Summary
Decidual leukocytes play key roles in maternal-fetal tolerance and immunity. Here, we present detailed methods to purify, culture, and functionally analyze human placental dNK, dTreg, dTem, and dMɸ from decidua parietalis, the maternal part of the placental membranes; decidua basalis, the maternal part of the placenta; and placental villi. These sites have high clinical relevance in the development of villitis and chorioamnionitis. This allows in-depth phenotypic and functional investigation of placental immune populations and their interactions with extravillous trophoblasts.
For complete details on the use and execution of this protocol, please refer to Ikumi et al.,1 Tilburgs et al.,2 Salvany-Celades et al.,3 Crespo et al.,4 van der Zwan et al.5
Subject areas: Cell Biology, Cell culture, Cell isolation, Cell separation/fractionation, Developmental biology, Immunology
Graphical abstract

Highlights
-
•
Protocol to obtain high yield and high viability human placental leukocytes
-
•
Method to obtain maternal leukocytes from 3 placental sites simultaneously
-
•
Isolation of maternal decidual leukocytes and fetal HLA-G+ EVT from a single placenta
-
•
Co-cultures and functional assays to accurately model maternal-fetal interactions
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Decidual leukocytes play key roles in maternal-fetal tolerance and immunity. Here, we present detailed methods to purify, culture, and functionally analyze human placental dNK, dTreg, dTem, and dMɸ from decidua parietalis, the maternal part of the placental membranes; decidua basalis, the maternal part of the placenta; and placental villi. These sites have high clinical relevance in the development of villitis and chorioamnionitis. This allows in-depth phenotypic and functional investigation of placental immune populations and their interactions with extravillous trophoblasts.
Before you begin
Institutional permissions
Laboratory experiments using human specimens and involving potentially infectious pathogens require approved institutional review board (IRB) and biosafety protocols. Inquire at your Institution for how to obtain the required approvals and do not start work before the approvals have been obtained.
Biological samples
Human placental tissues including placental membranes obtained after delivery between 18 and 42 weeks of gestation. Placental tissues from 5 - 18 weeks of gestation can also be used, however often using these earlier placental tissues, collection of placental membranes is difficult and only decidua basalis can be used for leukocyte isolation.
General laboratory preparation
Timing: 1–2 h
-
1.
All experiments should be performed in a class II biosafety (BSL2) cabinet.
-
2.
Set up humidified CO2 incubator at 37°C.
-
3.
Set up a shaking water bath at 37°C.
-
4.
Prepare all media and stock solutions.
Collection of placental tissues
Timing: 30 – 60 min
-
5.
After delivery collect the placenta in wash medium A (see Materials and equipment) in a clean and sterile container. Wash Medium A can be prepared ahead of time and stored up to 1 week at 4°C until use.
-
6.
Keep the placenta at room temperature until processing.
Note: The placental tissues should be stored at room temperature at all times. Refrigerated but not frozen tissues can be used but will result in low lymphocyte yields.
-
7.
Start processing the placenta within 5 h after delivery to obtain maximum leukocyte yield and viability.
Preparation of BSL2 cabinet for specimen dissection
Timing: 20 min
-
8.
Place a container with 10% bleach, biocidal or virucidal solution to collect liquid waste in the BSL2 cabinet.
-
9.
Place a second waste container with a biohazard bag to discard solid waste including contaminated plastics in the BSL2 cabinet.
-
10.
Place absorbent pads (linen savers), sterile dissection tools (sterile curved tip tweezers and sterile 6 ½” scissors), tube racks, sterile 50 mL tubes, sterile PBS, wash medium A and B in the BSL2 cabinet.
-
11.
Thaw collagenase and DNase solutions at room temperature.
-
12.
