (fig. S3, D to F). As expected from the decrease of NXK2-1+ MGE IPs (Fig. 1, A and B, and fig. S3, F and G) in E370K-vCOs, the IP-MGE trajectory is missing in mutant organoids.
Next, we performed differential expression (DE) analysis in progenitors (IPs and MGE-IPs) and neurons. All three populations of cells in E370K-vCOs down-regulated transcription factors (TFs), such as NKX2-1, NKX6-2, SIX3, and OCIAD2, and the secreted molecule SHH, all associated with ventral patterning. On the other hand, E370K-vCOs up-regulated dorsal TFs, such as PAX6, PTN, POU3F2, GLI3, and NEUROG2, and the secreted molecule WLS, confirming the dorsal identity of E370K ventral cells (Fig. 2, D to F). In particular, we found dysregulation of the WNT, NOTCH, and HEDGEHOG pathways, known to regulate cell differentiation and dorsoventral patterning (Fig. 2, D to G) (27–33). We also identified molecules associated with axon guidance: EFNA5, EPHB2, SEMA5B, and SLIT2. Moreover, ROBO3, DCC, FAT4, DLL1, and RND2, genes previously associated with periventricular heterotopia (PH) (10), were dysregulated in E370K neurons (Fig. 2F), suggesting that the cell fate switch can affect the dynamics of migrating neurons. Mutant neurons showed down-regulation of IN markers, such as DLX1, DLX2, and DLX5, while they up-regulated cortical markers such as NEUROG1, NEUROG2, and NEUROD4 (Fig. 2F), confirming the dorsal identity of E370K-vCOs.
The DE analysis revealed that E370K-vCOs dysregulate genes involved in pattern specification, regionalization, differentiation, and neurogenesis, as indicated by the enriched Gene Ontology (GO) terms (fig. S3, H to J), suggesting a specific contribution of LGALS3BP in human ventral forebrain development. The alteration in expression of secreted and transmembrane molecules, such as SHH, WLS, EPHB2, and SLIT2 in E370K-vCOs, showed that cell fate might be regulated in a non–cell-autonomous manner.
Cell fate changes result in migratory dynamics defects
Cortical neurons and INs migrate in different manners (12, 34, 35), and an altered cell fate could lead to migratory defects. In particular, the patient with the LGALS3BP variation presents PH (20).
To understand whether the cell fate changes observed in E370K-vCOs can lead to migratory defects and, therefore, to PH, we studied the migratory dynamics of E370K neurons with a dorsal identity. For this purpose, we generated human dorsoventral cerebral assembloids (dvCAs), a suitable human model system to study IN migration by resembling the ventral-dorsal forebrain axis in vitro (24, 36, 37). dvCAs express typical ventral forebrain markers such as NKX2-1 in the ventral region (vCAs) and dorsal forebrain markers such as TBR1 in the dorsal region (dCAs) (fig. S4, A to D).
We then generated dvCAs from isogenic CTRL and genetically edited iPSCs, LGALS3BP-E370K (E370K) (Fig. 3A). We generated control dvCAs (vCTRL-dCTRL) and dvCAs with a mutant ventral side (vE370K-dCTRL) to focus on the migration of ventral cells with the LGALS3BP variation (Fig. 3A).
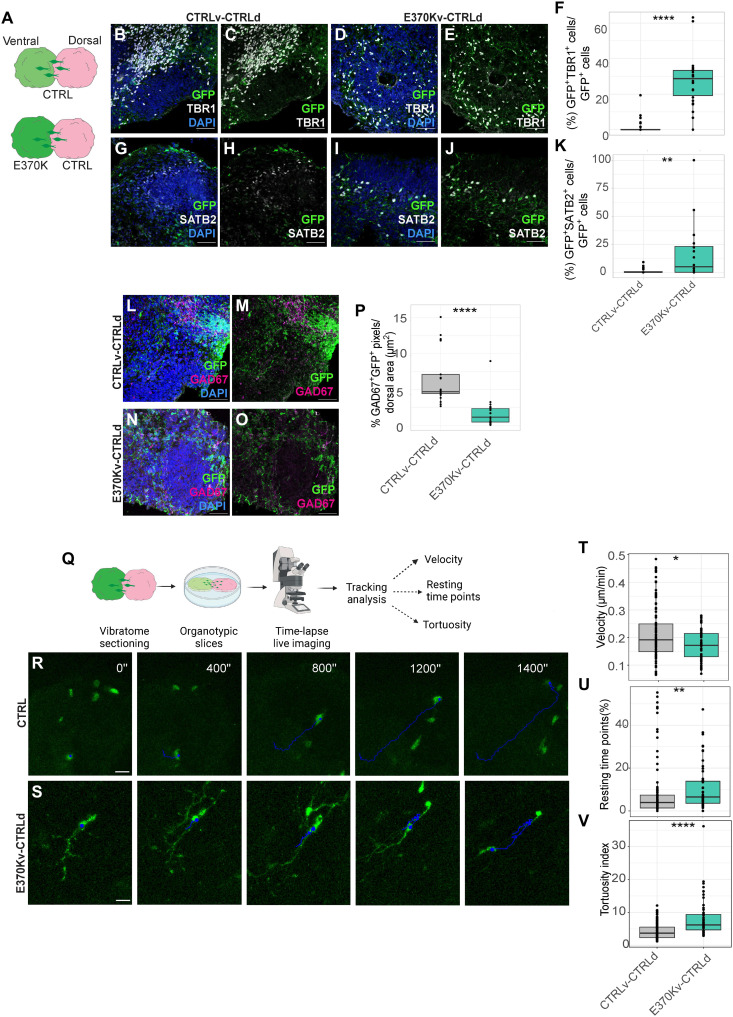
Fig. 3. Cell fate changes result in migratory dynamics defects.
(A) Schematic of the experimental setup showing dvCAs. (B to E) Micrograph of the dorsal region of D60 CTRLv-CTRLd (B) and (C) and E370Kv-CTRLd CAs (D) and (E) immunostained for green fluorescent protein (GFP) and TBR1. (F) Quantification of the percentage of GFP+ cells expressing TBR1 normalized by GFP+ cells in D60 CTRLv-CTRLd and E370Kv-CTRLd CAs. (G to J) Micrograph of the dorsal region of D60 CTRLv-CTRLd (G) and (H) and E370Kv-CTRLd CAs (I) and (J) immunostained for GFP and SATB2. (K) Quantification of the percentage of GFP+ cells expressing SATB2 normalized by GFP+ cells in D60 dvCAs. (L to O) Micrograph of the dorsal region of 60D CTRLv-CTRLd (L) and (M) and E370Kv-CTRLd (N) and (O) CAs immunostained for GFP and GAD67. (P) Quantification of the percentage of GFP+GAD67+ signal normalized by area (μm2) in D60 dvCAs. (Q) Schematic of the experimental setup for time-lapse live imaging in dvCAs. (R and S) Time-lapse live imaging of migrating GFP+ cells in organotypic slices of CTRLv-CTRLd (R) and E370Kv-CTRLd CAs (S). (T to V) Quantification of velocity (T), number of resting time points (U), and tortuosity index (V) of GFP+ cells in the dorsal side of dvCAs. Each individual data point represents one ventricle of dvCAs. Data are shown as means ± SEM. Statistical significance was based on the Mann-Whitney U test; *P < 0.05, **P < 0.01, and ****P < 0.0001. Scale bars, 50 μm (B) to (E), (G) to (J), and (L) to (O) and 80 μm (R) and (S).
To monitor migrating neurons, we first analyzed neurons migrating into the dorsal region by quantifying the number of cells migrating from the ventral [green fluorescent protein-positive (GFP+); in green] to the dorsal [red fluorescent protein-positive (RFP+); in magenta] region in dvCAs (fig. S4, E and F). We did not observe significant differences in the total number of migrated cells (fig. S4E) or significant changes in their distribution within the dorsal region of dvCAs (fig. S4, F and G). We analyzed the distribution of GFP+ cells that had migrated from the ventral to the dorsal region of dvCAs by subdividing the dorsal region (dCAs) into three equally distributed regions (bins in fig. S4, F and G). In CTRL dvCAs, GFP+ migrating cells were mainly distributed among bin 2 (~45%) and bin 3 (~55%) of CTRL-dCAs. In contrast, in vE370K-dCTRL CAs, most of the cells (70%) were found in bin 2 and only 10% in bin 3 of CTRL-dCAs, showing a different distribution trend in mutant cells when they migrate into the dorsal region, probably because of their altered molecular identity.
We then quantified the percentage of ventral E370K GFP+ cells that expressed TBR1 or SATB2 in CTRL-dCAs (Fig. 3, B to K) to assess whether ventral E370K neurons expressing cortical markers migrate to dCAs. Thirty percent of the total E370K GFP+ cells that migrated into the dorsal region expressed TBR1 (Fig. 3F), while 10% expressed SATB2 (Fig. 3K). On the contrary, CTRL GFP+ ventral cells expressing dorsal markers were absent in CTRL-dCAs, as expected. We additionally quantified TBR1+ cells in CTRL-vCAs and E370K-vCAs, as well as TBR1+GFP+ cells in CTRL-dCAs after clearing (fig. S4, H to M) (38). We confirmed the presence of both TBR1+ cells in the ventral E370K region of E370Kv-CTRLdCAs (fig. S4L) and GFP+TBR1+ cells that migrated from E370K-vCAs to CTRL-dCAs (fig. S4M).
Next, we examined whether the migrated ventral cells expressed interneuronal markers by quantifying the percentage of GFP+/GAD67+ signal (quantified in pixels) normalized by area (signal/area) (Fig. 3, L to P) in the dorsal region of dvCAs. As expected, we found a significant decrease of GFP+/GAD67+ signal/area for E370K ventral neurons that migrated into the dorsal region of dvCAs (Fig. 3P). These results suggest that E370K migrating ventral neurons exhibit a dorsal identity, rather than ventral, compared to CTRL migrating INs. Moreover, we assessed whether migrating mutant cells coexpressed dorsal and ventral neuronal markers. For this purpose, we analyzed whether E370K-GFP neurons coexpressed GAD67 and TBR1, or SATB2 and CALB1 in the dorsal region of vE370K-dCTRL dvCA (fig. S4, N to Q). We found that migrating E370K ventral neurons did not simultaneously express dorsal and ventral markers, suggesting that there are two distinct neuronal populations.
Next, we sought to understand whether the observed difference in the cellular identity could influence the distribution of migrated E370K neurons. For this purpose, we compared the distribution of GFP+TBR1− and GFP+TBR1+ cells in the dorsal region of vE370K-dCTRL dvCA, finding no differences in their distribution (fig. S4R).
Moreover, we also did not find any difference in the distribution of GFP+TBR1− migrated cells in CTRL and vE370K-dCTRL dvCAs (fig. S4S). Together, these data confirm that the localization of E370K migrated cells is not affected. Therefore, considering that LGALS3BP can act extrinsically, we decided to assess whether the extracellular environment could affect IN distribution in the dorsal region of dvCAs. To understand whether LGALS3BP might influence the recruitment of neurons, we generated vCTRL-dE370K dvCA (where the LGALS3BP variation is present only in the dorsal side) and quantified the distribution of migrated CTRL cells in the mutant dorsal environment (fig. S4T). We observed that CTRL INs, when migrating into the dorsal mutant region, are mostly localized at the first and second bins. These data suggest a dual role of LGALS3BP in regulating IN migration: cell-autonomously and non–cell-autonomously by influencing their recruitment.
We showed that scRNA-seq data from mutant neurons revealed an alteration in secreted molecules and transmembrane proteins associated with axon guidance (Fig. 2, F and G), suggesting changes in migratory dynamics. To dissect the migratory dynamics and behavior of E370K neurons, we monitored the trajectories of GFP+ cells in the dorsal region of dvCAs. We performed time-lapse imaging of D60 dvCAs slices and tracked the GFP+ ventral cells during their migration within dCAs (Fig. 3, Q to V). We measured velocity (speed of migration) (Fig. 3T), resting time points (time cells spent without moving) (Fig. 3U), and tortuosity (the ability to move in a straight trajectory) (Fig. 3V) as previously described by Klaus et al. (10). For all three parameters, we observed a significant difference compared to CTRL. We found a decreased speed of migration and increased resting time points and tortuosity, as previously shown in the case of mutations of PH genes DCHS1 and FAT4 (10). Not only neuronal dynamics but also similar genes (ROBO3, GNG5, and DCC) were found altered in E370K ventral neurons and DCHS1 and FAT4 altered neurons (10), suggesting common signatures for neurons associated with PH (fig. S5, A and B). In support of their fate switch, the average speed of E370K ventrally generated neurons is similar to the speed of previously observed dorsally generated CTRL neurons analyzed by Klaus et al. (10) (fig. S5C). Together, these results showed that cell fate changes, observed in E370K cells, lead to a migratory dynamics alteration, specifically in directionality, sliding movement, and speed.
