Abstract
We isolated the LIP2 gene from the lipolytic yeast Yarrowia lipolytica. It was found to encode a 334-amino-acid precursor protein. The secreted lipase is a 301-amino-acid glycosylated polypeptide which is a member of the triacylglycerol hydrolase family (EC 3.1.1.3). The Lip2p precursor protein is processed by the KEX2-like endoprotease encoded by XPR6. Deletion of the XPR6 gene resulted in the secretion of an active but less stable proenzyme. Thus, the pro region does not inhibit lipase secretion and activity. However, it does play an essential role in the production of a stable enzyme. Processing was found to be correct in LIP2A (multiple LIP2 copy integrant)-overexpressing strains, which secreted 100 times more activity than the wild type, demonstrating that XPR6 maturation was not limiting. No extracellular lipase activity was detected with the lip2 knockout (KO) strain, strongly suggesting that extracellular lipase activity results from expression of the LIP2 gene. Nevertheless, the lip2 KO strain is still able to grow on triglycerides, suggesting an alternative pathway for triglyceride utilization in Y. lipolytica.
Several yeasts are able to utilize triglycerides (TGs) as their sole carbon source. One such yeast, nonpathogenic Yarrowia lipolytica, has potential for use as a model system for lipid utilization because classical and molecular genetic tools have been developed for this species (2, 3). Integrative and replicative vectors are available (16), and more recently, vectors for multiple integration have been developed (H. Wang and J.-M. Nicaud, unpublished data).
This yeast naturally secretes several proteins, depending on the growth conditions (21). For example, if the pH is higher than 6, it secretes an alkaline extracellular protease (AEP) (33, 38). Under optimal conditions, up to 1 g of AEP is secreted per liter (37). AEP is encoded by XPR2 (11, 27, 33). This gene codes for a 454-amino-acid (aa) prepro enzyme precursor containing a 15-aa signal sequence and a stretch of nine X-Ala or X-Pro dipeptides, followed by a 124-aa pro region that includes a glycosylation site (Asn123) and a Lys156-Arg157 processing site, and finally the mature form itself. The AEP precursor undergoes complex processing (27). A diaminopeptidase processes the stretch of nine dipeptides (X-Ala or X-Pro) (28, 29), and the endoprotease encoded by the XPR6 gene is required to cleave the pro region, releasing the mature form (14). The pro region is involved in both the inhibition of protease activity and the folding of the propeptide into a conformation compatible with secretion, and secretion at 28°C depends on the glycosylation of the pro region (15, 28).
Several enzymes are secreted by Y. lipolytica, and lipase and esterase activities have been detected and analyzed in various studies. Lipase secretion was first reported in 1948 by Peters and Nelson (41, 42), who described a single type of glucose-repressible activity. An extracellular and two cell-bound types of activity corresponding to lipase I (39 kDa) and lipase II (44 kDa) were described by Ota and coworkers (39, 47). The extracellular lipase required oleic acid as a stabilizer-activator, whereas the cell-bound lipases did not and differed in several properties from the extracellular enzyme (40). The rates of production of the extracellular and cell-bound enzymes were reported to depend on the carbon and nitrogen composition of the medium. Extracellular lipase was only detected in cultures grown with an organic nitrogen source (36, 48), and lipase levels were shown to be modulated by cell morphology. In minimal medium supplemented with N-acetylglucosamine or citrate buffer, both of which promote dimorphic growth, higher levels of cell-bound lipases were detected. However, no clear relationship was established between the dimorphic state and lipase production (35).
Recently, Ota and coworkers purified the 39-kDa extracellular lipase (called lipase A) and determined the N-terminal amino acid sequence (22). Destain and coworkers isolated Y. lipolytica strains overproducing an extracellular lipase. They used chemical mutagenesis to produce a first generation of mutants with levels of lipase production five times higher than that of the wild type. A second round of mutagenesis generated strains able to secrete 1,200 U of lipase per ml, 25 times the level of the wild-type strain. These mutant strains were used for large-scale fermentation (500 liters) to produce lipase (13). The secreted lipase was shown to have an apparent molecular mass of 38.5 kDa, giving three bands in isofocusing (pIs of 5.0, 5.2, and 5.4). The sequence of the first 49 aa of the N terminus was determined (12) and found to be identical to that of lipase A. This sequence is similar to that of cell-bound lipase I; however, the extracellular lipase and lipase I are considered to differ in amino acid composition (22).
This strongly suggests that several genes encoding lipase are expected in Y. lipolytica. This is supported by the lack of success of attempts to obtain mutants unable to grow on TG. The mutants isolated were affected only in lipase levels (32). Such multicomponent gene families have been observed in other yeasts. A lipase family of five genes, with 80% identity, has been isolated in Candida rugosa (4, 25), and two genes have been isolated in Geotrichum candidum (5).
In Y. lipolytica, two lipase genes of the carboxylesterase family have been identified by Dominguez and coworkers: LIP1, which codes for a 486-aa lipase (17), and LIP3, which codes for a 498-aa lipase (9). Both are similar to the lipases of the fungi Candida cylindracea and G. candidum and belong to the carboxylesterase family (30). These are probably intracellular or cell-bound lipases, because no clear signal sequence was identified.
Here, we report on the isolation and characterization of the LIP2 gene, which codes for the extracellular lipase Lip2p, a triacylglycerol hydrolase. We show that this gene is responsible for all of the extracellular lipase activity of Y. lipolytica. Lipase expression was repressed by glucose and induced by olive oil and oleic acid. We show that Lip2p processing is similar to AEP processing, involving cleavage of the signal sequence and processing by the endoprotease encoded by the XPR6 gene. We also demonstrate that glycosylation and processing by the XPR6 gene product are not essential for activity and suggest that they are required for stabilization of the protein.
MATERIALS AND METHODS
Strains and media.
