Abstract
Plant ecologists and molecular biologists have long considered the hypothesis of a trade-off between plant growth and defence separately. In particular, how genes thought to control the growth–defence trade-off at the molecular level relate to trait-based frameworks in functional ecology, such as the slow–fast plant economics spectrum, is unknown. We grew 49 phenotypically diverse rice genotypes in pots under optimal conditions and measured growth-related functional traits and the constitutive expression of 11 genes involved in plant defence. We also quantified the concentration of silicon (Si) in leaves to estimate silica-based defences. Rice genotypes were aligned along a slow–fast continuum, with slow-growing, late-flowering genotypes versus fast-growing, early-flowering genotypes. Leaf dry matter content and leaf Si concentrations were not aligned with this axis and negatively correlated with each other. Live-fast genotypes exhibited greater expression of OsNPR1, a regulator of the salicylic acid pathway that promotes plant defence while suppressing plant growth. These genotypes also exhibited greater expression of SPL7 and GH3.2, which are also involved in both stress resistance and growth. Our results do not support the hypothesis of a growth–defence trade-off when leaf Si and leaf dry matter content are considered, but they do when hormonal pathway genes are considered. We demonstrate the benefits of combining ecological and molecular approaches to elucidate the growth–defence trade-off, opening new avenues for plant breeding and crop science.
Keywords: Defence gene, growth–defence trade-off, intraspecific variation, plant defence, plant economics spectrum, plant functional trait, plant immunity, rice (Oryza sativa), silica, silicon
Slow-growing, late-flowering (live slow–die old) rice genotypes exhibit greater expression of defence genes than fast-growing, early-flowering genotypes, demonstrating a growth–defence trade-off.
Introduction
One of the most influential theories in plant biology is that defences against pathogens and herbivores are costly, which results in trade-offs between growth and defence (Simms and Rausher, 1987; Fagerström, 1989; Herms and Mattson, 1992). Plants have indeed developed a wide range of constitutive and induced defence mechanisms to protect themselves against pathogens and herbivores, and this comes with a cost (Agrawal, 2007; Savatin et al., 2014). Plant ecologists have long considered the costs of resistance to pathogens or herbivores by testing a decrease in plant growth and/or reproduction associated with increased resistance (Strauss et al., 2002; Moore et al., 2003; Paul-Victor et al., 2010; Garcia et al., 2021). Such an approach showed some success in identifying trade-offs between growth and defence, whether chemical or mechanical (Endara and Coley, 2011; Garcia et al., 2021). However, whether these trade-offs result from genetic variation or phenotypic plasticity remains unclear (Hahn and Maron, 2016; Züst and Agrawal, 2017; but see Vázquez-González et al., 2020; Cope et al., 2021). In addition, the emergence of trait-based ecology enabled the exploration of major spectrum shaping plant strategies, such as the leaf economics spectrum (LES) (Wright et al., 2004), but plant mechanical or chemical defences against pathogens and herbivores are still poorly captured in this spectrum.
The LES describes a major axis of cross-species leaf ecophysiology comprising key traits such as leaf lifespan, nitrogen (N), and phosphorus (P) concentrations, and photosynthetic rate (Wright et al., 2004). The spectrum runs from fast-growing species having traits associated with rapid resource acquisition and return on resource investment (acquisitive species), to slow-growing species having traits involved in resource conservation (Wright et al., 2004; Reich, 2014). An implicit assumption underpinning the LES is that slow-growing species (conservative strategies) are better defended against pathogens and herbivores, to avoid the loss of scarce nutrients and hard-to-get carbon (Coley et al., 1985; Endara and Coley, 2011). The implementation of the LES to study trade-offs between growth and defence or survival across different species has shown some success (Hallik et al., 2009; Züst and Agrawal, 2017). For instance, conservative Helianthus spp. have tougher, more succulent leaves with higher tannin activity compared to more resource-acquisitive species (Mason et al., 2016). However, these studies most of the time ignore trait variation within species and do not control for the role of phylogeny in shaping these patterns (Albert et al., 2011). Considering the intraspecific level is therefore often more appropriate to understand mechanisms underlying the co-variations and trade-offs among traits (Vasseur et al., 2012; Sartori et al., 2019). In addition, although some traits thought to be involved in stress tolerance and resource conservation are often considered by trait-based ecology [e.g. leaf dry matter content (LDMC); Wright and Cannon, 2001; Mason and Donovan, 2015; Blumenthal et al., 2020], others are generally ignored. For instance, mineral deposits made of silica in plant tissues can increase the resistance to herbivores (Massey and Hartley, 2009) and pathogens (Ahammed and Yang, 2021), especially in grasses including rice, but leaf silicon (Si) concentration is rarely considered in trait-based ecology (de Tombeur et al., 2023).
In parallel with ecological studies, molecular biologists have long studied the plant immune system to understand how plants recognize and respond to pathogens (Ferreira et al., 2006; Jones and Dangl, 2006; Karasov et al., 2017). These studies allowed a thorough mechanistic understanding of the growth–defence trade-off. In particular, some genes, proteins, and hormonal pathways have been identified as important in the plant immune system and plant defence but also to mediate plant growth parameters (Ning et al., 2017; van Butselaar and Van Den Ackerveken, 2020). For instance, in rice (Oryza sativa), the OsNPR1 gene, originally identified as a key regulator of the salicylic acid pathway and defence response (Sugano et al., 2010), also suppresses growth by interfering with the auxin pathway (Li et al., 2016). Similarly, the rice protein IPA1, depending on its phosphorylation status, was shown to either activate the transcription of yield genes or to activate the defence gene WRKY45, leading to enhanced disease resistance (Jiao et al., 2010; Wang et al., 2018). These studies have been pivotal in improving our mechanistic understanding of the growth–defence trade-off (He et al., 2022), but they are mostly based on one single genotype and the use of mutants, and multi-genotypic comparative studies are scarce. More generally, these studies are at the molecular level and we still ignore whether they are reflected in plant ecological strategies and associated with the LES framework, and more generally with trait-based ecology.
Here, we combine ecological and molecular approaches to study the trade-off between growth and defence/immunity at the intraspecific level. To do so, we grew 49 genotypes of temperate rice (Oryza sativa subsp. japonica.) in similar conditions and under optimal growing conditions, and we measured growth- and defence-related functional traits together with defence-related gene expression. We first built a phenotypic space based on several key traits directly involved in plant metabolism and/or the LES framework (leaf N, P, and S concentrations, chlorophyll fluorescence as a proxy for photosynthetic efficiency). Concentrations of Si—a proxy for silica-based defences—and LDMC were considered as mechanical defence/stress-resistance traits, and the leaf carbon (C):N ratio was used as a proxy for leaf nutritional quality (Agrawal and Fishbein, 2006). These traits were added to the phenotypic space to analyse the relationships between mechanical defence and LES strategies. The main axes of trait variation—and especially the one contrasting fast-growing versus slow-growing genotypes—were then compared with the expression of genes involved in basal immunity to further test the hypothesis of a growth–defence trade-off. We hypothesized that slow-growing genotypes will have higher levels of leaf mechanical defensive traits (LDMC and leaf [Si]), and will express genes involved in plant defence more strongly.