Neuronal migration is a complex process, regulated by intracellular and extracellular mechanisms. We hypothesized that ventrally originated E370K neurons, because of their acquired dorsal identity, might not respond properly to the extracellular cues and, therefore, show defects in their migratory behavior. We identified genes associated with axon guidance and neuronal migration (including semaphorins, Eph/ephrins, and adhesion molecules) that are enriched in dorsal and/or ventral organoids (fig. S5D). These data suggest that the cell fate switch from ventral to dorsal leads to the expression of genes (associated with axon guidance or migration) typically enriched in dorsal neurons, affecting migratory behavior and dynamics of mutant ventrally originated neurons.
Extrinsic effect of LGALS3BP on ventral progenitor fate and neuronal specification
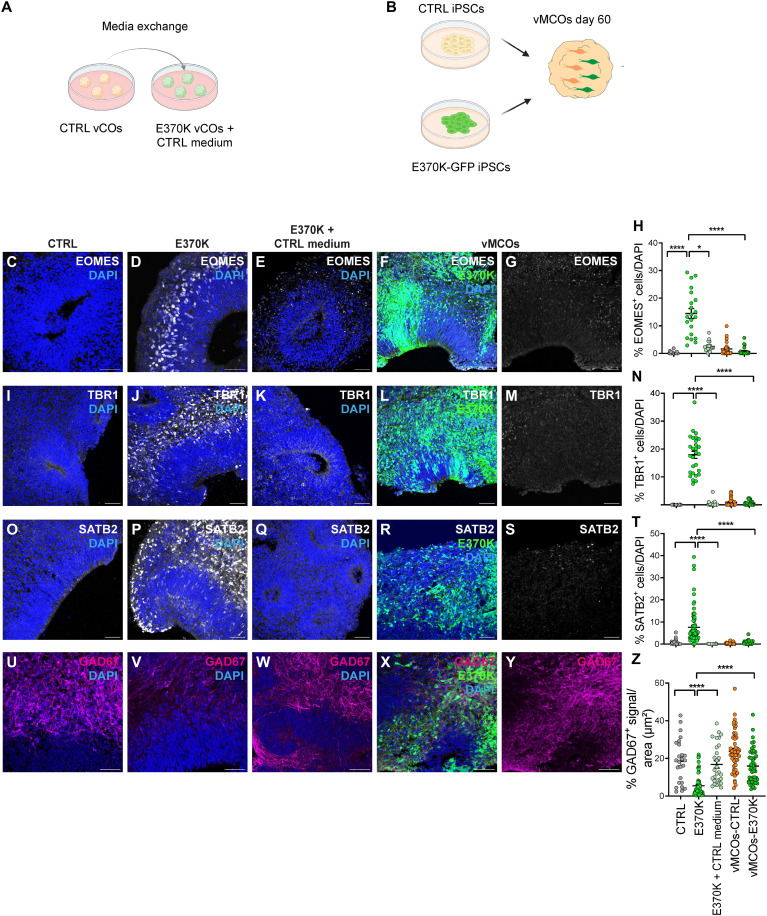
LGALS3BP is a secreted glycoprotein whose functions have been mainly characterized in cancer studies, being LGALS3BP a tumor biomarker (28, 39–45). Given its secretion, we expected that LGALS3BP would function in a non–cell-autonomous manner. We, therefore, assessed whether the dorsal identity acquired by vCOs generated from mutant cells could be rescued by providing a more physiological extracellular environment. To this end, we used two different approaches: On one hand, we treated E370K-vCOs with culture media from CTRL-vCOs (Fig. 4A), and on the other hand, we generated ventral mosaic cerebral organoids (vMCOs), where both isogenic CTRL iPSCs and GFP+ E370K iPSCs (Fig. 4B) were mixed together in a ratio of 1:1. We observed a rescue of the phenotype in both cases (Fig. 4, C to Z). Specifically, upon exposure to CTRL medium or cell-to-cell contact with CTRL cells, we observed a significant reduction in the expression of the cortical IP marker EOMES (Fig. 4, C to H), as well as the deep layer TBR1 (Fig. 4, I to N) and upper layer SATB2 cortical neuronal markers (Fig 4, O to T). We also observed a significant increase in the inhibitory neuronal marker GAD67 (Fig. 4, U to Z) compared to E370K-vCOs. These findings highlight the extracellular function of LGALS3BP, supporting the important role of the extracellular environment in brain development, suggesting that secreted molecules and proteins regulate cell proliferation, delamination, and distribution, as previously shown by Kyrousi et al. (20), as well as cell identity.
Fig. 4. Extrinsic effect of LGALS3BP on ventral progenitor fate and neuronal specification.
(A and B) Experimental setup for media exchange in vCOs (A) and for the generation of vMCOs (B). (C to G) Micrograph of D60 CTRL (C), E370K (D), E370K-vCOs fed with CTRL CM (E), and vMCOs (F) and (G) immunostained for EOMES. (H) Quantification of the percentage of EOMES+ cells in vCOs and vMCOs normalized by DAPI in the analyzed GE unit. (I to M) Micrograph of D60 CTRL (I), E370K (J), E370K-vCOs fed with CTRL CM (K), and vMCOs (L) and (M) immunostained for TBR1. (N) Quantification of the percentage of TBR1+ cells in vCOs and vMCOs normalized by DAPI in the analyzed GE unit. (O to S) Micrograph of D60 CTRL (O), E370K (P), E370K-vCOs fed with CTRL CM (Q), and vMCOs (R) and (S) immunostained for SATB2. (T) Quantification of the percentage of SATB2+ cells in vCOs and vMCOs normalized by DAPI in the analyzed GE unit. (U to Y) Micrograph of D60 CTRL (U), E370K (Y), E370K-vCOs fed with CTRL CM (W), and vMCOs (X) and (Y) immunostained for GAD67. (Z) Quantification of the percentage of GAD67+ signal in vCOs and vMCOs normalized by area [for (U) to (W)] and by GFP+ area [for (X) and (Y)] in the analyzed GE unit. Each individual data point represents one GE unit (ventricle). Data are shown as means ± SEM Significance was based on the Kruskal-Wallis multiple comparison test; *P = 0.029 and ****P < 0.001. Scale bars, 50 μm.
We observed a stronger rescue in vMCOs compared to E370K-vCOs treated with CTRL culture media, where we still observed an increase of EOMES+ cells compared to CTRL (Fig. 4, E and H). For this reason, we decided to dissect the transcriptional signatures of vMOs-E370K, performing scRNA-seq analysis on vMCOs (Fig. 5A). The 4898 identified cells were clustered into five main clusters (Fig. 5, B and C), including progenitors expressing TOP2A; IP expressing NKX2-1, ASCL1, and PAX6; and neurons expressing DLX5 and MAP2 (Fig. 5D) (7, 11).
Fig. 5. Extrinsic effect of LGALS3BP on ventral progenitor fate and neuronal specification.
(A) Schematic of the experimental setup for the generation and processing of vMCOs. After 60 days of culture, vMCOs are dissociated into single cells, and CTRL- and GFP-labeled E370K cells are sorted through fluorescence-activated cell sorting (FACS) and processed for scRNA-seq. The image was partially generated using BioRender. (B) UMAP visualization of scRNA-seq data of D60 vMCOs (n = 4898 cells from a pool of five organoids). (C) UMAP visualization of scRNA-seq clusters of D60 vMCOs. Each color cluster represents a different cell population. (D) Feature plot depicting the expression of progenitor markers (TOP2A and PAX6), IPs markers (NKX2-1, ASCL1, and GLI3), and neuron markers (DLX5, MAP2, TBR1, NEUROG1, and SLC17A6) in D60 vMCOs. (E) Volcano plot showing the genes rescued in progenitors in vMCOs, plotting the negative log10 P.adj of all genes against their log2 fold change (vMCOs-E370K versus vMCOs-CTRL). Rescued genes are labeled in gray, significantly up-regulated in red, and down-regulated in blue (P.adj < 0.01). (F) Volcano plot showing the genes rescued in IP and MGE IP in vMCOs, plotting the negative log10 P.adj of all genes against their log2 fold change (vMCOs-E370K versus vMCOs-CTRL). Rescued genes are labeled in gray, significantly up-regulated in red, and down-regulated in blue (P.adj < 0.01). (G) Volcano plot showing the genes rescued in neurons in vMCOs, plotting the negative log10 P.adj of all genes against their log2 fold change (vMCOs-E370K versus vMCOs-CTRL). Rescued genes are labeled in gray, significantly up-regulated in red, and down-regulated in blue (P.adj < 0.01).
The identified ventral telencephalic cells were aligned on a developmental pseudo-time axis showing the trajectory from progenitors to neurons, as seen in vCOs (fig. S6, A and B). As with to vCOs, we could not detect major changes in the pseudo-differentiation trajectory (fig. S6B). Using Monocle 3 (https://cole-trapnell-lab.github.io/monocle3/) (26), we identified pseudo-time trajectories in both CTRL and E370K cells obtained from vMCOs (fig. S6C). The trajectories were populated by both CTRL and E370K cells and resembled the ones found in CTRL-vCOs, suggesting a partial rescue of E370K cells due to exposure (proximity) to CTRL cells in the vMOs.
Specifically, the cell proportions of progenitors, IPs, and MGE IPs were increased compared to E370K-vCOs (figs. S3G and S6D). In E370K-vMCOs, we observed a 30% increase of cells in progenitors, 25% in IPs, and 50% in MGE IPs compared to E370K-vCOs, indicating a change in cell proportions when cells differentiate in a more physiological environment.
Cortical genes such as TBR1, NEUROG1, GLI3, and SLC17A6 are also expressed in vMCOs, but at lower levels compared to E370K-vCOs (Figs. 2C and 5D and figs. S3A and S6E). As for E370K-vCOs, E370K progenitors, IP, and MGE-IP obtained from vMCOs express a higher level of dorsal genes, such as PAX6 and GLI3, compared to CTRL. Gene expression of cortical markers such as TBR1, NEUROG1, and SLC17A6 is decreased in E370K neurons generated from vMCOs, showing a partial rescue of the neuronal identity affected in mutant vCOs (Figs. 2C and 5D and figs. S3A and S6E).
Next, we sought to understand to which extent vMCOs could rescue the E370K phenotype at the transcriptional level. Therefore, we specifically focused on genes that were differentially expressed in E370K-vCOs versus CTRL-vCOs, but those were not differentially expressed in E370K-vMCOs versus CTRL-vMCOs (Fig. 5, E to G). We identified that 85% of the genes that were DE in E370K progenitors were rescued in E370K cells in vMCOs; for IP and neurons, 62 and 68% of DE genes were rescued in E370K-vMCOs, respectively.
These results suggest a non–cell-autonomous regulation of neuronal cell fate and specification. The ventral molecular identity rescue observed in E370K cells was guided by the presence of CTRL neighboring cells. Both progenitors, IPs, and MGE-IPs, retained their dorsal identity, down-regulating NKX2-1 and SHH; moreover, IPs and MGE IPs still express PAX6 (Fig. 5, E and F, and fig. S6E), suggesting an extrinsic control over interneuronal specification. This is in line with the progressive specification hypothesis, which states that INs are restricted to a particular subtype at birth, but their definitive identities are established later in development and are shaped by the extracellular (cortical) environment (46, 47).
