Abstract
The shufflon, a multiple DNA inversion system in plasmid R64, consists of four invertible DNA segments which are separated and flanked by seven 19-bp repeat sequences. The product of a site-specific recombinase gene, rci, promotes site-specific recombination between any two of the inverted 19-bp repeat sequences of the shufflon. To analyze the molecular mechanism of this recombination reaction, Rci protein was overproduced and purified. The purified Rci protein promoted the in vitro recombination reaction between the inverted 19-bp repeats of supercoiled DNA of a plasmid carrying segment A of the R64 shufflon. The recombination reaction was enhanced by the bacterial host factor HU. Gel electrophoretic analysis indicated that the Rci protein specifically binds to the DNA segments carrying the 19-bp sequences. The binding affinity of the Rci protein to the four shufflon segments as well as four synthetic 19-bp sequences differed greatly: among the four 19-bp repeat sequences, the repeat-a and -d sequences displayed higher affinity to Rci protein. These results suggest that the differences in the affinity of Rci protein for the 19-bp repeat sequences determine the inversion frequencies of the four segments.
Several multiple-inversion systems which function to create biological diversity have been identified in prokaryotes (for a review, see reference 14). The shufflon of plasmid R64 consists of four DNA segments, A, B, C, and D, which are separated and flanked by seven 19-bp repeat sequences (17, 18) (Fig. 1A). The site-specific recombination between any inverted 19-bp repeat sequences mediated by the rci product results in the inversion of a DNA segment(s) independently or in groups.
FIG. 1.
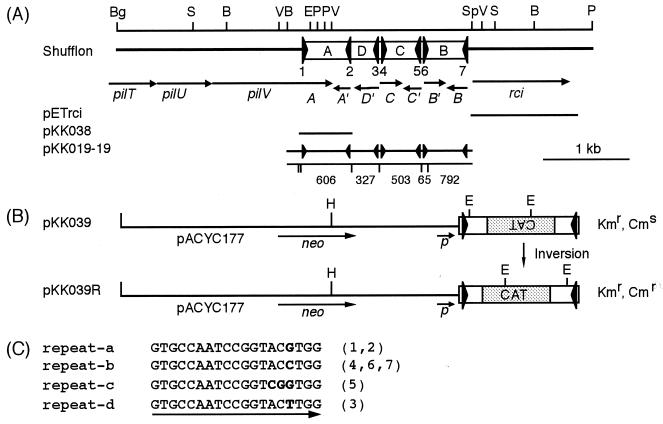
(A) Gene organization of pilTUV, shf, and rci regions of plasmid R64. This figure represents an arrangement (corresponding to pKK010-85) of many isomers of the shufflon subjected to multiple DNA inversions. At the top, a restriction map is shown. B, BstBI; Bg, BglII; E, EcoRI; P, PstI; S, SspI; Sp, SphI; V, EcoRV. The large triangles represent the 19-bp repeat sequences. Open reading frames deduced from the nucleotide sequence are represented by arrows. The lines indicate the DNA portions present in various plasmids. Below pKK019-19, HaeIII sites and the lengths (in base pairs) of DNA fragments produced by HaeIII and SspI digestion of pKK019-19 are indicated by vertical lines and numbers, respectively (see Fig. 5A). (B) DNA substrate for Rci-mediated recombination. The open bars represent segment A of the R64 shufflon. The large triangles represent the 19-bp sequences. The stippled bars indicate a DNA fragment carrying the promoterless CAT gene. The lines represent the pACYC177 sequence. (C) Four types of seven 19-bp repeat sequences. The numbers in parentheses correspond to the numbers of the 19-bp repeat sequences represented in panel A. Nucleotides that differ from the other sequences are indicated by boldface letters.