The Y. lipolytica strains used in this study were PO1d and derivatives (Table 1). Escherichia coli strain DH5α was used for DNA propagation. The media and techniques used to grow and handle Y. lipolytica were described by Barth and Gaillardin (3), and those used for E. coli were described by Sambrook et al. (44). The compositions of the media used were as follows (per liter): YPD, 10 g of yeast extract (Difco), 10 g of Bacto Peptone (Difco), and 10 g of glucose; YPDH, 10 g of yeast extract, 10 g of Bacto Peptone, 10 g of glucose, and 10 g of olive oil; YTDH, 10 g of yeast extract, 10 g of Bacto Tryptone (Difco), 10 g of glucose, and 50 g of olive oil. The minimal medium, YNB, contained 1.7 g of yeast nitrogen base without ammonium and without amino acids (Difco), 4 g of ammonium chloride, and various carbon sources as follows: YNBD, 10 g of glucose; YNBcas, YNBD with 5 g of Casamino Acids; YNBO, 10 g of oleic acid; YNBH, 10 g of olive oil; YNBTo, 10 g of triolein (To); YNBT, 10 g of tributyrin (T). Uracil (0.1 g/liter) and leucine (0.3 g/liter) were added to the media as required. Stock solutions of fatty acid (20% oleic acid, 0.5% Tween 40), oil (10% olive oil, 1% Tween 40), and tributyrin (20% tributyrin, 1% Tween 20) were subjected to sonication three times for 1 min each on ice. Agar (2%; Pastagar B; Institut Pasteur) was added for solid media. Media were buffered with 50 mM phosphate buffer, pH 6.8.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain (host strain) or primer | Plasmid, genotype, or sequence (5′→3′)c | Reference, source or location (bp) |
|---|---|---|
| E. coli strains | ||
| JME225 (DH5α) | pKS-LIP2W29a | This work |
| JME257 (DH5α) | JMP5 (ura3d1) | This work |
| JME259 (DH5α) | JMP6 (ura3d4/pPOX2-LIP2) | This work |
| JME276 (DH5α) | JMP3 (ura3d4) | This work |
| JME260 (DH5α) | JMP8 (ura3d1/pPOX2-LIP2) | This work |
| JME298 (DH5α) | JMP13 (ura3d1/pLIP2-LIP2) | This work |
| JME300 (DH5α) | JMP14 (ura3d4/pLIP2-LIP2) | This work |
| JME337 (DH5α) | pIMR83 | 14 |
| Y. lipolytica strains | ||
| W29 | MAT | 3 (CLIB 89, ATCC 20460)a |
| POld | MATaura3-302 leu2-270 xpr2-322 | 3 (CLIB 139) |
| E150 | MATaura3-302 leu2-270 xpr2-322 his1 | 3 (CLIB 122) |
| Y. lipolytica recombinant strainsb | ||
| JMY184 = POld-6-15 | JMP6/pPOX2-LIP2A | This work |
| JMY279 = POld-6-17 | JMP6/pPOX2-LIP2A | This work |
| JMY277 = POld-lip2 KO | plip2 KO lip2 KO | This work |
| JMY278 = POld-6-15-xpr6 KO | pxpr6 KO/pPOX2-LIP2Axpr6 KO | This work |
| JMY281 = POld-14-1 | JMP14/pLIP2-LIP2A | This work |
| JMY282 = POld-14-2 | JMP14/pLIP2-LIP2A | This work |
| JMY283 = E150-6-2 | JMP6/pPOX2-LIP2A | This work |
| Primers | ||
| YlipSA | GCi CCi CCi AGR GAR TGi CC | 4313 |
| YlipATG | CCC GGG CCG CAG TGG CCA CAA TGA AGC TTT CCA CCA TCC TTT TCA CAG CC | 3733 |
| YlipSTOP | GTT GGC GGC CGC GAA TTC GGA TCC TAA ATA AAC GAT ATT CAT TTA TTA AAG TAG ATA GTT GAG G | 4806 |
CLIB, Collection de Levures d'Intérêt Biotechnologique, Thiverval-Grignon, France.
Transformants were named according to the strain and plasmid used, and transformants were numbered as follows: for example; POld1-6-1 (strain-plasmid-clone number)
Underlined sequences correspond to introduced restriction enzyme sites as follows: for YlipATG, HindIII; for YlipSTOP, NotI, EcoRI, and BamHI. The locations of the various oligonucleotides are based on the position in the EMBL sequence of the described LIP2 locus.
cDNA sequencing and LIP2 gene isolation.
The wild-type strain, PO1d, was grown in YPDH for 12 h. When cultures reached an optical density at 600 nm of 10, cells were harvested by centrifugation and washed twice with cold water and total RNA was extracted as previously described (1). mRNA was purified using the Quick Prep Micro mRNA kit (Pharmacia) and a cDNA library was constructed using the Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.) in accordance with the manufacturer's instructions. A 5′ rapid amplification of cDNA ends (RACE)-PCR amplification was performed with primer YlipSA and the linker primer AP1, using 50 ng of cDNA, giving a 620-bp fragment. This PCR product was isolated after electrophoresis in an agarose gel and was introduced into the EcoRV site of the pBluescript II KS+ plasmid (Stratagene, La Jolla, Calif.). The insert was then sequenced. New primers corresponding to the determined sequence were synthesized and used for a second round of 5′ and 3′ RACE amplification. PCR fragments were sequenced directly after gel purification. pKS-LIP2W29a, containing the LIP2 cDNA, was constructed as follows. The cDNA was amplified by PCR using the YlipATG and reverse YlipSTOP primer pair (Table 1; see Fig. 2B). The PCR product was isolated and introduced into pKS+. Plasmids were checked by sequencing of the inserts.
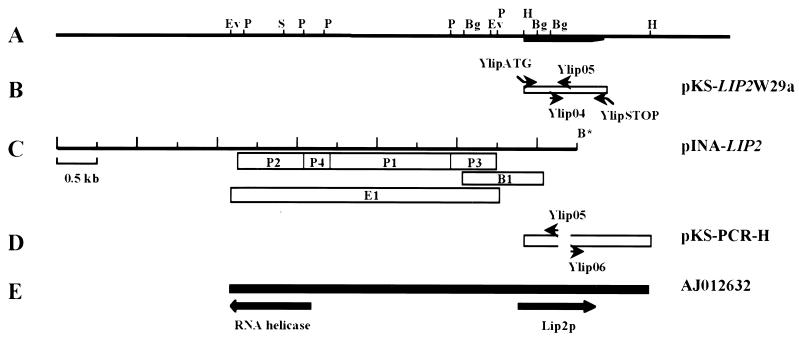
FIG. 2.
Sequencing of the LIP2 gene coding for the Y. lipolytica extracellular lipase. (A) Partial restriction map of the LIP2 locus. Abbreviations: EcoRV, Ev; PstI, P; BglII, Bg; HindIII, H. The location of the LIP2 ORF is indicated by the thick line. (B) Location of the cDNA sequence and schematic representation of primers used for gene library screening and the oligonucleotide pair YlipATG-YlipSTOP used for amplification of the coding region of the lipase gene. The recombinant plasmid containing the LIP2 gene was called pKS-LIP2W29a. (C) Schematic representation of the 6.5-kb insert contained within pINA-LIP2 and location of the PstI (P1 to P4), BglII (B1), and EcoRV (E1) subclones used for sequencing of the 5′ region of the LIP2 gene. (D) Sequence of the 1.4-kb HindIII genomic fragment. Genomic DNA was digested with HindIII, ligated, and amplified by PCR with the Ylip05-Ylip06 primer pair. The PCR fragment was cloned at the EcoRV site of pBluescript, giving pKS-PCR-H. (E) Location of the sequenced EcoRV-HindIII 5,304-bp region containing part of the DEAD box RNA helicase gene and the LIP2 gene (AJ012632). All scales are the same.
Sequence determination and analysis of the LIP2 gene.
The six pools of the Xuan library (51) were tested by PCR, and it was found that a clone containing the lipase gene was present in pool 2. pINA-LIP2 was isolated by colony hybridization using the 5′ RACE-derived 620-bp fragment as a probe against pool 2 clones. PstI (P1 to P4), BglII (B1), and EcoRV (E1) fragments (see Fig. 2C) were subcloned into the PstI, BamHI, and EcoRV sites of the Bluescript KS+ plasmid, respectively, and used for sequencing. We performed divergent PCR by digesting 100 ng of total genomic DNA with HindIII and ligating the digested product overnight at 16°C. We used 10 ng of the ligation product as a template for PCR with the primer pair Ylip05-Ylip06 (see Fig. 2D). The 1.4-kb PCR fragment was cloned into the EcoRV site of pBluescript KS+, giving pKS-PCR-H (see Fig. 2D). Template preparation, sequencing, and nucleotide sequence analysis were performed as previously described (26). Proteins were compared using the GCG-gap program (version 9.1-UNIX, Sept. 97) and the BLOSUM 62 matrix (Genetics Computer Group, University of Wisconsin, Madison).
Southern blot analysis.