Set up 50 mL conical tubes containing 30 mL PBS for the collection of placental tissue pieces during dissection.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45 - Pacific Orange; HI30 (1:50) | Invitrogen | RRID:AB_10376143; Cat #MHCD4530 |
| CD14 - PerCP; HCD14 (1:50) | BioLegend | RRID:AB_2564058; Cat #301847 |
| CD56 - PE; HCD56 (1:50) | BioLegend | RRID:AB_604093; Cat #318305 |
| CD3 - PerCP; UCHT1 (1:50) | BioLegend | RRID:AB_893300; Cat #300427 |
| CD4 - PerCP; SK3 (1:50) | BioLegend | RRID:AB_2563326; Cat #344624 |
| CD8 - Pacific Orange; 3B5 (1:50) | Invitrogen | RRID:AB_10372066; Cat #MHCD0830 |
| CD45RA - Alexa700; HI100 (1:100) | BioLegend | RRID:AB_493762; Cat #304119 |
| CD25 - PE; 2A3 (1:30) | BD Biosciences | RRID:AB_2783790; Cat #341011 |
| PD1 – PE/Cy7; EH12.1 (1:50) | BD Biosciences | RRID:AB_10611585; Cat #561272 |
| TIGIT - APC; MBSA43 (1:20) | eBioscience | RRID:AB_2573305; Cat #17-9500-42 |
| FOXP3 - Pacific Blue; 259D (1:30) | BioLegend | RRID:AB_940354; Cat #320215 |
| HELIOS - Alexa488; 22F6 (1:30) | BioLegend | RRID:AB_10645334; Cat #137213 |
| IFNγ - APC; B27 (1:50) | BioLegend | RRID:AB_315443; Cat #506510 |
| TNFα - Pacific Blue; Mab11 (1:50) | BioLegend | RRID:AB_528965; Cat #502920 |
| CD107a - PerCP/Cy5.5; H4A3 (1:100) | BioLegend | RRID:AB_1227508; Cat #328616 |
| IgG1 - PerCP/Cy5.5; MOPC-21 (1:50) | BioLegend | RRID:AB_893664; Cat #400150 |
| KIR2DL1 – PE/Cy7; Clone HP-MA4 (1:50) | BioLegend | RRID:AB_11415800; Cat #39512 |
| 7AAD (1:20) | Thermo Fisher | Cat #A1310 |
| Biological samples | ||
| Human placental samples | Local hospital and clinics | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Percoll | GE Healthcare | Cat #17-5445-02 |
| DNase I | Sigma | Cat #DN25 |
| Collagenase IV | Sigma-Aldrich | Cat #C5138 |
| Recombinant human EGF | PeproTech | Cat #AF-100-15 |
| Human gonadotropic hormone (HcG) | Sigma-Aldrich | Cat #C1063 |
| Ionomycin | Sigma | Cat #I9657 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat #P1585 |
| New born calf serum | GIBCO | Cat #16010159 |
| Human AB serum | Corning | Cat #35060CI |
| Fetal bovine serum (FBS) | Seradigm | Cat #1500-500 |
| LPS | Sigma-Aldrich | Cat #916374 |
| Selenium, transferrin, 100× | Gibco | Cat #41400-045 |
| 10 × PBS | Fisher | Cat #BP399-4 |
| Penicillin and streptomycin | Gibco | Cat #15140-122 |
| RPMI 1640 | Gibco | Cat #11875-093 |
| X-vivo Medium | Lonza | Cat #04-380Q |
| DMEM/F12 | Gibco | Cat #11320-033 |
| Human fibronectin | Corning | Cat #354008 |
| Compensation Beads, Anti-Mouse Ig K/ Negative Control Compensation Particles Set | BD Biosciences | Cat #552843 |
| Critical commercial assays | ||
| FOXP3 fix and perm kit | Invitrogen | Cat #00-5523-00 |
| Software and algorithms | ||
| BD FACSDIVA software | BD Biosciences | http://www.bdbiosciences.com/us/instruments/research/software/flowcytometry-acquisition/bd-facsdivasoftware/m/111112/features |
| FlowJo software | FlowJo | https://www.flowjo.com/ |
| Other | ||
| Cell dissociation sieve | Millipore Sigma | CD1-1KT (mesh size 60) |
| Cell strainers 40 μm, 70 μm, 100 μm | Thermo Fisher | Cat #22363547; Cat #22363548; Cat #22363549 |
| 15 mL Centrifuge tubes | Thermo Fisher | Cat #14-955-238 |
| 50 mL Conical centrifuge tubes | Thermo Fisher | Cat #14-955-240 |
| Curved tip tweezers | ||
| 6 ½” scissors | ||
| Cell sorter | BD Aria/Sony Nano/ Bigfoot | |
| Flow cytometer analyzer | BD Fortessa/Cytek Aurora | |
| −20°C freezer | ||
| −80°C freezer | ||
| 4°C fridge | ||
| Liquid nitrogen storage tank | ||
| BSL2 biosafety cabinet | ||
| Shaking water bath 37°C | ||
| Centrifuge | ||
| 37°C 5% CO2 Incubator | ||