The R64 shufflon is located in the C-terminal region of the pilV gene, which is the last gene of the pil operon. The R64 pil operon functions in the formation of conjugative thin pili which are required only for liquid matings. From the deduced amino acid sequences of several pil gene products, the R64 thin pilus was shown to belong to the type IV pilus family (13). DNA inversions in the shufflon give rise to seven pilV genes in which the N-terminal regions are constant while the C-terminal regions are variable. Recently, it was revealed that the pilV gene product is a minor component of the R64 thin pilus (30). The R64 shufflon is believed to determine recipient specificity in liquid matings through the switching of the seven C-terminal segments of the PilV protein (16). Shufflons with three or four invertible segments are present in all IncI1 and IncI2 plasmids (15). Recently, a variant shufflon consisting of a single invertible DNA segment was found in the pathogenicity island of the Salmonella enterica serovar Typhi genome (31).
The rci gene encodes a basic protein belonging to the integrase (Int) family of site-specific recombinases (19). The seven 19-bp repeat sequences separating the four segments are not identical and can be classified into four types (repeat-a, -b, -c, and -d) (Fig. 1C). The in vivo inversion frequencies of three segments (A, B, and C), which were cloned separately, were found to differ greatly (8). Recombination also occurred between inverted synthetic 19-bp repeat sequences that were inserted into pBR322 (8). The inversion frequencies of DNA segments flanked by various repeat sequences indicate that four types of repeat sequences determine the inversion frequency. The Rci protein is a recombinase that functions solely to catalyze DNA inversion between any two inverted 19-bp repeat sequences of the shufflon, and it cannot promote recombination between the direct 19-bp repeat sequences.
In this study, Rci protein was purified from Escherichia coli cells overexpressing the rci gene. Purified Rci protein promoted the in vitro inversion of a DNA segment between the inverted 19-bp repeat sequences. This reaction was enhanced by the addition of host protein HU. Specific binding of the Rci protein to DNA fragments carrying the 19-bp repeat sequences was observed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli JM83 Δ(lac-proAB) rpsL thi ara φ80dlacZΔM15 (29) was used. E. coli BL21(DE3) carrying the T7 RNA polymerase gene under the control of the lac promoter (26) was used to overproduce the Rci protein. pUC19 (29), pUC118 (29), pACYC177 (4), and pET11a (26) were used as vectors for cloning and expression.
Media.
Luria-Bertani (LB) and M9 glucose media were prepared as previously described (24). Solid media contained 1.5% agar. Antibiotics were added to liquid or solid media when necessary at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol (CM), 50 μg/ml; and kanamycin (KM), 50 μg/ml.
Construction of plasmids.
Recombinant DNA techniques were carried out as described previously (24). The rci gene was inserted into a T7 expression vector, pET11a (26). At first, an NdeI site was generated at the ATG initiation codon of the rci gene of pKK024 (19) by site-directed mutagenesis to generate pKK024N. After conversion of the PstI site to a BamHI site in pKK024N, the 1.4-kb NdeI-PstI fragment of pKK024N containing the rci gene was inserted into the NdeI-BamHI sites of pET11a to construct pETrci (Fig. 1A).
A tester plasmid for assay of the Rci activity was constructed as follows. A 606-bp HaeIII fragment of pKK010-85 (17) carrying segment A of the R64 shufflon was inserted into the ScaI site of pACYC177 to generate pKK038 (Fig. 1A). A 775-bp HincII fragment of pCM1 carrying a promoterless CM resistance (Cmr) gene (5) was inserted into the EcoRV site of pKK038 in the orientation opposite to that of the bla promoter to generate pKK039 (Fig. 1B).
A 2.2-kb BstBI-SphI fragment of pKK010-85 (17) carrying the entire R64 shufflon was inserted into the AccI-SphI sites of pUC19 to construct pKK019-19 (Fig. 1A).
The oligonucleotides corresponding to both strands of repeat-a, -b, -c, and -d sequences were annealed and inserted into the KpnI and PstI sites of pUC118 to generate pAG001, pAG002, pAG003, and pAG004, respectively. pAG005, carrying a deletion between the SmaI and HincII sites of pUC118, was also constructed.
Overproduction and purification of Rci protein.