Genomic DNA was prepared from cells grown overnight in 5 ml of YPD as previously described (3). DNA was digested, subjected to electrophoresis in a 1.2% agarose gel, and transferred to a Hybond-N nitrocellulose membrane (Amersham). DNA probes were radiolabeled using the Megaprime kit (Amersham). After hybridization, membranes were placed for 1 h against a Molecular Dynamics PhosphorImager screen and scanned using a Storm 860 PhosphorImager (Molecular Dynamics).
Disruption of LIP2.
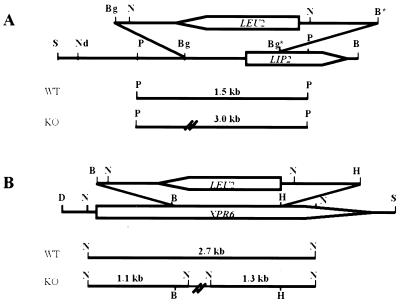
To construct a plasmid containing the lipase gene with its promoter, we inserted into SalI-BamHI-digested pKS+ a 2.7-kb SalI-HindIII fragment from pINA-LIP2 containing the LIP2 promoter together with the 1-kb HindIII-BamHI fragment from pKS-LIP2W29a carrying the lipase gene. The resulting plasmid was called pLIPpr-LIP2. To disrupt the LIP2 gene, a 1,091-bp BglII fragment in pLIPpr-LIP2 containing part of the promoter and half of the open reading frame (ORF) was deleted and replaced with the 2.7-kb BglII-BamHI Y. lipolytica LEU2 gene isolated from pINA240 (see Fig. 5A). The resulting plasmid, plip2::LEU2, was digested with NdeI and BamHI to yield a linear fragment suitable for one-step gene disruption (43) (see Fig. 5A) and used to transform PO1d. Southern blots of PstI-digested DNA from Leu+ transformants were probed with LIP2. In the disrupted strain, the 3-kb PstI fragment was replaced with a 1.5-kb fragment (data not shown). The resulting strain, containing a lip2::LEU2 deletion, was called JMY277.
FIG. 5.
Disruption of the LIP2 and XPR6 genes. Physical maps of the lipase (LIP2, A) and endoprotease (XPR6, B) genes and disruption cassettes are shown. ORFs are represented by arrow boxes indicating the direction of transcription. (A) For LIP2 gene disruption, the BglII fragments were replaced with a BglII-BamHI LEU2 gene and the NdeI-BamHI linear fragment was used for transformation. (B) For XPR6 gene disruption, the BamHI-HindIII fragment was replaced with a BamHI-HindIII LEU2 gene and the DraI-SalI linear fragment was used for transformation. The sizes of the PstI (A) and NcoI (B) fragments obtained by Southern blotting of the wild-type (WT) and disrupted (KO) strain DNAs using LIP2 (A) and XPR6 (B) probes are shown. Restriction sites: B, BamHI; Bg, BglII; D, DraI; H, HindIII; N, NcoI; Nd, NdeI; P, PstI; S, SalI.
Disruption of XPR6.
pIMR83 contains an xpr6A976::URA3 disruption cassette in which a 976-bp BamHI-HindIII fragment has been deleted and replaced with the 1.7-kb Y. lipolytica URA3 gene (14). The derivative plasmid, pxpr6Δ976::LEU2 (JME337), was constructed by replacing the 1.7-kb BamHI-HindIII URA3 fragment with the 2.7-kb BamHI-HindIII LEU2 fragment from pINA1192 (8). The DNA was digested with DraIII and SphI prior to transformation of the JMY184 strain (PO1d-6-15) and selection of the Leu+ transformants. Gene disruption was checked by Southern blotting of NcoI-digested DNA from putative disruptants with an XPR6 probe. Strain JMY278 had the two expected 1.1- and 1.3-kb NcoI fragments replacing the 2.7-kb wild-type signal (see Fig. 6B; data not shown).
FIG. 6.
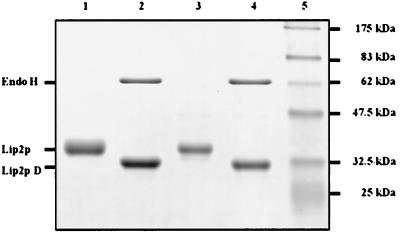
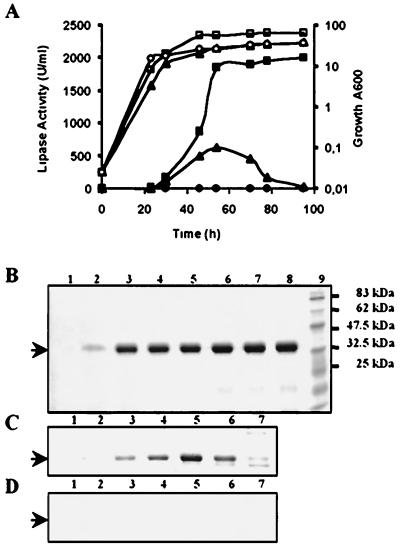
Analysis of the extracellular lipase from JMY184 and JMY278 by SDS-PAGE. The equivalent of 5 μl of the supernatant (52 h) from strains JMY184 (LIP2A; mature form) (Fig. 4B, lane 5) and JMY278 (xpr6 KO; pro form) (Fig. 4C, lane 5) was treated with endo H. Untreated (lanes 1 and 3) and deglycosylated (lanes 2 and 4) proteins were separated by SDS–12% PAGE and stained with Coomassie blue. The positions of endo H and the untreated (Lip2p) and deglycosylated (Lip2p D) lipases are indicated on the left. Prestained molecular mass markers were used as standards (lane 5).
Plasmid construction.
We recently constructed vectors JMP3 and JMP5 for gene expression and amplification in the yeast Y. lipolytica (Wang and Nicaud, unpublished data). They are derived from the amplification vectors previously described (3, 24). These vectors contain the ura3d1 (nondefective allele allowing single-plasmid integration) or ura3d4 (defective allele allowing multicopy plasmid integration) marker for selection in Y. lipolytica, a polylinker for insertion of the expression cassette, an 812-bp zeta region, and, inserted into the middle of the zeta region at the NotI site, pHSS6, which confers replication and kanamycin resistance in E. coli. The zeta region corresponds to the long terminal repeat of the Y. lipolytica retrotransposon Ylt1, which has been shown to be present in the genome in about 60 copies (46).
Using JMP3 (defective) and JMP5 (nondefective) as parental vectors, we constructed the expression vectors JMP6, JMP8, JMP13, and JMP14 (Table 1). The promoter region from LIP2 was rescued from pINA-LIP2 by PCR and digested with HindIII. The resulting 1,361-bp LIP2 promoter region was ligated together with the HindIII-EcoRI LIP2 fragment and JMP3 or JMP5 digested with HpaI and EcoRI. The POX2 promoter used (49) was a 2,163-bp ClaI-HindIII fragment produced by PCR. The resulting POX2 promoter was ligated together with the HindIII-EcoRI LIP2 fragment and JMP3 or JMP5 digested with ClaI and EcoRI. The resulting expression cassettes from the various plasmids described here were sequenced before transformation.
Transformation of Y. lipolytica.