LGALS3BP can revert the molecular identity of mutant ventral progenitors and INs
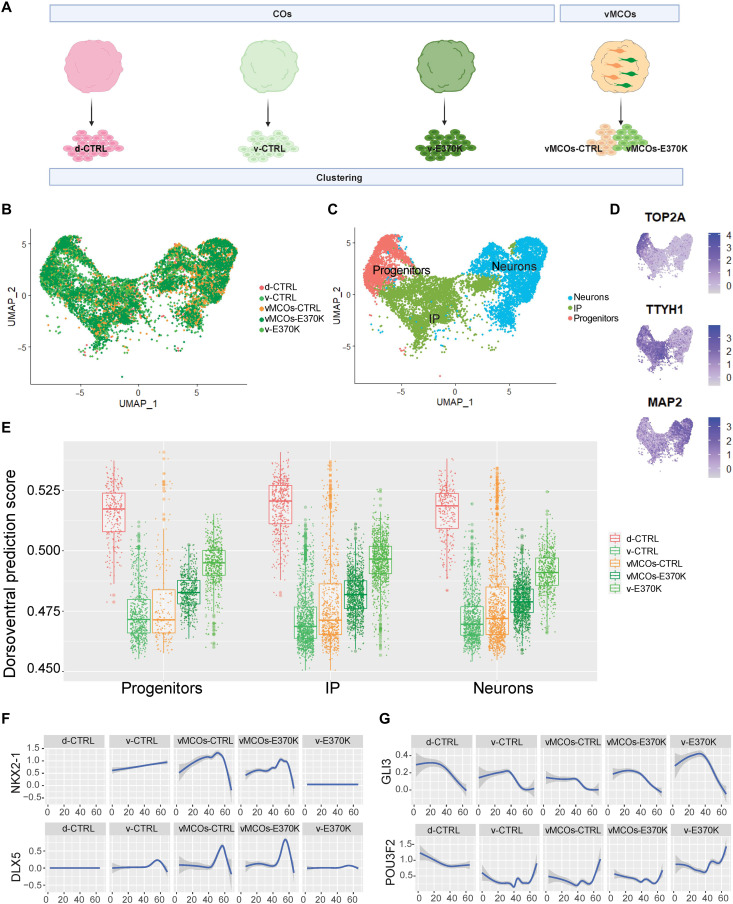
To investigate the molecular identity of E370K-vCOs and vMOs, we built a dorsoventral model that predicts the dorsal or ventral molecular identity of cells based on their transcriptomic features, defining the dorsoventral score (DV) (fig. S7A). We performed cluster analysis of scRNA-seq transcriptome data from CTRL-dCOs and CTRL-vCOs, E370K-vCOs, and CTRL/E370K-vMCOs (Fig. 6A) and identified three main clusters: progenitors expressing TOP2A, IPs expressing TTYH1, and neurons expressing MAP2 (Fig. 6, B to D, and fig. S7B).
Fig. 6. LGALS3BP can revert the molecular identity of mutant ventral progenitors and neurons.
(A) Schematic of the experimental setup for the cluster analysis of D60 CTRL-dCOs (d-CTRL), CTRL-vCOs (v-CTRL), E370K-vCOs (v-E370K), and vMCOs (vMCOs-CTRL and vMCOs-E370K). scRNA-seq datasets are clustered together and analyzed. (B) UMAP visualization of scRNA-seq data of D60 CTRL-dCOs (d-CTRL), CTRL-vCOs (v-CTRL), E370K-vCOs (v-E370K), and vMCOs (vMCOs-CTRL and -E370K). (C) UMAP visualization of scRNA-seq clusters of D60 CTRL-dCOs (d-CTRL), CTRL-vCOs (v-CTRL), E370K-vCOs (v-E370K), and vMCOs (vMCOs-CTRL and -E370K). Each color cluster represents a different cell population. (D) Feature plot depicting the expression of progenitor marker (TOP2A), IP marker (TTYH1), and neuronal marker (MAP2). (E) Boxplot showing the DV score of progenitors, IPs, and neurons in all conditions. In all three populations, v-E370K cells show a higher DV score compared to v-CTRL cells, and vMCOs-E370K cells show a lower DV score compared to v-E370K cells. Boxplots show median and interquartile range. Each dot in the plot represents one cell. (F) Expression level of ventral (NKX2-1 and DLX5) genes along the pseudo-time axis in each condition, showing a lower expression in v-E370K compared to v-CTRL and an increased expression in vMCOs-E370K compared to v-E370K. (G) Expression level of ventral (GLI3 and POU3F2) genes along the pseudo-time axis in each condition, showing a higher expression in v-E370K compared to v-CTRL and a decreased expression in vMCOs-E370K compared to v-E370K.
As expected, the dorsoventral prediction model revealed the highest DV score for CTRL-dCOs and the lowest for CTRL-vCOs (Fig. 6E, and fig. S7, C and D). All the other conditions showed blended dorsoventral identity and were distributed according to extrinsic short-distance exposure. E370K-vCOs cells mapped midway through the dorsoventral trajectory, while cells from vMCOs were mapped similarly. As expected, CTRL cells from vMCOs were closer to CTRL cells from vCOs, while E370K cells from vMCOs were closer to E370K-vCOs. These data strongly indicate that E370K cells acquire a dorsal molecular identity, and this can be reverted by exposure to CTRL cells. Last, CTRL cells from vMCOs showed mostly a ventral score with some outlier cells with a higher DV score, suggesting that CTRL cells have also been affected by the extracellular environment generated by the neighboring E370K cells (Fig. 6E and Fig. S7, C and D).
Gene expression is affected in both CTRL and E370K cells from vMCOs. NKX2-1 and DLX5, which are almost undetectable or have low expression levels in E370K-vCOs, are expressed along the pseudo-differentiation axis in E370K cells from vMCOs (Fig. 6F). GLI3 and POU3F2, which are mostly expressed in CTRL-dCOs, are highly expressed in E370K-vCOs, but lower in E370K cells from vMCOs, without showing great changes in CTRL cells from vMCOs (Fig. 6G).
To map CO single-cell transcriptome data to the developing mouse brain, we then used VoxHunt (https://quadbiolab.github.io/VoxHunt/) (48) obtained from in situ hybridization data (Allen Developing Mouse Brain Atlas) (49). As suspected, transcriptome data from CTRL-dCOs and vCOs map, respectively, to the mouse cortex (pallium) and ventral forebrain (subpallium). Data from E370K-vCOs map not only to the ventral forebrain but also to the mouse cortex, confirming their dorsal molecular identity. CTRL and E370K cells from vMCOs mostly map to the ventral forebrain; however, E370K cells still show a correlation with the mouse pallium (fig. S7E). Together, these findings showed that the molecular identity of progenitors, IPs, and neurons is influenced by extrinsic factors.
Extrinsic function of LGALS3BP in progenitor specification and neuronal migration dynamics
During brain development, neurogenesis and cell migration are influenced by extrinsic factors released into the ECM, including via EVs (50–52). LGALS3BP can be secreted into the extracellular environment directly or via EVs. This secreted glycoprotein is expressed at the surface of EVs (39, 40, 53) and was previously described as an EV marker (54). Moreover, LGALS3BP is enriched in cancer cell lines (i.e., neuroblastoma, ovarian carcinoma, and melanoma) (39, 41, 44, 55), in plasma from patients with cancer (44) or with pulmonary disease (54), and in cerebrospinal fluid from patients with neurological disorders (56). Our previous work confirmed that LGALS3BP is secreted in EVs in human cerebral organoids and modulates the extracellular space and dorsal progenitor cell fate (20).
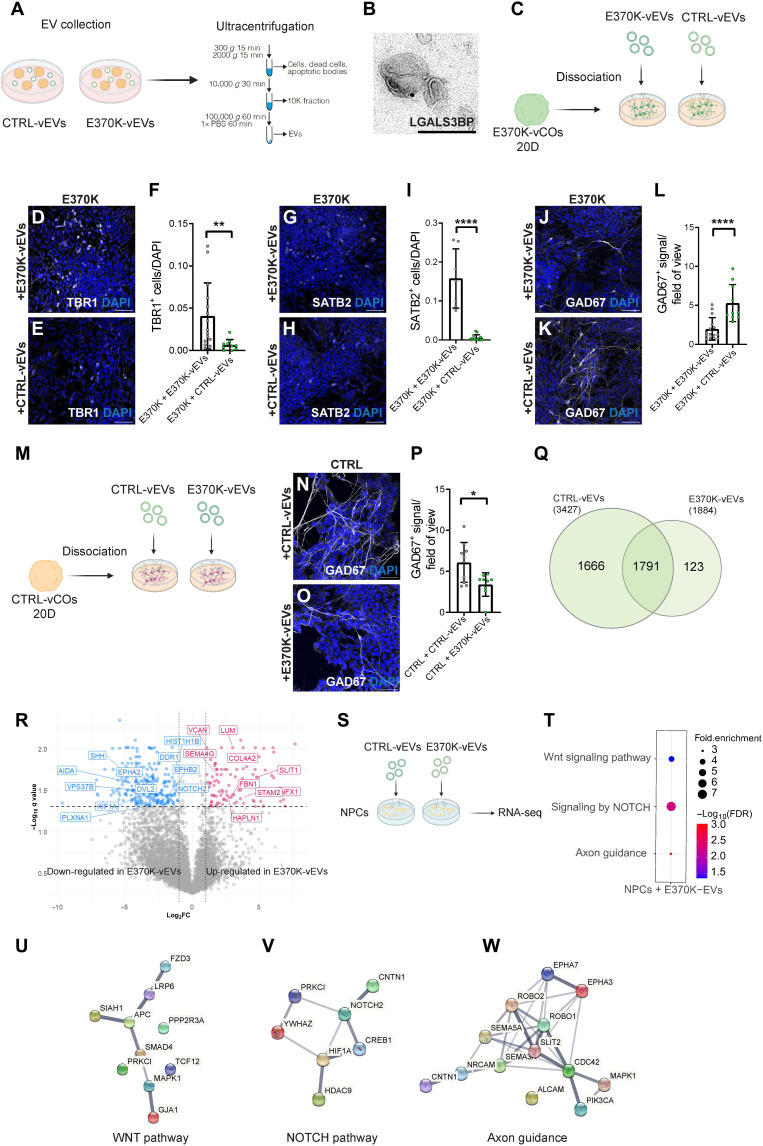
We, therefore, hypothesized that EV-associated LGALS3BP might regulate the cell fate switch and neuronal dynamics alteration observed in E370K-vCOs and dvCAs in a non–cell-autonomous manner. To this end, we isolated EVs from the conditioned media of vCOs by ultracentrifugation (Fig. 7A) and confirmed that LGALS3BP is present in EVs via immunoelectron microscopy (Fig. 7B).
Fig. 7. Extrinsic function of LGALS3BP in progenitor specification and neuronal migration.
(A) Schematic of isolation of EVs from vCOs. (B) LGALS3BP immunoelectron micrographs in EVs collected from COs. (C) Schematic of 20D E370K-vCO dissociation and EV treatment. (D and E) Micrograph of E370K cells treated with E370K-vEVs (D) and CTRL-vEVs (E) immunostained for TBR1. (F) Quantification of TBR1+ cells in (D) and (E) normalized by DAPI. (G and H) Micrograph of E370K cells treated with E370K-vEVs (G) and CTRL-vEVs (H) immunostained for SATB2. (I) Quantification of SATB2+ cells in (G) and (H) normalized by DAPI. (J and K) Micrograph of ventral E370K cells treated with E370K-vEVs (J) and CTRL-vEVs (K) immunostained for GAD67. (L) Quantification of GAD67+ signal in (J) and (K) normalized by area. (M) Schematic of 20D CTRL-vCO dissociation and EV treatment. (N and O) Micrograph of ventral CTRL cells treated with CTRL-vEVs (N) and E370K-vEVs (O) immunostained for GAD67. (P) Quantification of GAD67+ signal in (N) and (O) normalized by area. (Q) Venn diagram of proteins detected in CTRL-vEVs and E370K-EVs. (R) Volcano plot of DE proteins in E370K-vEVs, plotting the negative log10 q values (FDR) of all proteins against their log2 fold change. (S) Schematic of treatment of NPCs with CTRL-vEVs and E370K-vEVs. (T to W) GO terms of up-regulated pathways (T), and gene networks of WNT (U), NOTCH (V), and axon guidance (W) pathways in NPCs after E370K-vEVs exposure. Every dot in the plots refers to an analyzed field of view. Significance was based on the unpaired t test; *P < 0.05, **P < 0.01, and ****P < 0.001. Scale bars, 100 nm (B) and 50 μm (D), (E), (G), (H), (J), (K), (N), and (O).