Materials and equipment
Wash Medium A
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | 445 mL | |
| Fetal Bovine Serum (FBS) | 10% | 50 mL |
| Penicillin and Streptomycin 100× | 1× | 5 mL |
| Total | 500 mL |
Can be stored for up to 1 week at 4°C
Wash Medium B
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | 445 mL | |
| Fetal Bovine Serum (FBS) | 10% | 50 mL |
| Penicillin and Streptomycin 100× | 1× | 5 mL |
| DNase I | 0.1 mg/ mL | 5 mg |
| Total | 500 mL |
Can be stored for up to 1 week at 4°C
Tissue Digestion Enzyme Cocktail A
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | 500 mL | |
| Collagenase IV | 1 mg/ mL | 500 mg |
| DNase | 0.1 mg/ mL | 50 mg |
| Total | 500 mL |
Can be stored or up to 3 months at −20°C
100% Percoll
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll | 90% | 900 mL |
| 10× PBS | 10% | 100 mL |
Can be stored for up to 1 month at 4°C
70% Percoll
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll 100% | 70% | 70 mL |
| RPMI 1640 | 30% | 30 mL |
Can be stored for up to 1 week at 4°C
50% Percoll
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll 100% | 50% | 50 mL |
| RPMI 1640 | 50% | 50 mL |
Can be stored for up to 1 week at 4°C
45% Percoll
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll 100% | 45% | 45 mL |
| 1× PBS | 55% | 55 mL |
Can be stored for up to 1 week at 4°C
Cell culture Medium A
| Reagent | Final concentration | Amount |
|---|---|---|
| X-vivo Medium | 44.5 mL | |
| Human Serum | 5% | 5 mL |
| Penicillin and Streptomycin 100× | 1× | 0.5 mL |
| Total | 50 mL |
Can be stored for up to 1 week at 4°C
Cell culture Medium B
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F12 | 44 mL | |
| Insulin-Transferrin-Selenium, Ethanolamine, 100× (ITS-X) | 1× | 0.5 mL |
| Penicillin and Streptomycin 100× | 1× | 0.5 mL |
| Human chorionic gonadotropin (HcG) (2.5 × 106 units/mL) | 400 units/mL | 8 μL |
| Epidermal growth Factor (EGF) (100 μg/mL) | 5 ng/mL | 2.5 μL |
| New born Calf serum | 10% | 5 mL |
| Total | 50 mL |
Should be made fresh on the day of use.
Freeze Medium A
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI | 4.9 mL | |
| FBS | 40% | 4 mL |
| Penicillin and Streptomycin 100× | 1× | 100 μL |
| DMSO | 10% | 1 mL |
| Total | 10 mL |
Should be made fresh on the day of use.
Step-by-step method details
Tissue dissection
Timing: 1 h
This step details how to dissect and collect decidua parietalis, decidua basalis and villous tissues.
-
1.Remove the amnion by placing the placenta with the fetal surface (umbilical cord side) facing upwards.
-
a.Manually peel off the amnion from the chorion towards the umbilical cord (Figure 1A).
-
b.Remove and discard the amnion completely.
-
a.
-
2.Collect decidua parietalis by gently scraping the decidua parietalis from the chorion using sterile curved tip forceps (Figure 1B).
-
a.Avoid necrotic tissue and blood clots.
-
b.Collect 20–50 mL of tissue in a labelled 50 mL conical tube containing 1× PBS.
-
a.
-
3.Obtain decidua basalis and villous tissues by turning over the placenta to have the maternal surface (basal plate with cotyledons) facing upwards.
-
a.Carefully cut thin pieces of approximately L 2 × W 1 × H 0.5 cm3 from the placenta basal plate using sterile scissors (Figure 1C).
-
b.Avoid necrotic tissue and blood clots.
-
c.Then carefully remove villi from the decidua basalis with scissors.
-
d.Collect villi in a labelled 50 mL conical tube containing 1× PBS.
-
e.Remove all villi until decidua basalis is a clean 1–2 mm thin membrane (Figures 1D–1G).
-
f.Collect 20–50 mL villi and 20–50 mL decidua basalis in labelled 50 mL conical tube containing 1× PBS.