E. coli BL21(DE3) cells harboring pETrci were grown in M9 glucose medium containing 5 μg of thiamine/ml, 0.1% Casamino Acid, and 100 μg of ampicillin/ml at 37°C. When the A600 of the culture reached approximately 0.4, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.4 mM (final concentration). After 2 h, the cells were collected by centrifugation, washed with 50 mM Tris-HCl (pH 7.4), and resuspended in 80 ml of ice-cold buffer Y (50 mM Tris-HCl [pH 7.4], 0.8 M KCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, and 10% glycerol) supplemented with 1 mg of lysozyme/ml. After 30 min at 0°C, the suspension was frozen at −80°C. The frozen cells were thawed at room temperature and lysed by sonication. The following purification steps were performed at 4°C. The lysate was centrifuged at 100,000 × g for 90 min, and the supernatant was loaded onto a 25-ml phosphocellulose P11 (Whatman) column equilibrated with buffer Y. The column was eluted with 50 ml of buffer Y with a linear gradient (from 0.8 to 1.8 M) of KCl. Every fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The Rci-containing fractions were concentrated by ammonium sulfate precipitation. The pellet was dissolved in 2 ml of buffer Y and loaded onto a Sephacryl S-200HR (Pharmacia LKB) column (1.6 by 60 cm) equilibrated with buffer G (50 mM Tris-HCl [pH 7.4], 0.5 M KCl, and 1 mM EDTA). The Rci-containing fractions were concentrated, dialyzed against buffer Y containing 50% glycerol, and stored at −80°C.
The N-terminal amino acid sequence of the purified Rci protein was determined by Edman degradation in a model 477A/120A protein sequencer (Applied Biosystems).
HU protein and anti-HUα antiserum.
HU was partially purified from E. coli JM83 cells as described previously (12). Purified HUα and anti-HUα antiserum were kindly provided by Sumiko Inouye.
In vitro recombination assay.
Recombination assays were carried out at 37°C in 20 μl of reaction mixture containing 30 mM Tris-HCl (pH 7.6), 15 mM NaCl, 80 mM KCl, 4 mM spermidine, 0.7 mM EDTA, 7% glycerol, 4.4 nM supercoiled pKK039 DNA, and 68 nM Rci protein. The reactions were terminated by the addition of 2 μl of 2% SDS, and then the product DNA was collected by ethanol precipitation. To estimate inversion frequencies, the fraction of rearranged molecules was determined by two methods. First, the product DNA was digested with EcoRI and HindIII and subjected to electrophoresis on a 0.7% agarose gel. DNA fragments in the gel were stained with ethidium bromide and visualized under UV light. The gel image was scanned and then analyzed with NIH Image (ver. 1.58) software. The inversion frequency was determined as the ratio of the density of the new band to the sum of those of the new and original bands. Second, the product DNA was introduced into E. coli JM83. The cells were grown for 18 h on LB plates containing both CM and KM or only KM. The ratio of the number of CM- and KM-resistant (Cmr Kmr) colonies to that of KM-resistant (Kmr) colonies was determined.
Rci-binding assay.
Purified Rci protein and DNA fragments (0.1 pmol) were mixed in a buffer solution (20 μl) containing 50 mM Tris-HCl (pH 8.0), 0.2 M KCl, 5 mM spermidine, 0.3 mg of bovine serum albumin, and 10% glycerol. After incubation at 37°C for 10 min, the reaction mixtures were immediately loaded onto a 4% polyacrylamide gel in TBE buffer (90 mM Tris-borate [pH 8.0] and 2.5 mM EDTA) and electrophoresed at 4°C. Following electrophoresis, DNA fragments in the gel were stained with ethidium bromide and visualized under UV light.
RESULTS
Overproduction of the Rci protein.
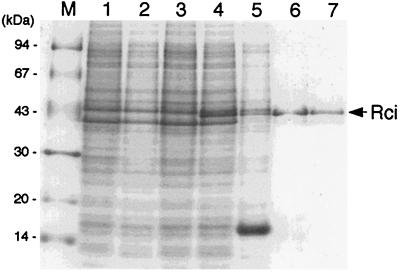
To overproduce the Rci protein, the rci coding sequence was inserted into an expression vector, pET11a, generating pETrci. Then pETrci was introduced into E. coli BL21(DE3) cells carrying the T7 RNA polymerase gene under the control of the lac promoter. Following IPTG induction, the overproduction of a 44-kDa protein was observed (Fig. 2, lane 4) which was not produced in the absence of induction (lane 3) or in cells harboring the control plasmid pET11a (lanes 1 and 2). The apparent molecular mass of the overproduced protein is consistent with the value of 44.3 kDa calculated from the predicted amino acid sequence (19).