Materials and methods
Plant material
We selected temperate rice genotypes (Oryza sativa subsp. japonica) from a set obtained from the European Rice Germplasm Collection (Courtois et al., 2012). This population was characterized for genome-wide analysis and phenotypic data were published elsewhere (Biscarini et al., 2016; Volante et al., 2017; Frontini et al., 2021). These phenotypic data were used in a clustering analysis, and 49 lines were chosen in order to maximize phenotypic and genotypic diversity. Traits used in this analysis were root biomass, days to flowering, leaf area, panicle and node height, leaf chlorophyll concentration, flavonoid concentration, yield, hundred grain weight, and number of tillers per metre. The list of the 49 rice genotypes can be found in Supplementary Table S1.
Growth conditions
The experiment was conducted in an experimental field of the CEFE (Montpellier, France) from June to September 2021, in outdoor conditions (mean daily temperature from 18.7 °C to 28.1 °C). We used a randomized complete block design using four blocks, with each genotype replicated once in each block (i.e. four replicates per genotype, a total of 196 pots). Plants were grown in 8.8-litre plastic pots (15 cm diameter; 50 cm depth) filled with a mixture of 50% (volume based) quartz sand and 50% soil (62% sand, 27% silt, and 11% clay) and amended with 3.5 g l−1 of NPK fertilizer (Basacote High K 6M NPK 13-5-18; Compo Expert) and 5.9 mg l−1 of Fe fertilizer (Ferveg 6; 6% Fe EDDHA). Plants were watered every day with a drip irrigation system with about 150 ml of tap water to avoid water stress.
Plant trait measurements
Traits were measured at the beginning of the flowering stage to standardize measurements among individuals. Daily maximum and minimum temperatures were used to calculate the age at flowering in growing degree-days (GDD), as the sum of GDD from germination to the appearance of the first panicle and considering a base temperature of 10 °C (Lee et al., 2015). Meteorological data were recorded using a Davis Vantage Pro2 weather station installed in the experimental field at CEFE.
The chlorophyll fluorescence, hereafter Y(II), was measured with a MINIPAM II (pulse amplitude modulation) fluorometer (Walz, Effeltrich, Germany). A mature N-1 leaf (below the flag leaf) was light-adjusted for approximately 5 min to reach a steady state photosynthesis level, prior to measurements of the effective quantum yield of photochemical energy conversion (yield). The relative effective quantum yield of photochemical energy conversion at steady-state photosynthesis was calculated as: yield=(Fmʹ−Fs)/Fmʹ (Genty et al., 1989), where Fs and Fmʹ are the fluorescence at steady-state photosynthesis and maximum fluorescence in the light, respectively. The chlorophyll fluorescence gives an estimate of the efficiency of photosystem II involved in photosynthesis. The same leaf was then collected, rehydrated overnight, weighed, and dried at 60 °C for 72 h to calculate the LDMC as the ratio between leaf dry weight and leaf fresh weight (% DW). Leaf N and C concentrations were determined on the same leaf after grinding, using a CN elemental analyser (Fisons Instruments—CHN model EA 1108).
Three additional N-1 adult leaves were then sampled on other tillers that had developed their flag leaves and begun to flower. Leaves were then dried at 60 °C for 72 h, and ground to quantify concentrations of P, Si, and S with a portable X-ray fluorescence spectrometer (Reidinger et al., 2012). Dried leaf material was ball-milled (Retsch MM400 Mixer mill, Germany) and ground material was pressed at 10 tons into pellets using a manual hydraulic press with a 13 mm die (Specac, UK). Elemental analysis was performed using a P-XRF instrument (Nitron XL3t900 GOLDD analyser: Thermo Scientific, UK) held in a test stand (SmartStand, Thermo Scientific). The P-XRF instrument was calibrated using Si-spiked synthetic methyl cellulose (Sigma-Aldrich, product no. 274429) and Certified Reference Materials of NCS DC73349 ‘Bush branches and leaves’ obtained from China National Analysis Center for Iron and Steel. To avoid signal loss by air absorption, the analyses were performed under a helium atmosphere (Reidinger et al., 2012). A reading of each side of the pellet was made, approximately 1 h apart, to account for u-drift in the instrument (i.e. variation in readings between consecutive runs using identical parameters; Johnson, 2014). The two readings were averaged to obtain P, Si, and S concentrations.
At harvest, aboveground biomass was dried at 60 °C for 72 h. The relative growth rate (RGR, in g g−1 GDD−1) was calculated as follows:
where W2 is the aboveground plant biomass at harvest, and t2–t1 is the number of GDD from germination to harvest. We used the mean weight of all the plants pulled out after germination for W1 (0.05 g).
Gene expression
At the beginning of the flowering stage, the last fully developed leaf (flag leaf) of the tallest tiller was sampled for RNA extraction on each plant. Only the middle part of the leaves was taken. Once collected, the leaf was transferred to a tube and placed immediately in liquid nitrogen.
For RNA extraction, we used protocols described in Delteil et al. (2012). Briefly, frozen leaf material was ground in liquid nitrogen. Approximately 500 mg of powder was treated with 1 ml of TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). RNA samples (5 µg) were denatured for 5 min at 65 °C with oligo(dT) 18 (3.5 mM) and deoxynucleoside triphosphate (dNTP) (1.5 mM). They were later subjected to reverse transcription for 45 min at 37 °C with 200 U of reverse transcriptase M-MLV (Promega, Madison, WI, USA) in the appropriate buffer.
