Abstract
The luminescence (lux) operon (luxICDABEG) of the symbiotic bacterium Vibrio fischeri is regulated by the transcriptional activator LuxR and two acyl-homoserine lactone (acyl-HSL) autoinducers (the luxI-dependent 3-oxo-hexanoyl-HSL [3-oxo-C6-HSL] and the ainS-dependent octanoyl-HSL [C8-HSL]) in a population density-responsive manner called quorum sensing. To identify quorum-sensing-regulated (QSR) proteins different from those encoded by lux genes, we examined the protein patterns of V. fischeri quorum-sensing mutants defective in luxI, ainS, and luxR by two-dimensional polyacrylamide gel electrophoresis. Five non-Lux QSR proteins, QsrP, RibB, AcfA, QsrV, and QSR 7, were identified; their production occurred preferentially at high population density, required both LuxR and 3-oxo-C6-HSL, and was inhibited by C8-HSL at low population density. The genes encoding two of the QSR proteins were characterized: qsrP directs cells to synthesize an apparently novel periplasmic protein, and ribB is a homolog of the Escherichia coli gene for 3,4-dihydroxy-2-butanone 4-phosphate synthase, a key enzyme for riboflavin synthesis. The qsrP and ribB promoter regions each contained a sequence similar to the lux operon lux box, a 20-bp region of dyad symmetry necessary for LuxR/3-oxo-C6-HSL-dependent activation of lux operon transcription. V. fischeri qsrP and ribB mutants exhibited no distinct phenotype in culture. However, a qsrP mutant, in competition with its parent strain, was less successful in colonizing Euprymna scolopes, the symbiotic host of V. fischeri. The newly identified QSR genes, together with the lux operon, define a LuxR/acyl-HSL-responsive quorum-sensing regulon in V. fischeri.
Quorum sensing is a regulatory mechanism by which gram-negative bacteria control gene expression in response to population density (13, 31). Quorum sensing involves acyl-homoserine lactone (acyl-HSL) signal molecules, produced by members of the LuxI family of acyl-HSL synthases, and proteins of the LuxR family of transcriptional activators, which mediate the response to local concentrations of acyl-HSLs (37, 40). The first acyl-HSL, 3-oxo-hexanoyl-HSL (3-oxo-C6-HSL) and the luxI and luxR genes were first identified in the marine bioluminescent bacterium Vibrio fischeri (20, 23). At present, LuxI/LuxR quorum-sensing systems have been identified in over 25 species of gram-negative bacteria from diverse habitats, including both marine and terrestrial bacteria and several pathogens of plants and animals (13, 37, 65).
Quorum sensing controls various different activities in these different bacteria, including luminescence, the production of extracellular enzymes, plasmid transfer, antibiotic synthesis, and biofilm formation (13, 31, 65). In some species, quorum sensing coordinates the expression of several unlinked genetic loci. For example, in the opportunistic human pathogen Pseudomonas aeruginosa, two LuxI/LuxR homolog pairs, LasI/LasR and RhlI/RhlR, regulate more than 40 genes via a complex quorum-sensing network (39, 52, 68). The coordinated production of multiple proteins of diverse function suggests that quorum sensing is an adaptational response to conditions of high population density, such as those encountered in association with plant and animal hosts (13, 52, 65).
In V. fischeri, quorum sensing controls luminescence; LuxR and 3-oxo-C6-HSL direct a population density-responsive transcriptional activation of the lux operon (luxICDABEG; genes for 3-oxo-C6-HSL synthase and light production) (reviewed in reference 14). Luminescence, which plays a defining role in the symbiosis formed by this bacterium with certain squids and fishes (16, 56), is dependent also on 3′:5′-cyclic AMP and GroESL, which influence the production and activity of LuxR, respectively. Furthermore, various physiological factors, including oxygen, carbon source, and iron, control luminescence, apparently by influencing the quorum-sensing mechanism (reviewed in reference 13). The complexity of control over lux operon expression and the apparent integration of quorum sensing with cellular physiology suggest that luminescence is part of a coordinated adaptational response. Except for luminescence, however, the nature of the quorum-sensing response in V. fischeri, its extent and functional significance, is presently unknown.
To begin gaining insight into these issues, we sought to identify LuxI/LuxR quorum-sensing-regulated (QSR) genes “downstream” of the lux operon. To do so, we used two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) and quorum-sensing mutants of V. fischeri defective in production of acyl-HSLs and LuxR. In this study, we report the identification of several non-Lux QSR proteins and their genes. The newly identified QSR genes, together with the lux operon, define a LuxR/acyl-HSL-responsive quorum-sensing regulon in V. fischeri. At least one member of the regulon, qsrP, apparently is involved in the ability of V. fischeri to colonize its sepiolid squid host, Euprymna scolopes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli SM10-λpir (provided by C. Gardel and J. Mekalanos) and Novablue were grown on Luria-Bertani (LB) medium (58) at 37°C with antibiotics as appropriate (ampicillin, 100 μg ml−1; chloramphenicol, 30 μg ml−1). V. fischeri MJR1, a spontaneous rifampin-resistant derivative of MJ-1 (57), was isolated by plating a saturated culture of MJ-1 on LBS agar (12) containing 100 μg of rifampin per ml. Strains of V. fischeri were maintained on LBS agar with the appropriate antibiotics (chloramphenicol, 3 μg ml−1; naladixic acid, 20 μg ml−1; neomycin, 200 μg ml−1; and rifampin, 100 μg ml−1). For protein isolation, cells were grown with aeration in 3-ml cultures of liquid LBS without antibiotics at 27°C to an absorbance of 660 nm (A660) of 0.4 (mid-exponential phase), 0.8 (late exponential phase), or 1.2 (early stationary phase). Cells from 1-ml volumes were pelleted at 4°C, washed twice with an equal volume of ice-cold artificial seawater (17), and stored frozen as a pellet at −75°C until use.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| Novablue | endA hsdR17(rK− mK+) supE thi recA gyrA relA lac [F′ proA+B+lacIqZΔM15::Tn10 (Tcr)] | Novagen |
| SM10-λpir | thi thr leu supE tonA lacY recA::RP4-2-Tc::Mu Kmr λ::pir | 50 |
| V. fischeri | ||
| ES114 | Wild type | 4 |
| ESR1 | ES114, Rfr | 34 |
| MJ-1 | Wild type | 57 |
| MJR1 | MJ-1, Rfr | This study |
| MJ-100 | MJ-1, Nxr | 18 |
| MJ-208 | MJ-100, ΔluxR | 42 |
| MJ-211 | MJ-100, ΔluxI (nonpolar) | 42 |
| MJ-215 | MJ-211, ainS | 42 |
| MJ-216 | MJ-100, ainS::neo | 33 |
| MJ-10X | MJR1, qsrP::p300-10XM | This study |
| ES-10X | ESR1, qsrP::p300-10XE | This study |
| MJ-ribBX | MJR1, ribB::p300-12XM | This study |
| Plasmids | ||
| pT7Blue | Cloning vector; Apr | Novagen |
| pGP704 | R6K ori RP4 mob Apr | 50 |
| pSMC300 | pGP704; Δbla Cmr | This study |
| p300-10XE | pSMC300; qsrP nt 119–269 from ESR1 | This study |
| p300-10XM | pSMC300; qsrP nt 119–269 from MJ-100 | This study |
| p300-12XM | pSMC300; ribB nt 194–450 from MJ-100 | This study |
nt, nucleotides.
Culture density was determined by measuring the A660. Under the conditions used, an A660 unit corresponded to approximately 109 cells ml−1. Synthetic autoinducers were added to culture tubes as solutions in chloroform to yield a final concentration of 100 nM. The chloroform was removed by evaporation with a stream of sterile air prior to addition of the medium and bacteria. Strains MJ-10X and ES-10X were tested for their ability to utilize various carbon and energy sources in VFM, a previously described minimal salts medium (15), containing glucose, glycerol, ribose, mannose, or N-acetylglucosamine (20 mM).
Protein isolation for 2-D PAGE and cell fractionation.
