Abstract
Platelets and their parent cell, the megakaryocyte (MK), are increasingly recognized for their roles during infection and inflammation. The MK residing in the bone marrow or arising from precursors trafficked to other organs for development go on to form platelets through thrombopoiesis. Infection, by direct and indirect mechanisms, can alter the transcriptional profile of MKs. The altered environment, whether mediated by inflammatory cytokines or other signaling mechanisms results in an altered platelet transcriptome. Platelets released into the circulation, in turn, interact with each other, circulating leukocytes and endothelial cells and contribute to the clearance of pathogens or the potentiation of pathophysiology through such mechanisms as immunothrombosis.
In this article we hope to identify key contributions that explore the impact of an altered transcriptomic landscape during severe, systemic response to infection broadly defined as sepsis, and viral infections, including SARS-CoV2. We include current publications that outline the role of MKs from bone-marrow and extra-medullary sites as well as the circulating platelet. The underlying diseases result in thrombotic complications that exacerbate organ dysfunction and mortality. Understanding the impact of platelets on the pathophysiology of disease may drive therapeutic advances to improve the morbidity and mortality of these deadly afflictions.
Keywords: Transcriptome, Sepsis, Virus, COVID-19, Megakaryocyte, Platelet
1. Introduction
Platelets are abundant, anucleate cells that are released from the large, multi-nucleated parent megakaryocyte (MK). This phenomenon was described by Wright as occurring in the bone marrow however more recent reports have shown this can occur at extra-medullary sites including the lung [[1], [2], [3]]. Platelets were long considered cell fragments due to their lack of nucleus and genomic DNA. As such, it was believed these cells were only capable performing functions related to an inscribed and fixed proteome. Importantly, platelets contain MK-derived mRNA and the translational machinery to perform activation-dependent protein synthesis [4]. Therefore, platelets are transcriptionally though not translationally inert.
Because of the anucleate nature of platelets, the translational machinery and mRNA transcripts must be invested from the parent MK or taken up by circulating platelets as occurs with “tumor educated platelets” whereby platelets take up spliced RNA via microvescicles [5,6].
The initial efforts to interrogate MK-derived transcripts in platelets were made using cDNA libraries that led to the identification of heritable, monogenic platelet disorders. The first description of the platelet transcriptome was performed by Gnatenko et al. in 2003 using microarray techniques and identified approximately 2000 genes. Interestingly, these initial studies found >30 % of the identified transcriptome had unassigned functions. Additional investigations utilizing serial gene analysis of gene expression (SAGE) then sequenced 25,000 tags and found greater than half (51 %) of them were non-mitochondrial tags and corresponded to 2300 transcripts. Thus, had similar complexity to nucleated cells though contained unique enrichment of transcripts specific to platelets as compared to other tissues [7]. The most striking observation was that there was strong correlation between transcript abundance and protein expression [8]. Additional studies have since found the correlations between platelet transcriptome and proteome to be more controversial [9]. The application of RNA-sequencing platforms vastly increased the ability to detect platelet transcripts with an estimated ∼9500 protein coding mRNAs identified and allows the detection of low abundance mRNAs and novel transcripts. Furthermore, miRNAs, lncRNAs and other non-coding RNAs have been identified, thus shedding light on the complexity of the transcriptional landscape of platelets [10]. More recent work has utilized single cell RNA sequencing to identify and sequence MK precursors and MKs from non-bone marrow tissues such as the lung and spleen [2,3,11].
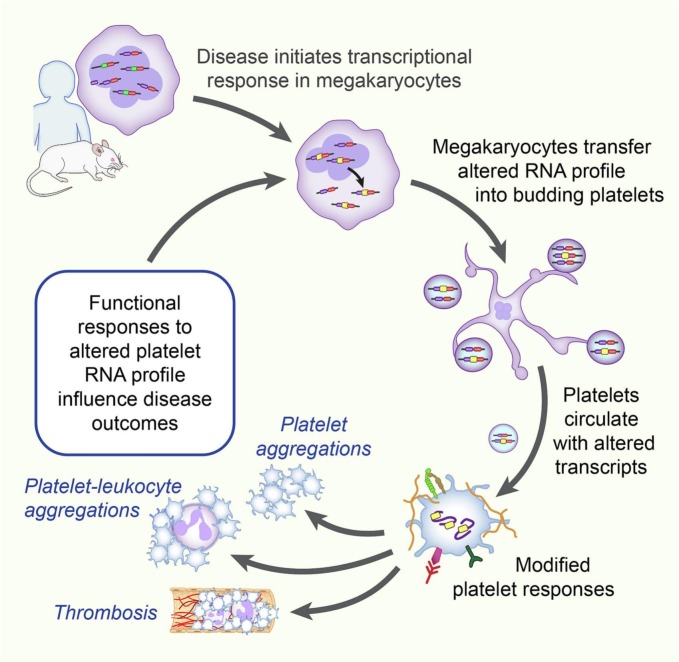
Megakaryocytes invest RNAs into platelets prior to platelets entering circulation (Fig. 1 ).
Fig. 1.
Inflammation and infection induces alterations at the transcriptome level in megakaryocytes. Bone marrow resident or other tissue resident MK invest an altered transcript profile into platelets during thrombopoiesis. Platelets are released into the circulation with an altered RNA profile and may undergo activation-dependent translation. Platelets have altered function in the setting of infection and inflammation that influences disease outcomes through such mechanisms as macro-, microthrombosis or formation of heterotypic aggregates.
The importance of the platelet transcriptome can be attributed, in part, to RNA content in platelets as well as the total blood mass of platelets. Platelets contain a total of 2.20 femtograms of RNA per platelet, 1000 times less than circulating leukocytes. However, platelets are the second most abundant blood cell in circulation. Therefore, the RNA contribution from platelets is orders of magnitude greater than leukocytes [12]. Investigation of the platelet transcriptome in disease has been a natural progression in understating the contribution of these cells to hematologic maladies, cardiovascular disease, and more recently inflammation and infectious diseases [13].
Platelets express surface receptors involved in immune responses such as Toll-like receptors (TLRs), CLEC2, GPVI, FcγRIIA that aid in recognition of circulating pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP), cell-cell interactions, and binding of complement and circulating antibodies, respectively [14,15]. Additionally, platelets secrete proteins that mediate inflammation and interact with leukocytes prompting transcriptional changes in other cells [[16], [17], [18]]. This review will focus on the alterations in the MK and platelet transcriptome in viral infections and infectious syndromes.
2. Transcriptional changes in infection
Systemic inflammation and infection by specific pathogens (bacterial, viral, and parasitic) have been reported to impact the transcriptomic profile of MKs and platelets resulting in alterations in canonical platelet function and the phenotype of circulating platelets [[19], [20], [21], [22]]. We will discuss the global transcriptional alterations and platelet functional responses in sepsis, influenza, dengue virus and SARS-CoV2 in the following sections.