Gene expression analysis was performed using the Fluidigm Biomark analysis protocol. Briefly, 2 µl of cDNA samples were diluted 10 times (samples around 3 µM) and pre-amplified following the ‘Pre-amplification of cDNA for Gene Expression with delta Gene Assays’ protocol provided by the manufacturer (Fluidigm). The reactions were cleaned up using Exonuclease I (Exo I, 4U μl−1). Gene expression analysis was performed with the 96.96 IFC Machine using the Delta Gene Assays and the protocol provided by Fluidigm on 1/10 diluted pre-amplified samples (5 µl gene assay mix and sample assays used for running the plate). Amplification was performed as follows: 95 °C for 1 min; 30 cycles of 96 °C for 5 s and 60 °C for 1 min; finally, 95 °C for 1 min. Gene expression was measured for 10 genes that were transcriptionally regulated in the leaf defence process in rice: OsMT2, NPR1, SPL7, POX223, PBZ1, RBBI2, PR1B, WRKY45, OsCHI, and PR5 (Supplementary Table S2; Delteil et al., 2012). We also included a gene of the GH3 family, GH3.2, because of its involvement in biotic and abiotic stresses (Fu et al., 2011; Du et al., 2012; Kong et al., 2019). Gene expression was calculated by the 2DDCt method (Livak and Schmittgen, 2001) using three references genes, EF1a, EF4a, and UBQ5, with the R package ‘fluidigr’.
Statistical analyses
First, we characterized the phenotypic space of the 49 genotypes using a principal component analysis (PCA) and considering three traits underlying the LES (leaf [N], leaf [P], Y(II)), to which we added traits related to leaf defence/stress resistance (LDMC, leaf [Si], leaf C:N ratio). Leaf [S] was added to the PCA because of its alignment with the LES framework and direct involvement in photosynthesis and growth (component of proteins) (Fratte et al., 2021). RGR and life history (age at flowering) were also added to the PCA, as quantitative supplementary variables (no influence on PCA results) to consider only leaf traits in the analysis. We then ran a second PCA with gene expression, considering the genes mentioned above. PCAs were run with the package FactoMineR on genotype-mean values and log-transformed data (Lê et al., 2008). Beyond PCA analyses, bivariate relations within traits and within genes were tested through standardized major axis (SMA) regressions (Warton et al., 2006), using the package SMATR (Warton et al., 2012).
To test potential relationships between the phenotypic space (first PCA) and the gene expression space (second PCA), genotype scores on the first two dimensions of both PCAs were extracted and potential co-variation between genotype scores on the four axes (two axes by PCAs) were tested through SMA regressions, as indicated above. We also tested potential relationships between axes of the trait PCAs and the expression of genes taken individually. All analyses were conducted in R (R Core Team, 2021).
Results
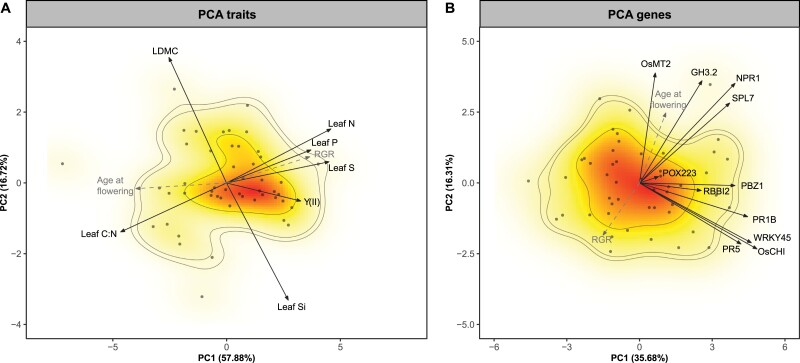
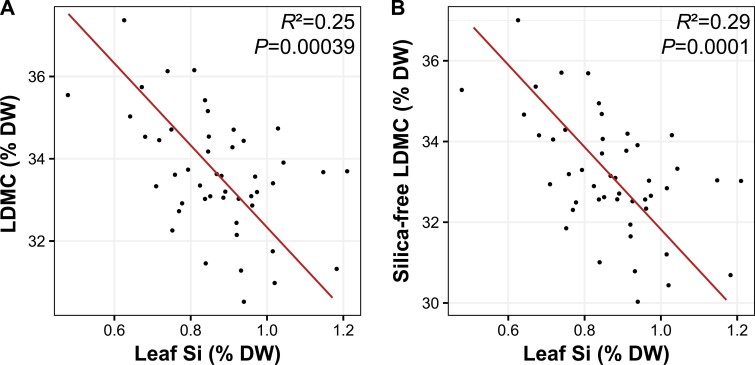
The first two principal components (PCs) describing the phenotypic space of rice functional traits explained 75% of the total variance (Fig. 1A; Supplementary Table S3). PC1 opposed genotypes with high foliar nutrient concentrations (leaf [N], [P], and [S]) and chlorophyll fluorescence to genotypes with higher leaf C:N ratio. The RGR and age at flowering were both well represented (Fig. 1A) and correlated (Supplementary Fig. S1) with this first axis, which overall describes a slow–fast continuum, with fast-growing, early-flowering genotypes with high nutrient concentrations contrasting with late-flowering genotypes with slower growth rates. The five traits mentioned above, RGR, and age at flowering were all significantly correlated with each other (Supplementary Table S4; Supplementary Fig. S2). Fast-growing genotypes also tended to have higher leaf [Si] but lower LDMC, although these two traits were better represented by PC2 (Fig. 1A) and negatively related to each other (either LDMC was corrected or not by silica weight; Fig. 2). Nevertheless, leaf [Si] was positively related to leaf [N], leaf [S], and Y(II), and negatively related to age at flowering and C:N ratio (Supplementary Table S4; Supplementary Fig. S2). Genotype-mean values of traits can be found in Supplementary Table S5.
Fig. 1.
Principal component analyses (PCAs) based on correlation matrices of the main functional traits (A) and gene expression (B) for 49 rice genotypes. The relative growth rate (RGR) and age at flowering are supplementary quantitative variables for visualization (grey, dashed lines), and have no influence on the PCA results. The colour gradient indicates regions of highest (red) to lowest (white) occurrence of probability of genotypes in the traits/genes space, and contour lines show 0.50, 0.90, and 0.95 quantiles (see Díaz et al., 2016 for the method). The results of the PCAs are presented in Supplementary Tables S3, S6. Both PCAs were run on log-transformed data. LDMC, leaf dry matter content; Y(II), chlorophyll fluorescence.
Fig. 2.
Relationship between leaf silicon concentrations and leaf dry matter content. (A) Standardized major axis regression line and statistics of the bivariate relationship between leaf silicon (Si) concentrations and leaf dry matter content (LMDC) among 49 rice genotypes. (B) To avoid spurious correlations, we also examined the relationship between leaf [Si] and silica-free LDMC. Silica-free LDMC was calculated as follows: (leaf dry weight−silica weight)/(leaf fresh weight−silica weight). The weight of silica in the leaves used for LDMC was calculated using leaf [Si]. Si was converted into silica by multiplying by 2.14 and assuming a 10% mean water content and a 5% content of other elements (Blecker et al., 2006). Both relationships were very similar, with a slightly greater explanatory power for the relationship between leaf [Si] and corrected LDMC.