All preparative procedures were conducted at 4°C or on ice. Frozen cell pellets were resuspended in 80 μl of sample buffer 1 (SB1; 40 mM Tris-HCl [pH 8.0], 200 mM dithiothreitol, 0.3% sodium dodecyl sulfate) and boiled for 2 min. After cooling, 8 μl of sample buffer 2 (0.5 M Tris-HCl [pH 8.0], 50 mM MgCl2, 1 mg of DNase I per ml, 0.25 mg of RNase A per ml) was added to the mixture, which was incubated for 30 min. Proteins were precipitated by the addition of acetone to 80% (vol/vol), collected by centrifugation, and resuspended in 80 μl of SB1. After a second acetone precipitation, the pellet was air-dried for 30 min, resuspended in 60 μl of SB1 and 240 μl of sample buffer 3 (9.9 M urea, 100 mM dithiothrietol, 4% Triton X-100, 2.2% polyampholytes [40% [wt/vol], pH 3 to 10; Genomic Sciences, Inc.]), and stored at −75°C until use. For cell fractionation, cells were separated into outer membrane, inner membrane, and cytoplasmic/periplasmic (i.e., soluble) fractions as described for Vibrio cholerae (50). Cells were treated with chloroform to isolate periplasmic proteins (1).
2-D PAGE.
Proteins were separated essentially as described by O'Farrell (51) using the 2-D Investigator system from Millipore and reagents from Genomic Sciences, Inc. First-dimension gels contained 4.1% acrylamide, 9.5 M urea, 5 mM CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 2% Triton X-100, and 5.8% polyampholytes. A 10-μl amount of sample was loaded at the basic end of the gel for analytical gels (50 μl for preparative gels) and electrophoresed for 18,000 volt-hours. After completion of the run, gels were incubated in equilibration buffer (0.375 M Tris-HCl [pH 9.2], 50 mM dithiothreitol, 0.01% bromophenol blue) for 2 min or stored frozen in 10% glycerol at −75°C.
Second-dimension separation was performed with continuous gels of 12.5% acrylamide (Duracryl) at 4°C. Analytical gels were stained with silver as described previously (52) and stored in 10% glycerol. Preparative gels were electroblotted to a polyvinylidene difluoride membrane (Millipore) and stained with Coomassie brilliant blue. Individual protein spots were excised for microsequence analysis.
Peptide sequencing.
For amino-terminal sequencing, conducted at the Massachusetts Institute of Technology Biopolymers Laboratory, excised proteins were subjected to automated Edman degradation on an Applied Biosystems model 477A protein sequencer with an on-line model 120 phenylthiohydantoin amino acid analyzer. For internal sequencing, conducted at the Harvard Microchemistry Facility, protein bound to the polyvinylidene difluoride membrane was digested in situ with endoproteinase LysC (Wako) (24). The resulting peptide mixture was separated by microbore high-performance liquid chromatography (HPLC) using a Zorbax C18 reverse-phase column (1.0 by 150 mm) on a Hewlett-Packard 1090 HPLC/1040 diode array detector. Optimum fractions from the chromatogram were chosen based on differential UV absorption at 205, 277, and 292 nm, peak symmetry, and resolution. Peaks were further screened for length and homogeneity by matrix-assisted laser desorption time-of-flight mass spectrometry on a Finnigan Lasermat 2000, and selected fractions were subjected to automated Edman degradation. Details of strategies for the selection of peptide fractions and their microsequencing have been described previously (44). The internal peptide sequences were: 10-PK12, NVPQIGDTYK; 10-PK39, DRVSIPPIYVELSDNRFSGLHVSGMK; 10-PK51, YGNLFFELIK; 12-PK32, KGVTTGVSATDR; and 12-PK78, LAELTPAGVLCEVTN.
Isolation of qsrP and ribB, the genes encoding QSR 10 and QSR 12.
QSR 10 was extracted from gels and subjected to internal proteolytic digestion. Three of the resulting peptides, 10-PK12, 10-PK39, and 10-PK51, were separated by reversed-phase HPLC and sequenced. Primers corresponding to regions of peptides 10-PK12 and 10-PK39 (10-12F, 10-12R, 10-39F, and 10-39R) were combined as appropriate in two separate PCRs using MJ-100 chromosomal DNA as the template. From the reaction with 10-12F and 10-39R as the primers, a single product of 209 bp was obtained. Attempts to use the 209-bp portion as a probe to isolate the complete gene for QSR 10 from a plasmid-borne genomic library of MJ-100 DNA (15) were unsuccessful, possibly because of underrepresentation of this DNA region in the library. As an alternative, we used PCR to amplify the DNA flanking the 209-bp region and subsequently PCR amplified the intact gene using primers designed from the flanking DNA sequence. To facilitate PCR amplification of the DNA flanking the 209-bp fragment, MJ-100 chromosomal DNA was digested to completion with CfoI, the 4-bp recognition sequence of which is not present in the fragment. The chromosomal fragments were then circularized by ligation, and primers 10-210R and 10-210F were used to amplify the regions flanking the 209-bp fragment, which were joined at the CfoI sites, making them contiguous on the circular molecule. The resulting 760-bp fragment was cloned and sequenced. From the sequences adjacent to the CfoI site, primers 10-826F and 10-826R were designed and used to PCR-amplify the intact qsrP gene.
To isolate the ribB gene, QSR 12 was digested, and two internal fragments were sequenced. Primers corresponding to regions of peptides 12-PK32 and 12-PK78 (12-32F, 12-32R, 12-78F, and 12-78R) were used to amplify a portion of the genome that codes for the protein. From the reaction using 12-32F and 12-78R as the primers, a single product of 215 nucleotides was obtained. Sequencing of the resultant 215-nucleotide fragment confirmed that it was part of the coding region for QSR 12. The intact gene was then isolated from a plasmid-borne library of MJ-1 chromosomal DNA in E. coli (15), using PCR analysis as a screen. Plasmids from 2,000 colonies representing the library were initially pooled in groups of 50, and DNA from these 40 groups served as the template for individual PCRs using primers 12-32F and 12-78R. One of the reactions generated a 215-bp fragment. Each plasmid from the pool of 50 for that reaction was then screened individually to identify a single positive clone, which contained approximately 7.5 kb of chromosomal insert DNA. The sequence of the region containing the QSR 12 open reading frame was obtained using a primer walk strategy.
PCR amplification, cloning, and sequencing of DNA.
PCR amplifications were performed in an Idaho Technology Rapidcycler using 50-μl glass capillary tubes. The following oligonucleotides were used as primers (* denotes degenerate primers; Y is T or C; R is A or G; H is A, T, or C; D is A, T, or G; and N is A, T, C, or G): 10-12F*, AAYGTNCCNCARATHGGNGAYACRTA; 10-12R*, TAYGTRTCNCCDATYTGNGGNACRTT; 10-39F*, ATHTAYGTNGARCTNAGYGAYAAYCG; 10-39R*, CGRTTRTCRCTNAGYTCNACRTADAT; 12-32F*, ACIACIGGDGTWAGYGCWACIGAYAG; 12-32R*, CTRTCIGTWGCTCTWACHCCIGTIGT; 12-78F*, GCWGGDGTWTTDTGYGARGTWACIAA; 12-78R*, TTIGTWACYTCRCAHAAWACHCCWGC; 10-210F, CTGTAGAGGCTTGCTCTATTGGATCT; 10-210R, TCAGAAGAAGC-TCTCTACGACCCTTG; 10-826F, ATCAAGAAACCTCGACGAGATAAACG; 10-826R, GTCCTAAAGAGGAAATGCTAAGTGGT; R6KR, GGCTTTTAAAGCTTTTAAGGTTTAACGG; SP12-101R, CGACAGTCCCTTCTGTATGTCCTC; and P12S-2, CACTTATTATCGAAACATCTATCATTA. Standard conditions were used except for the initial reactions with degenerate primers, which used 4 mM MgCl2 in the buffer and 30 cycles, with denaturation at 94°C for 2 s, annealing at 40°C for 10 s, and elongation at 50°C for 10 s. PCR products were purified from agarose gels with a QIAquick gel extraction kit (Qiagen) and cloned into pT7-Blue using the Perfectly Blunt cloning kit (Novagen).
DNA sequencing, conducted at the CRC DNA Sequencing Facility, University of Chicago, used the dideoxy nucleotide chain termination method of Sanger et al. (58) with dye terminator labeling.
Construction of qsrP and ribB mutants.