FIG. 2.
Expression by the T7 RNA polymerase system and purification of the Rci protein. E. coli BL21(DE3) cells harboring pET11a (lanes 1 and 2) or pETrci (lanes 3 and 4) were grown in M9 medium and treated with 0.4 mM IPTG (lanes 2 and 4) or untreated (lanes 1 and 3). The proteins were analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue. Lane M, marker proteins; lanes 1 to 4, total cell protein; lane 5, soluble proteins of IPTG-induced cells harboring pETrci; lane 6, peak fraction of phosphocellulose column chromatography; lane 7, peak fraction of gel filtration chromatography. The numbers on the left indicate the molecular masses of marker proteins. The location of the Rci protein is indicated.
Purification of the Rci protein.
Rci protein was purified from the IPTG-induced BL21(DE3) cells harboring pETrci. Approximately 60% of the overproduced Rci protein was recovered in the soluble fraction. The soluble proteins extracted from the induced cells (Fig. 2, lane 5) were loaded onto a phosphocellulose column and eluted with a linear gradient of KCl. Rci protein was eluted between 1.2 and 1.4 M KCl (lane 6). The fractions containing the Rci protein were then loaded onto a gel filtration column. The peak fractions (lane 7) were pooled and used as the purified Rci protein. SDS-PAGE followed by silver staining revealed that the purity of the Rci protein was over 95% (data not shown). Approximately 2 mg of purified Rci protein was obtained from a 2-liter culture. The Rci protein retained its activity for at least several months when stored at −80°C.
The molecular mass of native Rci protein was analyzed by gel filtration on Sephacryl S-200HR in G buffer. Rci protein was eluted as a sharp peak at a position corresponding to that of ovalbumin (44 kDa), suggesting that Rci protein is a monomer in G buffer.
The N-terminal amino acid sequence of the purified Rci protein was determined to be NH2-Pro-Ser-Pro-Arg-Ile-Arg-Lys-Met-Ser. This is identical to the sequence predicted from the nucleotide sequence of the rci gene (19), with the exception of an initial methionine residue.
The Rci protein mediates recombination in vitro.
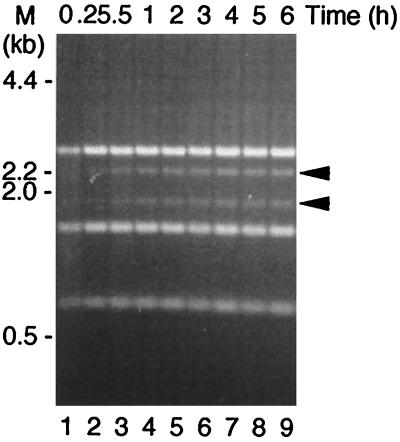
To demonstrate Rci-mediated in vitro DNA recombination, plasmid pKK039 (Fig. 1B) was constructed as a substrate. After incubation of supercoiled pKK039 DNA with the Rci protein, the reaction was stopped by the addition of SDS. The DNA products were digested with EcoRI and HindIII and subjected to agarose gel electrophoresis (Fig. 3). During the time course of the recombination reaction, two new DNA bands were produced, indicating that segment A containing the chloramphenicol transacetylase (CAT) gene in pKK039 was inverted in vitro by the activity of the Rci protein. To roughly estimate the DNA inversions, the gel image shown in Fig. 3 was scanned, and the inversion frequency was determined (Fig. 4A). The fraction of recombined molecules increased rapidly without any time lag. The addition of HU to the reaction mixture resulted in a 30 to 50% increase of the inversion rate (Fig. 4A). These results were consistent with the in vivo observation that the inversion frequency of the shufflon was low in a hupA hupB double mutant (28).
FIG. 3.