The first two PCs describing gene expression of rice genotypes explained about 52% of the total variance (Fig. 1B; Supplementary Table S6). PC1 supported almost all genes (NPR1, OsCHI, PBZ1, PR5, SPL7, WRKY45, and PR1B), with the exception of OsMT2 and GH3.2, which were better represented on PC2; POX223, which was better represented on PC4; and RBBI2, which was better represented on PC5 (Fig. 1B; Supplementary Table S6). Genes described by PC1 tended to be significantly correlated with each other (Supplementary Table S7). However, two groups of less-related genes were identified (SPL7, NPR1, and GH3.2 versus OsCHI, WRKY45, PR5, and PR1B) in the PCA (Fig. 1B) and through the correlation coefficients (Supplementary Table S7). The RGR and age at flowering were both better supported by PC2 (Fig. 1B; Supplementary Table S6), with RGR associated with lower gene expression (mostly opposed to GH3.2, NPR1, and SPL7 expression) and age at flowering with higher expression of these same genes. Genotype-mean values of gene expression can be found in Supplementary Table S8.
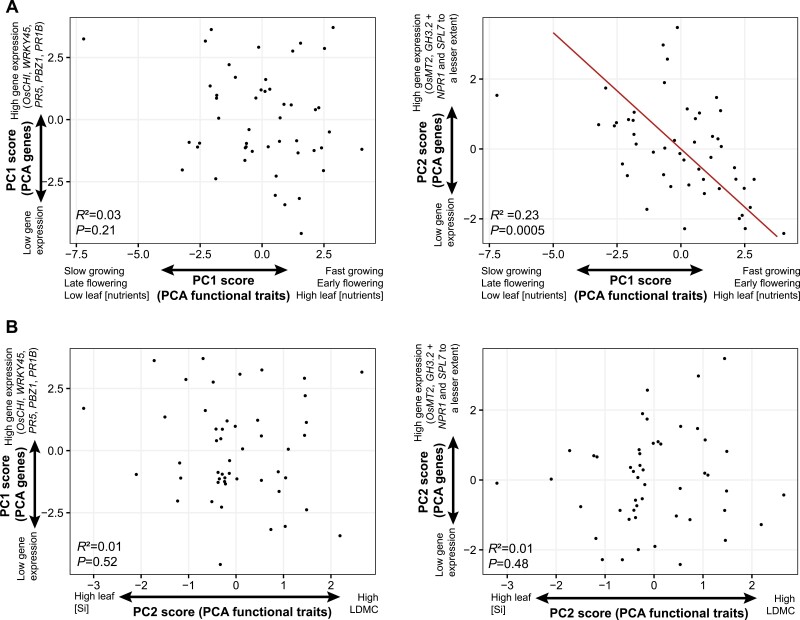
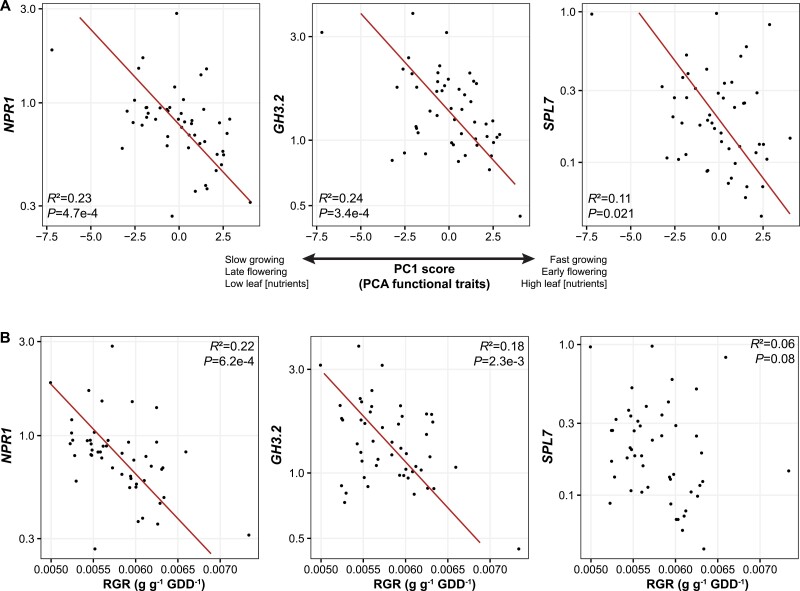
Genotype scores on PC1 of the PCA considering functional traits (slow–fast continuum) were not correlated with those on PC1 of the PCA considering gene expression (Fig. 3A). However, they were negatively correlated with those on PC2 of the PCA considering gene expression (Fig. 3A), on which OsMT2, GH3.2, and, to a lesser extent, NPR1 and SPL7 were represented. The same observation was made when RGR was considered instead of genotype scores on PC1 of the PCA considering traits (Supplementary Fig. S3). Considering each gene individually, genotype scores on the slow–fast axis were negatively correlated with the expression of NPR1, GH3.2, and SPL7 (Fig. 4A), but not with the expression of other genes (data not shown). The same observation was made when RGR was considered instead of PCA genotype scores (PC1), at least for NPR1 and GH3.2 (Fig. 4B).
Fig. 3.
Relationships between PCA (shown in Fig. 1) axis scores. (A) Relationships between genotype scores on PC1 of the PCA considering functional traits (slow–fast continuum) and both PCs of the PCA considering gene expression. (B) Relationships between genotype scores on PC2 of the PCA considering functional traits (Si–LDMC axis) and both PCs of the PCA considering gene expression. Standardized major axis regression lines and statistics of bivariate relationships are given.
Fig. 4.
Relationships between the expression of NPR1, GH3.2, and SPL7 and the genotype scores on PC1 of the PCA considering functional traits (slow–fast continuum) (A) and the genotypes relative growth rates (RGR) (B). Standardized major axis regression lines and statistics of bivariate relationships are given. y-axes are on a logarithmic scale.
Genotype scores on PC2 of the PCA considering rice functional traits (leaf [Si] versus LDMC axis) were not related to scores on PCs of the PCA genes (Fig. 3B), nor to gene expression considered individually.
Discussion
The present study showed that ‘live slow–die old’ rice genotypes grown in optimal conditions and without biotic stress exhibited greater expression of OsNPR1, OsSPL7, and OsGH3.2 (Figs 3, 4), which are thought to suppress growth and promote immunity in rice (Fu et al., 2011; Du et al., 2012; Ning et al., 2017; Liu et al., 2019), while fast genotypes exhibited lower expression of these genes. This result shows evidence of a growth–immunity trade-off in rice. Interestingly, this trade-off was less supported when traits considered as physical defences—leaf [Si] and leaf dry matter content (LDMC)—were considered, since these two traits were not aligned with the slow–fast continuum (Fig. 1). In fact, leaf [Si] tended to be higher among ‘live fast–die young’ genotypes. Our results also showed that, although genes involved in immunity tended to co-vary among each other, two independent molecular pathways for resistance appeared to dominate among the 49 rice genotypes (OsNPR1 versus OsWRKY45 (sub)pathways) (Fig. 1).