Because we observed a rescue of the E370K phenotype in E370K-GFP cells in vMCOs, we hypothesized that LGALS3BP might regulate interneuronal molecular identity and specification in an EV-dependent manner. To investigate whether physiological EV content can restore the ventral identity of mutant vCOs, we treated cells dissociated from D20 E370K-vCOs with E370K-vEVs (ventral EVs) (to resemble the mutant condition) and with CTRL-vEVs (to attempt a rescue) for 1.5 weeks (Fig. 7C).
We observed a rescue consisting of a decrease in TBR1+ (Fig. 7, D to F) and SATB2+ cells (Fig. 7, G to I) as well as an increase in GAD67 expression (Fig. 7, J to L) in E370K cells after exposure to CTRL-vEVs (Fig. 7, D to L). These data suggest that cargos transported in EVs generated by cells with physiological LGALS3BP, as well as secreted in the mutant environment, are sufficient to inhibit dorsalization. These results emphasize that the dorsal identity acquired by E370K ventral cells is mediated by EVs.
From the dorsoventral model applied to scRNA-seq data from vMCOs (Fig. 6E and Fig. S7C), in all the analyzed clusters, a portion of CTRL cells from vMCOs showed a high DV score, suggesting that the molecular identity of CTRL cells can also be influenced by the mutant extracellular environment. To dissect whether E370K-EVs can affect the specification of CTRL cells, we treated cells dissociated from D20 CTRL-vCOs with CTRL-vEVs (to resemble the CTRL condition) and with E370K-vEVs (to investigate the extrinsic pathological function of mutant EVs) for 1.5 weeks (Fig. 7M). We observed that CTRL cells exposed to E370K-EVs significantly reduced the expression of GAD67, suggesting that E370K-EVs might inhibit ventralization (Fig. 7, N to P). However, no expression of TBR1 or SATB2 was detected in cells dissociated from CTRL-vCOs, suggesting that specific levels of LGALS3BP and time-dependent developmental stages of extrinsic exposure can be crucial for the proper specification and molecular identity of cells.
We then performed proteomic analysis of EVs collected from CTRL and E370K-vCOs (fig. S8A). E370K-vEVs showed a different protein composition with a lower number of detected proteins, suggesting alterations in EV cargo loading (Fig. 7Q and Fig. S8B). As expected, we found LGALS3BP being expressed in EVs derived from both CTRL and E370K-vCOs, with a lower level in mutant EVs, also confirmed by Western blot analysis (fig. S8, C to E). We, therefore, hypothesize that the LGALS3BP patient variant can lead to changes in the number and/or type of proteins loaded onto EVs. To this end, we looked at the DE proteins in E370K-EVs (Fig. 7R). We found dysregulation of proteins STAM2 and VPS37B, involved in EV biogenesis (57), suggesting defects in mutant EV biogenesis. Previous studies have shown that the glycosylation state of EV-associated glycoproteins can affect their recruitment, as well as that of other proteins, in EVs. Specifically, altering glycosylation in melanoma cells and in the produced EVs affects the trafficking of LGALS3BP into EVs (53, 55). Moreover, the secreted molecule SHH [involved in dorsoventral patterning inducing ventralization (58)] is down-regulated in E370K-vEVs (Fig. 7R and Fig. S8, F and G), suggesting that the low level of SHH carried by EVs into the extracellular space might influence the differentiation of neighboring cells, resulting in a dorsal cell fate. Signaling protein DVL2, known to down-regulate the Wnt pathway (59), is down-regulated in mutant EVs. The decrease of DVL2 in mutant EVs could lead to an activation of the WNT pathway in neighboring cells, resulting in the transcription of dorsal genes (30, 60). EVs secreted by E370K-vCOs also showed dysregulated proteins involved in cell migration and motility—PLXNA1, SEMA3G, KIF1A, EPHA2, EPHB2, and SLIT1—and proteins of the ECM—HAPLN1, LUM, and VCAN—known to modulate cell proliferation and differentiation (Fig. 7R and fig. S8, F and H) (17, 61, 62). We showed that E370K-vCOs present alterations in the secretion of proteins involved in EV biogenesis, cellular migration, differentiation, and dorsoventral patterning, which could explain the extrinsic effect on these processes.
Therefore, we hypothesize that EVs might regulate progenitor cell fate and motility, resulting in the dorsal identity observed in ventral E370K neurons. To this end, we treated CTRL-NPCs in 2D acutely (12 hours) with EVs isolated from CTRL and E370K-vCOs (Fig. 7S) and identified up-regulated genes in the recipient cells. The RNA-seq analysis performed on NPCs revealed up-regulation of the WNT and NOTCH pathways (Fig. 7, T to V) after treatment with E370K-vEVs, showing a similar profile to E370K neural progenitors isolated from vCOs and analyzed by scRNA-seq (Fig. 2G). These results suggest that mutant EVs can change the transcriptomic profile of NPCs (fig. S8I) and activate pathways involved in cell dorsoventral patterning and cell-cell communication, resulting in an alteration in their fate (27, 32, 33, 63). Moreover, upon treatment with E370K-vEVs, NPCs showed up-regulation of genes involved in axon guidance (Fig. 7, T and W), as it was found in E370K neurons isolated from vCOs and analyzed by scRNA-seq (Fig. 2V). Together, these results propose a non–cell-autonomous regulation of neuronal differentiation and migration dynamics mediated via EVs.
We showed that vCOs with a variation in the ECM protein LGALS3BP manifest a dorsal identity and migratory defect that could lead to the development of neurological disorders. It has been shown that EVs can also transport ECM remodeling cargos, similar to matrix metalloproteinases, which regulate cell growth, differentiation, and cell migration (64). An altered deposition of collagens mediated via EVs can change the ECM composition and might lead to neurological diseases, including epilepsy (17). To better correlate the dysregulated network of proteins altered in E370K-EVs with neurological diseases, we identified proteins whose genes are associated with CMs, autism spectrum disorder (ASD), and epilepsy. We identified multiple proteins altered in E370K-EVs compared to CTRL-EVs that are mostly associated with microtubule organization, morphogenesis, and synaptic activity, highlighting the key role of EV-mediated signaling in neurodevelopmental disorders (NDDs) (fig. S8, J to L).
DISCUSSION
Defects in IN specification and migration might cause an excitatory/inhibitory imbalance in neuronal circuits, characteristic of neurological disorders, such as epilepsy and ASD. For example, some patients with ASD have reduced GABAergic neurons in the cortex (65, 66), and organoids derived from patients with ASD show dysregulation in genes involved in GABAergic IN differentiation and migration (67, 68). Other studies performed in cerebral organoids with mutations in ASD-associated genes revealed alterations in both GABAergic and excitatory neuron developmental trajectories (69, 70). Recent studies highlighted the high heterogeneity of human INs (5, 71). As LGALS3BP is highly expressed in human (but not in mouse) neural progenitors during brain development, it is an interesting candidate protein that could contribute to the higher heterogeneity of human neural progenitors and neurons. We previously described the extracellular function of LGALS3BP in regulating proliferation, NPC delamination, neuronal distribution, and migration in COs (20). In this work, we showed that vCOs with a mutation in the LGALS3BP gene exhibit alterations in dorsoventral patterning resulting in a population of cells with a dorsal identity. Therefore, we propose an extrinsic regulation of LGALS3BP in ventral progenitor specification, mainly mediated by EVs. We showed that the extracellular environment influences molecular identity and neuronal specification in vMCOs. In vMCOs, control, and mutant progenitors share similar transcriptomic features; however, after the IP transition, they reveal their intrinsic program identity (ventral for CTRL and more dorsal for E370K). Final neuronal identity is refined at later stages of development in interaction with the local extracellular environment. We showed that the proximity of CTRL cells has partially reverted the dorsal identity of E370K cells. We propose that LGALS3BP regulates IN specification by influencing the cellular cross-talk mediated by EVs. We showed that EVs carry secreted molecules that can drive dorsoventral patterning. EV treatment could significantly reduce the expression of dorsal markers in mutant cells and restore the expression of ventral genes, suggesting that CTRL-vEVs might be important for the induction of ventralization during brain development. On the contrary, mutant EVs inhibited ventralization in CTRL cells via a reduction of the expression of inhibitory neuronal markers. Therefore, we proposed a previously unidentified mechanism in dorsoventral patterning mediated by vEVs, and in particular, we showed a role of EV-associated LGALS3BP in inducing ventralization during early neurodevelopment. We identified fewer proteins in mutant vEVs compared to CTRL-vEVs. Because we showed that E370K-vEVs dysregulate proteins associated with the ESCRT-0 and ESCRT-II (57) transport machinery, we could speculate that the LGALS3BP variation might influence EV biogenesis. It has been shown that the glycosylation pattern of EV-associated proteins regulates the recruitment of specific proteins into the vesicles (53, 55). Further studies will focus on a detailed investigation of the glycosylation pattern of EV-associated LGALS3BP both in control and mutant COs. Moreover, treatment with E370K-vEVs changed the transcriptomic profile of NPCs, activating WNT and NOTCH pathways, involved in cell fate decision and dorsoventral regionalization (27, 29–33).
Previous studies in mice showed that the dorsoventral patterning during brain development is regulated by Pax6, Gli3, and Shh (72–74). Gli3 and Pax6 mutant mice show a ventral identity acquired by dorsal pallial tissue expressing ectopic Dlx2 (73, 74). Mice carrying a deletion in the Gli3 gene show ectopic ventral telencephalic identity in the dorsal forebrain, probably given by an alteration in the Wnt pathway (74). In the extracellular space, LGALS3BP binds Galectin-3 (LGALS3), forming a complex that interacts with membrane tetraspanins CD9 and CD82, activating Wnt/β-catenin signaling (19, 75). This pathway triggers the expression of Wnt target genes in the dorsal forebrain (30). One possible scenario is that the E370K variation activates Wnt/β-catenin signaling in vCOs. The activation of the signaling pathway might lead to the expression of PAX6 and repression of NKX2-1, resulting in the expression of other cortical dorsal genes (EOMES, TBR1, and SATB2) that define the dorsal identity of E370K-vCOs. Moreover, Wnts can act as paracrine molecules, affecting neighboring cells. In another possible scenario, the reduced protein level of LGALS3BP, observed in mutant vEVs, might result in less LGALS3BP binding CD9 and CD82. We propose that the activation of Wnt/β-catenin signaling through membrane tetraspanins might function in a dose-response manner, meaning that, in mutant cells, the lower level of EV-associated LGALS3BP is not sufficient to repress the dorsalization. The dose-response effect might also explain the partial rescue of the dorsal phenotype observed in mutant vCOs. The E370K mutation is heterozygous, which means that the wild-type form of LGALS3BP is still present in mutant organoids. However, when adding CM from CTRL organoids or generating a more physiological environment in vMOs, the additional LGALS3BP wild-type form can overcome the mutant form, restoring the ventral identity. On the other hand, E370K-vEVs were not sufficient not only to induce dorsalization when added to CTRL cells but only to inhibit ventralization, suggesting that dorsalization is also regulated in a dose-response manner. However, it has been shown that LGALS3 regulates negatively Wnt/β-catenin pathways in neuronal stem cells in the postnatal SVZ (76), suggesting that LGALS3BP might regulate the dorsoventral patterning via other pathways, such as SHH and NOTCH, as suggested from our data.
We described the migratory behavior of E370K cells, finding a significant decrease in velocity that becomes more similar to radially migrating excitatory neurons as shown previously (10). These cells reflect a more tortuous migratory behavior as it was previously described in migrating neurons derived from an individual with PH (with a mutation in DCHS1 and FAT4) (10). Ventrally derived neurons in E370K-COs show similar molecular signatures to altered neurons found in DCHS1 and FAT4 patient organoids, suggesting shared dysregulated pathways in ectopic neurons accumulating below the white matter of patients with PH. Future studies with cells derived from PH patients with different mutations will be essential to understand whether these are common signatures for all PH neurons, providing additional tools to target a specific subpopulation of affected neurons.
We observed that the migratory dynamics of neurons are altered. This could be justified by the dorsal identity acquired by E370K neurons that could result in a different response to extrinsic attractive/repulsive stimuli. However, LGALS3BP has already been shown to play a role in many processes, including dorsal progenitor delamination, regulation of ECM composition, and expression/localization of cytoskeletal and guidance molecules, suggesting that the changes in migration could also be independent of the cell fate switch.