Restriction analysis of the products of in vitro recombination by Rci protein. Reactions were carried out with 68 nM Rci protein using 4.4 nM supercoiled pKK039 DNA as a substrate. The reaction mixtures were incubated at 37°C for the indicated times. The product DNAs were digested with EcoRI and HindIII and subjected to electrophoresis on a 0.7% agarose gel. The locations of molecular length markers are indicated on the left. DNA bands from the recombination product are indicated by arrowheads on the right.
FIG. 4.
Time course of Rci-mediated recombination in vitro. Reactions were carried out without (open circles) or with (solid circles) 64 nM HU protein. The inversion frequency (percentage) was determined from the gel image shown in Fig. 3 (A) or by transforming E. coli JM83 cells with reaction products (B).
The DNA products were also used to transform E. coli JM83 cells to determine the inversion frequency. The inversion frequency estimated by the ratio of Cmr Kmr colonies to Kmr colonies was similar to that estimated by gel analysis (Fig. 4B). These results suggest that a major portion of the recombined molecules was biologically active.
When pKK039R, in which segment A including a CAT gene was inverted (Fig. 1B), was used as the substrate, the in vitro inversions were also observed (data not shown). In both pKK039 and pKK039R, however, the in vitro reaction reached a plateau at about 20% inversion product, although DNA inversion of segment A easily reached 50%-50% equilibrium in vivo (8). It is unlikely that the saturation was a result of relaxation of the DNA substrate, since most of the DNA substrate remained in supercoiled form even after prolonged incubation with the Rci protein. The reason for the inefficiency of the in vitro system is unknown. Up to 40% inversion product was formed in the presence of 34% glycerol.
The Int family of site-specific recombinases shares four invariant residues (22) that are equivalent to Arg154, His234, Arg237, and Tyr270 of the Rci protein. A mutant Rci protein in which the conserved tyrosine was replaced (RciY270F) was also purified. When the RciY270F protein was used in the in vitro reaction, recombination products were not detected by gel analysis or by the transformation assay (data not shown). These results indicate that tyrosine-270 of the Rci protein plays a key role in the recombination reaction, as was found in the other Int family recombinases.
Effects of different reaction conditions.
The effects of different reaction conditions on Rci-mediated in vitro recombination were examined and are summarized in Table 1. We observed 30% stimulation of the inversion by the addition of 64 nM HU protein to the standard reaction mixture. Since higher concentrations of HU protein did not increase the inversion, 15 HU dimers per DNA molecule were demonstrated to be sufficient to promote inversion (data not shown). Changes in various parameters of the reaction conditions had similar effects on inversion frequencies with or without HU protein.
TABLE 1.
Requirements for in vitro recombination
| Parameter | Value | Relative activity (%)a
|
|
|---|---|---|---|
| −HU | +HU | ||
| Substrates | |||
| Supercoiled DNAb | 100 | 100 | |
| Relaxed DNA | 5 | 1 | |
| Linear DNA | <0.9 | <0.2 | |
| Components | |||
| Standardb | 100 | 100 | |
| +MgCl2 | 5 mM | 93 | 102 |
| +EDTA | 5 mM | 79 | 76 |
| +ATP | 2 mM | 79 | 85 |
| −Spermidine | 25 | 54 | |
| KCl concn | 40 mM | 83 | 58 |
| 200 mM | 6 | 1 | |
| Other | |||
| Standardb | 100 | 100 | |
| pH | 6.8 | 73 | 83 |
| 8.0 | 97 | 115 | |
| 8.5 | 99 | 94 | |
| Temp | 25°C | 66 | 62 |
| 30°C | 82 | 109 | |
| 42°C | 131 | 147 | |
| DNA concn | 2.2 nM | 187 | 152 |
| 8.8 nM | 28 | 38 | |
| Rci concn | 34 nM | 60 | 30 |
| 136 nM | 228 | 218 | |
| 272 nM | 38 | NDc | |
Recombination activity was determined as the yield of recombination product, the ratio of the number of Cmr Kmr colonies to that of Kmr colonies. Relative activity is expressed as the percentage of the recombination activity relative to the activity under the standard conditions (100% relative activity).