Understanding how life history strategies and the ‘slow–fast continuum’ align with key traits remains a key challenge in trait-based ecology. Here, we found strong evidence for this in a crop species, with fast-growing acquisitive and early-flowering genotypes contrasting with late-flowering genotypes with slower growth rates (Fig. 1), in accordance with other studies using non-crop species (e.g. Vasseur et al., 2012; Sartori et al., 2019). Interestingly, we found that leaf [S] was well-aligned with the main axis of trait variation and correlated with leaf [N], and [P], in line with interspecific trends and consistent with its role in protein synthesis (Fratte et al., 2021). More surprisingly, traits considered as mechanical defences or involved in stress tolerance, such as LDMC and leaf [Si], were not aligned with this axis. Species with high LDMC are generally more resistant to stresses and have slower growth rates and decomposition rates (Blumenthal et al., 2020). Its non-alignment with the LES traits was therefore somewhat surprising (e.g. Wang et al., 2022), and suggests that C allocation to growth is not necessarily accompanied by less investment in structural tissues in rice.
Regarding Si, associating its concentration with either plant growth or defence is challenging because both have been demonstrated and the cost of silicification remains unclear, making potential links between leaf [Si] and key traits hard to capture (Massey et al., 2007; de Tombeur et al., 2023). That said, leaf [Si] was correlated positively with leaf [N] and chlorophyll fluorescence, confirming global interspecific trends and suggesting a potential role of Si in photosynthesis (Detmann et al., 2012; de Tombeur et al., 2023). Finally, the results also suggest a trade-off between LDMC and leaf [Si] among rice genotypes (Fig. 2)—in contrast to global interspecific trends where both traits are positively correlated (de Tombeur et al., 2023)—perhaps as different solutions to minimize the impact of some biotic and abiotic stresses and/or improving leaf mechanical properties. Interestingly, a negative relationship between leaf [Si] and leaf mass per area (LMA) has already been reported in rice (de Tombeur et al., 2021), in line with the present result and reinforcing the linkages and potential trade-offs between silicification and leaf morphological traits such as LDMC, LMA, and leaf thickness (de Tombeur et al., 2023).
Since rice is an important crop and the most advanced model species for monocotyledonous, understanding its basal immunity level is important for several purposes (Chen and Ronald, 2011). Similar to salicylic acid concentration (Silverman et al., 1995), basal immunity levels can be used as a proxy for disease resistance against several pathogens in rice (Vergne et al., 2010; Grand et al., 2012), but this has never been investigated in such a large intraspecific range of cultivars. Unlike in Arabidopsis, in rice defence signalling after pathogen infection branches into two gene subpathways controlled by two gene regulators, OsNPR1 and OsWRKY45 (Nakayama et al., 2013; Mutuku et al., 2015; van Butselaar and Van Den Ackerveken, 2020). Knockdown experiments have demonstrated that both transcriptional regulators are essential but antagonistic for induced resistance in rice. For instance, some immunity-related gene activation relies on OsWRKY45, but is suppressed by OsNPR1 (Ning et al., 2017). Our results confirmed these two independent pathways at the intraspecific level (Fig. 1), suggesting different strategies of rice to activate defence in the japonica subgroup. Previously differences in the expression of OsWRKY45 between indica and japonica subgroups were reported with different impact on rice resistance to Xanthomonas oryzae pv. oryzae (Deng et al., 2018), but this regulation was not described in the japonica subgroup. Moreover, the link between these two pathways and trade-off regulation between growth and immunity has also been reported (van Butselaar and Van Den Ackerveken, 2020). For instance, IPA1 (the IDEAL PLANT ARCHITECTURE 1 gene in rice) gene can regulate either growth or immunity via OsWRKY45 (Wang et al., 2018).
Although the non-alignment of LDMC and leaf [Si] with the slow–fast axis shown in the rice phenotypic space does not support the hypothesis of a growth–defence trade-off, such a hypothesis was strongly supported by genetic data (Figs 3, 4). In particular, OsNPR1, SPL7, and GH3.2 were more constitutively expressed among live slow–die old genotypes (Fig. 4), having more ‘conservative’ strategies. First, OsNPR1 is well-known to both inhibit growth and enhance resistance to pathogens (Sugano et al., 2010; Li et al., 2016; Ning et al., 2017; van Butselaar and Van Den Ackerveken, 2020). Second, SPL7 overexpression can reduce tiller numbers in rice (Dai et al., 2018) and, more generally, this gene plays a critical role in plant growth and balancing reactive oxygen species during biotic and abiotic stress (Hoang et al., 2019; Liu et al., 2019). Finally, GH3.2 can mediate basal resistance by suppressing indole-3-acetic acid amido synthetase (the major form of auxin in rice) accumulation (Fu et al., 2011), and is involved in growth regulation and abiotic stress resistance (Du et al., 2012; Kong et al., 2019). Overall, this result supports the linkage between genes identified as key regulators of the growth–immunity trade-off and traits underlying the LES/slow–fast continuum framework. It is important to note, however, that plants have been grown without fungal infection or herbivores in this study. The fact that constitutive expression of immunity-related genes is a proxy of resistance against blast fungus in rice (Grand et al., 2012) suggests that the live fast–die young genotypes identified here will be more susceptible to infection, but future studies should now consider the susceptibility of these same genotypes to pathogens and herbivores. In addition to this, our results raise several other avenues for future research.
First, combining ecological and molecular approaches should be made more consistently and in different environmental conditions for a better mechanistic understanding of trait co-variation and functional trait frameworks. Ultimately, specific genes could be identified as key players in the growth–immunity trade-offs at the phenotypic level (Strauss et al., 2002; Endara and Coley, 2011; Garcia et al., 2021), but also more generally for trait co-variations highlighted in trait-based ecology (e.g. Díaz et al., 2016). Second, how environmental parameters and trade-offs in resource allocation shape species distribution and/or population genetic variation in natural ecosystems is still not well understood (but see Cope et al., 2021), and could gain from studies combining molecular and trait-based approaches. Finally, one of the key challenges in crop science is to breed varieties resistant to pathogens and herbivores, while being productive and fast-growing (Karasov et al., 2017; Ning et al., 2017; van Butselaar and Van Den Ackerveken, 2020; He et al., 2022). On the one hand, our study clearly identified a growth–immunity trade-off that could be difficult to overcome by artificial selection. On the other hand, our multidimensional approach also identified phenotypic axes of variation that are independent of this trade-off (in particular the LDMC–leaf [Si] axis) and could be considered in breeding strategies, after assessing the benefits of these mechanical defences/stress-resistance traits. This kind of phenotype-based optimization has already been suggested in the context of plant adaptation to abiotic stresses to, for instance, overcome the drought-resistance–early growth trade-off in rice (e.g. increased growth with unchanged drought resistance through larger and thicker leaves; Rebolledo et al., 2013; Luquet et al., 2016). Overall, functional space approaches, such as those widely used in ecology (Mouillot et al., 2021), can open new avenues for crop science and plant breeding, beyond traditional trait-by-trait approaches.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Relationships between genotype scores on both PCs of the PCA considering functional traits (Fig. 1A) and genotypes relative growth rate (RGR) and age at flowering.