-
a.
-
4.
For first trimester decidual tissue, macroscopically separate decidual tissue from villi. Avoid necrotic tissue. Collect 10–30 mL of tissue in a 50 mL conical tube containing 1× PBS.
-
5.
Continue to process decidua parietalis, decidua basalis and villous tissues separately.
Figure 1.
Tissue dissection
(A) Remove the amnion by placing the placenta with the fetal surface facing upwards and manually peeling off the amnion from the chorion, peeling towards the umbilical cord. Remove and discard the amnion completely.
(B) Decidua parietalis collection is done by gently scraping the decidua parietalis from the chorionic membrane using sterile curved tip forceps.
(C) To collect decidua basalis and villous tissues, turn over the placenta to have the maternal side basal plate with cotyledons facing upwards and carefully cut thin pieces from the placenta basal plate with sterile scissors.
(D–E) carefully remove villi from decidua basalis with scissors until (F–G) decidua basalis is a clean 1–2 mm thin membrane. Collect 20–50 mL of each tissue in a 50 mL conical containing 1× PBS.
Tissue washing and digestion
Timing: 2.5 h
This step details how to wash and then digest the placental tissues.
-
6.Wash the tissues without centrifugation by adding sterile 1× PBS into each of the conical tubes,
-
a.Wait until the tissue settles in the lower half of the tube (for approximately 1 min)
-
b.Carefully discard the supernatant by either pouring it out or using a serological pipette.
-
c.Fill up the tubes with PBS and wash all tissues several times until the PBS is clear and there is no blood visible.
-
d.If more than 20 mL of tissue was collected during dissection, split the tissue in 2 or more tubes while washing.
-
a.
-
7.
Cut the tissue into small pieces (1–2 mm2) inside the 50 mL conical tube using scissors.
-
8.
Wash the tissues without centrifugation by adding sterile 1× PBS into each of the conical tubes until the PBS is clear as described in step 6.
-
9.
After the last wash step, let the tissue settle for 2 min to the bottom of the 50 mL conical tube until no debris are visible in the PBS supernatant, measure and write down tissue volume.
-
10.
Divide the tissue into 50 mL conical tubes to obtain 10 mL of tissue per tube. Fill up each tube with serum-free RPMI and centrifuge at 650 g for 1 min.
-
11.Carefully discard the supernatant. Add serum free RPMI to obtain a total RPMI and tissue volume of 20 mL. Add 10 mL of the enzyme cocktail A (see materials and equipment) per tube.
-
a.Enzyme cocktail A aliquots can be prepared and stored for up to 3 months at −20°C until ready to use.
-
a.
-
12.
Wrap the tubes with parafilm to ensure they are well sealed to prevent spillage and place the tubes horizontally in a shaking 37°C water bath at 50 rpm for 75–90 min.
Filtration and density gradient centrifugation
Timing: 3 h
This step details how to filter placental tissues after digestion and how to load suspensions on Percoll gradients for density gradient centrifugation.
-
13.Remove the tubes from the water bath and remove the parafilm.
-
a.Fill up the tubes with wash medium B (see materials and equipment). Wash Medium B can be prepared ahead of time and stored for up to 1 week at 4°C until use.
-
b.Let the tissue settle to the bottom of the tube for approximately 1 min
-
c.Filter the supernatant through a metal dissociation sieve placed in a 15 cm petri dish.
-
d.Gently stir the solution through the sieve with the back of a transfer pipette to strain the supernatant into the petri dish.
-
e.Fill up the tube with wash medium B to resuspend the remaining tissue and repeat steps 13b-d once.
-
a.
-
14.Transfer the remaining tissue into the metal dissociation sieve,
-
a.add 10–30 mL wash medium B.
-
b.gently stir the solution through the sieve with the back of a transfer pipette to strain the supernatant into the petri dish.
-
c.Discard the remaining tissue fragments.
-
a.
CRITICAL: When stirring the tissue and liquid in the strainer do not attempt to grind the remaining tissue through the filter as this will result in impurities.
-
15.
Filter the supernatants from the petri dish progressively through 100 μm, 70 μm and then 40 μm cell strainers into 50 mL conical tubes.
-
16.
Centrifuge filtered supernatants at 650 g for 7 min, discard the supernatant and gently resuspend the cell pellet by pipetting up and down with a sterile bulb pipette.