3. Sepsis
Sepsis and septicemia, colloquially known as “blood poisoning”, is the clinical syndrome identified as a systemic inflammatory response to infection. The clinical presentation of sepsis is heterogenous and is caused by many pathogens [23]. Individuals may present with severe physiologic derangements such as hyper- or hypothermia, tachycardia and hypotension with minimal prodrome while others may present with smoldering, protracted symptoms and develop end-organ dysfunction over days rather than hours [23,24]. The variation in presentation is multi-factorial and due to underlying co-morbidities, pathogen burden, environmental influences and variations in gene expression [24,25]. The need for clear diagnostic criteria must be used to study well-defined populations for mechanistic and therapeutic research.
The link between sepsis and thrombocytopenia has been well-recognized [26]. An abrupt decline in platelet count can occur in the absence of bacteremia suggesting both a direct and indirect impact on platelets. Disseminated intravascular coagulopathy (DIC) and more recently sepsis-induced coagulopathy (SIC) is the commonly identified cause of severe thrombocytopenia that occurs during sepsis [27]. However, platelets are consumed through additional processes including thrombin-induced apoptosis, autophagy, splenic sequestration, losses from vascular leak and others [[28], [29], [30]].
4. Megakaryocytes and sepsis
Cytokines induced during inflammation and direct infection may directly impact megakaryopoiesis, and in-turn, platelet production [31,32]. Thrombocytopenia that occurs during sepsis may result in increased megakaryopoiesis. Interestingly, elevated levels of thrombopoietin (TPO) are seen during sepsis and promote megakaryopoiesis [3,30]. Additionally, other pro-inflammatory cytokines released in sepsis, including TNF-α, IL-1β, IL-6, also may contribute to increased MK development [[33], [34], [35]]. However some cytokines such as, type 1 interferons, which are induced in viral and bacterial infection, and have direct anti-viral responses, may have suppressive effects on hematopoetic stem cells, including megakaryocytes and MK precursors. Specifically, Wang et al., demonstrated that IFN-A directly suppressed TPO-induced megakaryopoiesis by suppressing phosphorylation of c-Mpl and STAT 5 [36].
Recent studies highlight how sepsis alters the MK transcriptome and dynamically transfer these altered transcripts to platelets [37]. Increased platelet activation and functional responses marked by increases in surface expression of P-selectin, increased circulating platelet-leukocyte aggregates and integrin activation are commonly observed in septic patients [19,37,38]. Platelets isolated from septic patients and mice subjected to cecal ligation and puncture (CLP) model of sepsis express >500 differentially expressed transcripts compared to platelets from healthy donors and untreated mice. Of these, ITGA2B, which codes for the platelet-specific αIIb integrin, was one of the most upregulated genes [19]. Interestingly, ITGA2B expression was upregulated in autologous, bone marrow (BM) MKs from mice 24-48 h after CLP whereas platelets had increased ITGA2B expression 72 h following CLP. This suggests the increase in platelet ITGA2B was invested from the parent MK and was caused by the systemic response to infection. In a similar CLP model, increases in granzyme B, a serine protease involved in apoptosis, was observed in BM MKs and closely following by increased platelet mRNA and protein expression. Similar increases in platelet granzyme B were observed in a cohort of pediatric patients with sepsis [39].
Regulation of transcription at the level of the MK during sepsis is complex. Investigations have begun to focus on micro-RNAs (miRNA) and their role in regulation of gene expression. Recent work implicated a down-regulation in Dicer, one of the key miRNA processing enzymes, and mir-26b resulting in increased P-selectin expression in MEG-01s, an immortalized megakaryoblastic cell line. These cells were treated with LPS and resulted in significant gene alterations involved in inflammatory pathways, lipid homeostasis, responses to calcium, and angiogenesis [38]. Alterations in platelet miRNA from septic individuals in a similar pattern were observed. It has commonly been viewed that MKs reside in the bone marrow where they are the source of platelets released into the circulation. Provoking studies performed by several investigators have shown that MKs found in extra-medullary sites are responsible not only for a significant degree of thrombopoiesis but also express genes that direct the extra-medullary MKs to immune function [3,11]. Lung-derived MKs are enriched in pathways involved in innate immunity. Recently, Valet et al. reported that the acute inflammation occurring in sepsis causes megakaryocyte-erythrocyte progenitors (MEPs) to migrate to the spleen and differentiate into MKs [3]. RNAseq of spleen MKs and BM MKs suggest that spleen MKs have an immune-like transcriptional signature with higher expression of CD40L compared to BM MKs. This increase in CD40L expression was also observed in platelets arising from MKs in the spleen. Intriguingly, these differences in transcriptional signature were seen in both septic and homeostatic conditions, which caused researchers to suggest that differences in MK transcriptional signature is dependent on the tissue from where they were derived and not on sepsis itself [3]. Taken together, these observations suggest that sepsis indeed affects the transcript levels of MKs and newly released platelets, either by directly affecting BM MKs, induce the migration of MKs to other organs, or inducing a resident population of precursor cells to develop into MKs. The observations made by Valet et al. may also be attributed to differences in the murine model of sepsis employed and further work is required to dissect and understand the expression profile of MKs and platelets that arise from extramedullary sites.
Similar to findings in bacterial sepsis MK and platelet transcriptomic variations are observed during viral infections. Key anti-viral genes such as interferon-induced transmembrane protein 3 (IFITM3), are upregulated in CD34+-derived MKs co-cultured with dengue virus. Moreover platelets from patients infected with the influenza or dengue virus also express increased IFITM3 mRNA [20]. IFITM-3 aids in the restriction of viral infections by regulating the fusion of viruses to endocytic vessels and directing them to lysosomes [40,41]. Campbell et al. utilized a lentivirus overexpression model in the MEG-01 cell line to restrict dengue virus infection. Furthermore, mutations in IFITM3 (rs12252-C) led to human MKs being more susceptible to the virus [20]. Thus, the upregulation of IFTIM3 in MKs with exposure to dengue virus suggests a role in immune function. The role of IFITM3 in platelets requires further elucidation.