Fig. S2. Bivariate relationships between traits used to build the phenotypic space (Fig. 1A).
Fig. S3. Relationships between genotype scores on both PCs of the PCA considering gene expression (Fig. 1B) and genotype relative growth rates.
Table S1. List of the 49 rice genotypes used in this study, and countries of origin.
Table S2. IDs and origins of marker genes used for expression analysis.
Table S3. Results of the principal component analysis based on a correlation matrix of the main plant traits for 49 rice genotypes, as shown in Fig. 1A (PC1 and PC2).
Table S4. Standardized major axis statistics of bivariate relationships between the main plant traits considered in this study.
Table S5. Genotype-mean values of traits used in the principal component analysis describing the phenotypic space of rice (Fig. 1A; Supplementary Table S3).
Table S6. Results of the principal component analysis based on a correlation matrix of gene expression involved in plant defence for 49 rice genotypes, as shown in Fig. 1B (PC1 and PC2).
Table S7. Standardized major axis statistics of bivariate relationships between gene expression considered in this study.
Table S8. Genotype-mean values of relative gene expression used in the PCA (Fig. 1B; Supplementary Table S6).
Acknowledgements
We are grateful to Maëva Tremblay, Elodie Certenais, Ana Elkaïm, Thierry Mathieu, Pauline Durbin, Hubert Vo Van, Fabien Lopez, and David Degueldre for their invaluable help both in the field and in the lab. We also thank the TE platform team of the LabEx CeMEB for all the help provided to conduct the experiment.
Contributor Information
Felix de Tombeur, CEFE, Univ Montpellier, CNRS, EPHE, IRD, Montpellier, France; School of Biological Sciences and Institute of Agriculture, The University of Western Australia, Perth, Australia.
Rémi Pélissier, PHIM Plant Health Institute, Univ Montpellier, Institut Agro, INRAE, CIRAD, Montpellier, France.
Ammar Shihan, CEFE, Univ Montpellier, CNRS, EPHE, IRD, Montpellier, France.
Koloina Rahajaharilaza, Faculty of Sciences, DS Life and Environmental Sciences, University of Antananarivo 101, Antananarivo, Madagascar; CIRAD, UMR AGAP Institut, F-34398 Montpellier, France.
Florian Fort, CEFE, Univ Montpellier, Institut Agro, CNRS, EPHE, IRD, Univ Valéry, Montpellier, France.
Lucie Mahaut, CEFE, Univ Montpellier, CNRS, EPHE, IRD, Montpellier, France.
Taïna Lemoine, CEFE, Univ Montpellier, CNRS, EPHE, IRD, Montpellier, France.
Sarah J Thorne, School of Biosciences, University of Sheffield, Sheffield, UK.
Sue E Hartley, School of Biosciences, University of Sheffield, Sheffield, UK.
Delphine Luquet, CIRAD, UMR AGAP Institut, F-34398 Montpellier, France; UMR AGAP Institut, Univ Montpellier, CIRAD, INRAE, Institut Agro, Montpellier, France.
Denis Fabre, CIRAD, UMR AGAP Institut, F-34398 Montpellier, France; UMR AGAP Institut, Univ Montpellier, CIRAD, INRAE, Institut Agro, Montpellier, France.
Hans Lambers, School of Biological Sciences and Institute of Agriculture, The University of Western Australia, Perth, Australia.
Jean-Benoît Morel, PHIM Plant Health Institute, Univ Montpellier, Institut Agro, INRAE, CIRAD, Montpellier, France.
Elsa Ballini, PHIM Plant Health Institute, Univ Montpellier, Institut Agro, INRAE, CIRAD, Montpellier, France.
Cyrille Violle, CEFE, Univ Montpellier, CNRS, EPHE, IRD, Montpellier, France.
Ros Gleadow, Monash University, Australia.
Author contributions
RP, AS, KR, FF, DL, J-BM, EB, and CV planned and designed the research. FdT, RP, AS, KR, FF, and ST performed experiments. FdT and RP analysed the data and wrote the first version of the manuscript, with inputs from all the authors. FdT and RP contributed equally.
Conflict of interest
All authors declare that they have no conflicts of interest.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 101021641 (project SiliConomic granted to FdT), and was supported by the European Research Council (ERC) Starting grant ‘Ecophysiological and biophysical constraints on domestication in crop plants’ (grant ERC-StG-2014-639706-CONSTRAINTS) awarded to CV. This work was also supported by the ANR MUSE (ANR-16-IDEX-0006 AMUSER).