-
17.
Add wash medium A in a volume equal to the tissue volume measured in step 9.
-
18.Add a 1:1 volume of Percoll 50% solution (see Materials and equipment) to the cell suspension to achieve a final Percoll concentration of 25%.
-
a.All Percoll solutions can be prepared ahead of time and stored for up to 1 week at 4°C until ready to use.
-
a.
-
19.
Layer the Percoll gradients in 50 mL conical tubes using 10 mL of 70% Percoll, 15 mL of 45% Percoll, 20 mL of 25% Percoll containing the cells and 5 mL PBS as described in: Figure 2 and Methods Videos S1, S2, and S3.
Figure 2.
Percoll gradient
(A) The Percoll gradient is loaded by making layers of 10 mL 70% Percoll (1.082 g/mL), 15 mL 45% Percoll (1.053 g/mL), and 20 mL 25% Percoll (1.029 g/mL) with the isolated cell fractions in suspension and 5 mL PBS.
(B) After density gradient centrifugation the lymphocyte enriched fraction is collected from the 1.082 / 1.053 g/mL interface and macrophage enriched fractions from the 1.053 / 1.029 g/mL interface.
(C) The pellet and the proteins, fats and debris are discarded; (C) a photo of a Percoll gradient after density gradient centrifugation.
CRITICAL: Practice to make Percoll gradients with perfectly separated layers; fuzzy layers result in diminished cell yields. Use bulb transfer pipettes to carefully layer the different concentrations of Percoll on top of one another without mixing. Touching the tip of the pipette to the wall of the tube helps to control the loading (see attached Methods Videos S1, S2, and S3). Loading Percoll gradients using 4°C Percoll solutions generates better results.
-
20.
Gently load the tubes in a centrifuge and centrifuge the samples at 800 g for 30 min with no brake.
-
21.
Once the centrifugation is complete, there will be three distinct cell fractions visible in each tube (Figure 2).
-
22.
Using a sterile bulb pipette carefully collect the top layer containing debris and discard this fraction.
-
23.
Using a sterile bulb pipette carefully collect the macrophage band at the 45%/25% interface and transfer it into a sterile conical tube.
-
24.
Using a sterile bulb pipette carefully collect the lymphocyte band at the 70%/45% interface and transfer it into a sterile conical tube.
-
25.
For both macrophage and lymphocyte fractions, transfer a maximum of 20 mL of cells into one 50 mL conical tube and fill up the tubes with wash medium A and centrifuge at 800 g for 7 min.
CRITICAL: With the transfer of cells, some Percoll from the gradient is transferred to the 50 mL conical tube. Not sufficiently diluting this Percoll with wash medium will result in excessive cell loss, as cells will not deposit properly. After centrifuging the cells, take care to notice if there are cells attached to the walls of the tube and make sure to collect them in the bottom of the tube while resuspending.
-
26.
Carefully decant your supernatant and resuspend cell pellets in 0.5–1 mL wash medium B. Count cells.
Note: The cell pellet now contains the isolated and purified cells for immune-phenotyping or cell sorting by flow cytometry.
-
27.
For cell sorting, bring cells up to a concentration of about 2 × 106 cell per mL add anti-CD45, CD14, CD56, CD8 and CD4 antibodies listed for 30 min on ice and in the dark.
-
28.
Add 10 mL wash medium B and centrifuge cells at 800 g for 7 min. Discard supernatant and carefully resuspend cell pellets in 0.5–1 mL wash medium B.
-
29.
Filter cells through a 40 μm cell strainer and transfer to a 5 mL sterile FACS tube with cap.
-
30.
Add a viability dye e.g., 7AAD prior to cell sorting and sort live CD45+CD14-CD56+ single dNK cells, CD45+CD14-CD56-CD4+ and CD45+CD14-CD56-CD8+ T cells according to the gating strategy depicted in Figure 3.
-
31.
After cell sorting, centrifuge all cell fractions at 650 g for 7 min, discard supernatant and resuspend purified lymphocytes at 1 × 106 cells per mL in cell culture medium A. For dNK cells add 2.5 ng/mL IL-15 and for T cells add 50 Units IL-2.
-
32.