Y. lipolytica strains were transformed by the lithium acetate method as described previously (24). Expression vectors (5 μg) were digested with NotI and subjected to electrophoresis. The bands corresponding to the expression cassettes were extracted from the gel and used for transformation. Typically, 0.05 or 2 μg of expression cassettes was used for nondefective (transformation control) and defective vectors, respectively. Transformants were selected on YNBcas and appeared after 3 days at a frequency of 105 transformants per μg of DNA for the nondefective vector (defective vectors: 5 to 15 days, 10 to 30 transformants).
Protein analysis.
Protein concentration was determined as described by Bradford (7) using the Bio-Rad protein assay system with bovine serum albumin as the standard. Proteins were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) as described by Laemmli (23), using prestained low-range protein markers (Bio-Rad) as molecular mass standards. Cell culture protein amounts equivalent to an optical density at 600 nm of 0.2 or to 5 μl of cell supernatant were loaded per lane.
Lipase activity detection and assay.
For detection of lipase activity on agar plates, plates containing emulsions of either T or To were used. A halo (T) or a clearing zone (To) developed within 24 to 48 h of incubation at 28°C. Lipase activity was routinely measured by titrimetric assay as follows. The substrate emulsion was prepared with olive oil (50 ml; Sigma, Paris, France) and gum arabic (50 ml; 10%, wt/vol). The solution was emulsified in a Waring blender (three pulses of 2 min each at maximum speed). The supernatant of the cell culture (20 μl), pure or diluted, depending on the quantity of lipase, was added to 5 ml of substrate emulsion and 2 ml of 50 mM phosphate buffer, pH 6.8 (Na2HPO4-KH2PO4). Samples were incubated for 20 min at 37°C with shaking (200 rpm). The reaction was stopped with 4 ml of acetone-ethanol (50/50, vol/vol) containing 0.09% thymolphthalein (Prolabo) as an indicator. Enzymatic activity was determined by titration of the fatty acid released with 50 mM sodium hydroxide. All lipase activity assays were performed at least in duplicate. One unit of lipase is the amount of enzyme that catalyzes the release of 1 μmol of fatty acid per min at 37°C.
N-terminal amino acid sequence of the lipase.
The sequence of the N-terminal 7 aa of the lipase present in the cell culture supernatant was determined. The lipase was blotted onto a polyvinylidene difluoride membrane after SDS-PAGE, and the blotted protein, detected by staining of the membrane with Coomassie brilliant blue R-250, was used directly for Edman degradation in a protein sequencer. Automated Edman sequencing was performed using a Perkin-Elmer Applied Biosystems Procise 494A sequencer with the reagents and methods of the manufacturer.
Lipase modification.
Deglycosylation of extracellular lipase using endoglycosidase H (endo H; NEB-Ozyme) was performed in accordance with the manufacturer's instructions. A 50-μl volume of cell culture supernatant (52 h) from lipase-overproducing strains JMY184 (PO1d-6-15) and JMY278 (PO1d-6-15-xpr6 KO) was treated with 0.2% (wt/vol) SDS and 0.08 U of endo H per mg of protein at 37°C for 2 h (native condition) or heated for 5 min at 85°C and treated for 24 h (denaturing conditions).
Antibody preparation and detection on Western blots.
Polyclonal antiserum directed against Lip2p was raised in a rabbit. Before use, the antibodies were immunopurified with the nitrocellulose membrane band containing the blotted Lip2p as previously described (19). For Western blot analysis, proteins were resolved by SDS–10% PAGE and transferred onto a nitrocellulose membrane (Schleicher & Schuell). Antigen-antibody complexes were detected using the ECL Kit (Amersham Pharmacia Biotech).
Nucleotide sequence accession number.
The nucleotide sequence of the LIP2 gene will appear in the EMBL nucleotide sequence database under accession no. AJ012632.
RESULTS
Isolation and sequencing of the LIP2 gene.
We previously isolated the LECE gene coding for a 40-kDa extracellular triacylglycerol hydrolase from the yeast Candida ernobii (44). To isolate the corresponding gene from Y. lipolytica, we first compared the predicted amino acid sequences of secreted lipases from the yeasts C. ernobii and Saccharomyces cerevisiae (P47145) and from the fungi Rhizomucor miehei (P19515) (6), Rhizopus delemar (P21811) (18), Fusarium heterosporum (S77816) (31), and Penicillium camembertii (P25234) (52). Alignment of these protein sequences led to the identification of the highly conserved motif GHSLGG/AA, which contains the serine involved in the active site (Fig. 1). We designed the degenerate antisense oligonucleotide YlipSA (Table 1) for reverse transcription-PCR, based on the conserved core sequence, using available DNA sequences and on the basis of Y. lipolytica codon usage.
FIG. 1.
Alignment of the predicted amino acid sequences of secreted lipases from the yeasts C. ernobii (C. e) and S. cerevisiae (S. c; P47145) and from the fungi R. miehei (R. m; P19515), R. delemar (R. d; P21811), F. heterosporum (F. h; S77816), and P. camembertii (P. c; P25234). Conserved amino acids are highlighted. Potential N glycosylation sites in the Y. lipolytica sequence are in boldface type. Asterisks indicate amino acids involved in the active site.
Lipase secretion in Y. lipolytica has been reported to be induced in rich complex media with olive oil as the carbon source (36). Therefore, a cDNA library was constructed using mRNA isolated from strain PO1d grown in the induction medium YPDH, used for 5′ RACE-PCR, and sequenced as described in Materials and Methods. The complete cDNA sequence was determined, and it corresponds to coordinates 3737 to 4806 of the sequence with EMBL accession no. AJ012632. Plasmid pINA-LIP2 was isolated from pool 2 of the Xuan library (51). It contains a 7.1-kb genomic insert that carries the promoter and part of the LIP2 gene (Fig. 2C). Indeed, restriction analysis of pINA-LIP2 and location of the HindIII and BglII sites (from the cDNA sequence) showed that only the promoter and part of the ORF were present in the EcoRV-BamHI region. This region was sequenced using six subclones (Fig. 2C). We used a divergent PCR strategy and genomic DNA to obtain the 3′ end of the gene, which was absent from pINA-LIP2. First, we determined which restriction enzymes could be used by probing a Southern blot of genomic DNA digested with various restriction enzymes with the 5′ RACE fragments, detecting 1.4-, 1.66-, and 3.7-kb fragments for the HindIII, PstI, and SphI digestions, respectively (data not shown). The 1.4-kb HindIII fragment was amplified, cloned, and sequenced. The 5,304-bp EcoRV-HindIII sequence will appear in the EMBL database under accession no. AJ012632.
We obtained the complete lipase gene, a 1,118-bp fragment carrying the lipase ORF and the terminator right up to the polyadenylation site by PCR using as a template either the cDNA library or genomic DNA from wild-type strain W29 and cloning the amplified fragments into pKS, giving rise to pKS-LIP2W29a (Fig. 2B). No sequence differences were observed between fragments of genomic and cDNA origin.
Sequence analysis.