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Agrawal AA. 2007. Macroevolution of plant defense strategies. Trends in Ecology and Evolution 22, 103–109. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Fishbein M.. 2006. Plant defense syndromes. Ecology 87, 132–149. [DOI] [PubMed] [Google Scholar]
- Ahammed GJ, Yang Y.. 2021. Mechanisms of silicon-induced fungal disease resistance in plants. Plant Physiology and Biochemistry 165, 200–206. [DOI] [PubMed] [Google Scholar]
- Albert CH, Grassein F, Schurr FM, Vieilledent G, Violle C.. 2011. When and how should intraspecific variability be considered in trait-based plant ecology? Perspectives in Plant Ecology, Evolution and Systematics 13, 217–225. [Google Scholar]
- Biscarini F, Cozzi P, Casella L, et al. 2016. Genome-wide association study for traits related to plant and grain morphology, and root architecture in temperate rice accessions. PLoS One 11, e0155425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecker SW, McCulley RL, Chadwick OA, Kelly EF.. 2006. Biologic cycling of silica across a grassland bioclimosequence. Global Biogeochemical Cycles 20, doi: 10.1029/2006GB002690. [DOI] [Google Scholar]
- Blumenthal DM, Mueller KE, Kray JA, Ocheltree TW, Augustine DJ, Wilcox KR.. 2020. Traits link drought resistance with herbivore defence and plant economics in semi-arid grasslands: the central roles of phenology and leaf dry matter content. Journal of Ecology 108, 2336–2351. [Google Scholar]
- Chen X, Ronald PC.. 2011. Innate immunity in rice. Trends in Plant Science 16, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley PD, Bryant JP, Chapin FS.. 1985. Resource availability and plant antiherbivore defense. Science 230, 895–899. [DOI] [PubMed] [Google Scholar]
- Cope OL, Keefover-Ring K, Kruger EL, Lindroth RL.. 2021. Growth-defense trade-offs shape population genetic composition in an iconic forest tree species. Proceedings of the National Academy of Sciences, USA 118, e2103162118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois B, Frouin J, Greco R, et al. 2012. Genetic diversity and population structure in a European collection of rice. Crop Science 52, 1663–1675. [Google Scholar]
- Dai Z, Wang J, Yang X, Lu H, Miao X, Shi Z.. 2018. Modulation of plant architecture by the miR156f–OsSPL7–OsGH3.8 pathway in rice. Journal of Experimental Botany 69, 5117–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tombeur F, Cooke J, Collard L, Cisse D, Saba F, Lefebvre D, Burgeon V, Nacro HB, Cornelis J-T.. 2021. Biochar affects silicification patterns and physical traits of rice leaves cultivated in a desilicated soil (Ferric Lixisol). Plant and Soil 460, 375–390. [Google Scholar]
- de Tombeur F, Raven JA, Toussaint A, et al. 2023. Why do plants silicify? Trends in Ecology and Evolution 38, 275–288. [DOI] [PubMed] [Google Scholar]
- Delteil A, Blein M, Faivre-Rampant O, Guellim A, Estevan J, Hirsch J, Bevitori R, Michel C, Morel JB.. 2012. Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defence. Molecular Plant Pathology 13, 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Liu H, Zhou Y, Zhang Q, Li X, Wang S.. 2018. Exploring the mechanism and efficient use of a durable gene-mediated resistance to bacterial blight disease in rice. Molecular Breeding 38, 18. [Google Scholar]
- Detmann KC, Araújo WL, Martins SCV, Sanglard LMVP, Reis JV, Detmann E, Rodrigues FÁ, Nunes-Nesi A, Fernie AR, Damatta FM.. 2012. Silicon nutrition increases grain yield, which, in turn, exerts a feed-forward stimulation of photosynthetic rates via enhanced mesophyll conductance and alters primary metabolism in rice. New Phytologist 196, 752–762. [DOI] [PubMed] [Google Scholar]
- Díaz S, Kattge J, Cornelissen JHC, et al. 2016. The global spectrum of plant form and function. Nature 529, 167–171. [DOI] [PubMed] [Google Scholar]
- Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L.. 2012. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. Journal of Experimental Botany 63, 6467–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endara MJ, Coley PD.. 2011. The resource availability hypothesis revisited: a meta-analysis. Functional Ecology 25, 389–398. [Google Scholar]
- Fagerström T. 1989. Anti-herbivory chemical defense in plants: a note on the concept of cost. The American Naturalist 133, 281–287. [Google Scholar]
- Ferreira RB, Monteiro S, Freitas R, Santos CN, Chen Z, Batista LM, Duarte J, Borges A, Teixeira AR.. 2006. Fungal pathogens: the battle for plant infection. Critical Reviews in Plant Sciences 25, 505–524. [Google Scholar]
- Fratte MD, Pierce S, Zanzottera M, Cerabolini BEL.. 2021. The association of leaf sulfur content with the leaf economics spectrum and plant adaptive strategies. Functional Plant Biology 48, 924–935. [DOI] [PubMed] [Google Scholar]
- Frontini M, Boisnard A, Frouin J, Ouikene M, Morel JB, Ballini E.. 2021. Genome-wide association of rice response to blast fungus identifies loci for robust resistance under high nitrogen. BMC Plant Biology 21, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S.. 2011. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiology 155, 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A, Martinez M, Diaz I, Santamaria ME.. 2021. The price of the induced defense against pests: a meta-analysis. Frontiers in Plant Science 11, 615122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR.. 1989. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochimica et Biophysica Acta – General Subjects 990, 87–92. [Google Scholar]
- Grand X, Espinoza R, Michel C, Cros S, Chalvon V, Jacobs J, Morel JB.. 2012. Identification of positive and negative regulators of disease resistance to rice blast fungus using constitutive gene expression patterns. Plant Biotechnology Journal 10, 840–850. [DOI] [PubMed] [Google Scholar]
- Hahn PG, Maron JL.. 2016. A framework for predicting intraspecific variation in plant defense. Trends in Ecology and Evolution 31, 646–656. [DOI] [PubMed] [Google Scholar]
- Hallik L, Niinemets U, Wright IJ.. 2009. Are species shade and drought tolerance reflected in leaf-level structural and functional differentiation in Northern Hemisphere temperate woody flora? New Phytologist 184, 257–274. [DOI] [PubMed] [Google Scholar]
- He Z, Webster S, He SY.. 2022. Growth–defense trade-offs in plants. Current Biology 32, R634–R639. [DOI] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ.. 1992. The dilemma of plants: to grow or defend. The Quarterly Review of Biology 67, 283–335. [Google Scholar]
- Hoang TV, Vo KTX, Rahman MM, Choi SH, Jeon JS.. 2019. Heat stress transcription factor OsSPL7 plays a critical role in reactive oxygen species balance and stress responses in rice. Plant Science 289, 110273. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Johnson J. 2014. Accurate measurements of low Z elements in sediments and archaeological ceramics using portable X-ray fluorescence (PXRF). Journal of Archaeological Method and Theory 21, 563–588. [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Karasov TL, Chae E, Herman JJ, Bergelson J.. 2017. Mechanisms to mitigate the trade-off between growth and defense. The Plant Cell 29, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Zhong H, Deng X, Gautam M, Gong Z, Zhang Y, Zhao G, Liu C, Li Y.. 2019. Evolutionary analysis of GH3 genes in six Oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. Japonica. Plants 8, 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F.. 2008. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software 25, 1–18. [Google Scholar]