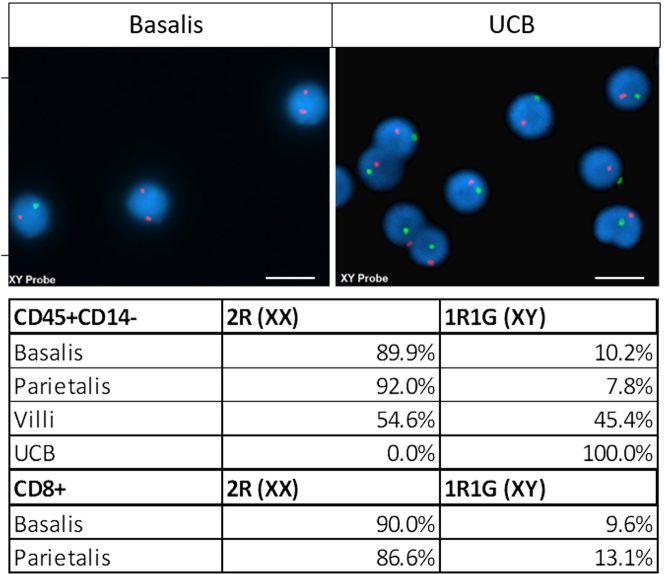
Upon FACS sort, purified leukocyte fractions were submitted to the Cincinnati Children’s Hospital Medical Centre (CCHMC) cytology core to determine maternal (XX) and male fetal (XY) cells (Figure 4).
Figure 3.
Gating strategy for cell sorting
Gating strategy of a representative term placenta decidua parietalis lymphocyte sample is shown. First, leukocytes are selected by gate R1 in the FSC-A / SSC-A plot. Second, doublets are excluded by gate R2 in the FSC-A / FSC-H plot and R3 in the SSC-A / SSC-H plot. CD45 and 7AAD are plotted and CD45+7AAD- live cells are selected by R4. Next CD45 and CD14 are plotted to select CD45+CD14+ macrophages and CD45+CD14- lymphocytes (R5). Lymphocytes (R5) are then plotted for CD45 and CD56 to select CD45+CD56+ dNK (sort gate I) and CD45+CD56- lymphocytes (R6). (F) CD45+CD56- lymphocytes (R6) are then plotted for CD4 and CD8 to select CD45+CD56-CD4+ and CD45+CD56-CD8+ T cells (sort gates II and III). CD3 and TCR staining are not used during live cell sorting to avoid T cell activation. Decidual macrophages can be sorted from the decidual macrophage preparation using R1 + R2 + R3 + R4 and then selecting CD45+CD14+ cells.
Figure 4.
Delineation of maternal and fetal CD45+CD14- lymphocytes
Representative microscopy images of Fluorescent In Situ Hybridization (FISH) for X (Red) and Y (Green) chromosomes in leukocytes isolated from the decidua basalis (left) and Umbilical Cord Blood (right). The table depicts percentages of female (2R, XX) and Male (1R1G, XY) cells in CD45+CD14- lymphocytes and CD8+ T cells isolated from the decidua basalis, decidua parietalis, villous tissue and cord blood. The scale bars indicate 10 μm.
Cryopreservation
Timing: 20 min
This step details how to cryo preserve decidual lymphocyte fractions.
-
33.
Sorted decidual lymphocyte fractions can be frozen for future research.
-
34.Following step 31, add 1 mL freeze medium A to 0.5–10 × 106 cells. Freeze Medium A should be made fresh on the day of use.
-
a.Transfer cells to sterile 1.2 mL cryo tube.
-
b.Slow freeze cells in cool cell at −80°C
-
c.After 2 days transfer vials to liquid nitrogen for long term storage.
-
a.
Note: freezing and thawing decidual lymphocyte fractions may result in the loss of over 50%–80% of viable cells. Particularly, freezing <2 × 106 decidual lymphocyte will result in high cell loss.
Co-culture of placental lymphocytes with sample matched fetal HLA-G+ EVT
Timing: 1 h
This step details how to set up co-cultures of placental lymphocyte fractions with sample matched fetal primary HLA-G+ EVT2,3,6.
-
35.
Isolate primary HLA-G+ EVT from the same placenta or use established HLA-G+ EVT-like cell lines according to the protocols described by Hamilton et al. 20237.
-
36.
Seed 5 × 104 HLA-G+ EVT in 100 μL Cell Culture Medium B on a collagen or fibronectin coated flat bottom 48 well culture plate.
-
37.