4.1. Alterations in platelet function during sepsis
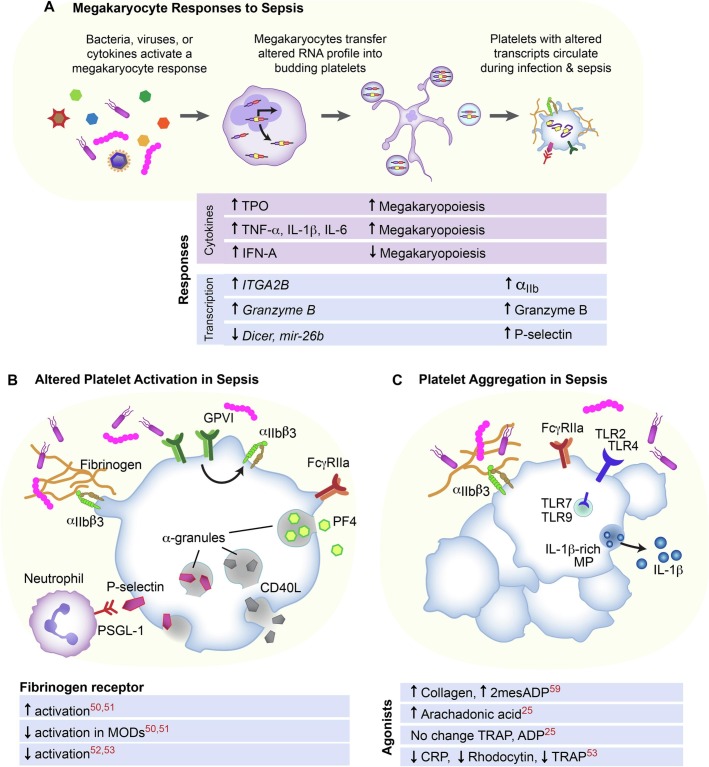
A platelet's most recognized role is a mediator of thrombosis, however platelets also play an important role as immune effectors providing protective immune functions or accelerating inflammatory outcomes [42,43]. Sepsis can impact both canonical platelet functions and immune functions of platelets. These alterations are driven by a number of factors including splicing precursor RNA and containing an altered transcriptome invested into budding platelets released into circulation [19,44]. Additionally, sepsis can drive direct platelet-pathogen interactions and the impact of platelets on other inflammatory cells (e.g. neutrophils, monocytes, or dendritic cells) [45] (Fig. 2).
Fig. 2.
Sepsis is a heterogenous clinical syndrome caused by multiple pathogens including bacteria and viruses that induce a host response. (A) This host response causes alterations in the MK and platelet transcriptome. The platelet transcriptome results in altered platelet activation, including alterations in sub-populations of septic patients (B). Platelet aggregation in response to agonists (C) in sepsis suggests altered function of specific receptors in sepsis.
4.2. The controversial role of platelet activation in sepsis
Platelets contain a number of surface receptors capable of binding to circulating proteins or other cells. Platelet activation occurs as the initial step in coagulation that allows amplification of internal signals resulting in increased cell-cell interaction and promotes the formation of a platelet plug. Classic platelet activation is induced by exposed collagen or soluble agonists that bind to G-protein coupled receptors (GPCR) leading to activation of adhesion receptors (αIIbβ3) which then results in ADP-mediated adhesion and aggregation. Investigations exploring the role and downstream signaling have broadly expanded the field to include a number of surface receptors beyond classic GPCRs. For example, immune receptors (FcR and FcγRIIA) contain ITAM regions that allow signal transduction between platelets and leukocytes as well as platelet adhesion [46]. Many of these surface receptors are present under resting conditions on platelets from healthy subjects. Flow cytometry assays designed to detect antibodies (PAC-1 and JonA, human and mouse, respectively) that bind to the activated conformation of the platelet fibrinogen receptor, αIIbβ3, is a well-established assessment of early platelet activation. Additional surface markers include those involved in degranulation such as CD62P (P-selectin, α granules) and CD63 (GP53, dense granules and lysosomes) [47]. Other assays to evaluate platelet activation include exposure to shear stress via GPVI dependent mechanisms [48]. In disease states such as dengue virus and sepsis circulating platelets have increased surface expression of αIIbβ3 and increased PAC-1 binding suggesting these platelets are in circulation with evidence of early activation [19,49]. Early work by Gawaz et al. identified increased fibrinogen receptor activity assessed by flow cytometry in two separate cohorts of septic patients compared to other critically ill patients without sepsis. The presence of multiple organ failure syndrome (MODs) abrogated the effect of increased activation [50,51]. Since that time additional reports have had conflicting findings. Yaguchi et al. found under basal conditions that platelets from septic patients had decreased activation as assessed by PAC-1 and fluorescent fibrinogen binding with no difference following agonist stimulation when compared to platelets from healthy volunteers [52]. More recent studies have extended those findings with no increased in GPIIb-IIIa receptor expression nor basal activation of the fibrinogen receptor in septic patients compared to healthy donors [53]. Differences in pathogen, source of infection, and time of hospitalization to enrollment may account for some of the conflicting reports.
4.3. Platelet aggregation in sepsis
Platelet-platelet binding is termed aggregation and occurs when integrin αIIbβ3 transitions into its active conformation (activation) to become a high affinity receptor for fibrinogen. The subsequent cross-linking of fibrinogen to two integrin receptors results platelet-platelet aggregates and a “platelet clump” begins to form. The process of integrin surface expression, binding fibrinogen, and progression of an αIIbβ3-mediated operation is metabolically demanding and could be impacted by derangements that limit a platelet's metabolic processes [54].
Platelets have a number of surface receptors that have immune functions and can potentiate or result in platelet activation and aggregation. Danger-associated molecular pattern (DAMP) and pathogen-associated molecular pattern (PAMP) molecules are detected by a number of platelet receptors including FcγRIIA, CLEC and Toll-like receptors (TLR). Platelets have been shown to contain protein and mRNA for TLRs 1, 2, 3, 4, 6, 7, 8, and 9 [[55], [56], [57]]. TLR4 and TLR2 are expressed on the platelet surface while TLR9 receptors translocate from T-granules to the plasma membrane upon activation with Type C CpG dinucleotides [58]. They recognize specific molecules from pathogens and have each been shown to potentiate or independently activate platelets [14].
Alterations in platelet aggregation are dependent on select agonists, for example, Nuhrenberg et al. found an increase in aggregation in response to arachidonic acid (AA) in platelets from septic patients requiring vasopressor support compared to patients with stable coronary disease. No difference in aggregation was seen in response to thrombin receptor associated protein 6 (TRAP) nor adenosine diphosphate (ADP) [22]. These findings are attributed to an increase in immature platelet fraction. Transcriptomic analysis of these platelets revealed an upregulation of genes involved in protein synthesis pathways and did not overlap with transcripts found in immature platelet fractions found in healthy donors. Campbell et al. showed increased platelet aggregation to collagen and 2mesADP in platelets from septic patients compared to healthy donors [59]with the hypothesis that increased IFITM3 causes increased fibrinogen endocytosis and platelet hyperreactivity. Conversely, Weiss et al. have shown decreased platelet aggregation in response to CRP-XL, rhodocytin, and TRAP with findings to suggest GPVI dysfunction and increased GPVI shedding [53]. These findings reproduce prior work that has shown decreased platelet aggregation in severe sepsis/septic shock regardless of platelet count to agonists ADP, AA, TRAP, collagen, and thromboxane A2 [52]. The heterogeneity of the septic patients in regard to source of infection and methods of assessing aggregation, e.g. washed platelets vs platelet-rich plasma, may account for some of the differences reported in the literature.