The standard reaction was carried out at 37°C for 2 h with 68 nM Rci protein (−HU) or with 68 nM Rci protein and 64 nM HU dimer (+HU) in 20 μl of 30 mM Tris-HCl (pH 7.6), 15 mM NaCl, 80 mM KCl, 4 mM spermidine, 0.7 mM EDTA, 22% glycerol, and 4.4 nM supercoiled pKK039 DNA. Under the standard conditions, the recombination frequencies were 17 and 22% without and with HU dimers, respectively.
ND, not determined.
Supercoiled DNA substrate was absolutely required for the inversion reaction. No recombination product was detected using linear DNA (which had been digested once with HindIII) as a substrate. When the substrate DNA had been relaxed by topoisomerase I, the inversion rate was severely decreased. Additions of magnesium ion, EDTA, or ATP had little effect on recombination, suggesting that neither an energy source nor divalent cations are necessary. Removal of spermidine from the reaction mixture decreased the reaction rate. In the absence of spermidine, HU exhibited 180% stimulation on recombination. The inversion reaction showed a broad pH optimum between 7.6 and 8.5. The inversion was sensitive to KCl concentration, with a maximal inversion activity at a KCl concentration of 80 mM. Standard reaction mixtures consisted of 68 nM Rci protein and 4.4 nM substrate DNA at the approximate molar ratio of 15:1. Reaction rates were nearly proportional to the Rci/DNA ratios when the ratios were smaller than 31:1. However, a higher ratio of Rci protein to DNA (62:1) reduced the rate of the inversion reaction.
Rci protein binds to various shufflon segments.
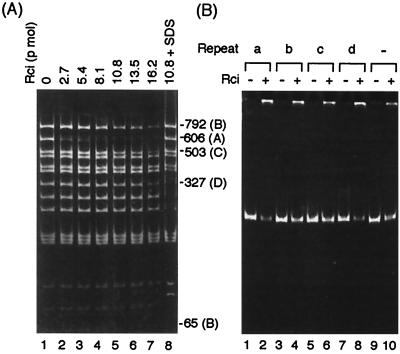
DNA of plasmid pKK019-19 carrying the entire R64 shufflon was digested with HaeIII and SspI to separate the segments A, B, C, and D (Fig. 1A). The DNA fragments were used in the binding assay (Fig. 5A). As the amount of Rci protein added to the reaction mixture was increased, the DNA fragments derived from segments A, B, C, and D disappeared from the gel and large complexes which could not enter the gel matrix were seen in the well. Segment A carrying two repeat-a sequences displayed the highest affinity for the Rci protein among the four segments. Higher concentrations of Rci protein were required for binding to DNA segments B, C, and D. The DNA molecules in these complexes were not covalently linked to the Rci protein, because the large complexes were replaced by bands that migrated at the position of free DNA after the addition of SDS to the reaction mixture before electrophoresis (Fig. 5A, lane 8). Regeneration by SDS of three fragments containing segment D was insufficient for an unknown reason.
FIG. 5.
(A) Binding of Rci protein to the four shufflon segments. DNA fragments of pKK019-19 (0.1 pmol) digested with HaeIII and SspI (see Fig. 1A) were incubated with the indicated amounts of Rci protein and electrophoresed on a polyacrylamide gel as described in Materials and Methods. (B) Binding of Rci protein to four 19-bp sequences. The names of the 19-bp sequences are indicated at the top. −, control without a 19-bp sequence. PCR-generated fragments carrying the 19-bp sequences (0.1 pmol) were incubated without (−) or with (+) 6.8 pmol of Rci protein. A binding assay was performed as described in Materials and Methods except that bovine serum albumin was omitted from the reaction mixtures.
In this binding assay, the A and C segments carried two repeat sequences while the others had only one repeat sequence. For direct comparison of the affinity of Rci for the four repeat sequences, four plasmids, pAG001 through pAG004, carrying synthetic repeat-a, -b, -c, and -d sequences in pUC118, were constructed. The 314-bp DNA fragments containing each of the four 19-bp repeat sequences were produced by PCR amplification of these plasmids and used for the Rci-binding assay. With the addition of Rci protein, the number of free DNA fragments in the gel decreased and a large complex was formed that remained in the well of the gel (Fig. 5B). The DNA fragments carrying repeat-a and -d sequences displayed higher affinity for the Rci protein (Fig. 5B, lanes 2 and 8), while the DNA fragments carrying repeat-b and -c sequences displayed lower affinity (Fig. 5B, lanes 4 and 6).