To construct a suicide plasmid, pSMC300, effective in V. fischeri, which is naturally resistant to ampicillin, the ampicillin resistance locus of pGP704 (50) was replaced by a gene encoding resistance to chloramphenicol. pGP704, which contains the R6K origin of replication and the mob region of pRP4, was digested with PstI, and the 1.1-kb cat gene, supplied as a control insert in the pCR-Script cloning kit (Stratagene), was blunt-end cloned in place of the 5′ end of the gene for β-lactamase to form pSMC300. This vector can be conjugated from E. coli SM10-λpir to V. fischeri, but it cannot replicate independently; it therefore confers resistance to chloramphenicol only if it has recombined into the chromosome. To promote recombination between the vector and qsrP, a 150-bp PCR fragment corresponding to nucleotides 119 to 269 of the coding regions from MJ-100 and ESR1 was cloned into the XbaI and SacI sites of pSMC300 to yield p300-10XM and p300-10XE, respectively. For ribB, a 257-bp PCR fragment that corresponds to nucleotides 194 to 450 of the coding region was cloned into the XbaI and SphI sites of pSMC-300 to yield p300-12XM. Conjugation of the vectors from SM10-λpir to V. fischeri strains MJR1 and ESR1 was performed as described previously (18). Transconjugants were selected for their ability to grow on LBS containing rifampin and chloramphenicol.
Disruption of the qsrP and ribB genes was confirmed by PCR analysis. For the qsrP mutants MJ-10X and ES-10X, primers R6KR, which spans the HindIII site of the R6K origin and reads towards the multiple cloning site of pSMC300, and 10-826R were used to PCR-amplify MJ-10X and ES-10X chromosomal DNA. Both reactions yielded the 700-bp product anticipated if the plasmid disrupted the qsrP coding region. PCR amplification with primers 10-826F and 10-826R, which bracket the qsrP coding region, yielded no product with MJ-10X and ES-10X DNA as the template. For the ribB mutant MJ-ribBX, primers R6KR and SP12-101R, the latter of which corresponds to DNA sequence 26 nucleotides downstream of the sequence cloned into the suicide plasmid, was used to PCR-amplify MJ-ribBX DNA. The reaction yielded the 420-bp fragment anticipated if the vector disrupted the ribB gene. PCR of the mutant chromosomal DNA using primers SP12-101R and P12S-2 yielded no product.
Colonization of E. scolopes.
Colonization of the light organs of juvenile E. scolopes was performed in general according to previously published procedures (49). Adult animals were maintained and mated and egg clutches were handled as previously described (6). Within 24 h of hatching, juvenile aposymbiotic squids were placed individually in 10 ml of filter-sterilized artificial seawater in 30-ml glass scintillation vials, and bacterial cells grown to mid-exponential phase in VFM medium with 20 mM ribose as the sole carbon source were added to a final concentration of 104 cells ml−1 per strain. After a 3-h exposure to the bacteria and at 12-h intervals, the squid were transferred to vials containing fresh filter-sterilized artificial seawater. Luminescence was used as an indicator of development of the symbiosis (49). At 48 h postinoculation, the animals were rinsed in filter-sterilized artificial seawater and homogenized to release the bacteria within the light organs. Dilutions of the homogenate were spread-plated on LBS agar containing rifampin (LBSrif). For platings from each animal, approximately 200 colonies were transferred from LBSrif to LBSrif containing chloramphenicol to distinguish between ESR1 (chloramphenicol sensitive) and ES-10X (chloramphenicol resistant). A similar plating procedure was used for the in vitro growth competition experiment, with 50 colonies screened; the ratio of the mutant to the parent strain was determined from cultures grown to an A660 of 0.6.
Nucleotide sequence accession numbers.
The nucleotide sequences of the qsrP and ribB genes from MJ-100 and qsrP from ESR1 have been deposited in GenBank under accession numbers AF233626, AF233627, and AF233628, respectively. Preliminary sequence data for V. cholerae RibB were obtained from The Institute for Genomic Research website at http://www.tigr.org.
RESULTS
Identification of non-Lux QSR proteins.
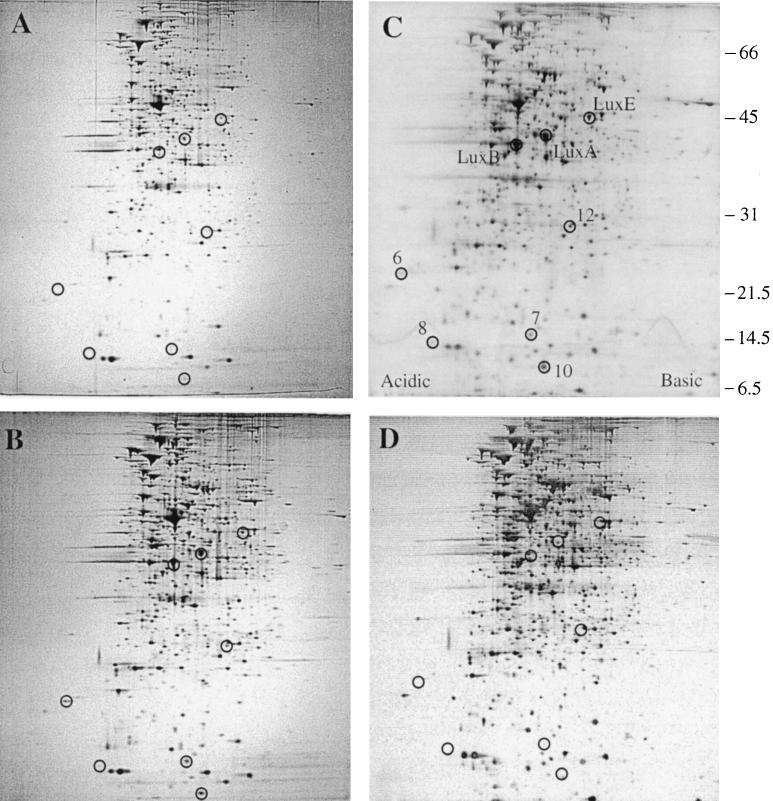
To identify V. fischeri QSR proteins distinct from those encoded by the lux operon, we used 2-D PAGE and quorum-sensing mutants of V. fischeri MJ-100. MJ-100, derived from the wild-type fish-symbiotic strain MJ-1, was chosen for this work because, like MJ-1, it strongly expresses luminescence and produces a high level of acyl-HSL in laboratory culture. In contrast, the luminescence of the squid-symbiotic strain ES114 is weak in laboratory culture, due apparently to underproduction of 3-oxo-C6-HSL (4, 17, 35). We first analyzed the cellular proteins of MJ-100 to determine what proteins V. fischeri might produce in a population density-dependent manner. MJ-100 was grown to mid-exponential phase (A660 = 0.4), at which point luminescence is uninduced or just beginning to induce, to late exponential phase (A660 = 0.8), at which point luminescence induction is strongly under way, and to early stationary phase (A660 = 1.2), at which point MJ-100 is fully induced for luminescence. Visual comparison of gels revealed eight proteins not apparent or weakly detectable at mid-exponential phase that increased in their abundance during growth of MJ-100 through late exponential phase to early stationary phase (Fig. 1A, B, and C).
FIG. 1.
Production of QSR proteins in V. fischeri. Whole-cell proteins were analyzed by 2-D PAGE (Materials and Methods) for (A) MJ-100 (parent strain) at mid-exponential phase (A600 = 0.4), (B) MJ-100 at late exponential phase (A660 = 0.8), (C) MJ-100 at early stationary phase (A660 = 1.2), and (D) MJ-215 (ΔluxI ainS) at early stationary phase (A660 = 1.2). (C) The positions of LuxA, LuxB, LuxE, and the newly identified QSR proteins QSR 6 (AcfA), QSR 7 (unidentified), QSR 8 (QsrV), QSR 10 (QsrP), and QSR 12 (RibB) are circled and designated. The positions of molecular size standards are indicated at the right (in kilodaltons).
To determine whether these eight proteins were under an acyl-HSL-type of quorum-sensing control, we then compared the proteins produced by MJ-100 with those produced by MJ-215 at early stationary phase. MJ-215 is defective in the two V. fischeri autoinducer synthase genes luxI and ainS and therefore makes neither 3-oxo-C6-HSL nor C8-HSL (33, 38, 42, 60). MJ-215 failed to produce detectable amounts of seven of the proteins and produced the eighth to a lesser extent than MJ-100 (Fig. 1D). These results demonstrate that V. fischeri produces at least eight proteins in a population density-dependent manner and that the production of these proteins is apparently subject to acyl-HSL-mediated control. We therefore designated these proteins QSR proteins.