5. COVID-19
The SARS-CoV2 virus that erupted in 2019 swept across the world and resulted in >6 million deaths at the time of this publication. The primary cause of hospitalization and death was due to severe hypoxemic respiratory failure or a complication there-of.
5.1. Role of thromboembolic disease in COVID-19
We quickly learned that patients who suffered from COVID-19 had macrothrombi and microthrombi that were associated with end-organ failure [19,60,61]. Platelets were enriched in these small thrombi as well as neutrophils undergoing neutrophil extracellular trap (NET) formation. Multiple investigators have now shown platelets and leukocytes, micro-immunothrombi, were found in the capillaries and small vessels within multiple tissues on autopsy of patients who were presumed to have died from COVID-19 [62,63]. The role of immunothrombi and the biologic processes that drive immunothrombosis are resultant from a complex interplay of dysregulated coagulation, platelet and neutrophil activation [63].There are conflicting reports on the presence and contribution of pro-coagulant platelets to the thrombotic pathology in COVID-19 [64,65]. While there has been marked variability in detection and reporting of both venous and arterial thromboembolic events in COVID-19 the incidence has ranged from <10 % to 63 % of patients requiring support with extracorporeal membrane oxygenation (ECMO) with the greater incidence occurring in those with more severe disease [66]. Because the frequency of thromboembolic events despite standard prophylactic measures many clinical trials investigated anticoagulation and antiplatelet therapies such as the Accelerating COVID-19 Therapeutic Interventions and Vaccines-4 (ACTIV-4), however many have closed due to limited enrollment or futility at interim analysis (https://www.nih.gov/research-training/medical-research-initiatives/activ/covid-19-therapeutics-prioritized-testing-clinical-trials). At the time of review there have been two large clinical trials testing antiplatelet therapies, RECOVERY and REMAP-CAP. RECOVERY with 7351 patients in the aspirin compared to usual care 7541 patients did not show improved survival at 28 days. However, there was a modest decreased in hospital length of stay and thrombotic events [67]. Similarly, REMAP-CAP compared aspirin vs P2Y12 inhibitor vs usual care did not improve organ support-free days however there was a 5 % reduction in mortality for critically ill patients [68]. Both studies support the clinical relevance of platelet dysfunction in the pathology of COVID-19.
6. Transcriptomic changes in platelets in COVID-19
Similar to viral infections, such as dengue and influenza, the platelet transcriptome is altered during SARSCoV-2 infection [20,69]. RNA-seq performed on platelets isolated from COVID-19 patients and healthy matched donors, showed a clustering of transcriptome-wide gene expression from COVID-19 patients compared to healthy donors by a primary cluster analysis (PCA). This suggests a systemic shift in the transcriptional landscape expressed within platelets [69,70]. Such changes in the transcriptome provide a more complete picture of the drivers that cause platelet hyperactivity which contributes to the thromboembolic phenomenon seen in COVID-19. Though many biological processes at the transcriptome level are disparate in COVID-19 platelets, four main pathways have been identified as significantly altered across the literature: immune regulation, secretion, stress response and programmed cell death pathways. Direct viral invasion of MKs and platelets via the ACE2 receptor have been mixed though several groups have noted the ACE2 receptor is not present on MKs nor platelets. The ability of the virus to infect via an alternate receptor has been described in subsequent studies [71,72].
7. Platelet function in COVID-19
7.1. Immune regulation and interferon response
Indeed, enrichment analysis of the platelet transcriptome and three integrated single cell RNA-seq data sets demonstrated that IFITM3 and other genes in the antigen presenting pathways such as IFIT1, IFIT3, IFI27, IFI6, and IFITM2 were enriched in COVID-19 samples [69,73]. Though recent studies suggests platelets can engage in immune-like activities, further studies are needed to determine if platelets restrict viral infection such as COVID-19 through proteins like IFITM3. Single-cell sequencing of small circulating MKs from COVID-19 patients show an increase in interferon (IFN)-stimulated genes including IFITM3 [74]. Though the role of IFITM3 in platelets needs to be further elucidated, an antiviral role of IFITM3 in MKs has been described in other viral infections [20]. Additional findings of MKs in COVID-19 infection will be discussed in greater detail below.
Interestingly, Garma and colleagues found an increase of expression in genes associated with an antiviral response and in genes associated to the prothrombotic response and platelet activation. Interestingly, one of the genes upregulated was the CD40LG that codes for CD40L [73]. CD40L can be found in platelet α granules and contribute to platelet activation by enhancing ROS production [75,76]. Platelets treated with TRAP and CD40L have enhanced mito-ROS production which may contribute to the increases of basal ROS observed in COVID 19 platelets, however this possibility has not been explored [75]. Furthermore, CD40L expression on activated platelets plays a key role in dendritic cell maturation, B-cell isotype switching and CD8+ T-cell responses [77] (Fig. 3).
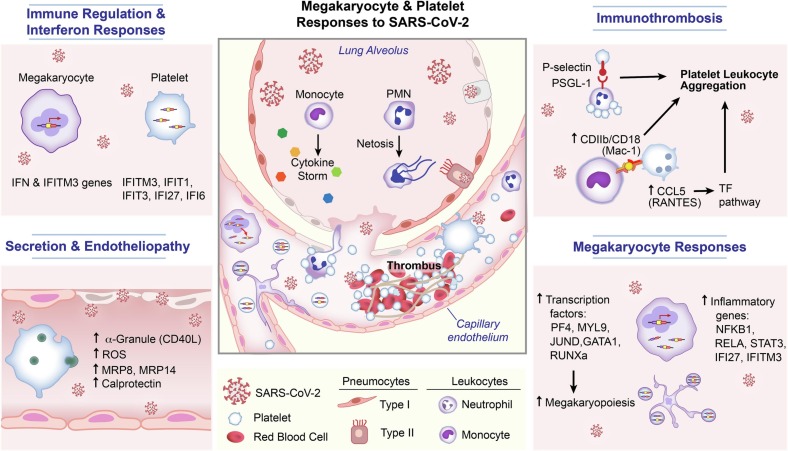
Fig. 3.
SARS-CoV2 enhances transcription factors that cause increased megakaryopoiesis and induce immune responses in megakaryocytes and enhancing transcriptional changes in the MK and platelet. The virus may also directly enter platelets or MKs and induces changes that impact interactions with circulating leukocytes and endothelial cells.