In conclusion, we propose a non–cell-autonomous regulation of the secreted glycoprotein LGALS3BP in IN progenitor fate decision and dorsoventral patterning during brain development. Using ventral cerebral organoids and dvCAs, we showed that a mutation in the ECM component LGALS3BP, identified in an individual with neurodevelopmental disorders, alters cell fate specification of ventral progenitors and that these mechanisms might be regulated by EVs.
MATERIALS AND METHODS
iPSC culture
iPSCs reprogrammed from NuFF3-RQ human newborn foreskin feeder fibroblasts (GSC-3404, GlobalStem) (77) were cultured on Matrigel (Corning)–coated plates (Thermo Fisher Scientific, Waltham, MA, USA) in mTesR1 basic medium supplemented with 1× mTesR1 supplement (STEMCELL Technologies, Vancouver, Canada) at 37°C, 5% CO2, and ambient oxygen level. Passaging was done using Accutase (STEMCELL Technologies) treatment.
CRISPR genome editing for generation of mutant iPSC lines
For CRISPR genome editing for the generation of mutant iPSCs, one control iPSC line was used to generate an isogenic control and mutant lines as described by Kyrousi et al. (20).
Generation of labeled iPSC line
The GFP- and RFP-labeled iPSC lines were generated using the piggyBac (PB) transposase (1 μg) and PB-GFP (1 μg) (78) and PB-RFP (1 μg) nucleofection (79). Dissociated iPSCs were transfected with the Amaxa Nucleofector 2b (program B-016). GFP and RFP colonies were picked and cultured on Matrigel (Corning/VWR International, 354234)–coated plates in mTeSR1 basic medium (STEMCELL Technologies, 85850) supplemented with 1× mTeSR1 supplement (STEMCELL Technologies, 85850) at 37°C and 5% CO2.
Generation of dvCAs and vCOs
dvCAs and vCOs were generated according to Bagley et al. (80). Embryoid bodies (EBs) were guided to generate ventral and dorsal identities. iPSCs from isogenic control and mutant lines were dissociating into single cells using Accutase (Sigma-Aldrich, A6964), and approximately 9000 cells were transferred to one well of an ultralow-attachment 96-well plate (Corning). Five days later, during the neuronal induction, to induce brain regional patterning, EBs were treated individually with Smoothened Agonist (SAG) (1:10,000) (Millipore, 566660) and IWP-2 (1:2000) (Sigma-Aldrich, I0536) for ventral identity and with cyclopamine A (1:500) (Calbiochem, 239803) for dorsal identity. After 7 days, one ventral EB and one dorsal EB were embedded together into the same Matrigel (Corning/VWR International, 354234) droplet to form a fused organoid. The ventrally patterned EBs were embedded separately, each in one drop of Matrigel. After this point, the generation of vCOs followed methods according to Lancaster and Knoblich (81).
Exchange of culture media from CTRL-vCOs and E370K-vCOs
E370K-vCOs were fed with culture media collected from CTRL-vCOs starting from D12 of the protocol for the generation of vCOs. The CTRL culture medium was added to E370K-vCOs every day until the day of the analysis. CTRL-vCOs were fed with fresh media every day. At D60, vCOs were collected for immunohistochemistry analysis.
Generation of vMCOs
Ventral MCOs were generated according to Bagley et al. (24). iPSCs from isogenic control and from GFP-labeled E370k line were dissociating into single cells using Accutase (Sigma-Aldrich, A6964), and approximately 4500 cells of each line were mixed together and transferred to one well of an ultralow-attachment 96-well plate (Corning), in a ratio of 1:1. The protocols continue as described in the “Generation of dvCAs and vCOs” section.
Immunohistochemistry
For immunohistochemistry, sections were postfixed using 4% paraformaldehyde (PFA) for 10 min and permeabilized with 0.3% Triton X-100 for 5 min. After postfixation and permeabilization, sections were blocked with 0.1% Tween 20 and 10% normal goat serum (Biozol, VEC-S-1000). Primary and secondary antibodies were diluted in blocking solution. Nuclei were visualized using 4′,6-diamidino-2-phenylindole (0.5 mg/ml; Sigma-Aldrich, D9542). Immunostained sections were analyzed using a Leica TCS SP8 confocal microscope (Leica, Germany). For nuclear antibodies, before the postfixation step, sections were incubated in a freshly made 10 mM citric buffer (pH 6) in a microwave for 1 min at 720 W and for 10 min at 120 W and then left it to cool down for 20 min at room temperature (RT). Used primary and secondary antibodies are shown in tables S1 and S2.
Quantifications
Cell quantifications were performed with the ImageJ software and analyzed with RStudio or GraphPad. For the analysis of EOMES, we analyzed 96 CTRL and 53 E370K ventricles; for TBR1, we analyzed 100 CTRL and 107 E370K ventricles; and for SATB2, we analyzed 48 CTRL and 76 E370K ventricles, from three different batches. For analysis of those that are not nuclear markers, such as GAD67 and GFP, we used the “Analyze particles” tool of ImageJ software. After setting parameters for threshold intensity and size, this function calculates the particles (+signal) positive for GAD67. We used the “Area Fraction” parameter that calculates the percentage of pixels positive for GAD67 and GFP per area. For data visualization, every dot in the plot refers to analyzed ventricles per vCOs, dvCAs, or vMCOs from at least three different organoids and assembloids generated in at least two independent batches.
Human COs 3D immunohistochemistry and tissue clearing
The dvCA 3D immunohistochemistry and tissue clearing were performed following Masselink et al. (38). For immunohistochemistry, fused organoids were fixed in 4% PFA overnight at 4°C. They were incubated on a shaker for 2 days in phosphate-buffered saline (PBS)–TxDBN solution [10% PBS10X, 2% TX100, 20% dimethyl sulfoxide, 5% bovine serum albumin (BSA), and 0.05% NaN]. Primary and secondary antibodies were diluted in PBS-TxDBN solution and incubated for 4 days. For the following dehydration step, the organoids were transferred sequentially in 30, 50, and 70% as well as 2× 99.7% in 1-Propanol (Sigma-Aldrich, catalog no. W292818):1× PBS (pH 9) solutions. Dehydration is performed at 4°C on a gyratory rocker for at least 4 hours per step. For the refractive index matching, the organoids were transferred into ethyl cinnamate (Sigma-Aldrich, catalog no. W243000) solution and incubated on a gyratory rocker at RT for at least 1 hour before recording. The imaging was performed using a Leica TCS SP8 confocal microscope (Leica, Germany). The cells were counted using Imaris software and analyzed with RStudio.
Binning analysis in cleared dvCAs
The binning analysis in cleared dvCAs was performed by dividing the dorsal region into three equally distributed bins. This assay was used to get an overview of migration and distribution of ventrally generated neurons in the dorsal region of assembloids.
Human COs 3D time-lapse imaging
For 3D time-lapse imaging, slices of dvCAs were prepared and imaged as described previously (82). dvCAs were sliced at 300-μm thickness on a vibratome (Leica VT1200S) in ice-cold Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Invitrogen) supplement with sodium bicarbonate, glucose, and 10% antibiotics, oxygenated with 100% O2 for 20 min before cutting. The slices were placed on a cell culture insert (Millicell) and further cultured in normal organoid medium. The slices were kept in an atmosphere with 5% CO2 at 37°C. Live imaging was performed for 48 hours using a Leica TCS SP8 confocal microscope (Leica, Germany), taking an image every 20 min. The cell movement was tracked using ImageJ software and the Manual Tracking Plugin, and the movement parameters were calculated and analyzed in RStudio. The selected parameters are standard dynamics measurements previously shown in vivo (83, 84) and in vitro (10).
scRNA-seq library preparation and data analysis
Five 60-day-old vCOs and vMCOs were randomly selected from each condition. Single cells were dissociated using the StemPro Accutase Cell Dissociation Reagent (Life Technologies), filtered through 30 and 20 μM filters (Miltenyi Biotec) and cleaned of debris using a Percoll (Sigma-Aldrich, P1644) gradient. For vMCOs, single cells were fluorescence-activated cell sorting (FACS) sorted to collect control and GFP-labeled E370K single cells. Single cells were resuspended in ice-cold PBS supplemented with 0.04% BSA at a concentration of 1000 cells/μl. Single cells were loaded onto a Chromium Next GEM Single Cell 3′ chip (Chromium Next GEM Chip G Single Cell Kit, 16 rxns; 10x Genomics, PN-1000127) with the a Chromium Next GEM Single Cell 3′ GEM Library and Gel Bead Kit v3.1 (Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1, 4 rxns; 10x Genomics, PN-1000128), and cDNA libraries were generated with the Single Index Kit T Set A, 96 rxns (10x Genomics, PN-1000213) according to the manufacturer’s instructions. Libraries were sequenced using Illumina NovaSeq 6000 in 28/8/91 bp mode (SP flow cell); quality control and Unique Molecular Identifier (UMI) counting were performed by the Max-Planck für molekulare Genetik (Germany). Downstream analysis was performed using the R package Seurat (version 3.2). Cells with more than 2500 or less than 200 detected genes or with mitochondrial content higher that 10% were excluded, as well as genes that were not expressed in at least three cells. We excluded clusters with “glycolysis” identity based on GO terms of cluster-specific markers genes (21, 85). Normalization of gene expression was done using a global-scaling normalization method (“LogNormalize,” scale.factor = 10000), and the 2000 most variable genes were selected (selection method, “vst”) and scaled (mean = 0 and variance = 1 for each gene) before principal components analysis (PCA). The “FindNeighbors” and “FindClusters” functions were used for clustering with resolution of 0.5 and uniform manifold approximation and projection (UMAP) for visualization. Clusters were grouped on the basis of the expression of known marker genes and differentially expressed genes identified with the “FindAllMarkers” function. The PCA was used to determine the pseudo-differentiation axis of telencephalic cells. For pseudotime analysis, we used the Monocle 3 algorithm (26), which identifies the overall trajectory of gene expression changes.
FACS analysis
Single cells obtained after dissociation of 60-day-old vMCOs were collected for FACS analysis (see the “scRNA-seq library preparation and data analysis” section) to sort control and GFP-labeled E370K for scRNA-seq analysis. FACS analysis was performed at a FACSAria (BD) in BD FACSFlow medium, with a nozzle diameter of 100 μm. For each run, 10,000 cells were analyzed.
Dorsoventral identity model
The model implemented in the bmrm R package was used to classify the identity of cells with genes showing minimal expression (10,407), as described by Oberst et al. (86). The model was trained with a subset of dorsal and ventral control cells, and the 30 most-weighted genes were used for fold cross-validation of additional dorsal and ventral control cells and prediction of dorsal and ventral mutant cells. The same method was applied to build a model to classify control and mutant cells, selecting for 100 most-weighted genes.
EV collection and analysis
EVs were collected from conditioned media from ventral and dorsal COs by the following steps: conditioned media centrifugation at 300g for 15 min, supernatant centrifugation at 2000g for 10 min, supernatant centrifugation at 10,000g for 30 min, supernatant centrifugation at 100,000g for 90 min and pellet wash with 1× PBS, and centrifugation at 100,000g for 90 min. Alternatively, the miRCURY Exosome Cell/Urine/CSF Kit (QIAGEN, 76743) was used to isolate EVs from the conditioned medium according to the manufacturer’s instructions.