To determine whether any of the eight QSR proteins were different from those encoded by the lux operon, we examined their amino-terminal sequences. We extracted the eight proteins from gels (Materials and Methods), had their amino termini sequenced, and then compared those sequences with the published amino-terminal sequences of V. fischeri Lux proteins (2). Three of the eight proteins were found to be LuxA, LuxB, and LuxE, whereas four of the other proteins (QSR 6, QSR 8, QSR 10, and QSR 12) were distinct from Lux proteins (Table 2). The amino terminus of the fifth protein (QSR 7) apparently was blocked, precluding acquisition of its amino-terminal sequence. However, the estimated size of QSR 7, about 15 kDa, was smaller than that of any of the Lux proteins, the smallest of which, LuxI, is approximately 25 kDa (23). These results demonstrate that V. fischeri produces five QSR proteins distinct from those encoded by the lux operon. Cell fractionation experiments (Materials and Methods) localized these proteins to the outer membrane (QSR 7), the periplasm (QSR 10), and the cytoplasm (QSR 12). QSR 6 was solubilized from the envelope fraction by N-lauroylsarcosine (Materials and Methods), consistent with its being associated with either the periphery of the outer membrane or the cytoplasmic membrane. QSR 8, though readily observed in whole-cell extracts on 2-D PAGE, was not detected in any of the cell fractions.
TABLE 2.
N-terminal peptide sequences of V. fischeri QSR proteins
| Peptide or oligonucleotide | Sequencea | Approx. size (kDa) |
|---|---|---|
| LuxA | MKFGN | 40 |
| LuxB | –KFGL | 37 |
| LuxE | ––V–TEY | 44 |
| QSR 6 (AcfA) | [A]PRVGL | 22 |
| QSR 7 (not identified) | NH2 terminus apparently blocked | 15 |
| QSR 8 (QsrV) | –[D]NAPVKGGFT | 14 |
| QSR 10 (QsrP) | KNTYS | 12 |
| QSR 12 (RibB) | ––KLNQ | 24 |
Brackets are used for low-confidence amino acid designations.
Requirement for LuxR and 3-oxo-C6-HSL.
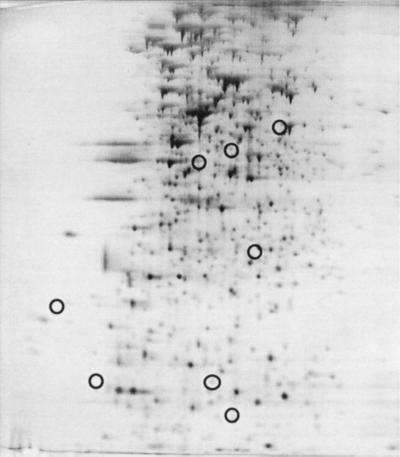
To determine whether the newly identified QSR proteins required LuxR for their production, we examined the protein pattern of MJ-208, a luxR deletion mutant. MJ-208 is unable to induce lux operon transcription regardless of the presence of 3-oxo-C6-HSL (42). MJ-208 failed to produce LuxA, LuxB, and LuxE and also produced none of the five non-Lux QSR proteins (Fig. 2). Therefore, LuxR is required for the production of the newly identified QSR proteins, as it is for production of the Lux proteins.
FIG. 2.
Protein pattern of V. fischeri MJ-208 (ΔluxR). Cells were grown to the early stationary phase (A660 = 1.2). For identification of circled proteins, see Fig. 1C.
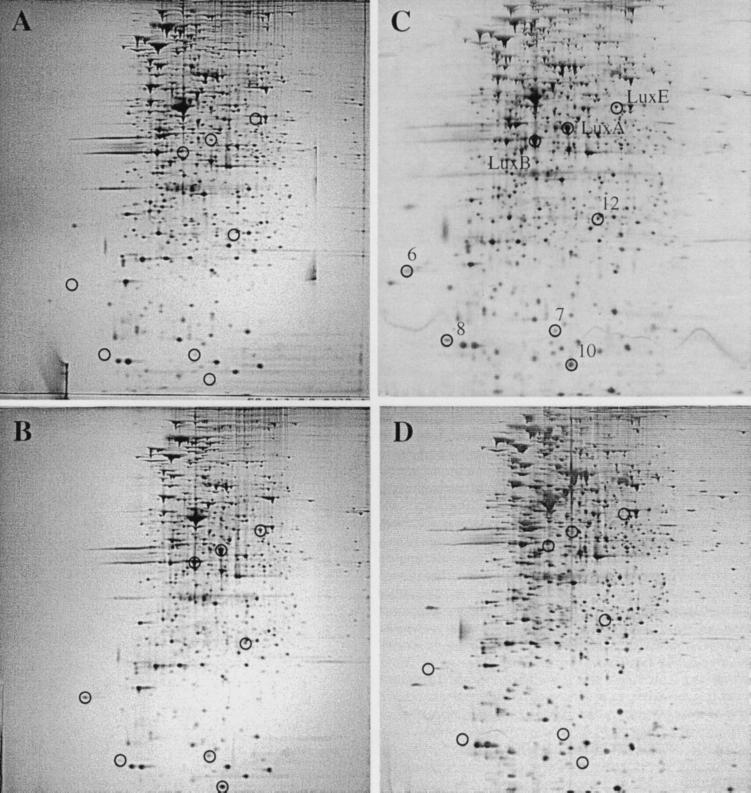
The results presented above for the double autoinducer synthase mutant MJ-215 (ΔluxI ainS) (Fig. 1D) are consistent with a requirement for either 3-oxo-C6-HSL or C8-HSL or both, via LuxR for the production of the QSR proteins. To determine which of the V. fischeri acyl-HSLs fulfills that requirement, we took two approaches. First, we examined the protein patterns of the single autoinducer synthase mutants MJ-211 (ΔluxI) and MJ-216 (ainS). MJ-211, which is unable to synthesize 3-oxo-C6-HSL but does synthesize C8-HSL, failed to produce substantial amounts of LuxA, LuxB, LuxE, or any of the five non-Lux QSR proteins (Fig. 3A). In contrast, MJ-216, which is unable to synthesize C8-HSL but does synthesize 3-oxo-C6-HSL, produced all eight of these proteins (Fig. 3B). Next, we compared the protein patterns of MJ-215 grown to early stationary phase in the presence of exogenously added acyl-HSLs (100 nM). In the presence of 3-oxo-C6-HSL, production of the three Lux and the five non-Lux QSR proteins was restored (Fig. 3C), whereas the presence of C8-HSL did not restore the production of these proteins (Fig. 3D). These results demonstrate that 3-oxo-C6-HSL is required for production of the QSR proteins. Furthermore, they demonstrate that C8-HSL is neither sufficient for the production of detectable levels of these proteins nor necessary for their high-level production.
FIG. 3.
Protein patterns of V. fischeri acyl-HSL synthase mutants. The single acyl-HSL synthase mutants (A) MJ-211 (ΔluxI) and (B) MJ-216 (ainS) were grown to early stationary phase (A660 = 1.2) in the absence of added acyl-HSL. The double acyl-HSL synthase mutant MJ-215 (ΔluxI ainS) was grown in the presence of exogenously added (C) 3-oxo-C6-HSL (100 nM) or (D) C8-HSL (100 nM). (C) LuxA, LuxB, LuxE, and the newly identified QSR proteins QSR 6 (AcfA), QSR 7 (unidentified), QSR 8 (QsrV), QSR 10 (QsrP), and QSR 12 (RibB) are circled and designated.
Absence of positive regulation of QSR proteins by C8-HSL.
We previously hypothesized that C8-HSL activates the production of non-Lux quorum-regulated proteins in V. fischeri via LuxR or another regulatory protein (42). The results presented above provide a test of that hypothesis by comparison of the protein patterns of MJ-216 (ainS), which makes no C8-HSL, and MJ-100. For cells grown to the early stationary phase, the protein pattern of MJ-216 was essentially indistinguishable from that of MJ-100 (Fig. 3B and 1C). A further test of the hypothesis is provided by the results of the double autoinducer synthase mutant MJ-215 (ΔluxI ainS) grown in the presence and absence of exogenously added C8-HSL to the early stationary phase. Regardless of the presence or absence of C8-HSL, the protein patterns of MJ-215 appeared identical (Fig. 3D and 1D). Thus, neither the absence nor the presence of C8-HSL had a discernible effect on the proteins produced by V. fischeri at high population density. Either no proteins in V. fischeri are produced under positive control by C8-HSL or they are not revealed by the 2-D PAGE conditions used here.
Negative regulation of QSR proteins by C8-HSL.