7.2. Platelet secretion and endothelial cell interaction
In COVID-19 patients, systemic inflammation, hypercoagulability and endothelial dysfunction have become hallmarks of severe disease [[78], [79], [80]]. Platelets' ability to interact with endothelial cells (EC) and contribute to EC's proinflammatory state makes them a likely candidate to contribute to the endothelial dysfunction observed in severe COVID-19 patients. Barret et al. explored this by incubating ECs with platelet releasate from COVID-19 patients. Interestingly, gene set enrichment analysis (GSEA) of EC transcriptome revealed increases in pathway associated with dysregulation of cell-to-cell tight junction, proinflammatory processes and coagulation, suggesting that platelet and EC crosstalk could be a key contributing factor to endotheliopathy [79]. Integrated platelet and EC RNA sequence analysis and subsequent gene ontology pathway analysis demonstrated an upregulation of the platelet degranulation pathway. Further work has shown abnormalities in alpha-granule and dense granule release through an increase in immune receptor activation in those with severe disease [63,81,82]. Focused RNAseq analysis of platelet genes that are upregulated during COVID-19, revealed that the s100A8/A9 transcripts that code for MRP8 and MRP14 respectively were upregulated. MRP8 and MRP14 MRP8/14 heterodimerize and produce calprotectin, an inflammation associated protein, found in platelet granules and released from activated platelets [83]. Furthermore, calprotectin derived from COVID-19 platelets activates ECs and may add to the hypercoagulable phenotype described in COVID-19 [79,84].
7.3. Cell-cell interaction
Platelets are able to form heterotypic aggregates with circulating leukocytes during infectious conditions [45,85]. Platelets express other surface receptors, namely P-selectin (CD62P) which binds to P-selectin glycoprotein ligand 1 (PSGL1) and activated leukocyte CD11b/CD18 (Mac-1) on the surface of neutrophils via platelet GPIb [86,87]. Notably, platelets express P-selectin on their surface which allows platelets to interact with endothelial cells and neutrophils while in circulation. Furthermore, shedding of P-selectin is dependent on endothelial expression of PSGL-1 [88]. Activated platelets may also release CCL5 (RANTES) from their α granules [87]. CCL5 is a chemokine that attracts monocytes and to which platelets may bind via PSGL-1 and P-selectin. Platelets then influence the transcription and down-stream expression of tissue factor (TF) mRNA in monocytes. Inhibition of platelet-monocyte aggregates by selectively inhibiting P-selectin or with a non-P-selectin selective antibody, abciximab (targeting integrin αIIbβIIIa) abolished the effect of increased TF expression [17,89]. Similar findings have been shown with activated platelets and leading to increased TF mRNA in healthy endothelial cells [90]. Moreover, increased circulating platelet-leukocytes have been reported by multiple groups in acute COVID-19 infection [69].
7.4. Stress response
Reactive oxygen species (ROS) have recently emerged as pivotal regulators of platelet function [76,91,92]. Upon platelet activation, signaling pathways can trigger endogenous ROS production that aids in amplifying platelet activation and in some instances alters mitochondrial function [75,76]. Though changes in platelet ROS production occur physiologically upon platelet activation, an oxidative stress like the one triggered by viral infections can lead to platelet toxicity and hyperactivation [69,73,79]. While mitochondrial dysfunction may be the main cause of increase of basal ROS levels, COVID-19 may increase ROS in patient platelets as shown by Manne et al. 2020, increases in other transcriptional factors may also contribute to rising basal ROS levels [69,75,76]. Interestingly, using Ingenuity Pathway Analysis of the transcriptome of platelets from COVID-19 patients, Manne et al. found upregulation in genes related to mitochondrial dysfunction pathways and stress response, suggesting these responses are occurring with MK and not solely due to platelet activation in circulation [69]. Considering platelet mitochondrial dysregulation may lead to thrombotic complications such as ischemic stroke, mitochondrial pathway dysregulation was further explored [69,[93], [94], [95]]. Two main characteristics of mitochondrial dysfunction were measured, reactive oxygen species and phosphatidylserine (PS) exposure. Interestingly, though there was a rise in basal ROS in COVID-19 patients compared to the healthy donors, no increase in PS exposure, a marker of mitochondrial-dependent apoptosis, was observed [69].
7.5. SARS-CoV2 enhances pathways involved in programmed cell death
In a small, single-center series, Denorme et al. showed no increase in basal annexin V surface expression in platelets from patients with COVID-19 compared to healthy donors. These platelets when exposed agonist failed to increase PS exposure [65]. Though no difference in surface expression of apoptosis markers (PS exposure) was seen between platelets from COVID-19 patients compared to the healthy donors, increases in gene expression in programmed cell death pathways was observed in platelets from patients with acute COVID-19. Koupenova et al. performed transcriptome analysis of platelets and compared patients with acute COVID-19 infection to patient with myocardial infarction to evaluate pathways related to acute viral infection and not related to the prothrombotic phenotype often described in COVID-19 patients. When comparing the platelet transcriptome from patients with acute COVID-19 to platelets from patients with myocardial infarction the most upregulated pathways aside from IFN I pathways, were the programmed cell death pathways, more specifically apoptosis and necroptosis. Further functional assays demonstrated that markers for necroptosis and apoptosis such as caspase 1 and 3 were upregulated in patients with severe COVID-19 [72]. Additional work interrogating the platelet proteome in severe, non-severe COVID-19 infection and healthy donors found an upregulation in differentially expressed proteins involved in programmed cell death in individuals with COVID-19 compared to healthy donors. Confirming findings that the alterations in the platelet transcriptome impacts protein expression in circulating platelets [96].
Similarly, findings in cohorts of adult and pediatric patients have identified increased expression of genes and proteins involved in autophagy and pyroptosis. The latter has been identified as contributing to NETs and increased patient mortality [29,97].
7.6. SARS-CoV2 effects on megakaryocytes
Considering platelet transcripts are acquired during thrombopoiesis, transcriptional analysis of MKs in COVID-19 patients can provide further information on the causes of the platelet phenotype observed during COVID-19. Single cell RNA-seq (scRNASeq) on MKs revealed increases transcription factors related to megakaryopoiesis (PF4, MYL9, JUND, GATA1, RUNXa and histone associated genes) and inflammatory response (NFKB1, RELA, STAT3, IFI27 and IFITM3). This suggests that MKs have been infected, are maturing faster and possibly contributing to increased production of platelets with altered transcriptomes [72,98,99]. Moreover, the single-nucleotide polymorphism rs12252 which truncates the N-terminal of IFITM3 has been associated with increased COVID-19 severity and mortality in an age dependent manner [[100], [101], [102]]. The relationship between MK IFITM3 expression and mortality in severe COVID-19 has not been established.
New data shows both bone marrow and lung MKs may become infected with the SARS-CoV2 virus and promote increased viral dissemination [74]. Moreover, analysis of both primary human MKs and established MK cell line revealed increase in s100A8/A9 expression when MKs were treated with COVID19 [74,79]. Interestingly, treatment of MKs with another corona virus (COV-Oc43) did not increase s100A8/A9 expression [74]. This suggests that increases in s100A8/A9 expression maybe specific to the SARS-CoV2 coronavirus though if it is upregulated in other infectious syndromes needs further investigation (Fig. 3).
8. Conclusion
As technology continues to evolve and allows further dissection and more complete understanding of the RNA profile of MKs and platelets, the more we are able to understand how MKs and platelets are altered by and contribute to disease. It is clear through numerous investigations that MKs play direct roles in pathogen interactions and are both indirectly and directly impacted by infection of the host species. Furthermore, platelets are now well-established as immune effector cells with contributions to bacterial clearance, secretion of antimicrobial proteins and viral clearance.