For immune-electron microscopy, aliquots of EV suspensions were analyzed by I. Miinalainen at Biocenter Oulu/EM laboratory, Finland (87). Vesicles were deposited on Formvar carbon-coated, glow-discharged grids and incubated in a blocking serum containing 1% BSA in PBS, and LGALS3BP primary antibodies and secondary gold conjugates (Zymed, San Francisco, CA, USA) were diluted in 1% BSA in PBS. The blocking efficiency was controlled by performing the labeling procedure in the absence of primary antibody.
vCOs dissociation
The D20 vCOs (24) were dissociated into single cells using the StemPro Accutase Cell Dissociation Reagent (Life Technologies) and filtered through 30 μM filters (Miltenyi Biotec). A total of 20,000 cells were plated onto each well of a 24-well plate coated with poly-l-ornithine (10 μg/ml) (Sigma-Aldrich, P4957)/laminin (10 μg/ml) (Sigma-Aldrich, L2020) and kept in neural differentiation medium (NDM) + DMEM/F12 + GlutaMAX and Neurobasal medium in a ratio 1:1 supplemented with 1:100 N2 supplement (100×), 1:100 B-27 supplement (50×), 0.5% of MEM nonessential amino acid solution (100×), 0.5% GlutaMAX supplement, 50 μM 2-mercaptoethanol (50 mM), antibiotic antimycotic solution (100×), and insulin (2.5 μg/ml) for 24 hours. The day after, EVs collected from vCOs (see the “EV collection and analysis” section) were resuspended in NDM [Neurobasal medium supplemented with 1:100 N2 supplement (100×), 1:100 B-27 supplement (50×), 0.5% of MEM nonessential amino acid solution (100×), and antibiotic antimycotic solution (100×)]. Brain-derived neurotrophic factor (20 ng/ml), glial cell line–derived neurotrophic factor (20 ng/ml), dibutyryl cyclic adenosine monophosphate (1 μM), ascorbic acid (200 μM), and laminin (2 μg/ml) were freshly added. NDM including EVs was added to the cells for 1.5 weeks. The medium was exchanged every 2 days.
Sample preparation for mass spectrometry
Purified EVs were lysed in radioimmunoprecipitation assay (RIPA) buffer [150 mM NaCl, 50 mM tris (pH 8), 0.1% sodium deoxycholate, 0.1% SDS, and 0.1% NP-40]. Ten micrograms of protein for each sample was subjected to the modified FASP protocol (88). Briefly, the protein extract was loaded onto the centrifugal filter CO 10 kDa (Merck Millipore, Darmstadt, Germany), and the detergent was removed by washing five times with 8 M urea (Merck, Darmstadt, Germany) and 50 mM tris (Sigma-Aldrich, USA) buffer. Proteins were reduced by adding 5 mM dithiothreitol (DTT) (Bio-Rad, Canada) at 37°C for 1 hour in the dark. To remove the excess of DTT, the protein sample was washed three times with 8 M urea and 50 mM tris. Subsequently, protein thiol groups were blocked with 10 mM iodoacetamide (Sigma-Aldrich, USA) at RT for 45 min. Before proceeding with the enzymatic digestion, urea was removed by washing the protein suspension three times with 50 mM ammonium bicarbonate (Sigma-Aldrich, Spain). Proteins were digested first by Lys-C (Promega, USA) at RT for 2 hours and then by trypsin (Premium Grade, MS Approved, SERVA, Heidelberg, Germany) at RT overnight; both enzymes were added at an enzyme-protein ratio of 1:50 (w/w). Peptides were recovered by centrifugation followed by two additional washes with 50 mM ammonium bicarbonate and 0.5 M NaCl (Sigma-Aldrich, Switzerland). The two filtrates were combined; the recovered peptides were lyophilized under vacuum. Dried tryptic peptides were desalted using C18 tips (Thermo Fisher Scientific, Pierce, USA), following the manufacturer’s instructions. Briefly, the peptides dissolved in 0.1% (v/v) formic acid (Thermo Fisher Scientific, USA) were loaded onto the C18 tip and washed 10 times with 0.1% (v/v) formic acid; subsequently, the peptides were eluted by 95% (v/v) acetonitrile (Merck, Darmstadt, Germany) and 0.1% (v/v) formic acid. The desalted peptides were lyophilized under vacuum. The purified peptides were reconstituted in 0.1% (v/v) formic acid for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
MS data acquisition
Desalted peptides were loaded onto a 25-cm, 75-μm ID C18 column with an integrated nanospray emitter (Odyssey/Aurora, ionopticks, Melbourne) via the autosampler of the Thermo Easy-nLC 1000 (Thermo Fisher Scientific) at 60°C. Eluting peptides were directly sprayed onto the timsTOF Pro (Bruker Daltonics). Peptides were loaded in buffer A [0.1% (v/v) formic acid] at 400 nl/min, and percentage of buffer B (80% acetonitril and 0.1% formic acid) was ramped from 5 to 25% over 90 min followed by a ramp to 35% over 30 min, then 58% over the next 5 min, 95% over the next 5 min, and maintained at 95% for another 5 min. Data acquisition on the timsTOF Pro was performed using timsControl. The mass spectrometer was operated in data-dependent PASEF mode with one survey TIMS-MS and 10 PASEF MS/MS scans per acquisition cycle. Analysis was performed in a mass scan range from 100 to 1700 m/z (mass/charge ratio) and an ion mobility range from 1/K0 = 0.85 to 1.30 V s cm−2 using equal ion accumulation and ramp time in the dual TIMS analyzer of 100 ms each at a spectra rate of 9.43 Hz. Suitable precursor ions for MS/MS analysis were isolated in a window of 2 Th for m/z < 700 and 3 Th for m/z > 700 by rapidly switching the quadrupole position in sync with the elution of precursors from the TIMS device. The collision energy was lowered as a function of ion mobility, starting from 45 eV for 1/K0 = 1.3 V s cm−2 to 27 eV for 0.85 V s cm−2. Collision energies were interpolated linearly between these two 1/K0 values and kept constant above or below these base points. Singly charged precursor ions were excluded with a polygon filter mask, and further m/z and ion mobility information was used for “dynamic exclusion” to avoid resequencing of precursors that reached a “target value” of 14,500 arbitrary units. The ion mobility dimension was calibrated linearly using three ions from the Agilent ESI LC-MS tuning mix (m/z, 1/K0: 622.0289, 0.9848 V s cm−2; 922.0097, 1.1895 V s cm−2; 1221.9906, 1.3820 V s cm−2).
Raw data analysis of MS measurements
Raw data were processed using the MaxQuant computational platform (version 1.6.17.0) (89) with standard settings applied for ion mobility data (90). In short, the peak list was searched against the UniProt database of human database (75,069 entries, downloaded in July 2020) with an allowed precursor mass deviation of 10 parts per million (ppm) and an allowed fragment mass deviation of 20 ppm. MaxQuant by default enables individual peptide mass tolerances, which was used in the search. Cysteine carbamidomethylation was set as static modification, and methionine oxidation, deamidation and N-terminal acetylation as variable modifications. The match-between-run option was enabled, and proteins were quantified across samples using the label-free quantification (LFQ) algorithm in MaxQuant, generating LFQ intensities. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (91), partner repository with the dataset identifier PXD028438.
Bioinformatic analysis
For the proteomic characterization in EVs, 3427 proteins were quantified. Proteins that were consistently detected in two of the three technical replicates per each condition were retained. Downstream analysis was performed using R. The LFQ values were log2-transformed. Missing values were imputed using the R package DEP (version 1.15.0) and replaced by random values of a left-shifted Gaussian distribution (shift of 1.8 units of the SD and a width of 0.3). DE analysis was performed on the imputed data using Student’s t test. Proteins with log2 fold change values (log2FC) ≥ 1 and ≤−1 and with a false discovery rate (FDR)–corrected q < 0.05 were considered as differentially expressed. The gene-disease association analysis was performed using DisGeNET (www.disgenet.org/search) (92).
Western blots
For Western blot, EVs were treated in RIPA buffer containing protease and phosphatase inhibitors (Roche, Basel, Switzerland). We used SDS–polyacrylamide gel electrophoresis with a 12.5% acrylamide gel. We used 50 mM DTT to separate the proteins and added 50 mM DTT to dissociate proteins and avoid protein aggregates. Then, the proteins were transferred to a nitrocellulose membrane (GE HealthCare, Chalfont St. Giles, Buckinghamshire, UK). Then, we colored the membranes with a Ponceau Red solution (Serva, 33427.01) to verify the quality of the transfer. Next, we incubated the membranes with primary antibodies (LGALS3BP, rabbit ab217572, Abcam; SHH, mouse, sc-365112, Santa Cruz Biotechnology; LUM, rabbit, ab168948, Abcam) overnight in rotation at 4°C. The membranes were washed three times with Tris-buffered saline with 0.1% Tween (TBS-T) for 10 min each and incubated with secondary antibodies for 1 hour at RT. After three washes in TBS-T, we treated the membrane with the ECL Western Blotting Detection solution (Millipore, Billerica, MA, USA) for protein detection. Bands were quantified using ImageJ software.
Bulk RNA-seq
RNA-seq was performed on 10 ng of total RNA collected from three independent wells of NPCs from a 24-well plate. NPCs were not treated with EVS or treated for 12 hours with EVs collected by ultracentrifugation from 25 ml of conditioned medium collected from 28 to 37 days in culture COs (control ventral, EPM1 ventral, control dorsal, and EPM1 dorsal COs). NPCs were lysed in 1 ml of TRIzol (QIAGEN) per well, and RNA was isolated using the RNA Clean & Concentrator kit (Zymo Research) including digestion of remaining genomic DNA according to producer’s guidelines. RNA was further processed according to Cernilogar et al. (93). Briefly, cDNA synthesis was performed with the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech, catalog no. 634888) according to the manufacturer’s instruction. cDNA was fragmented to an average size of 200 to 500 base pairs (bp) in a Covaris S220 device (5 min; 4°C; peak power 175; duty factor 10; cycle burst 200). Fragmented cDNA was used as input for library preparation with the MicroPlex Library Preparation Kit v2 (Diagenode, catalog no. C05010012) and processed according to the manufacturer’s instructions. Libraries were quality controlled by Qubit and Agilent DNA Bioanalyzer analysis. Deep sequencing was performed on a HiSeq 1500 system according to the standard Illumina protocol for 50-bp paired-end reads with v3 sequencing reagents.
RNA-seq analysis
Paired-end reads were aligned to the human genome version GRCh38 using STAR v2.6.1d (94) with default options “--runThreadN 32 --quantMode TranscriptomeSAM GeneCounts --outSAMtype BAM SortedByCoordinate.” Reads-per-gene counts were imported in R v4.1.0. Bioconductor package DESeq2 v1.32.0 (95) was used for DE analysis. Only genes with read counts > 1 were considered. Significantly changed genes were determined through pairwise comparisons using the DESeq2 results function (log2 fold change threshold = 1, adjusted P value < 0.05). Heatmaps with differentially expressed genes were plotted with pheatmap v1.0.12 and RColorBrewer v1.1-2 using rlog-normalized expression values. The bulk RNA-seq data used in this study have been deposited in the Gene Expression Omnibus under accession number GSE197252.
Enrichment analysis
GO term analysis of differentially expressed genes in mutant patterned human COs was tested using the FUMA algorithm (96) by inserting the DE gene lists from all the cell populations into the GENE2FUNC software (FDR < 0.05) or STRING.
Data visualization
Data visualization was performed using RStudio and GraphPad. Schematics in the figures were partially generated with BioRender (https://biorender.com/).
Acknowledgments
We thank M. Ködel, A. Krontira, P. Lopez, C. Cruceanu, F. Cernilogar, S. Moser, F. Di Matteo, V. Pravata, G. Berto, K. Christofi, and L. Nguyen for technical help and critical discussion.
Funding: This project is supported by Max Planck Society, ERA-Net E-Rare (HETEROMICS|01GM1914), ERA-Net Neuron (nEUrotalk|01EW1907), and DFG (CA 1205/4-1|RU 1232/7-1).