- Lee KJ, Kim DI, Choi DH, Lee BW.. 2015. Rice grain-filling characteristics under elevated air temperature in a temperate region. Journal of Crop Science and Biotechnology 18, 231–236. [Google Scholar]
- Li X, Yang D-L, Sun L, Li Q, Mao B, He Z.. 2016. The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-amido synthase expression. Plant Physiology 172, 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi Z, Zhang X, et al. 2019. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nature Plants 5, 389–400. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luquet D, Rebolledo C, Rouan L, Soulie C, Dingkuhn M.. 2016. Heuristic exploration of theoretical margins for improving adaptation of rice through crop-model assisted phenotyping. In: Yin X, Struik P, eds. Crop systems biology. Cham: Springer, 105–127. [Google Scholar]
- Mason CM, Bowsher AW, Crowell BL, Celoy RM, Tsai CJ, Donovan LA.. 2016. Macroevolution of leaf defenses and secondary metabolites across the genus Helianthus. New Phytologist 209, 1720–1733. [DOI] [PubMed] [Google Scholar]
- Mason CM, Donovan LA.. 2015. Does investment in leaf defenses drive changes in leaf economic strategy? A focus on whole-plant ontogeny. Oecologia 177, 1053–1066. [DOI] [PubMed] [Google Scholar]
- Massey FP, Ennos AR, Hartley SE.. 2007. Grasses and the resource availability hypothesis: the importance of silica-based defences. Journal of Ecology 95, 414–424. [Google Scholar]
- Massey F, Hartley S.. 2009. Physical defences wear you down: progressive and irreversible impacts of silica on insect herbivores. Journal of Animal Ecology 78, 281–291. [DOI] [PubMed] [Google Scholar]
- Moore JP, Taylor JE, Paul ND, Whittaker JB.. 2003. Reduced leaf expansion as a cost of systemic induced resistance to herbivory. Functional Ecology 17, 75–81. [Google Scholar]
- Mouillot D, Loiseau N, Grenié M, et al. 2021. The dimensionality and structure of species trait spaces. Ecology Letters 24, 1988–2009. [DOI] [PubMed] [Google Scholar]
- Mutuku JM, Yoshida S, Shimizu T, Ichihashi Y, Wakatake T, Takahashi A, Seo M, Shirasu K.. 2015. The WRKY45-dependent signaling pathway is required for resistance against Striga hermonthica parasitism. Plant Physiology 168, 1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama A, Fukushima S, Goto S, et al. 2013. Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biology 13, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Liu W, Wang GL.. 2017. Balancing immunity and yield in crop plants. Trends in Plant Science 22, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Paul-Victor C, Züst T, Rees M, Kliebenstein DJ, Turnbull LA.. 2010. A new method for measuring relative growth rate can uncover the costs of defensive compounds in Arabidopsis thaliana. New Phytologist 187, 1102–1111. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/ [Google Scholar]
- Rebolledo MC, Luquet D, Courtois B, Henry A, Soulié J-C, Rouan L, Dingkuhn M.. 2013. Can early vigour occur in combination with drought tolerance and efficient water use in rice genotypes? Functional Plant Biology 40, 582–594. [DOI] [PubMed] [Google Scholar]
- Reich PB. 2014. The world-wide ‘fast-slow’ plant economics spectrum: a traits manifesto. Journal of Ecology 102, 275–301. [Google Scholar]
- Reidinger S, Ramsey MH, Hartley SE.. 2012. Rapid and accurate analyses of silicon and phosphorus in plants using a portable X-ray fluorescence spectrometer. New Phytologist 195, 699–706. [DOI] [PubMed] [Google Scholar]
- Sartori K, Vasseur F, Violle C, et al. 2019. Leaf economics and slow-fast adaptation across the geographic range of Arabidopsis thaliana. Scientific Reports 9, 10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatin DV, Gramegna G, Modesti V, Cervone F.. 2014. Wounding in the plant tissue: the defense of a dangerous passage. Frontiers in Plant Science 5, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman P, Seskar M, Kanter D, Schweizer P, Métraux J-P, Raskin I.. 1995. Salicylic acid in rice. Plant Physiology 108, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms EL, Rausher MD.. 1987. Costs and benefits of plant resistance to herbivory. The American Naturalist 130, 570–581. [Google Scholar]
- Strauss SY, Rudgers JA, Lau JA, Irwin RE.. 2002. Direct and ecological costs of resistance to herbivory. Trends in Ecology and Evolution 17, 278–285. [Google Scholar]
- Sugano S, Jiang C-J, Miyazawa S-I, Masumoto C, Yazawa K, Hayashi N, Shimono M, Nakayama A, Miyao M, Takatsuji H.. 2010. Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Molecular Biology 74, 549–562. [DOI] [PubMed] [Google Scholar]
- van Butselaar T, Van Den Ackerveken G.. 2020. Salicylic acid steers the growth–immunity tradeoff. Trends in Plant Science 25, 566–576. [DOI] [PubMed] [Google Scholar]
- Vasseur F, Violle C, Enquist BJ, Granier C, Vile D.. 2012. A common genetic basis to the origin of the leaf economics spectrum and metabolic scaling allometry. Ecology Letters 15, 1149–1157. [DOI] [PubMed] [Google Scholar]
- Vázquez-González C, Sampedro L, Rozas V, Zas R.. 2020. Climate drives intraspecific differentiation in the expression of growth-defence trade-offs in a long-lived pine species. Scientific Reports 10, 10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne E, Grand X, Ballini E, Chalvon V, Saindrenan P, Tharreau D, Nottéghem JL, Morel JB.. 2010. Preformed expression of defense is a hallmark of partial resistance to rice blast fungal pathogen Magnaporthe oryzae. BMC Plant Biology 10, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volante A, Desiderio F, Tondelli A, et al. 2017. Genome-wide analysis of japonica rice performance under limited water and permanent flooding conditions. Frontiers in Plant Science 8, 1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JJ, Zhou L, Shi H, et al. 2018. A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang Z, Huang H, Wang H, Peñuelas J, Sardans J, Niinemets U, Niklas KJ, Li Y, Xie J, Wright IJ.. 2022. Leaf water content contributes to global leaf trait relationships. Nature Communications 13, 5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton DI, Duursma RA, Falster DS, Taskinen S.. 2012. smatr 3 – an R package for estimation and inference about allometric lines. Methods in Ecology and Evolution 3, 257–259. [Google Scholar]
- Warton DI, Wright IJ, Falster DS, Westoby M.. 2006. Bivariate line-fitting methods for allometry. Biological Reviews of the Cambridge Philosophical Society 81, 259–291. [DOI] [PubMed] [Google Scholar]
- Wright IJ, Cannon K.. 2001. Relationships between leaf lifespan and structural defences in a low-nutrient, sclerophyll flora. Functional Ecology 15, 351–359. [Google Scholar]
- Wright IJ, Westoby M, Reich PB, et al. 2004. The worldwide leaf economics spectrum. Nature 428, 821–827. [DOI] [PubMed] [Google Scholar]
- Züst T, Agrawal AA.. 2017. Trade-offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Annual Review of Plant Biology 68, 513–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.