Sources of funding
The preparation of this manuscript was supported by National Institutes of Health (1K08HL153953-01, EAM).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Diana Lim for preparation of the figures, critical comments, and consultation regarding effective display of the images.
References
- 1.Wright J.H. The origin and nature of the blood plates. Boston Med. Surg. J. 1906;154:643–645. [Google Scholar]
- 2.Lefrancais E., et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544(7648):105–109. doi: 10.1038/nature21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valet C., et al. Sepsis promotes splenic production of a protective platelet pool with high CD40 ligand expression. J. Clin. Invest. 2022;132(7) doi: 10.1172/JCI153920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weyrich A.S., et al. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc. Natl. Acad. Sci. U. S. A. 1998;95(10):5556–5561. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best M.G., et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson R.J., et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118(13):3680–3683. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittrich M., et al. Analysis of SAGE data in human platelets: features of the transcriptome in an anucleate cell. Thromb. Haemost. 2006;95(4):643–651. [PubMed] [Google Scholar]
- 8.Gnatenko D.V., et al. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101(6):2285–2293. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- 9.Londin E.R., et al. The human platelet: strong transcriptome correlations among individuals associate weakly with the platelet proteome. Biol. Direct. 2014;9:3. doi: 10.1186/1745-6150-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray P.F., et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pariser D.N., et al. Lung megakaryocytes are immune modulatory cells. J. Clin. Invest. 2021;131(1) doi: 10.1172/JCI137377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teruel-Montoya R., et al. MicroRNA expression differences in human hematopoietic cell lineages enable regulated transgene expression. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0102259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macaulay I.C., et al. Platelet genomics and proteomics in human health and disease. J. Clin. Invest. 2005;115(12):3370–3377. doi: 10.1172/JCI26885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portier I., Campbell R.A. Role of platelets in detection and regulation of infection. Arterioscler. Thromb. Vasc. Biol. 2021;41(1):70–78. doi: 10.1161/ATVBAHA.120.314645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rayes J., Watson S.P., Nieswandt B. Functional significance of the platelet immune receptors GPVI and CLEC-2. J. Clin. Invest. 2019;129(1):12–23. doi: 10.1172/JCI122955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celi Alessandro P.G., Roberto Lorenzet, Antonio De Blasi, Neal Ready, Furie Barbara C., Bruce Furie. P-selectin induces the expression of tissue factor on monocytes. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8767–8771. doi: 10.1073/pnas.91.19.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hottz E.D., et al. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136(11):1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer B.F., et al. Novel anti-bacterial activities of beta-defensin 1 in human platelets: suppression of pathogen growth and signaling of neutrophil extracellular trap formation. PLoS Pathog. 2011;7(11) doi: 10.1371/journal.ppat.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton E.A., et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;134(12):911–923. doi: 10.1182/blood.2019000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell R.A., et al. Human megakaryocytes possess intrinsic antiviral immunity through regulated induction of IFITM3. Blood. 2019;133(19):2013–2026. doi: 10.1182/blood-2018-09-873984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D., et al. ELF4 facilitates innate host defenses against Plasmodium by activating transcription of Pf4 and Ppbp. J. Biol. Chem. 2019;294(19):7787–7796. doi: 10.1074/jbc.RA118.006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nuhrenberg T.G., et al. Impact of high platelet turnover on the platelet transcriptome: results from platelet RNA-sequencing in patients with sepsis. PLoS One. 2022;17(1) doi: 10.1371/journal.pone.0260222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer M., et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellinger R.P., et al. Surviving Sepsis Campaign guidelines for management of severe sepsis and septic shock. Intensive Care Med. 2004;30(4):536–555. doi: 10.1007/s00134-004-2210-z. [DOI] [PubMed] [Google Scholar]
- 25.Rautanen A., et al. Genome-wide association study of survival from sepsis due to pneumonia: an observational cohort study. Lancet Respir. Med. 2015;3(1):53–60. doi: 10.1016/S2213-2600(14)70290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent J.L., Yagushi A., Pradier O. Platelet function in sepsis. Crit. Care Med. 2002;30(5 (Suppl)):S313–S317. doi: 10.1097/00003246-200205001-00022. [DOI] [PubMed] [Google Scholar]
- 27.Iba T., et al. New criteria for sepsis-induced coagulopathy (SIC) following the revised sepsis definition: a retrospective analysis of a nationwide survey. BMJ Open. 2017;7(9) doi: 10.1136/bmjopen-2017-017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraemer B.F., et al. Bacteria differentially induce degradation of Bcl-xL, a survival protein, by human platelets. Blood. 2012;120(25):5014–5020. doi: 10.1182/blood-2012-04-420661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwertz H., et al. Human platelets display dysregulated sepsis-associated autophagy, induced by altered LC3 protein-protein interaction of the Vici-protein EPG5. Autophagy. 2022;18(7):1534–1550. doi: 10.1080/15548627.2021.1990669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Assinger A., et al. Platelets in sepsis: an update on experimental models and clinical data. Front. Immunol. 2019;10:1687. doi: 10.3389/fimmu.2019.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cremer M., et al. Low immature platelet fraction suggests decreased megakaryopoiesis in neonates with sepsis or necrotizing enterocolitis. J. Perinatol. 2013;33(8):622–626. doi: 10.1038/jp.2013.21. [DOI] [PubMed] [Google Scholar]
- 32.Assinger A. Platelets and infection - an emerging role of platelets in viral infection. Front. Immunol. 2014;5:649. doi: 10.3389/fimmu.2014.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu T., et al. Bifunctional effect of the inflammatory cytokine tumor necrosis factor alpha on megakaryopoiesis and platelet production. J. Thromb. Haemost. 2022;20(12):2998–3010. doi: 10.1111/jth.15891. [DOI] [PubMed] [Google Scholar]
- 34.Beaulieu L.M., et al. Interleukin 1 receptor 1 and interleukin 1beta regulate megakaryocyte maturation, platelet activation, and transcript profile during inflammation in mice and humans. Arterioscler. Thromb. Vasc. Biol. 2014;34(3):552–564. doi: 10.1161/ATVBAHA.113.302700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams N., et al. The role of interleukin 6 in megakaryocyte formation, megakaryocyte development and platelet production. CIBA Found. Symp. 1992;167:160–170. doi: 10.1002/9780470514269.ch10. (discussion 170-3) [DOI] [PubMed] [Google Scholar]