Four Rci proteins with mutations at the invariant residues (R154Q, H234L, R237Q, and Y270F) were partially purified. These mutant proteins retained the ability to bind to the shufflon segments, whereas they lacked in vivo recombination activity (data not shown).
DISCUSSION
In vitro DNA recombination systems using the integrase family recombinases, including λ and HP1 integrases, P1 Cre protein, 2μm plasmid Flp protein, and E. coli XerCD proteins, have been constructed and characterized (3, 9, 11, 20, 23, 27). In this paper, the Rci protein of plasmid R64 was purified and the Rci-mediated recombination system was reconstituted in vitro. The Rci-mediated recombination reaction required neither an energy source nor divalent cations, as was found for the other Int family recombination systems. Negatively supercoiled DNA substrate was required for Rci protein, as is the case for the λ and HP1 integrases, which require a supercoiled attP substrate in attP × attB reactions. In contrast, Cre and Flp proteins are able to recombine supercoiled, linear, or relaxed DNA substrate (1, 6). In vitro recombination was observed only between inverted repeat-a sequences but not between direct repeat-a sequences (data not shown), as was observed in the in vivo system (8).
The HU protein had a stimulatory effect on Rci-mediated recombination in vitro, consistent with in vivo observations using a hupA hupB strain (28). However, the requirement for HU in the in vitro system is not as marked as that in the in vivo system. Addition of HU protein to the in vitro system increased the recombination frequency by only 30 to 50%. Possible contamination of the Rci preparation by HU protein was ruled out by Western blot analysis using anti-HU antiserum (data not shown). Spermidine may replace the function of HU in the in vitro recombination reaction, since HU displayed a marked stimulatory effect in the absence of spermidine (Table 1).
The bacterial histonelike protein HU is an abundant nonspecific DNA binding protein (7). HU protein induces DNA bending upon binding to DNA. HU is known to be required for various DNA transaction systems, including the stimulation of replication initiation at oriC, Mu DNA transposition, Hin inversion, and repression by GalR of the gal promoters (2, 10, 21, 25). In these systems, only a small number of HU dimers are required for stimulation. Similarly, in the Rci-mediated in vitro recombination reaction, a small number of HU dimers (15 HU dimers per DNA molecule or less) was sufficient for inversion stimulation. The binding of HU protein to DNA might facilitate assembly and/or stabilization of the Rci-DNA complex at the recombination sites.
Rci protein specifically bound to the shufflon segments carrying the 19-bp repeat sequences and the DNA fragments carrying the synthetic 19-bp repeat sequences. However, a stoichiometric Rci-DNA complex which would display slower migration during PAGE was not formed. The Rci-DNA complex was too large to enter the gel. A large complex was also formed under high-ionic-strength conditions (900 mM KCl [data not shown]). These results suggest that the Rci protein aggregates into a large complex with the shufflon segment. The Rci protein displayed higher affinity for repeat-a and -d sequences than for repeat-b and -c sequences. Shufflon segment A, which contains two repeat-a sequences, displayed higher affinity than segment D, which contains only one repeat-d sequence, suggesting that the Rci protein may bind cooperatively to the two repeat-a sequences in segment A. In the previous study, segment A displayed the highest in vivo inversion frequency among the four segments (8). The differences in the affinities of Rci protein for the four segments may explain their respective in vivo inversion frequencies. It is most likely that differences in the 14th, 15th, and 16th nucleotides of the four repeat sequences (Fig. 1C) affect the affinity of Rci protein for the four segments, and consequently their recombination abilities.