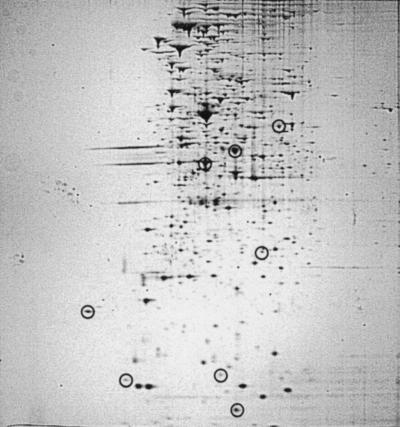
In contrast to a positive role, C8-HSL has been demonstrated to negatively modulate lux operon transcription. That negative modulation apparently operates by a competitive inhibition by C8-HSL of the interaction between 3-oxo-C6-HSL and LuxR, inhibiting production of the Lux proteins at low population density (21, 42, 43). Consistent with that inhibitory activity, MJ-216 (ainS) induces luminescence at a substantially lower population density than MJ-100 (43). We therefore asked whether C8-HSL might operate similarly on the newly identified QSR proteins, inhibiting their production at low population density. To examine that possibility, the protein patterns of MJ-100 and MJ-216 grown to mid-exponential phase (A660 = 0.4) were compared. For MJ-216, LuxA, LuxB, and LuxE and each of the five non-Lux QSR proteins were readily observed (Fig. 4), whereas all eight proteins were less abundant or not detected from MJ-100 at this population density (Fig. 1A). Thus, in the absence of C8-HSL, production of the QSR proteins is enhanced at low population density. Therefore, C8-HSL apparently operates in vivo to inhibit production of both the Lux proteins and the newly identified QSR proteins.
FIG. 4.
Production of QSR proteins at low population density by V. fischeri. Cells of MJ-216 (ainS) were grown to mid-exponential phase (A660 = 0.4). For identification of circled proteins, see Fig. 1C or 3C.
Characteristics of qsrP, the gene encoding QSR 10.
To gain insight into the functions of the newly identified QSR proteins, we isolated the genes for QSR 10, QSR 12, QSR 6, and QSR 8. In this report, we describe the genes for QSR 10 and QSR 12, which were given primary attention because of the abundance and ease of isolation of these proteins and because QSR 10 apparently is produced by V. fischeri in symbiosis with E. scolopes (see below). We note parenthetically here that the deduced translation products for the genes specifying QSR 6 and QSR 8, provisionally designated acfA and qsrV, respectively, exhibited sequence similarity to AcfA from Vibrio cholerae (53) and to hypothetical proteins F130 from E. coli and 1709 from Haemophilus influenzae, respectively. The characteristics of acfA and qsrV will be presented elsewhere.
The gene for QSR 10, designated qsrP, for quorum-sensing-regulated periplasmic, was isolated from a genomic library of MJ-100 DNA by a PCR amplification approach, with primers based on internal sequences of the protein (Materials and Methods). Neither the qsrP nucleotide sequence nor its deduced amino acid sequence exhibited significant similarity with known genes or gene products; apparently, qsrP is a novel gene. The 129-amino-acid QsrP precursor protein has a calculated molecular size of 14,746 Da. Consistent with the periplasmic location of QsrP (QSR 10), a 19-amino-acid sequence typical of prokaryotic leader peptides (67) is absent from the amino terminus of the mature protein, which begins at K20 of the deduced translation product. The mature 110-amino-acid protein has a calculated molecular weight of 12,660 and a calculated isoelectric point of 4.9, attributes consistent with the mobility of the protein on 2-D PAGE (Fig. 1C).
The qsrP coding region was bounded by a promoter with similarity to the lux operon promoter, including the presence of a probable lux box, and by a strong putative transcriptional terminator. The qsrP lux box, a 20-bp region of dyad symmetry, was found centered at 73 nucleotides upstream of the start of the qsrP coding region (Fig. 5A). This sequence matches 14 of the 20 nucleotides of the lux operon lux box (Table 3), which serves as the binding site for the LuxR/3-oxo-C6-HSL transcriptional activator complex (2, 22, 62). The lux operon promoter lacks a −35 region (23), and the location where a −35 region would occur is overlapped by the lux box (9, 24). With the positioning of the qsrP lux box as a guide, a similarly located −10 Pribnow box (TAATAT) can be discerned in the qsrP promoter region, 17 nucleotides downstream from the end of the qsrP lux box. As also seen for the lux operon promoter, no −35 region was apparent. A likely ribosome-binding site (GGA) was identified 5 bases upstream of the ATG translation initiation codon. Following the QsrP coding region, 23 bases after its translational stop codon, a probable rho-independent transcriptional terminator was identified. The stem-and-loop structure consists of a perfect 11-base stem and a 5-base loop, followed by a poly(T) region. Thus, the qsrP gene appears to be monocistronic, and its promoter region contains a lux box, which is consistent with direct transcriptional control of qsrP by LuxR/3-oxo-C6-HSL.
FIG. 5.
Nucleotide sequences of the promoter regions of the V. fischeri qsrP (A) and ribB (B) genes. Putative lux boxes, −10 Pribnow boxes, and ribosome-binding sites (rbs) are indicated. For gsrP, the arrow between amino acid residues A19 and K20 of the deduced protein indicates the leader peptide cleavage site. For ribB, a −35 sequence and a second −10 sequence are also underlined.
TABLE 3.
Alignment of lux boxes for V. fischeri QSR genes
| Gene | Strain | lux box sequence | Source or reference |
|---|---|---|---|
| luxI | MJ-1 | ACCTGTAGGA TCGTACAGGT | 24 |
| ATCC | ACCTGTAGGA TCGTACAGGT | 9 | |
| ES114 | AGCTGTAGGA TGGTACAGGT | 35 | |
| H905 | GGCTGTAGGA TAGTACAGGT | 36 | |
| EM30 | GCCTGTAGGA TCGTACAGGT | 36 | |
| ainS | MJ-1 | TAATGAGTTA TCAATCAATA | 33 |
| qsrP | MJ-1a | ACCTGTAATA AGTTACAGGA | This study |
| ES114b | ACCTGTAATA AACGACAGGA | This study | |
| ribB | MJ-1a | ACCTGCAATA ATTTACAGTA | This study |
| Consensusc | ACCTGTAGGA T GTACAGGT |
From Nalr derivative strain MJ-100.
From Rifr derivative strain ESR1.
Greater than 50% identity; underlining denotes an identical nucleotide at that position in seven or more of the nine listed sequences.
Comparison of qsrP from MJ-100 and ESR1.
To examine the role of QsrP in the bioluminescent symbiosis of V. fischeri with the sepiolid squid E. scolopes, we isolated qsrP from strain ESR1, a derivative of the squid-symbiotic strain ES114. ESR1 was chosen for this work because it is fully competent to establish symbiosis with the squid (34). In contrast, the fish-symbiotic strain MJ-1 and its derivatives, reported on above, are not effective for analysis of V. fischeri genes involved in squid symbiosis; they fail to colonize E. scolopes or do so inconsistently (10; unpublished data). Motivating our interest in QsrP as a possible symbiosis determinant is the observation that a protein similar in size to MJ-100 QsrP, but with a more basic isoelectric point, is one of the more abundant proteins revealed by 2-D PAGE of proteins from V. fischeri cells taken directly from light organs of adult E. scolopes. Like the Lux proteins of ESR1, this protein is not produced by squid-symbiotic strains grown in culture (unpublished data), indicating that it, like the Lux proteins, is subject to a quorum-sensing control apparently specific to the symbiotic state.
The qsrP gene was isolated from ESR1 by PCR amplification from chromosomal DNA with oligonucleotides 10-826F and 10-826R. Sequence analysis of the cloned PCR product revealed that the DNA flanking the QsrP coding region was essentially identical in MJ-100 and ESR1, whereas the QsrP coding region itself differed in the two strains. Specifically, of 234 nucleotides preceding the lux box, 233 were identical in the two strains. Furthermore, 17 of the 20 lux box nucleotides were the same in the two strains (Table 3). For the QsrP coding region, however, only 84% of the nucleotides and 78% of the deduced amino acids were identical. Notable differences in the coding region included the addition of amino acid residues R and S between D63 and F64 of the MJ-100 sequence. The mature QsrP protein from ESR1 had a calculated molecular weight of 12,908 and a calculated pI of 7.0. The higher calculated isoelectric point of the ESR1 protein is consistent with the position of the putative QsrP protein on 2-D PAGE of proteins from V. fischeri cells taken from the E. scolopes light organ.