- 36.Wang Q., et al. Interferon-alpha directly represses megakaryopoiesis by inhibiting thrombopoietin-induced signaling through induction of SOCS-1. Blood. 2000;96(6):2093–2099. [PubMed] [Google Scholar]
- 37.Shannon O. The role of platelets in sepsis. Res. Pract. Thromb. Haemost. 2021;5(1):27–37. doi: 10.1002/rth2.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szilagyi B., et al. Reduced miR-26b expression in megakaryocytes and platelets contributes to elevated level of platelet activation status in sepsis. Int. J. Mol. Sci. 2020;21(3) doi: 10.3390/ijms21030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freishtat R.J., et al. Sepsis alters the megakaryocyte-platelet transcriptional axis resulting in granzyme B-mediated lymphotoxicity. Am. J. Respir. Crit. Care Med. 2009;179(6):467–473. doi: 10.1164/rccm.200807-1085OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hachim M.Y., et al. Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front. Immunol. 2020;11:1372. doi: 10.3389/fimmu.2020.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suddala K.C., et al. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 2019;15(1) doi: 10.1371/journal.ppat.1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrell C.N., et al. Emerging roles for platelets as immune and inflammatory cells. Blood. 2014;123(18):2759–2767. doi: 10.1182/blood-2013-11-462432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho-Tin-Noe B., Demers M., Wagner D.D. How platelets safeguard vascular integrity. J. Thromb. Haemost. 2011;9(Suppl. 1):56–65. doi: 10.1111/j.1538-7836.2011.04317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rondina M.T., et al. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J. Thromb. Haemost. 2011;9(4):748–758. doi: 10.1111/j.1538-7836.2011.04208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dib P.R.B., et al. Innate immune receptors in platelets and platelet-leukocyte interactions. J. Leukoc. Biol. 2020;108(4):1157–1182. doi: 10.1002/JLB.4MR0620-701R. [DOI] [PubMed] [Google Scholar]
- 46.Arman M., Krauel K. Human platelet IgG Fc receptor FcgammaRIIA in immunity and thrombosis. J. Thromb. Haemost. 2015;13(6):893–908. doi: 10.1111/jth.12905. [DOI] [PubMed] [Google Scholar]
- 47.Goodall A.H., Appleby J. In: Methods in Molecular Biology: Platelets and Megakarycytes, Vol 1: Functional Assays. Gibbins J.M., Mahaut-Smith M.P., editors. Humana Press; Totowa, N.J: 2004. Flow-cytometric analysis of platelet-membrane glycoprotein expression and platelet activation; pp. 225–253. [DOI] [PubMed] [Google Scholar]
- 48.Nieswandt B., Pleines I., Bender M. Platelet adhesion and activation mechanisms in arterial thrombosis and ischaemic stroke. J. Thromb. Haemost. 2011;9(Suppl. 1):92–104. doi: 10.1111/j.1538-7836.2011.04361.x. [DOI] [PubMed] [Google Scholar]
- 49.Hottz E.D., et al. Dengue induces platelet activation, mitochondrial dysfunction and cell death through mechanisms that involve DC-SIGN and caspases. J. Thromb. Haemost. 2013;11(5):951–962. doi: 10.1111/jth.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gawaz M., et al. Platelet activation and interaction with leucocytes in patients with sepsis or multiple organ failure. Eur. J. Clin. Investig. 1995;25(11):843–851. doi: 10.1111/j.1365-2362.1995.tb01694.x. [DOI] [PubMed] [Google Scholar]
- 51.Gawaz M., et al. Platelet function in septic multiple organ dysfunction syndrome. Intensive Care Med. 1997;23(4):379–385. doi: 10.1007/s001340050344. [DOI] [PubMed] [Google Scholar]
- 52.Yaguchi A., et al. Platelet function in sepsis. J. Thromb. Haemost. 2004;2(12):2096–2102. doi: 10.1111/j.1538-7836.2004.01009.x. [DOI] [PubMed] [Google Scholar]
- 53.Weiss L.J., et al. Acquired platelet GPVI receptor dysfunction in critically ill patients with sepsis. Blood. 2021;137(22):3105–3115. doi: 10.1182/blood.2020009774. [DOI] [PubMed] [Google Scholar]
- 54.Jarvis G.E. In: Methods in Molecular Biology: Platelets and Megakaryocytes, Vol 1: Functional Assays. Gibbins J.M., Mahaut-Smith M.P., editors. vol. 1. Humana; Totwa, NJ: 2004. Platelet aggregation in whole blood. [Google Scholar]
- 55.Ebermeyer T., et al. Platelet innate immune receptors and TLRs: a double-edged sword. Int. J. Mol. Sci. 2021;22(15) doi: 10.3390/ijms22157894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leroy J., et al. Fungal chitin reduces platelet activation mediated via TLR8 stimulation. Front. Cell. Infect. Microbiol. 2019;9:383. doi: 10.3389/fcimb.2019.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cognasse F., et al. The inflammatory role of platelets via their TLRs and Siglec receptors. Front. Immunol. 2015;6:83. doi: 10.3389/fimmu.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thon J.N., et al. T granules in human platelets function in TLR9 organization and signaling. J. Cell Biol. 2012;198(4):561–574. doi: 10.1083/jcb.201111136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell R.A., et al. IFITM3 regulates fibrinogen endocytosis and platelet reactivity in nonviral sepsis. J. Clin. Invest. 2022;132(23) doi: 10.1172/JCI153014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ackermann M., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanff T.C., et al. Thrombosis in COVID-19. Am. J. Hematol. 2020;95(12):1578–1589. doi: 10.1002/ajh.25982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Middleton E.A., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136(10):1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicolai L., et al. Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation. 2020;142(12):1176–1189. doi: 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Althaus K., Zlamal J., Bakchoul T. Antibody-mediated platelet activation in COVID-19: a coincidence or a new mechanism of the dysregulated coagulation system? J. Thromb. Haemost. 2021;19(5):1171–1173. doi: 10.1111/jth.15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denorme F., et al. COVID-19 patients exhibit reduced procoagulant platelet responses. J. Thromb. Haemost. 2020;18(11):3067–3073. doi: 10.1111/jth.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenner W.J., Gorog D.A. Incidence of thrombotic complications in COVID-19: on behalf of ICODE: the International COVID-19 Thrombosis Biomarkers Colloquium. J. Thromb. Thrombolysis. 2021;52(4):999–1006. doi: 10.1007/s11239-021-02475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Group, R.C Aspirin in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2022;399(10320):143–151. doi: 10.1016/S0140-6736(21)01825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Investigators, R.-C.W.C.f.t.R.-C, et al. Effect of antiplatelet therapy on survival and organ support-free days in critically ill patients with COVID-19: a randomized clinical trial. JAMA. 2022;327(13):1247–1259. doi: 10.1001/jama.2022.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manne B.K., et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136(11):1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ji W., et al. Transcriptional landscape of circulating platelets from patients with COVID-19 reveals key subnetworks and regulators underlying SARS-CoV-2 infection: implications for immunothrombosis. Cell. Biosci. 