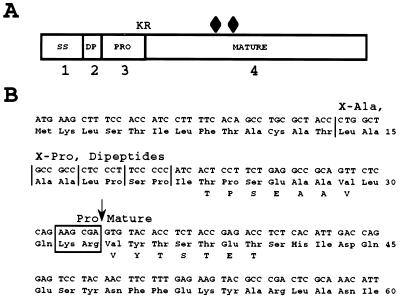
Two ORFs were identified in the 5,304-bp sequence (Fig. 2E). One corresponded to the NH2 terminus of a protein with a sequence similar to the first 324 aa of the human putative DEAD box RNA helicase (45). The second ORF was 1,005 bp long and encoded a 334-residue polypeptide similar in sequence to the yeast and fungal secreted lipases. The comparison results gave levels of identity (similarity) of 30.3% (39.2) to the yeast C. ernobii, 28.6% (38.2) to the S. cerevisiae, 27.4% (34.1) to the fungus R. miehei, 31.5% (38.6) to the R. delemar, 29% (36) to the F. heterosporum, and 26.8% (33.3) to the P. camembertii lipases, as shown in Fig. 1. These lipases belong to the triacylglycerol hydrolase family (EC 3.1.1.3). The LIP2 gene has a codon adaptation index of 0.426, indicating that it is a moderately expressed gene. The polypeptide encoded by the Y. lipolytica LIP2 gene contains several processing motifs (Fig. 3A). The prepro protein presents 13 aa, followed by four X-Ala or X-Pro dipeptides, possible substrates of the diamino peptidase (29); a short proregion of 12 aa and a Lys-Arg dipeptide, a putative substrate of the KEX2-like endopeptidase encoded by the XPR6 gene in Y. lipolytica (14); and the mature lipase of 301 residues. The signal sequence cleavage site is most probably located after aa 17, according to SignalP prediction (34), after the second potential X-Ala dipeptide, giving a 17-aa signal sequence, longer than that of AEP (28).
FIG. 3.
Processing of Y. lipolytica Lip2p. (A) Schematic representation of lipase processing. Shown are the putative 13-aa signal sequence (SS), followed by a stretch of four dipeptides (DP), a short 12-aa pro region (PRO) including the Lys-Arg (KR) cleavage site of the KEX2-like XPR6 endoprotease (14), and the mature 301-aa lipase. The diamonds indicate the positions of the potential signals for asparagine-linked glycosylation (Asn-X-Thr/Ser). (B) sequence of the preproregion and the first 27 N-terminal aa of the mature lipase encoded by the LIP2 gene from Y. lipolytica strain W29. The DNA sequence is shown together with the predicted amino acid sequence (three-letter code) and the N-terminal amino acid sequences (one-letter code) of the lipases secreted by the JMY184 (LIP2A; mature form) and JMY278 (xpr6 KO; pro form) strains. The vertical arrow indicates the confirmed pro-mature lipase processing site, and the vertical lines represent the four X-Ala and X-Pro dipeptides which act as possible substrates for dipeptidyl aminopeptidase activity.
The mature polypeptide contains the three conserved amino acids Ser162, Asp230, and His289, which have been shown to be part of the active site, and the conserved GHSLGGA motif (Fig. 1). A disulfide bridge may form between Cys76 and Cys120. Two potential N glycosylation sites were also identified (N113IS and N134NT).
Analysis of the nucleotide sequence and comparison with the cDNA sequence indicated that polyadenylation occurred at position 4806, 15 bp after the polyadenylation motif, AATAAA. The region between the lipase gene and the adjacent gene is 2,764 bp, larger than the mean intergenic region of the S. cerevisiae genome, although this is a general feature of intergenic regions in Y. lipolytica.
Lipase secretion, expression vectors, and strain construction.
The lipase gene was expressed under the control of the POX2 or LIP2 promoter (Table 1). The expression vectors were constructed using three-way ligation as described in Materials and Methods. Plasmids JMP6, JMP8, JMP13, and JMP14 were introduced into a strain in which Ylt1 was absent (PO1d) or present (E150). For each transformation, 20 to 50 clones on YNBT plates were tested for lipase production with the defective vectors and 5 clones were tested for nondefective vectors. The strains used in this study are listed in Table 1. No difference in the size of the halo on YNBT was observed between strains transformed with nondefective vectors (JMP8 and JMP14) and the untransformed strain (data not shown). For the defective vectors, most (85%) of the transformants had larger halos and the rest (15%) corresponded to cases of URA3 conversion (data not shown).
The copy number of integrated cassettes with the defective vectors was similar for all of the strains used, 10 copies, on average. However, the types of integration differed: mainly dispersed in the zeta-free strain (PO1d) and in tandem in strains containing zeta (E150). The copy number, the type of integration, and the stability of integrated expression cassettes will be presented elsewhere. We further studied lipase regulation and secretion using strains JMY184 and JMY279, which contained the multicopy cassette with the POX2 promoter driving the LIP2 gene, and JMY281 and JMY282, which contained the multicopy cassette with the LIP2 promoter driving LIP2. The four strains presented similar growth kinetics on both glucose and olive oil media (data not shown), but JMY281 and JMY282 secreted 25 to 50% of the amounts of lipase secreted by JMY184 and JMY279. Both the POX2 and LIP2 promoters gave low levels of expression in glucose media and were induced by oleic acid and olive oil (see below and data not shown).
Overproduction of lipase does not affect Lip2p processing and secretion.
Lipase secretion was compared in the wild-type (PO1d) and LIP2A strains (JMY184 and JMY281; lipase gene expressed under control of the POX2 and LIP2 promoters, respectively). Typically, for the PO1d strain, lipase activity was detected at 20 U/ml in YNBH, 50 U/ml in YPDH, and about 100 U/ml in YTDH. In contrast, it was detected at 150 U/ml (200 U/ml) in YNBH, 400 U/ml (500 U/ml) in YPDH, and 1,500 U/ml (2,000 U/ml) in YTDH for JMY281 and JMY184, respectively. The kinetics of extracellular lipase production and protein accumulation in the culture supernatant during the growth of JMY184 in YTDH are presented in Fig. 4. Lipase production started 15 h after transfer into the inducing media when glucose was consumed and increased until 50 h of culture, when it reached a plateau. Activity and protein levels were stable over 40 h. In JMY281 and JMY184, 50- and 100-fold increases in lipase production were obtained, respectively. We investigated the nature of the Lip2p produced by LIP2A strain JMY184 using immunoblotting to analyze the supernatant and extracts of cells induced in YTDH using anti-Lip2p antibodies. Only the 38.5-kDa mature form was detected in both fractions (data not shown). The N-terminal amino acid sequence of the secreted lipase (seven amino acids, Fig. 3B) corresponded to the mature form, indicating that precursor processing was not limiting (see below).
FIG. 4.
Growth and lipase production of strains JMY184 (squares), JMY278 (triangles), and JMY277 (circles). (A) Growth (open symbols) and extracellular lipase production (closed symbols) in YTDH were monitored over time. Supernatant protein analysis during growth was done by SDS-PAGE and Coomassie blue staining for strains JMY184 (LIP2A) (B), JMY278 (LIP2A, xpr6 KO) (C), and JMY277 (lip2) (D). Lanes 1 to 8 correspond to the equivalent of 5 μl of supernatant sampled after 0, 22, 28, 44, 52, 70, 78, and 94 h of culture, respectively. Sizes of prestained molecular mass standards (lane 9) are indicated on the right. The arrows mark the 38.5-kDa position.
The XPR6 gene product is involved in Lip2p processing.
The XPR6 gene has been shown to encode a dibasic processing endoprotease similar to that encoded by the KEX2 gene in S. cerevisiae (14). In Y. lipolytica, Xpr6p is involved in AEP processing. XPR6 is not an essential gene, but its disruption results in reduced growth and a defect in the yeast-to-mycelium switch (smooth colonies) (14). To confirm the role of Xpr6p in Lip2p processing, a xpr6Δ976::LEU2 gene disruption cassette was introduced into strain JMY184 to give strain JMY278 (Fig. 5B). Small and smooth colonies corresponding to disrupted clones were formed by 60% of Leu+ transformants. Correct gene disruption was confirmed by Southern blotting.