Author contributions: Conceptualization: S.C. and F.P. Methodology: F.P., N.B., C.K., R.D.G., R.B., A.F.-E., and G.M. Investigation: S.C., F.P., N.B., and R.D.G. Visualization: F.P. Funding acquisition: S.C. Supervision: S.C. and D.J. Writing—original draft: F.P.; review and editing: S.C. and F.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Single-cell RNA-seq datasets have been deposited in the Gene Expression Omnibus under accession number GSE202874. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (91), partner repository with the dataset identifier PXD028438. The bulk RNA-seq data used in this study have been deposited in the Gene Expression Omnibus under accession number GSE197252.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.J. Guo, E. S. Anton, Decision making during interneuron migration in the developing cerebral cortex. Trends Cell Biol. 24, 342–351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Steinecke, C. Gampe, G. Zimmer, J. Rudolph, J. Bolz, EphA/ephrin A reverse signaling promotes the migration of cortical interneurons from the medial ganglionic eminence. Development 141, 460–471 (2014). [DOI] [PubMed] [Google Scholar]
- 3.E. Peyre, C. G. Silva, L. Nguyen, Crosstalk between intracellular and extracellular signals regulating interneuron production, migration and integration into the cortex. Front. Cell. Neurosci. 9, 129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Tatti, M. S. Haley, O. K. Swanson, T. Tselha, A. Maffei, Neurophysiology and regulation of the balance between excitation and inhibition in neocortical circuits. Biol. Psychiatry 81, 821–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.F. M. Krienen, M. Goldman, Q. Zhang, R. C. H. del Rosario, M. Florio, R. Machold, A. Saunders, K. Levandowski, H. Zaniewski, B. Schuman, C. Wu, A. Lutservitz, C. D. Mullally, N. Reed, E. Bien, L. Bortolin, M. Fernandez-Otero, J. D. Lin, A. Wysoker, J. Nemesh, D. Kulp, M. Burns, V. Tkachev, R. Smith, C. A. Walsh, J. Dimidschstein, B. Rudy, L. S. Kean, S. Berretta, G. Fishell, G. Feng, S. A. McCarroll, Innovations present in the primate interneuron repertoire. Nature 586, 262–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D. Džaja, A. Hladnik, I. Bǐanić, M. Baković, Z. Petanjek, Neocortical calretinin neurons in primates: Increase in proportion and microcircuitry structure. Front. Neuroanat. 8, 103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Yu, Z. Zeng, D. Xie, R. Chen, Y. Sha, S. Huang, W. Cai, W. Chen, W. Li, R. Ke, T. Sun, Interneuron origin and molecular diversity in the human fetal brain. Nat. Neurosci. 24, 1745–1756 (2021). [DOI] [PubMed] [Google Scholar]
- 8.S. A. Fietz, W. B. Huttner, Cortical progenitor expansion, self-renewal and neurogenesis—A polarized perspective. Curr. Opin. Neurobiol. 21, 23–35 (2010). [DOI] [PubMed] [Google Scholar]
- 9.V. Fernández, C. Llinares-benadero, V. Borrell, Cerebral cortex expansion and folding: What have we learned ? EMBO J. 35, 1021–1044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. Klaus, S. Kanton, C. Kyrousi, A. C. Ayo-Martin, R. Di Giaimo, S. Riesenberg, A. C. O’Neill, J. Gray Camp, C. Tocco, M. Santel, E. Rusha, M. Drukker, M. Schroeder, M. Götz, S. P. Robertson, B. Treutlein, S. Cappello, Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat. Med. 25, 561–568 (2019). [DOI] [PubMed] [Google Scholar]
- 11.S. Bajaj, J. A. Bagley, C. Sommer, A. Vertesy, S. N. Wong, V. Krenn, J. Lévi-Strauss, J. A. Knoblich, Neurotransmitter signaling regulates distinct phases of multimodal human interneuron migration. EMBO J. 40, e108714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. G. Silva, E. Peyre, M. H. Adhikari, S. Tielens, S. Tanco, P. Van Damme, L. Magno, N. Krusy, G. Agirman, M. M. Magiera, N. Kessaris, B. Malgrange, A. Andrieux, C. Janke, L. Nguyen, Cell-intrinsic control of interneuron migration drives cortical morphogenesis. Cell 172, 1063–1078.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. Bellion, J. P. Baudoin, C. Alvarez, M. Bornens, C. Métin, Nucleokinesis in tangentially migrating neurons comprises two alternating phases: Forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25, 5691–5699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.K. R. Long, W. B. Huttner, How the extracellular matrix shapes neural development. Open Biol. 9, 180216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.L. Lim, D. Mi, A. Llorca, O. Marín, Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O. Marín, J. L. R. Rubenstein, A long, remarkable journey: Tangential migration in the telencephalon. Nat. Rev. Neurosci. 2, 780–790 (2001). [DOI] [PubMed] [Google Scholar]
- 17.S. Amin, V. Borrell, The extracellular matrix in the evolution of cortical development and folding. Front. Cell Dev. Biol. 8, 604448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.P. Stampolidis, A. Ullrich, S. Iacobelli, LGALS3BP, lectin galactoside-binding soluble 3 binding protein, promotes oncogenic cellular events impeded by antibody intervention. Oncogene 34, 39–52 (2015). [DOI] [PubMed] [Google Scholar]
- 19.J. H. Lee, J. A. Bae, J. H. Lee, Y.-W. Seo, D. H. Kho, E. G. Sun, S. E. Lee, S. H. Cho, Y. E. Joo, K. Y. Ahn, I. J. Chung, K. K. Kim, Glycoprotein 90K, downregulated in advanced colorectal cancer tissues, interacts with CD9/CD82 and suppresses the Wnt/β-catenin signal via ISGylation of β-catenin. Gut 59, 907–917 (2010). [DOI] [PubMed] [Google Scholar]
- 20.C. Kyrousi, A. C. O’Neill, A. Brazovskaja, Z. He, P. Kielkowski, L. Coquand, R. Di Giaimo, P. D’ Andrea, A. Belka, A. Forero Echeverry, D. Mei, M. Lenge, C. Cruceanu, I. Y. Buchsbaum, S. Khattak, G. Fabien, E. Binder, F. Elmslie, R. Guerrini, A. D. Baffet, S. A. Sieber, B. Treutlein, S. P. Robertson, S. Cappello, Extracellular LGALS3BP regulates neural progenitor position and relates to human cortical complexity. Nat. Commun. 12, 6298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.S. Kanton, M. J. Boyle, Z. He, M. Santel, A. Weigert, F. Sanchís-Calleja, P. Guijarro, L. Sidow, J. S. Fleck, D. Han, Z. Qian, M. Heide, W. B. Huttner, P. Khaitovich, S. Pääbo, B. Treutlein, J. G. Camp, Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 22.T. J. Nowakowski, A. Bhaduri, A. A. Pollen, B. Alvarado, M. A. Mostajo-Radji, E. Di Lullo, M. Haeussler, C. Sandoval-Espinosa, S. J. Liu, D. Velmeshev, J. R. Ounadjela, J. Shuga, X. Wang, D. A. Lim, J. A. West, A. A. Leyrat, W. J. Kent, A. R. Kriegstein, Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.G. La Manno, K. Siletti, A. Furlan, D. Gyllborg, E. Vinsland, A. Mossi Albiach, C. Mattsson Langseth, I. Khven, A. R. Lederer, L. M. Dratva, A. Johnsson, M. Nilsson, P. Lönnerberg, S. Linnarsson, Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021). [DOI] [PubMed] [Google Scholar]
- 24.J. A. Bagley, D. Reumann, S. Bian, J. Lévi-Strauss, J. A. Knoblich, Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Y. Miura, M. Y. Li, F. Birey, K. Ikeda, O. Revah, M. V. Thete, J. Y. Park, A. Puno, S. H. Lee, M. H. Porteus, S. P. Pașca, Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 38, 1421–1430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. Trapnell, D. Cacchiarelli, J. Grimsby, P. Pokharel, S. Li, M. Morse, N. J. Lennon, K. J. Livak, T. S. Mikkelsen, J. L. Rinn, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.P. Hayward, T. Kalmar, A. M. Arias, Wnt/Notch signalling and information processing during development. Development 135, 411–424 (2008). [DOI] [PubMed] [Google Scholar]
- 28.M. Luo, Q. Zhang, Y. Hu, C. Sun, Y. Sheng, C. Deng, Lgals3bp: A potential plasma biomarker associated with diagnosis and prognosis in patients with sepsis. Infect. Drug Resist. 14, 2863–2871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L. R. Hernández-Miranda, J. G. Parnavelas, F. Chiara, Molecules and mechanisms involved in the generation and migration of cortical interneurons. ASN Neuro 2, e00031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. Chi, B. Fan, D. Feng, Z. Chen, Z. Liu, Y. Hui, X. Xu, L. Ma, Y. Fang, Q. Zhang, G. Jin, L. Liu, F. Guan, X. Zhang, The dorsoventral patterning of human forebrain follows an activation/transformation model. Cereb. Cortex 27, 2941–2954 (2017). [DOI] [PubMed] [Google Scholar]
- 31.S. W. Wilson, C. Houart, Early steps in the development of the forebrain. Dev. Cell 6, 167–181 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.E. C. Lai, Notch signaling: Control of cell communication and cell fate. Development 131, 965–973 (2004). [DOI] [PubMed] [Google Scholar]
- 33.B. De Strooper, W. Annaert, Where Notch and Wnt signaling meet: The presenilin hub. J. Cell Biol. 152, F17–F20 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.I. Y. Buchsbaum, S. Cappello, Neuronal migration in the CNS during development and disease: Insights from in vivo and in vitro models. Development 146, dev163766 (2019). [DOI] [PubMed] [Google Scholar]
- 35.C. G. Silva, E. Peyre, L. Nguyen, Cell migration promotes dynamic cellular interactions to control cerebral cortex morphogenesis. Nat. Rev. Neurosci. 20, 318–329 (2019). [DOI] [PubMed] [Google Scholar]
- 36.F. Birey, J. Andersen, D. Makinson, S. Islam, W. Wei, N. Huber, H. Christina Fan, K. R. Cordes Metzler, G. Panagiotakos, N. Thom, N. A. O’rourke, L. M. Steinmetz, J. A. Bernstein, J. Hallmayer, J. R. Huguenard, S. P. Pașca, Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Xiang, Y. Tanaka, B. Patterson, Y. J. Kang, G. Govindaiah, N. Roselaar, B. Cakir, K. Y. Kim, A. P. Lombroso, S. M. Hwang, M. Zhong, E. G. Stanley, A. G. Elefanty, J. R. Naegele, S. H. Lee, S. M. Weissman, I. H. Park, Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383–398.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.W. Masselink, D. Reumann, P. Murawala, P. Pasierbek, Y. Taniguchi, F. Bonnay, K. Meixner, J. A. Knoblich, E. M. Tanaka, Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development 146, dev166884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.E. Capone, A. Lamolinara, F. Pastorino, R. Gentile, S. Ponziani, G. Di Vittorio, D. D’agostino, S. Bibbò, C. Rossi, E. Piccolo, V. Iacobelli, R. Lattanzio, V. Panella, M. Sallese, V. De Laurenzi, F. Giansanti, A. Sala, M. Iezzi, M. Ponzoni, R. Ippoliti, S. Iacobelli, G. Sala, Targeting vesicular lgals3bp by an antibody-drug conjugate as novel therapeutic strategy for neuroblastoma. Cancers 12, 2989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. Capone, S. Iacobelli, G. Sala, Role of galectin 3 binding protein in cancer progression: A potential novel therapeutic target. J. Transl. Med. 19, 405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.C. Escrevente, N. Grammel, S. Kandzia, J. Zeiser, E. M. Tranfield, H. S. Conradt, J. Costa, Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLOS ONE 8, e78631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.X. Zhang, H. Ding, Z. Lu, L. Ding, Y. Song, Y. Jing, Q. Hu, Y. Dong, Y. Ni, Increased LGALS3BP promotes proliferation and migration of oral squamous cell carcinoma via PI3K/AKT pathway. Cell. Signal. 63, 109359 (2019). [DOI] [PubMed] [Google Scholar]
- 43.J. Gomes, P. Gomes-Alves, S. B. Carvalho, C. Peixoto, P. M. Alves, P. Altevogt, J. Costa, Extracellular vesicles from ovarian carcinoma cells display specific glycosignatures. Biomolecules. 5, 1741–1761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Y. Song, M. Wang, H. Tong, Y. Tan, X. Hu, K. Wang, X. Wan, Plasma exosomes from endometrial cancer patients contain LGALS3BP to promote endometrial cancer progression. Oncogene 40, 633–646 (2021). [DOI] [PubMed] [Google Scholar]