2022;12(1):15. doi: 10.1186/s13578-022-00750-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett T.J., et al. Platelets contribute to disease severity in COVID-19. J. Thromb. Haemost. 2021;19(12):3139–3153. doi: 10.1111/jth.15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koupenova M., et al. SARS-CoV-2 initiates programmed cell death in platelets. Circ. Res. 2021;129(6):631–646. doi: 10.1161/CIRCRESAHA.121.319117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garma L.D., Deng H., Goldschmidt E. Integrated analysis of transcriptomic data reveals the platelet response in COVID-19 disease. Sci. Rep. 2022;12(1):6851. doi: 10.1038/s41598-022-10516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu A., et al. Infection of lung megakaryocytes and platelets by SARS-CoV-2 anticipate fatal COVID-19. Cell. Mol. Life Sci. 2022;79(7):365. doi: 10.1007/s00018-022-04318-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chakrabarti S., et al. CD40 ligand influences platelet release of reactive oxygen intermediates. Arterioscler. Thromb. Vasc. Biol. 2005;25(11):2428–2434. doi: 10.1161/01.ATV.0000184765.59207.f3. [DOI] [PubMed] [Google Scholar]
- 76.Masselli E., et al. ROS in platelet biology: functional aspects and methodological insights. Int. J. Mol. Sci. 2020;21(14) doi: 10.3390/ijms21144866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elzey B.D., et al. Platelet CD40L at the interface of adaptive immunity. Thromb. Res. 2011;127(3):180–183. doi: 10.1016/j.thromres.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wichmann D., et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern. Med. 2020;173(4):268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barrett T.J., et al. Platelets amplify endotheliopathy in COVID-19. Sci. Adv. 2021;7(37):eabh2434. doi: 10.1126/sciadv.abh2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goshua G., et al. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020;7(8):e575–e582. doi: 10.1016/S2352-3026(20)30216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weiss L.J., et al. Uncoupling of platelet granule release and integrin activation suggests GPIIb/IIIa as therapeutic target in COVID-19. Blood Adv. 2023;7(11):2324–2338. doi: 10.1182/bloodadvances.2022008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schrottmaier W.C., et al. Adverse outcome in COVID-19 is associated with an aggravating hypo-responsive platelet phenotype. Front. Cardiovasc. Med. 2021;8 doi: 10.3389/fcvm.2021.795624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larsen S.B., et al. Calprotectin and platelet aggregation in patients with stable coronary artery disease. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0125992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conway E.M., et al. Understanding COVID-19-associated coagulopathy. Nat. Rev. Immunol. 2022;22(10):639–649. doi: 10.1038/s41577-022-00762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rondina M.T., et al. In vivo platelet activation in critically ill patients with primary 2009 influenza A(H1N1) Chest. 2012;141(6):1490–1495. doi: 10.1378/chest.11-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zarbock A., Polanowska-Grabowska R.K., Ley K. Platelet-neutrophil-interactions: linking hemostasis and inflammation. Blood Rev. 2007;21(2):99–111. doi: 10.1016/j.blre.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 87.McGregor L., Martin J., McGregor J.L. Platelet-leukocyte aggregates and derived microparticles in inflammation, vascular remodelling and thrombosis. Front. Biosci. 2006;11:830–837. doi: 10.2741/1840. [DOI] [PubMed] [Google Scholar]
- 88.Dole V.S., et al. PSGL-1 regulates platelet P-selectin-mediated endothelial activation and shedding of P-selectin from activated platelets. Thromb. Haemost. 2007;98(4):806–812. [PubMed] [Google Scholar]
- 89.Christersson C., Johnell M., Siegbahn A. Tissue factor and IL8 production by P-selectin-dependent platelet-monocyte aggregates in whole blood involves phosphorylation of Lyn and is inhibited by IL10. J. Thromb. Haemost. 2008;6(6):986–994. doi: 10.1111/j.1538-7836.2008.02956.x. [DOI] [PubMed] [Google Scholar]
- 90.Slupsky J.R., et al. Activated platelets induce tissue factor expression on human umbilical vein endothelial cells by ligation of CD40. Thromb. Haemost. 1998;80(6):1008–1014. [PubMed] [Google Scholar]
- 91.Arthur J.F., et al. ITAM receptor-mediated generation of reactive oxygen species in human platelets occurs via Syk-dependent and Syk-independent pathways. J. Thromb. Haemost. 2012;10(6):1133–1141. doi: 10.1111/j.1538-7836.2012.04734.x. [DOI] [PubMed] [Google Scholar]
- 92.Choo H.J., et al. Mitochondrial calcium and reactive oxygen species regulate agonist-initiated platelet phosphatidylserine exposure. Arterioscler. Thromb. Vasc. Biol. 2012;32(12):2946–2955. doi: 10.1161/ATVBAHA.112.300433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agbani E.O., Poole A.W. Procoagulant platelets: generation, function, and therapeutic targeting in thrombosis. Blood. 2017;130(20):2171–2179. doi: 10.1182/blood-2017-05-787259. [DOI] [PubMed] [Google Scholar]
- 94.Jobe S.M., et al. Critical role for the mitochondrial permeability transition pore and cyclophilin D in platelet activation and thrombosis. Blood. 2008;111(3):1257–1265. doi: 10.1182/blood-2007-05-092684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Denorme F., et al. Platelet necrosis mediates ischemic stroke outcome in mice. Blood. 2020;135(6):429–440. doi: 10.1182/blood.2019002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trugilho M.R.O., et al. Platelet proteome reveals features of cell death, antiviral response and viral replication in covid-19. Cell Death Dis. 2022;8(1):324. doi: 10.1038/s41420-022-01122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Su M., et al. Gasdermin D-dependent platelet pyroptosis exacerbates NET formation and inflammation in severe sepsis. Nat. Cardiovasc. Res. 2022;1(8):732–747. doi: 10.1038/s44161-022-00108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Battina H.L., et al. Interaction of the inflammatory response and megakaryocytes in COVID-19 infection. Exp. Hematol. 2021;104:32–39. doi: 10.1016/j.exphem.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Valdivia-Mazeyra M.F., et al. Increased number of pulmonary megakaryocytes in COVID-19 patients with diffuse alveolar damage: an autopsy study with clinical correlation and review of the literature. Virchows Arch. 2021;478(3):487–496. doi: 10.1007/s00428-020-02926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y., et al. Interferon-induced transmembrane protein 3 genetic variant rs12252-C associated with disease severity in coronavirus disease 2019. J. Infect. Dis. 2020;222(1):34–37. doi: 10.1093/infdis/jiaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alghamdi J., et al. Interferon-induced transmembrane protein-3 genetic variant rs12252 is associated with COVID-19 mortality. Genomics. 2021;113(4):1733–1741. doi: 10.1016/j.ygeno.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Y., et al. Interferon-induced transmembrane protein 3 gene polymorphisms are associated with COVID-19 susceptibility and severity: a meta-analysis. J. Inf. Secur. 2022;84(6):825–833. doi: 10.1016/j.jinf.2022.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]