The extracellular lipase activity of strain JMY278, which carried xpr6Δ976, was, at most, one-quarter of that of nondisrupted strain JMY184 (Fig. 4A). During growth, extracellular lipase accumulates in JMY184 (Fig. 4B) whereas in JMY278, it increases and then decreases, in parallel with protein levels (Fig. 4C). This decrease may result from the production of a less stable pro protein or degradation by a protease released by the cells.
The N-terminal sequences of the secreted lipases produced in YPDH by strains JMY184 and JMY278 after 52 h of cell growth (Fig. 4B and C, lanes 5) were determined. For JMY278, the amino acid sequence obtained was TPSEAAV. The Thr residue corresponds to aa 23 (Fig. 3B). For JMY184, the sequence was VYTSTET, corresponding to the N-terminal sequence of the mature lipase (aa 34), as determined by Destain (12). The specific activity of the lipases secreted at 52 h by JMY184 (Lip2p) and JMY278 (pro-Lip2p) were compared and found to be similar, indicating that pro-Lip2p was as active as Lip2p (data not shown).
Lip2p is a glycosylated protein.
Many extracellular enzymes produced by yeast are glycosylated. Two potential N glycosylation sites were detected in the mature form (Fig. 1). As a difference was observed between the experimentally determined (SDS-PAGE) molecular mass (38.5-kDa) and the calculated molecular mass (33,384 Da) of Lip2p, we investigated whether N glycosylation was responsible for this difference. Both lipases, Lip2p and pro-Lip2p, were treated with endo H under native and denaturing conditions. The decrease in the apparent molecular masses of both proteins shows that they were glycosylated and contained 10 to 15% sugar (Fig. 6, lanes 2 and 4). Lip2p, pro-Lip2p, and their corresponding deglycosylated forms presented similar specific activities (data not shown), indicating that glycosylation is not required for activity.
A single gene accounts for extracellular lipase activity.
Several lipase genes have been identified in the yeasts C. rugosa and G. candidum. We investigated if there were also several lipase genes in Y. lipolytica by digesting genomic DNA with five restriction enzymes (PstI, HindIII, BamHI, BglII, and ClaI) and probing at low stringency with the LIP2 gene. For each enzyme, a single band was observed (data not shown) indicating that the LIP2 gene was unique in Y. lipolytica. We also constructed strain JMY277, in which the LIP2 gene was deleted (lip2 knockout [KO], Fig. 5A). We compared lipase production in cell supernatant after 40 h of culture for PO1d (wild type) and JMY277 (lip2 KO) in three different media. In YNBD, for PO1d and JMY277, less than 0.5 U/ml was detected. Under inducing conditions, YNBH and YNBO, we detected 19 and 10 U/ml for PO1d and less than 0.5 U/ml for JMY277. Comparison of YNBH values show the loss of at least 97% of the extracellular lipase activity. Even in the rich YTDH medium, no lipase activity or extracellular protein was detected (Fig. 4D). Finally, during LIP2 gene isolation, a single and unique 5′ RACE-PCR fragment was isolated using the YlipSA primer. Thus, the extracellular lipase is encoded by the single LIP2 gene, in contrast to previous reports suggesting the presence of several extracellular lipase-encoding genes.
LIP2 is not essential for growth on TG media.
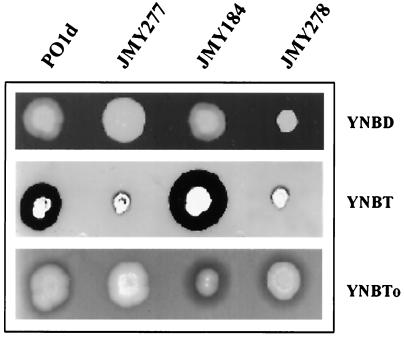
We tested whether LIP2 was essential for growth on TG media by comparing the growth on solid and liquid media of wild-type strain PO1d and modified strains JMY277 (lip2 KO), JMY184 (LIP2A), and JMY278 (xpr6 KO). In each case, we compared growth on YNBD, YNBT, and YNBTo (Fig. 7). No differences in growth were observed on YNBD plates, except for JMY278. Similarly, no growth differences were observed on YNBTo (Fig. 7, bottom) and YNBH (data not shown) plates. This was confirmed by the similar growth of these strains in liquid media (data not shown), demonstrating that LIP2 was not essential for growth on long-chain TGs. Surprisingly, JMY277 and JMY278 grew poorly on short-chain TGs (Fig. 7, middle) but normally in liquid media (data not shown). We have no explanation for this effect.
FIG. 7.
Growth and lipase activity of Y. lipolytica strains on plates. Cells growing exponentially in YNBD at 28°C were diluted, and 105 cells in 10 μl were used to inoculate YNBD, YNBT, and YNBTo plates. Lipase secretion was detected as a halo after 3 days of incubation.
Extracellular lipase activity was detected on YNBH and YNBTo plates and was particularly evident on YNBT plates. Halos were present on YNBTo plates for the wild-type strain and were larger for the amplified strain, JMY184. No halos were observed for JMY277 (Fig. 7, bottom), indicating the absence of any other extracellular lipase activity, which was confirmed by the absence of extracellular protein in this strain. This suggests that the growth of JMY277 on YNBH and YNBTo results from TG metabolism by another pathway, probably involving membrane-bound lipases I and II or the intracellular LIP1 and LIP3 gene products. This demonstrates that LIP2 is not essential for growth under these conditions.
DISCUSSION
Several lipases have been detected in Y. lipolytica, including intracellular, membrane-bound, and extracellular enzymes (for reviews, see references 2 and 21). The sequences of only two genes encoding lipases were determined by Dominguez and coworkers i.e., the LIP1 and LIP3 genes. These lipases are similar to the lipases of the fungi C. cylindracea and G. candidum and belong to the carboxylesterase family. They may be intracellular or membrane bound, because no clear signal sequence was detected (9). In this study, we isolated the LIP2 gene, which codes for the extracellular lipase Lip2p, which belongs to the triacylglycerol hydrolase family.
Lip2p is synthesized as a prepro protein with 13 aa followed by four dipeptides (X-Ala or X-Pro), a short 12-aa pro region ending in a Lys-Arg (KR) site, and finally the mature protein. This is very similar to the structure of the prepro AEP precursor of the AEP encoded by the XPR2 gene (28). This precursor has 13 aa followed by 10 dipeptides (X-Ala or X-Pro), a large 122-aa pro region ending in a KR site, and finally the mature protein. For Lip2p, SignalP predicted cleavage at Ala17, whereas for AEP, SignalP predicted cleavage at Ala15, the observed cleavage site (29). Whatever the location of the signal peptide cleavage site, two to three X-Ala or X-Pro dipeptides follow that are potential substrates for dipeptide aminopeptidase processing.