Binding of Rci protein to supercoiled substrate DNA was also confirmed. When the in vitro reaction mixture was analyzed by agarose gel electrophoresis followed by Western blotting, Rci protein was detected at the positions corresponding to those of supercoiled and open circular pKK039 DNAs (data not shown). The Rci protein was also shown to bind nonspecifically to supercoiled pACYC177 DNA under similar conditions. Binding of high concentrations of Rci protein to nonshufflon DNA fragments was also observed (Fig. 5A, lane 7). Nonspecific DNA binding of Rci and formation of the large Rci-DNA complex might explain the inhibition of in vitro recombination reactions by high Rci concentrations. Excess Rci protein may increase nonspecific binding, which could interfere with faithful recombination between the recombination sites. It is possible that addition of HU or spermidine to the in vitro system may increase production of faithful recombination complexes. Inhibition by high concentrations of integrase protein was also observed in the HP1 system (9).
Further elucidation of the shufflon recombination system would require more detailed investigations, including footprinting analyses of the Rci-DNA complex. Such work is currently in progress in our laboratory.
ACKNOWLEDGMENTS
We are grateful to S. Inouye for HU protein and anti-HU antiserum. We thank K. Takayama for critical reading of the manuscript.
This work was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Abremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 2.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arciszewska L, Grainge I, Sherratt D. Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J. 1995;14:2651–2660. doi: 10.1002/j.1460-2075.1995.tb07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Close T J, Rodriguez R L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982;20:305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- 6.Cox M M. DNA inversion in the 2μm plasmid of Saccharomyces cerevisiae. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 661–670. [Google Scholar]
- 7.Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyohda A, Funayama N, Komano T. Analysis of DNA inversions in the shufflon of plasmid R64. J Bacteriol. 1997;179:1867–1871. doi: 10.1128/jb.179.6.1867-1871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakimi J M, Scocca J J. Purification and characterization of the integrase from the Haemophilus influenzae bacteriophage HP1; identification of a four-stranded intermediate and the order of strand exchange. Mol Microbiol. 1996;21:147–158. doi: 10.1046/j.1365-2958.1996.6311351.x. [DOI] [PubMed] [Google Scholar]
- 10.Haykinson M J, Johnson R C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoess R H, Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci USA. 1984;81:1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R C, Simon M I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim S-R, Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komano T. Shufflons: multiple inversion systems and integrons. Annu Rev Genet. 1999;33:171–191. doi: 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- 15.Komano T, Kim S-R, Nisioka T. Distribution of shufflon among IncI plasmids. J Bacteriol. 1987;169:5317–5319. doi: 10.1128/jb.169.11.5317-5319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komano T, Kim S-R, Yoshida T, Nisioka T. DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J Mol Biol. 1994;243:6–9. doi: 10.1006/jmbi.1994.1625. [DOI] [PubMed] [Google Scholar]
- 17.Komano T, Kubo A, Kayanuma T, Furuichi T, Nisioka T. Highly mobile DNA segment of IncIα plasmid R64: a clustered inversion region. J Bacteriol. 1986;165:94–100. doi: 10.1128/jb.165.1.94-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komano T, Kubo A, Nisioka T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 1987;15:1165–1172. doi: 10.1093/nar/15.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubo A, Kusukawa A, Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988;213:30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- 20.Landy A. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 21.Lavoie B D, Shaw G S, Millner A, Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 22.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad P V, Horensky D, Young L J, Jayaram M. Substrate recognition by the 2 micron circle site-specific recombinase: effect of mutations within the symmetry elements of the minimal substrate. Mol Cell Biol. 1986;6:4329–4334. doi: 10.1128/mcb.6.12.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Skarstad K, Baker T A, Kornberg A. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 27.Vetter D, Andrews B J, Roberts-Beatty L, Sadowski P D. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci USA. 1983;80:7284–7288. doi: 10.1073/pnas.80.23.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada M, Kutsukake K, Komano T, Imamoto F, Kano Y. Participation of the hup gene product in site-specific DNA inversion in Escherichia coli. Gene. 1989;76:345–352. doi: 10.1016/0378-1119(89)90174-1. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X L, Morris C, Hackett J. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major ‘pathogenicity island’ of Salmonella typhi. Gene. 1997;202:139–146. doi: 10.1016/s0378-1119(97)00466-6. [DOI] [PubMed] [Google Scholar]