Mutants of MJR1, a spontaneous rifampin-resistant derivative of MJ-100, and of ESR1 defective in qsrP were constructed by a plasmid integration procedure (Materials and Methods). To ascertain whether the mutation in qsrP resulted in an obvious phenotype in laboratory culture, the mutants, MJ-10X and ES-10X, were examined for differences from their parent strains under a variety of growth conditions. Regardless of the growth condition or the attribute tested (Materials and Methods), however, the mutants grew and behaved very similarly to their parent strains.
Diminished symbiosis competence of a qsrP mutant.
With the construction of a qsrP mutant of ESR1, we were in a position to examine the involvement of QsrP in the ability of V. fischeri to colonize its symbiotic host, E. scolopes. First, to determine if the mutant had any obvious defect in its ability to grow compared with the parent, we used an in vitro growth competition assay. ES-10X and ESR1 were inoculated together at equal numbers into laboratory medium and allowed to grow for several doublings, and then the numbers of mutant and parent cells were quantified (Materials and Methods). A ratio of mutant to parent close to 1 was obtained (ES-10X/ESR1 = 1.17 in each of three replicates), indicating that the two strains were apparently competitively equal in vitro. Next, to determine whether the mutant exhibited any obvious defect in its ability to colonize the squid, juvenile aposymbiotic E. scolopes were presented separately with the qsrP mutant and its parent strain in a squid colonization assay (Materials and Methods). Separately, the mutant and the parent colonized equally well (ES-10X, 2.8 × 105 ± 1.0 × 105 CFU/light organ, n = 10; ESR1, 2.9 × 105 ± 1.3 × 105 CFU/light organ, n = 10, at 48 h postinoculation). These results indicate that the mutant had no obvious difference from the parent in its ability to grow or to colonize the squid.
In competition with the parent, however, the qsrP mutant was less successful in colonizing the squid. For the colonization competition assay, aposymbiotic juveniles of E. scolopes were presented with a 1:1 mix of ES-10X and ESR1, and the symbiosis was allowed to develop; the numbers of mutant and parent cells present in the E. scolopes light organs were then determined at 48 h postinoculation. Compared to the parent strain, lower numbers of the qsrP mutant were present in nearly all of the animals (Fig. 6). Of the 15 animals tested (three separate trials of 5 animals each), the parent dominated the colonizing population in 12 animals, and in half of those cases it constituted more than 80% of the symbiont population. For the other three animals, one had a ratio of parent to mutant near 50%, the expected average value if no difference existed in the competitive ability of the two strains, and two animals had a higher percentage of mutant than parent. These results indicate a distinct dominance of the parent strain over the mutant in the colonizing population, similar to that observed in competition assays between ESR1 and a katA mutant of V. fischeri (66). We nonetheless sought a statistical assessment of the results, using the sign-rank test (62), a two-tailed nonparametric test. The null hypothesis, equal ability of the parent and mutant strains to colonize, was rejected at a high level of confidence, P = 0.01. We interpret these results as indicating that the defect in qsrP confers a competitive disadvantage in the symbiosis. The QsrP protein therefore apparently contributes to the ability of V. fischeri to establish bioluminescent symbiosis with E. scolopes.
FIG. 6.
Colonization of juveniles of the sepiolid squid E. scolopes by a qsrP mutant of V. fischeri and its parent strain. Aposymbiotic, 1-day-old hatchling squid were presented with a 1:1 mixture of the mutant and parent strains. The percentage of the qsrP mutant in the population of V. fischeri cells colonizing the light organ was determined for each animal at 48 h postinoculation.
Isolation of ribB, the gene encoding QSR 12.
We next isolated the gene encoding QSR 12 (Materials and Methods). The deduced amino acid sequence of QSR 12 was 55 and 56% identical to RibB of E. coli (55) and H. influenzae (27), respectively, 65% identical to LuxH of Vibrio harveyi (64), and 67% identical to RibB of Vibrio cholerae (The Institute for Genomic Research [TIGR] database; J. F. Heidelberg, personal communication). The E. coli RibB protein is a 3,4-dihydroxy-2-butanone 4-phosphate synthase, which catalyzes a key step in riboflavin synthesis (55). We therefore designated the gene for QSR 12 ribB. In V. harveyi, luxH occurs downstream of luxG as the last gene of the lux operon (64), whereas in V. fischeri, ribB is not part of the lux operon, which ends after luxG with a strong bidirectional rho-independent terminator (63). A luxH gene has not been identified previously in V. fischeri.
The V. fischeri ribB coding region directs the synthesis of a 217-amino-acid protein of 23,616 Da with an estimated pI of 5.26. Like that of qsrP, the ribB coding region was bounded by a promoter region containing a lux box and by a probable transcriptional terminator. The lux box is centered 133 nucleotides upstream of the translational start (Fig. 5B) and is identical in 12 of the MJ-1 lux operon lux box nucleotides and 17 of the MJ-1 qsrP lux box nucleotides (Table 3). A possible −10 Pribnow box sequence (TAAAAT) starts 11 nucleotides downstream from the end of the ribB lux box, a spacing that is somewhat less than that between the lux box and −10 sequences in the promoter regions of the MJ-1 lux operon (19 nucleotides) and qsrP (17 nucleotides). However, the ribB promoter region also contains a −35 (TTGTCA) sequence and an additional −10 (TTAAAT) sequence, separated by 18 nucleotides, which occur between the lux promoter elements and the ribB coding region. The putative −35 region overlaps the lux box-associated −10 region by a single nucleotide (Fig. 5B). A likely ribosome-binding site (GGAG) is positioned 10 bases upstream of the proposed ATG translation initiation codon. Amino-terminal sequencing of the protein (Table 2) confirmed that the indicated Met residue was the translational start. The coding region is followed by a putative stem-and-loop rho-independent terminator, which begins 128 nucleotides after the ribB stop codon. Therefore, ribB, like qsrP, appears to be monocistronic, and its transcription appears to be regulated directly by LuxR/3-oxo-C6-HSL.
Construction and characterization of a ribB mutant.
Induction of luminescence presumably leads to a strongly increased demand for the luciferase substrate FMNH2, but how cells cope with that demand is not known and little is presently known about the generation of reduced flavin mononucleotide in V. fischeri (11, 32, 48, 69). To attempt to gain insight into these issues, we constructed a mutant of MJR1 defective in ribB, using a plasmid integration procedure (Materials and Methods), and examined the mutant for altered growth and luminescence in comparison with its parent strain. The ribB mutant MJ-ribBX, however, exhibited no obvious phenotype. MJ-ribBx grew normally, produced a high level of light, and induced luminescence in a fashion similar to that of MJR1, regardless of the presence or absence of exogenously supplied riboflavin. Therefore, RibB apparently is not required for and does not play a significant role in normal light production by V. fischeri, at least in routine laboratory culture.
To assess the role of RibB in symbiosis, we attempted to isolate ribB from the squid-symbiotic strain ESR1. However, the primers used to isolate ribB from the MJ-1 genomic library (Materials and Methods) and several combinations of primers used in sequencing of the MJ-1 ribB gene all gave negative results when used with ESR1 chromosomal DNA as the template. In contrast, all tested primer combinations were positive with MJR1 DNA. Either a ribB gene is absent from ESR1 or its sequence has diverged substantially from that in the fish-symbiotic strain MJ-1.
DISCUSSION
This study reports the identification in the bioluminescent symbiotic bacterium V. fischeri of five QSR proteins, QsrP, RibB, AcfA, QsrV, and QSR 7, distinct from those encoded by the luminescence operon luxICDABEG. Identification of the QSR proteins was facilitated by the availability of genetically defined mutants of V. fischeri defective in production of the quorum-sensing transcriptional activator LuxR and the quorum-sensing signals 3-oxo-C6-HSL and C8-HSL (33, 42, 43). The production of the QSR proteins occurs preferentially at high population density, requires both LuxR and 3-oxo-C6-HSL, and apparently is inhibited by C8-HSL at low population density. These regulatory attributes are the same as those controlling production of the proteins for luminescence. Consistent with LuxR/acyl-HSL-dependent control, the promoter regions of the qsrP and ribB genes contain a lux box, a regulatory site necessary for LuxR/3-oxo-C6-HSL-dependent activation of lux operon transcription. The genes for the newly identified QSR proteins, together with the genes for luminescence, define a LuxR/acyl-HSL-responsive quorum-sensing regulon in V. fischeri.