The two signal sequences had similar hydrophobicity characteristics (10, 34), indicating that Lip2p may be secreted via the signal recognition particle-dependent pathway (20). Lip2p maturation is dependent upon KEX2-like processing. In strain JMY278, which carries the xpr6 KO deletion, the secreted precursor started at Thr23 rather than Ile22, as expected from diaminopeptidase processing. Based on SignalP prediction, signal peptide cleavage after Ile22 is very unlikely. Although Lip2p was overproduced in JMY278, there was no evidence that X-Ala or X-Pro dipeptides were present at the N terminus of the precursor, whereas this was the case for the AEP precursor produced in an xpr6 KO strain. Thus, there is no direct evidence that dipeptidyl aminopeptidase processing occurs and the nature of the processing event removing Ile22 is unclear. Mature Lip2p and its corresponding deglycosylated form have similar specific enzymatic activities. Similarly, pro-Lip2p produced in strain JMY278 and its corresponding deglycosylated form also have specific activities similar to that of the mature form, but these pro proteins were shown to be less stable. Assuming that the pro protein observed in the xpr6 KO corresponds to the pro peptide in vivo, these results indicate that if the pro peptide is not removed: (i) secretion is not greatly affected, as lipase production in JMY278 is only one-quarter of that in JMY184; (ii) there is no evidence that the pro peptide inhibits enzymatic activity, as has been shown for AEP; and (iii) the precursor seems to be less stable, indicating that the pro peptide may destabilize the protein. We do not know whether N glycosylation is involved in the secretion of Lip2p, but it may contribute to the stability of the protein and its resistance to protease, as has been shown for the human gastric lipase (50). Wild-type strains secrete about 30 to 50 U of lipase per ml. Mutant strains producing 25 times more lipase than the wild type were isolated by two-step mutagenesis. These mutants produced 1,200 U/ml under optimized conditions in a 500-liter fermentor (13). We used the POX2 promoter to drive LIP2 gene expression and multicopy integration of this expression cassette and obtained stable transformants that produced 2,000 U/ml, under nonoptimized conditions, corresponding to about 0.5 g of lipase per liter of supernatant. The production of large quantities of protein will facilitate the detailed analysis of the biochemical characteristics of the enzyme and provide material for crystallization and structural analysis. The secreted lipase had an apparent molecular mass of 38.5 kDa, 5.2 kDa more than the expected size. This suggests that the two potential glycosylation sites (Asn113 and Asn134) are N glycosylated with core oligosaccharide.
Our PCR and DNA hybridization results show that unlike some yeasts, including C. rugosa (4, 25) and G. candidum (5), in which several highly homologous genes are present, Y. lipolytica probably contains a single gene encoding extracellular lipase. This is supported by the low residual extracellular lipase activity (less than 0.5 U/ml, the lower limit of detection of our assay) in strain JMY277, in which LIP2 was deleted. We cannot totally exclude the possibility that there is another extracellular lipase encoding a gene with a low level of sequence similarity that would not be detected by hybridization. If such a gene is present, it is only weakly expressed under the growth conditions tested.
A surprising result was that lip2 KO strain JMY277 grew with the same growth rate and cell yield as a LIP2 strain on TG media. This suggests either that a very low level of activity (0.5 U/ml) is sufficient to sustain normal growth or that TGs may enter the cells directly via an unknown process and be hydrolyzed by intracellular and/or cell-bound lipases. Preliminary results of partial sequencing indicate that a third carboxylesterase and two short sequences containing the motif GHSLGG/AA characteristic of the triacylglycerol lipase family are present in Y. lipolytica. Further analyses of these genes are in progress to determine the localization of the gene product and their function in the TG metabolism of this yeast.
ACKNOWLEDGMENTS
We thank J. Knight for editing the English version of the text and J.-C. Huet (Unité de Recherche de Biochimie et Structure des Protéines, INRA Jouy-en-Josas) for protein sequencing. We acknowledge E. Adamowicz for providing purified Lip2p. We are grateful to J. Angignard and S. Decollogne for anti-Lip2p antibody production.
This work was supported by the Institut National de la Recherche Agronomique, by the Centre National de la Recherche Scientifique, and by Mayoly-Spindler SA.
G.P. and H.W. contributed equally to this work.
REFERENCES
- 1.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 2.Barth G, Gaillardin C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev. 1997;19:219–237. doi: 10.1111/j.1574-6976.1997.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 3.Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf W K, editor. Nonconventional yeasts in biotechnology. Vol. 1. Berlin, Germany: Springer-Verlag; 1996. pp. 313–388. [Google Scholar]
- 4.Benjamin S, Pandey A. Candida rugosa lipases: molecular biology and versatility in biotechnology. Yeast. 1998;14:1069–1087. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1069::AID-YEA303>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 5.Bertolini M C, Laramee L, Thomas D Y, Cygler M, Schrag J D, Vernet T. Polymorphism in the lipase genes of Geotrichum candidum strains. Eur J Biochem. 1994;219:119–125. doi: 10.1111/j.1432-1033.1994.tb19921.x. [DOI] [PubMed] [Google Scholar]
- 6.Boel E, Huge-Jensen B, Christensen M, Thim L, Fiil N P. Rhizomucor miehei triglyceride lipase is synthesized as a precursor. Lipids. 1988;23:701–706. doi: 10.1007/BF02535672. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Casaregola S, Feynerol C, Diez M, Fournier P, Gaillardin C. Genomic organization of the yeast Yarrowia lipolytica. Chromosoma. 1997;106:380–390. doi: 10.1007/s004120050259. [DOI] [PubMed] [Google Scholar]
- 9.Choupina A, Gonzalez F, Morin M, Burguillo F, Ferminan E, Dominguez A. The lipase system of Yarrowia lipolytica. Curr Genet. 1999;35:297. [Google Scholar]
- 10.Cornette J L, Cease K B, Margalit H, Spouge J L, Berzofsky J A, DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987;195:659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- 11.Davidow L S, O'Donnell M M, Kaczmarek F S, Pereira D A, DeZeeuw J R, Franke A E. Cloning and sequencing of the alkaline extracellular protease gene of Yarrowia lipolytica. J Bacteriol. 1987;169:4621–4629. doi: 10.1128/jb.169.10.4621-4629.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Destain J. Ph.D. thesis. Gembloux, Belgium: Faculté Universitaire des Sciences Agronomiques de Gembloux; 1998. [Google Scholar]
- 13.Destain J, Roblain D, Thonart P. Improvement of lipase production from Yarrowia lipolytica. Biotechnol Lett. 1997;19:105–107. [Google Scholar]
- 14.Enderlin C S, Ogrydziak D M. Cloning, nucleotide sequence and functions of XPR6, which codes for a dibasic processing endoprotease from the yeast Yarrowia lipolytica. Yeast. 1994;10:67–79. doi: 10.1002/yea.320100107. [DOI] [PubMed] [Google Scholar]