- 45.M. L. Fogeron, H. Müller, S. Schade, F. Dreher, V. Lehmann, A. Kühnel, A. K. Scholz, K. Kashofer, A. Zerck, B. Fauler, R. Lurz, R. Herwig, K. Zatloukal, H. Lehrach, J. Gobom, E. Nordhoff, B. M. H. Lange, LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat. Commun. 4, 1531 (2013). [DOI] [PubMed] [Google Scholar]
- 46.B. Wamsley, G. Fishell, Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat. Rev. Neurosci. 18, 299–309 (2017). [DOI] [PubMed] [Google Scholar]
- 47.A. Kepecs, G. Fishell, Interneuron cell types are fit to function. Nature 505, 318–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J. S. Fleck, F. Sanchís-Calleja, Z. He, M. Santel, M. J. Boyle, J. G. Camp, B. Treutlein, Resolving organoid brain region identities by mapping single-cell genomic data to reference atlases. Cell Stem Cell 28, 1148–1159.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 49.C. L. Thompson, L. Ng, V. Menon, S. Martinez, C.-K. Lee, K. Glattfelder, S. M. Sunkin, A. Henry, C. Lau, C. Dang, R. Garcia-Lopez, A. Martinez-Ferre, A. Pombero, J. L. R. Rubenstein, W. B. Wakeman, J. Hohmann, N. Dee, A. J. Sodt, R. Young, K. Smith, T.-N. Nguyen, J. Kidney, L. Kuan, A. Jeromin, A. Kaykas, J. Miller, D. Page, G. Orta, A. Bernard, Z. Riley, S. Smith, P. Wohnoutka, M. J. Hawrylycz, L. Puelles, A. R. Jones, A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Taverna, W. B. Huttner, Neural progenitor nuclei IN motion. Neuron 67, 906–914 (2010). [DOI] [PubMed] [Google Scholar]
- 51.P. Sharma, P. Mesci, C. Carromeu, D. R. McClatchy, L. Schiapparelli, J. R. Yates, A. R. Muotri, H. T. Cline, Exosomes regulate neurogenesis and circuit assembly. Proc. Natl. Acad. Sci. U.S.A. 116, 16086–16094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.L. Peruzzotti-Jametti, J. D. Bernstock, C. M. Willis, G. Manferrari, R. Rogall, E. Fernandez-Vizarra, J. C. Williamson, A. Braga, A. van den Bosch, T. Leonardi, G. Krzak, Á. Kittel, C. Benincá, N. Vicario, S. Tan, C. Bastos, I. Bicci, N. Iraci, J. A. Smith, B. Peacock, K. H. Muller, P. J. Lehner, E. I. Buzas, N. Faria, M. Zeviani, C. Frezza, A. Brisson, N. J. Matheson, C. Viscomi, S. Pluchino, Neural stem cells traffic functional mitochondria via extracellular vesicles. PLOS Biol. 19, e3001166 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Y. Liang, W. S. Eng, D. R. Colquhoun, R. R. Dinglasan, D. R. Graham, L. K. Mahal, Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J. Biol. Chem. 289, 32526–32537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.I. K. Sundar, D. Li, I. Rahman, Proteomic analysis of plasma-derived extracellular vesicles in smokers and patients with chronic obstructive pulmonary disease. ACS Omega 4, 10649–10661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.B. S. Batista, W. S. Eng, K. T. Pilobello, K. D. Hendricks-Muñoz, L. K. Mahal, Identification of a conserved glycan signature for microvesicles. J. Proteome Res. 10, 4624–4633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. Costa, A. Pronto-Laborinho, S. Pinto, M. Gromicho, S. Bonucci, E. Tranfield, C. Correia, B. M. Alexandre, M. de Carvalho, Investigating LGALS3BP/90 K glycoprotein in the cerebrospinal fluid of patients with neurological diseases. Sci. Rep. 10, 5649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.F. Teng, M. Fussenegger, Shedding light on extracellular vesicle biogenesis and bioengineering. Adv. Sci. 8, 2003505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Rallu, R. Machold, N. Gaiano, J. G. Corbin, A. P. McMahon, G. Fishell, Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and hedgehog signaling. Development 129, 4963–4974 (2002). [DOI] [PubMed] [Google Scholar]
- 59.X. Jiang, O. Charlat, R. Zamponi, Y. Yang, F. Cong, Dishevelled promotes wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol. Cell 58, 522–533 (2015). [DOI] [PubMed] [Google Scholar]
- 60.C. R. Altmann, A. H. Brivanlou, Neural patterning in the vertebrate embryo. Int. Rev. Cytol. 203, 447–482 (2001). [DOI] [PubMed] [Google Scholar]
- 61.K. R. Long, B. Newland, M. Florio, N. Kalebic, B. Langen, A. Kolterer, P. Wimberger, W. B. Huttner, Extracellular matrix components HAPLN1, lumican, and collagen I cause hyaluronic acid-dependent folding of the developing human neocortex. Neuron 99, 702–719.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 62.N. Maeda, Proteoglycans and neuronal migration in the cerebral cortex during development and disease. Front. Neurosci. 9, 98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. Kumar, K. Reynolds, Y. Ji, R. Gu, S. Rai, C. J. Zhou, Impaired neurodevelopmental pathways in autism spectrum disorder: A review of signaling mechanisms and crosstalk. J. Neurodev. Disord. 11, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M. Nawaz, F. Fatima, Extracellular vesicles, tunneling nanotubes, and cellular interplay: Synergies and missing links. Front. Mol. Biosci. 4, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.N. A. J. Puts, E. L. Wodka, A. D. Harris, D. Crocetti, M. Tommerdahl, S. H. Mostofsky, R. A. E. Edden, Reduced GABA and altered somatosensory function in children with autism spectrum disorder. Autism Res. 10, 608–619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Ariza, H. Rogers, E. Hashemi, S. C. Noctor, V. Martínez-cerdeño, The number of chandelier and basket cells are differentially decreased in prefrontal cortex in autism. Cereb. Cortex 28, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.J. Mariani, G. Coppola, K. A. Pelphrey, J. R. Howe, F. M. Vaccarino, FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.H. Wang, Modeling neurological diseases with human brain organoids. Front. Synaptic Neurosci. 10, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.C. E. Villa, C. Cheroni, C. P. Dotter, J. G. Danzl, G. Testa, G. Novarino, CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 39, 110615 (2022). [DOI] [PubMed] [Google Scholar]
- 70.B. Paulsen, S. Velasco, A. J. Kedaigle, M. Pigoni, G. Quadrato, A. J. Deo, X. Adiconis, A. Uzquiano, R. Sartore, S. M. Yang, S. K. Simmons, P. Symvoulidis, K. Kim, K. Tsafou, A. Podury, C. Abbate, A. Tucewicz, S. N. Smith, A. Albanese, L. Barrett, N. E. Sanjana, X. Shi, K. Chung, K. Lage, E. S. Boyden, A. Regev, J. Z. Levin, P. Arlotta, Autism Genes Converge on Asynchronous Development of Shared Neuron Classes (Springer US, 2022), vol. 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R. N. Delgado, D. E. Allen, M. G. Keefe, W. R. Mancia Leon, R. S. Ziffra, E. E. Crouch, A. Alvarez-Buylla, T. J. Nowakowski, Individual human cortical progenitors can produce excitatory and inhibitory neurons. Nature 601, 397–403 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. Fuccillo, M. Rutlin, G. Fishell, Removal of Pax6 partially rescues the loss of ventral structures in Shh null mice. Cereb. Cortex 16, i96–i102 (2006). [DOI] [PubMed] [Google Scholar]
- 73.T. Theil, G. Alvarez-Bolado, A. Walter, U. Rüther, Gli3 is required for Emx gene expression during dorsal telencephalon development. Development 126, 3561–3571 (1999). [DOI] [PubMed] [Google Scholar]
- 74.S. Tole, C. W. Ragsdale, E. A. Grove, Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toesJ. Dev. Biol. 217, 254–265 (2000). [DOI] [PubMed] [Google Scholar]
- 75.T. Pikkarainen, T. Nurmi, T. Sasaki, U. Bergmann, S. Vainio, Role of the extracellular matrix-located Mac-2 binding protein as an interactor of the Wnt proteins. Biochem. Biophys. Res. Commun. 491, 953–957 (2017). [DOI] [PubMed] [Google Scholar]
- 76.O. Al-Dalahmah, J. Nicholson, S. Draijer, L. C. Soares, F. G. Szele, Galectin-3 diminishes Wnt signaling in the postnatal subventricular zone. Stem Cells 38, 1149–1158 (2020). [DOI] [PubMed] [Google Scholar]
- 77.A. Cárdenas, A. Villalba, C. de Juan Romero, E. Picó, C. Kyrousi, A. C. Tzika, M. Tessier-Lavigne, L. Ma, M. Drukker, S. Cappello, V. Borrell, Evolution of cortical neurogenesis in amniotes controlled by robo signaling levels. Cell 174, 590–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.F. Di Matteo, F. Pipicelli, C. Kyrousi, I. Tovecci, E. Penna, M. Crispino, A. Chambery, R. Russo, A. C. Ayo-Martin, M. Giordano, A. Hoffmann, E. Ciusani, L. Canafoglia, M. Götz, R. Di Giaimo, S. Cappello, Cystatin B is essential for proliferation and interneuron migration in individuals with EPM1 epilepsy. EMBO Mol. Med. 12, e11419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.F. Chen, J. LoTurco, A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods 207, 172–180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.J. A. Bagley, D. Reumann, S. Bian, J. A. Knoblich, Fused dorsal-ventral cerebral organoids model human cortical interneuron migration. bioRxiv 131250 [Preprint]. 26 April 2017. 10.1101/131250. [DOI]
- 81.M. A. Lancaster, N. S. Corsini, S. Wolfinger, E. H. Gustafson, A. W. Phillips, T. R. Burkard, T. Otani, F. J. Livesey, J. A. Knoblich, Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R. Stahl, T. Walcher, C. De Juan Romero, G. A. Pilz, S. Cappello, M. Irmler, J. M. Sanz-Aquela, J. Beckers, R. Blum, V. Borrell, M. Götz, Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell 153, 535–549 (2013). [DOI] [PubMed] [Google Scholar]
- 83.F. Lepiemme, J. Stoufflet, M. Javier-Torrent, G. Mazzucchelli, C. G. Silva, L. Nguyen, Oligodendrocyte precursors guide interneuron migration by unidirectional contact repulsion. Science 376, eabn6204 (2022). [DOI] [PubMed] [Google Scholar]
- 84.F. Lepiemme, C. G. Silva, L. Nguyen, Time lapse recording of cortical interneuron migration in mouse organotypic brain slices and explants. STAR Protoc. 2, 100467 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.A. Bhaduri, M. G. Andrews, W. Mancia Leon, D. Jung, D. Shin, D. Allen, D. Jung, G. Schmunk, M. Haeussler, J. Salma, A. A. Pollen, T. J. Nowakowski, A. R. Kriegstein, Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.P. Oberst, S. Fièvre, N. Baumann, C. Concetti, G. Bartolini, D. Jabaudon, Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature 573, 370–374 (2019). [DOI] [PubMed] [Google Scholar]
- 87.J. Van Deun, Q. Roux, S. Deville, T. Van Acker, P. Rappu, I. Miinalainen, J. Heino, F. Vanhaecke, B. G. De Geest, O. De Wever, A. Hendrix, Feasibility of mechanical extrusion to coat nanoparticles with extracellular vesicle membranes. Cells 9, 1797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.J. R. Wiśniewski, A. Zougman, N. Nagaraj, M. Mann, Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 89.S. Tyanova, T. Temu, J. Cox, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 90.N. Prianichnikov, H. Koch, S. Koch, M. Lubeck, R. Heilig, S. Brehmer, R. Fischer, J. Cox, Maxquant software for ion mobility enhanced shotgun proteomics. Mol. Cell. Proteomics 19, 1058–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Y. Perez-Riverol, A. Csordas, J. Bai, M. Bernal-Llinares, S. Hewapathirana, D. J. Kundu, A. Inuganti, J. Griss, G. Mayer, M. Eisenacher, E. Pérez, J. Uszkoreit, J. Pfeuffer, T. Sachsenberg, Ş. Yilmaz, S. Tiwary, J. Cox, E. Audain, M. Walzer, A. F. Jarnuczak, T. Ternent, A. Brazma, J. A. Vizcaíno, The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.J. Piñero, Á. Bravo, N. Queralt-Rosinach, A. Gutiérrez-Sacristán, J. Deu-Pons, E. Centeno, J. García-García, F. Sanz, L. I. Furlong, DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.F. M. Cernilogar, S. Hasenöder, Z. Wang, K. Scheibner, I. Burtscher, M. Sterr, P. Smialowski, S. Groh, I. M. Evenroed, G. D. Gilfillan, H. Lickert, G. Schotta, Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 47, 9069–9086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.K. Watanabe, E. Taskesen, A. Van Bochoven, D. Posthuma, Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2