We hypothesized previously that C8-HSL, via LuxR or another regulatory protein, represents a second quorum-sensing system in V. fischeri, one with target genes distinct from those regulated by 3-oxo-C6-HSL and LuxR (42). No protein whose production was dependent on C8-HSL was revealed, however, through 2-D PAGE analysis of MJ-216 (ainS) in comparison with its parent strain, or of MJ-215 (ΔluxI ainS) grown in the presence or absence of C8-HSL. If C8-HSL does positively regulate the production of proteins in V. fischeri, then the conditions necessary for their detection apparently differ from the conditions used in this study.
Conversely, the results of this study support and extend an inhibitory role in vivo for C8-HSL (43). The presence of C8-HSL in V. fischeri delays lux operon expression in a LuxR-dependent manner, apparently through a competitive inhibition of the interaction between 3-oxo-C6-HSL and LuxR (21, 43). In the present study, we used the observation that the ainS mutant MJ-216 induces luminescence at a lower population density than the parent strain (43). The mutation, by eliminating the ability of cells to synthesize C8-HSL, apparently relieves the inhibitory effect of C8-HSL, thereby allowing lux operon induction to occur at a lower concentration of 3-oxo-C6-HSL, i.e., at a lower population density. We therefore expected that MJ-216 would produce higher levels of the Lux proteins at a lower population density than its parent strain, MJ-100. 2-D PAGE analysis confirmed that expectation for the Lux proteins detected here, LuxA, LuxB, and LuxE, and revealed that higher levels of the five newly identified QSR proteins also were produced at a lower density. Thus, the new QSR proteins are subject to the same regulatory control, both positive and negative, as the proteins of the lux operon. We therefore believe that C8-HSL plays a global role in V. fischeri, inhibiting premature induction of QSR genes (i.e., induction at an inappropriately low population density). This in vivo inhibitory role is not unexpected. The inhibitory activity of C8-HSL supports the notion (see, for example, reference 13) that the production of QSR proteins is adaptive specifically at high population density. Furthermore, analogous mechanisms for inhibiting the expression of quorum-regulated genes exist in other bacteria, including LuxO in V. harveyi (3, 28), TraM in Agrobacterium tumefaciens (29), and RsaL in P. aeruginosa (8). In each of these other quorum-sensing systems, the inhibitor is a protein, whereas in V. fischeri the inhibitor is an acyl-HSL. Despite that difference, inhibition of QSR gene expression at low population density might be a common regulatory theme in bacterial quorum sensing.
The presence of a lux box in the promoter regions of qsrP and ribB is consistent with the requirement for LuxR and 3-oxo-C6-HSL for production of QsrP and RibB. LuxR and 3-oxo-C6-HSL presumably control transcription of these genes directly. The lux operon lux box, to which the qsrP and ribB lux boxes are similar (Table 3), is essential for the LuxR/3-oxo-C6-HSL-dependent activation of lux operon transcription (9, 22, 62). Identical or similar sequences have been identified in the lux operon promoters of various V. fischeri strains, the ainSR promoter region in V. fischeri MJ-1, and the promoters controlling expression of several genes in P. aeruginosa, traA and traI of the octopine-type Ti plasmid, and traI of the nopaline-type plasmid, respectively, of Agrobacterium tumefaciens, solI of Ralstonia solanacearum, and cepI of Burkholderia cepacia (26, 30, 33, 36, 39, 41, 45, 52, 68). Thus, both within V. fischeri (Table 3) and among various gram-negative bacteria, the lux box represents a conserved regulatory sequence, the presence of which infers direct quorum-sensing control of that gene or operon by a LuxR homolog and an acyl-HSL. The qsrP promoter region resembles that of the lux operon, with the lux box apparently complementing the lack of a ς70-type −35 region. That arrangement, by analogy with the lux operon (9, 24), suggests regulation of qsrP solely by LuxR/3-oxo-C6-HSL. In contrast, the ribB promoter region differs by also containing a likely −35 region adjacent to the lux box and a second appropriately spaced −10 region, suggesting both quorum-sensing and “housekeeping” regulation of ribB.
We had anticipated that comparisons between MJ-100 and MJ-215 might reveal all of the proteins of the lux operon, since the luxICDABEG genes are coordinately expressed from a single promoter under LuxR/acyl-HSL control. However, LuxI, LuxC, LuxD, and LuxG were not detected in our analysis. These proteins might comigrate on 2-D PAGE gels with more abundant proteins and thereby be masked, or differential stability might influence their abundance in the cell and, as a consequence, their detection by the 2-D PAGE conditions used here. Similar to our observations, a recent 2-D PAGE analysis of Photorhabdus luminescens lux genes luxCDABE on a recombinant plasmid identified only LuxA, LuxB, and LuxD (47). Regardless of the reason, failure to detect certain of the V. fischeri Lux proteins leads us to anticipate that a more direct approach may be necessary for the identification of additional QSR proteins and genes. Support for this notion is provided by the recent identification of 37 QSR genes in P. aeruginosa by transposon mutagenesis (68). We anticipate, therefore, that additional analysis will reveal many more QSR genes in V. fischeri.
Evidence presented here suggests that a major function of the quorum-sensing regulon in V. fischeri is to coordinate the expression of luminescence and the production of proteins involved in host association. Consistent with this notion is the high level of acyl-HSL detected in light organs of the sepiolid squid E. scolopes (5). With respect to luminescence, the ribB gene could contribute to light production through the involvement of RibB in synthesis of riboflavin, a precursor of the luciferase substrate reduced flavin mononucleotide (48, 55). However, no evidence for an essential role for RibB (QSR 12) in light production was revealed here by analysis of a V. fischeri ribB mutant in laboratory culture. Additional studies will be necessary to ascertain whether the defect in ribB is complemented in V. fischeri by another protein or is more obvious under growth conditions other than those used for routine laboratory culture. With respect to host association, QsrP, a novel protein, apparently is produced by V. fischeri in the symbiosis, and it may be involved in colonization of the squid light organ. By itself, a qsrP mutant colonized the light organs of juvenile E. scolopes to the same extent as its parent strain, whereas in competition with the parent, the mutant was less successful, suggesting that the defect in qsrP confers a competitive disadvantage in the symbiosis. Recently, a katA mutant of V. fischeri also was shown, through procedures similar to those used here, to exhibit a diminished ability to colonize E. scolopes in competition with its parent strain (66). The periplasmic location of QsrP suggests an involvement of the protein in acquisition of nutrients from the host or a response to the host environment. Also identified in this study is a second QSR protein, AcfA (QSR 6), the V. cholerae homolog of which is thought to contribute to the ability of V. cholerae to colonize the mouse intestinal epithelium (53). Furthermore, a possible role for QSR proteins in host colonization is not unexpected. Many QSR genes apparently mediate interactions between bacteria and their plant and animal hosts (13, 39, 65). Nonetheless, validation of the notion that the quorum-sensing regulon plays a major role in coordinating production of proteins involved in host association in V. fischeri must await the identification of additional QSR genes in this species.
Quorum sensing in V. fischeri serves as an important model for the response of gram-negative bacteria to population density and host association. Studies of the regulation of luminescence in V. fischeri, one of the first and presently the best understood quorum-sensing system, set the stage for the discovery of quorum sensing in other gram-negative bacteria (14, 40). The identification of non-Lux QSR genes and proteins in this species extends the significance of the V. fischeri model by making it apparent that luminescence is just one of several activities of a LuxR/acyl-HSL-controlled quorum-sensing regulon in this bacterium. Furthermore, the demonstration that a member of this regulon, qsrP, apparently is necessary for V. fischeri to efficiently colonize its sepiolid squid host highlights the value of V. fischeri as a mutualistic counterpoint to the several pathogenic bacteria whose host interactions are dependent on quorum sensing. Ongoing studies to characterize the roles of AcfA, QsrV, and QSR 7 and to identify additional QSR genes should provide further insight into the extent and significance of the quorum-sensing response in V. fischeri.
ACKNOWLEDGMENTS
We thank M. Claes for providing the E. scolopes hatchlings used in this study and for guiding the colonization assays, J. King for facilitating peptide sequencing at MIT, E. Russek-Cohen and M. Pascual for statistics advice, D. Caron for equipment and reagents used in an initial phase of this work, C. Gardel and J. Mekalanos for bacterial strains, and T. Marathe for technical assistance. We thank also the Biopolymers Laboratory at the Massachusetts Institute of Technology for amino-terminal protein sequencing, the Harvard Microchemistry Facility for internal protein sequencing, and the CRC DNA Sequencing Facility at the University of Chicago for DNA sequencing. Preliminary sequence data for V. cholerae RibB were obtained from The Institute for Genomic Research website at http://www.tigr.org.