- 15.Fabre E, Nicaud J M, Lopez M C, Gaillardin C. Role of the proregion in the production and secretion of the Yarrowia lipolytica alkaline extracellular protease. J Biol Chem. 1991;266:3782–3790. [PubMed] [Google Scholar]
- 16.Fournier P, Guyaneux L, Chasles M, Gaillardin C. Scarcity of ars sequences isolated in a morphogenesis mutant of the yeast Yarrowia lipolytica. Yeast. 1991;7:25–36. doi: 10.1002/yea.320070104. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez F. Ph.D. thesis. Salamanca, Spain: Universidad de Salamanca; 1996. [Google Scholar]
- 18.Haas M J, Allen J, Berka T R. Cloning, expression and characterization of a cDNA encoding a lipase from Rhizopus delemar. Gene. 1991;109:107–113. doi: 10.1016/0378-1119(91)90594-2. [DOI] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 20.He F, Beckerich J M, Gaillardin C. A mutant of 7SL RNA in Yarrowia lipolytica affecting the synthesis of a secreted protein. J Biol Chem. 1992;267:1932–1937. [PubMed] [Google Scholar]
- 21.Heslot H. Genetics and genetic engineering of the industrial yeast Yarrowia lipolytica. Vol. 43. Germany: Springer-Verlag Berlin; 1990. [Google Scholar]
- 22.Kuno H, Ota Y. The new method for the purification of extracellular lipases from Yarrowia (Saccharomycopsis) lipolytica and some properties of lipase A. J Fac Appl Biol Sci. 1996;35:191–197. [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Le Dall M T, Nicaud J M, Gaillardin C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr Genet. 1994;26:38–44. doi: 10.1007/BF00326302. [DOI] [PubMed] [Google Scholar]
- 25.Lotti M, Tramontano A, Longhi S, Fusetti F, Brocca S, Pizzi E, Alberghina L. Variability within the Candida rugosa lipases family. Protein Eng. 1994;7:531–535. doi: 10.1093/protein/7.4.531. [DOI] [PubMed] [Google Scholar]
- 26.Maftahi M, Gaillardin C, Nicaud J M. Sticky-end polymerase chain reaction method for systematic gene disruption in Saccharomyces cerevisiae. Yeast. 1996;12:859–868. doi: 10.1002/(SICI)1097-0061(199607)12:9%3C859::AID-YEA978%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Matoba S, Fukayama J, Wing R A, Ogrydziak D M. Intracellular precursors and secretion of alkaline extracellular protease of Yarrowia lipolytica. Mol Cell Biol. 1988;8:4904–4916. doi: 10.1128/mcb.8.11.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matoba S, Morano K A, Klionsky D J, Kim K, Ogrydziak D M. Dipeptidyl aminopeptidase processing and biosynthesis of alkaline extracellular protease from Yarrowia lipolytica. Microbiology. 1997;143:3263–3272. doi: 10.1099/00221287-143-10-3263. [DOI] [PubMed] [Google Scholar]
- 29.Matoba S, Ogrydziak D M. A novel location for dipeptidyl aminopeptidase processing sites in the alkaline extracellular protease of Yarrowia lipolytica. J Biol Chem. 1989;264:6037–6043. [PubMed] [Google Scholar]
- 30.Mileto D, Brocca S, Lotti M, Takagi M, Alquati C, Alberghina L. Characterization of the Candida rugosa lipase system and overexpression of the lip1 isoenzyme in a non-conventional yeast. Chem Phys Lipids. 1998;93:47–55. doi: 10.1016/s0009-3084(98)00028-0. [DOI] [PubMed] [Google Scholar]
- 31.Nagao T, Shimada Y, Sugihara A, Tominaga Y. Cloning and nucleotide sequence of cDNA encoding a lipase from Fusarium heterosporum. J Biochem (Tokyo) 1994;116:536–540. doi: 10.1093/oxfordjournals.jbchem.a124558. [DOI] [PubMed] [Google Scholar]
- 32.Nga B, Gaillardin C, Fournier P, Heslot H. Genetic analysis of lipase low-producing mutants of Yarrowia lipolytica. J Gen Microbiol. 1989;135:2439–2443. [Google Scholar]
- 33.Nicaud J M, Fabre E, Beckerich J M, Fournier P, Gaillardin C. Cloning, sequencing and amplification of the alkaline extracellular protease gene of Yarrowia lipolytica. J Biotechnol. 1989;12:285–298. [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Novotny C, Dolezalova L, Lieblova J. Dimorphic growth and lipase production in lipolytic yeasts. Folia Microbiol. 1994;39:71–73. doi: 10.1007/BF02814534. [DOI] [PubMed] [Google Scholar]
- 36.Novotny C, Dolezalova L, Musil P, Novak M. The production of lipases by some Candida and Yarrowia yeasts. J Basic Microbiol. 1988;4:221–227. [Google Scholar]
- 37.Ogrydziak D M. Yeast extracellular proteases. Crit Rev Biotechnol. 1993;13:1–55. doi: 10.3109/07388559309069197. [DOI] [PubMed] [Google Scholar]
- 38.Ogrydziak D M, Scharf S J. Alkaline extracellular protease produced by Saccharomycopsis lipolytica CX161-1B. J Gen Microbiol. 1982;128:1225–1234. doi: 10.1099/00221287-128-6-1225. [DOI] [PubMed] [Google Scholar]
- 39.Ota Y, Gomi K, Kato S, Sugiura T, Minoda Y. Purification and some properties of cell-bound lipase from Saccharomycopsis lipolytica. Agric Biol Chem. 1982;46:2885–2893. [Google Scholar]
- 40.Ota Y, Oikawa S, Morimoto Y, Minoda Y. Nutritional factors causing mycelial development of Saccharomycopsis lipolytica. Agric Biol Chem. 1984;48:1933–1940. [Google Scholar]
- 41.Peters I I, Nelson F E. Factors influencing the production of lipase by Mycotorula lipolytica. J Bacteriol. 1948;55:581–591. doi: 10.1128/jb.55.5.581-591.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters I I, Nelson F E. Preliminary characterization of lipase of Mycotorula lipolytica. J Bacteriol. 1948;55:593–600. doi: 10.1128/jb.55.5.593-600.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Savitsky K, Ziv Y, Bar-Shira A, Gilad S, Tagle D A, Smith S, Uziel T, Sfez S, Nahmias J, Sartiel A, Eddy R L, Shows T B, Collins F S, Shiloh Y, Rotman G. A human gene (DDX10) encoding a putative DEAD-box RNA helicase at 11q22-q23. Genomics. 1996;33:199–206. doi: 10.1006/geno.1996.0184. [DOI] [PubMed] [Google Scholar]
- 46.Schmid-Berger N, Schmid B, Barth G. Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J Bacteriol. 1994;176:2477–2482. doi: 10.1128/jb.176.9.2477-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiura T, Ota Y, Minoda Y. Partial characterisation of cell-bound lipase of Candida paralipolytica. Agric Biol Chem. 1976;40:2479–2480. [Google Scholar]
- 48.Ventura Pereira-Meirelles F, Miguez Rocha-Leao M H, Lippel Sant'Anna G. A stable lipase from Candida lipolytica: cultivation conditions and crude enzyme characteristics. Appl Biochem Biotechnol. 1997;63–65:73–85. doi: 10.1007/BF02920414. [DOI] [PubMed] [Google Scholar]
- 49.Wang H J, Le Dall M-T, Wache Y, Laroche C, Belin J-M, Gaillardin C, Nicaud J-M. Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol. 1999;181:5140–5148. doi: 10.1128/jb.181.17.5140-5148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wicker-Planquart C, Canaan S, Riviere M, Dupuis L. Site-directed removal of N-glycosylation sites in human gastric lipase. Eur J Biochem. 1999;262:644–651. doi: 10.1046/j.1432-1327.1999.00427.x. [DOI] [PubMed] [Google Scholar]
- 51.Xuan J-W, Fournier P, Declerck N, Chasles M, Gaillardin C. Overlapping reading frames at the LYS5 locus in the yeast Yarrowia lipolytica. Mol Cell Biol. 1990;10:4795–4806. doi: 10.1128/mcb.10.9.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi S, Mase T, Takeuchi K. Cloning and structure of the mono- and diacylglycerol lipase-encoding gene from Penicillium camembertii U-150. Gene. 1991;103:61–67. doi: 10.1016/0378-1119(91)90391-n. [DOI] [PubMed] [Google Scholar]