This research was supported by grant MCB 97-229772 from the National Science Foundation.
REFERENCES
- 1.Ames G F-L, Prody C, Kustu S. Simple, rapid, and qualitative release of periplasmic proteins by chloroform. J Bacteriol. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin T O, Devine J H, Heckel R C, Lin J-W, Shadel G S. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence. J Biolumin Chemilumin. 1989;4:326–341. doi: 10.1002/bio.1170040145. [DOI] [PubMed] [Google Scholar]
- 3.Bassler B L, Wright M, Silverman M R. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 4.Boettcher K J, Ruby E G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;176:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettcher K J, Ruby E G. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J Bacteriol. 1995;177:1053–1058. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claes M F, Dunlap P V. Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J Exp Zool. 2000;286:280–296. [PubMed] [Google Scholar]
- 7.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 8.De Kievit T, Seed P C, Nezezon J, Passador L, Iglewski B H. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2175–2184. doi: 10.1128/jb.181.7.2175-2184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devine J H, Shadel G S, Baldwin T O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc Natl Acad Sci USA. 1989;86:5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doino J A, McFall-Ngai M J. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- 11.Doudoroff M. Lactoflavin and bacterial luminescence. Enzymologia. 1938;4:239–243. [Google Scholar]
- 12.Dunlap P V. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J Bacteriol. 1989;171:1199–1202. doi: 10.1128/jb.171.2.1199-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunlap P V. N-Acyl-l-homoserine lactone autoinducers in bacteria: unity and diversity. In: Shapiro J A, Dworkin M, editors. Bacteria as multicellular organisms. New York, N.Y: Oxford University Press; 1997. pp. 69–106. [Google Scholar]
- 14.Dunlap P V. Quorum regulation of luminescence in Vibrio fischeri. J Mol Microbiol Biotechnol. 1999;1:5–12. [PubMed] [Google Scholar]
- 15.Dunlap P V, Callahan S M. Characterization of a periplasmic 3′:5′-cyclic nucleotide phosphodiesterase gene, cpdP, from the marine symbiotic bacterium Vibrio fischeri. J Bacteriol. 1993;175:4615–4624. doi: 10.1128/jb.175.15.4615-4624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunlap P V, Greenberg E P. Role of intercellular chemical communication in the Vibrio fischeri-monocentrid fish symbiosis. In: Dworkin M, editor. Microbial cell-cell interactions. Washington, D.C.: American Society for Microbiology; 1991. pp. 219–253. [Google Scholar]
- 17.Dunlap P V, Kita-Tsukamoto K, Waterbury J B, Callahan S M. Isolation and characterization of a visibly luminous variant of Vibrio fischeri ES114 from the sepiolid squid Euprymna scolopes. Arch Microbiol. 1995;164:194–202. [Google Scholar]
- 18.Dunlap P V, Kuo A. Cell density dependent modulation of the Vibrio fischeri luminescence system in the absence of autoinducer and LuxR protein. J Bacteriol. 1992;174:2440–2448. doi: 10.1128/jb.174.8.2440-2448.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunlap P V, Mueller U, Lisa T A, Lundberg K S. Growth of the marine luminous bacterium Vibrio fischeri on 3′:5′-cyclic AMP: correlation with a periplasmic 3′:5′-cyclic AMP phosphodiesterase. J Gen Microbiol. 1992;138:115–123. [Google Scholar]
- 20.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:24444–22449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 21.Eberhard A, Widrig C A, McBath P, Schineller J B. Analogs of the autoinducer of bioluminescence in Vibrio fischeri. Arch Microbiol. 1986;146:35–40. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 22.Egland K A, Greenberg E P. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 23.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engebrecht J, Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987;15:10455–10467. doi: 10.1093/nar/15.24.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez J, Andrews L, Mische S M. An improved procedure for enzymatic digestion of polyvinylidiene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:113–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 26.Flavier A B, Ganova-Raeva L M, Schell M A, Denny T P. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J Bacteriol. 1997;179:7089–7097. doi: 10.1128/jb.179.22.7089-7097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 28.Freeman J A, Bassler B L. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. 1999;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuqua C, Burbea M, Winans S C. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J Bacteriol. 1995;177:1367–1373. doi: 10.1128/jb.177.5.1367-1373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuqua C, Winans S C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR/LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giese A C. Studies on the nutrition of dim and bright variants of a species of luminous bacteria. J Bacteriol. 1943;46:323–331. doi: 10.1128/jb.46.4.323-331.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilson L, Kuo A, Dunlap P V. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray K M, Greenberg E P. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J Bacteriol. 1992;174:4384–4390. doi: 10.1128/jb.174.13.4384-4390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray K M, Passador L, Iglewski B H, Greenberg E P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenberg E G. Quorum sensing in gram-negative bacteria. ASM News. 1997;63:371–377. [Google Scholar]
- 38.Hanzelka B L, Parsek M R, Val D L, Dunlap P V, Cronan J E, Jr, Greenberg E P. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol. 1999;181:5766–5770. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassett D J, Ma J-F, Elkins J G, McDermott T R, Ochsner U A, West S E H, Huang C-T, Fredericks J, Burnett S, Stewart P S, McFeters G, Passador L, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 40.Hastings J W, Greenberg E P. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol. 1999;181:2667–2668. doi: 10.1128/jb.181.9.2667-2668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang I, Li P-L, Zhang L, Piper K R, Cook D M, Tate M E, Farrand S K. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo A, Blough N V, Dunlap P V. Multiple N-acyl-l-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J Bacteriol. 1994;176:7558–7565. doi: 10.1128/jb.176.24.7558-7565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo A, Callahan S M, Dunlap P V. Modulation of luminescence operon expression by N-octanoyl-l-homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol. 1996;178:971–976. doi: 10.1128/jb.178.4.971-976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane W S, Galat A, Harding M W, Schreiber S L. Complete amino acid sequence of the FK506 and rapamycin binding protein, FKBP, isolated from calf thymus. J Protein Chem. 1991;10:151–160. doi: 10.1007/BF01024778. [DOI] [PubMed] [Google Scholar]
- 45.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 46.Lewenza S, Conway B, Greenberg E P, Sokol P A. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 48.McElroy W D, Hastings J W, Sonnenberg V, Coulombre J. The requirement of riboflavin phosphate for bacterial luminescence. Science. 1953;118:385–386. doi: 10.1126/science.118.3066.385. [DOI] [PubMed] [Google Scholar]
- 49.McFall-Ngai M J, Ruby E G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 50.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Farrell P H. High-resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 52.Pesci E C, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 147–155. [Google Scholar]
- 53.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae toxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabilloud T. A comparison between low background silver diammine and silver nitrate protein stains. Electrophoresis. 1992;13:429–439. doi: 10.1002/elps.1150130190. [DOI] [PubMed] [Google Scholar]
- 55.Richter G, Volk R, Krieger C, Lahm H-W, Rothlisberger U, Bacher A. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J Bacteriol. 1992;174:4050–4056. doi: 10.1128/jb.174.12.4050-4056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruby E G. Lessons from a cooperative bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 57.Ruby E G, Nealson K H. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol Bull. 1976;141:574–586. doi: 10.2307/1540507. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Greenberg E P. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snedecor G W, Cochran W G. Statistical methods. 7th ed. Ames: The Iowa State University Press; 1980. [Google Scholar]
- 62.Stevens A M, Greenberg E P. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol. 1997;179:557–562. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swartzman E, Kapoor S, Graham A F, Meighen E A. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J Bacteriol. 1990;172:6797–6802. doi: 10.1128/jb.172.12.6797-6802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swartzman E, Miyamoto C, Graham A, Meighen E A. Delineation of the transcriptional boundaries of the lux operon of Vibrio harveyi demonstrates the presence of two new lux genes. J Biol Chem. 1990;265:3513–3517. [PubMed] [Google Scholar]
- 65.Swift S, Williams P, Stewart G S A B. N-Acylhomoserine lactones and quorum sensing in proteobacteria. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 291–313. [Google Scholar]
- 66.Visick K L, Ruby E G. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 68.Whiteley M, Lee K M, Greenberg E G. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zenno S, Saigo K. Identification of the genes encoding NAD(P)H-flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria: Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J Bacteriol. 1994;176:3544–3551. doi: 10.1128/jb.176.12.3544-3551.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]