Add 1 × 105 purified decidual or peripheral blood NK cells, T cells or macrophages in 100 μL cell culture medium A to the HLA-G+ EVT. Cell culture medium A and B should always be mixed 1:1 to maintain EVT and leukocyte viability.
-
38.
For negative controls include additional wells only containing HLA-G+ EVT or leukocyte fractions.
-
39.
For positive controls include additional wells with 1 μg/mL PMA and 1 μg/mL Ionomycin (NK cells and T cells; 1 μg/mL anti-CD3/CD28 beads (T cells) or 100 ng/mL LPS (macrophages).
Determination of Treg induction by HLA-G+ EVT using flow cytometry
Timing: 4 day incubation and 5–6 h processing
This step details how to determine Treg induction by HLA-G+ EVT using Flow cytometry of CD4+ T cells and EVT co-cultures.2,3
-
40.
After a 4-day (96 h) co-culture of HLA-G+ EVT and T cells, harvest T cells using a p200 pipette and transfer cells to a 5 mL FACS tube. Rinse the wells twice with 200 μL cell culture medium A to make sure all T cells are collected.
-
41.
Spin once at 7 min at 800 g and put on ice.
-
42.
Add antibodies for Treg cell surface antigens including but not limited to CD45, CD4, CD45RA, CD25, PD1 and TIGIT. Incubate 30 min on ice.
-
43.
Wash cells with 2 mL cell culture medium A and spin once at 7 min at 800 g.
-
44.
Use the ebiosciences FOXP3 fix and perm kit according to manufacturer instructions and add antibodies for intracellular detection of FOXP3 and HELIOS. Incubate 20 min on ice.
-
45.
Wash cells with 2 mL cell culture medium A and spin once at 7 min at 800 g.
-
46.
Add 0.2 mL cell culture medium A and acquire cells on 8+ parameter flow cytometer.
-
47.
Analyze cells according to strategy depicted in Figure 5.
Figure 5.
Co-culture of CD4+ T cells with EVT increases Treg populations
CD4+ T cells were cultured alone (A) or with primary HLA-G+ EVT (B) for 4 days and analyzed for the expression of CD25, FOXP3, PD1 and TIGIT. Co-culture with HLA-G+ EVT increased the frequency of FOXP3+ and PD1+ cells as well as the Mean Fluorescence Intensity of FOXP3. A representative example of HLA-G+EVT from term placenta decidua basalis is shown.
Cytokine secretion in response to HLA-G+ EVT
Timing: 4–24 h incubation and 1 h processing
This step details how to determine cytokine secretion by decidual immune cells in response to HLA-G+ EVT.2,3,8
-
48.
To detect cytokine secretion of placental NK cells, T cells and macrophages in response to HLA-G+ EVT, set up co-cultures as described in step 35–39.
-
49.
Incubate co-cultures for 4–24 h in an incubator at 37°C with 5% CO2.
-
50.
Collect 150 μL supernatant in a non-stick 0.7 mL tube and spin once at 800 g for 7 min to remove cell debris.
-
51.
Transfer two times 70 μL supernatant to clean non-stick 0.7 mL tubes and freeze directly in −80°C until use.
-
52.
Analyze supernatants by ELISA or multiplex assay.
Degranulation and intracellular cytokine responses to HLA-G+ EVT
Timing: 4–16 h incubation and 1 h processing
This step details how to determine degranulation and intracellular cytokines responses of decidual immune cells in response to HLA-G+ EVT.4,8
-
53.
To detect degranulation and cytokine production of NK and T cells in response to EVT, to each co-culture described in 35–37, add CD107a PerCP/Cy5.5 or IgG2a PerCP/Cy5.5 control antibodies at a final concentration of 250 ng/mL.
-
54.
Incubate co-cultures for 4–16 h in an incubator at 37°C with 5% CO2.
-
55.
Add monensin for the last 4–6 h of the co-culture to detect intracellular cytokines.
-
56.
Using a p200 pipette transfer lymphocytes to a 5 mL FACS tube. Rinse the wells twice with 200 μL cell culture medium A to make sure all lymphocytes are collected.
-
57.
Spin once at 7 min at 800 g and put on ice.
-
58.
Add antibodies for lymphocyte cell surface antigens including but not limited to CD45, CD4, CD8, CD45RA, CD56 and KIR2DL1. Incubate 30 min on ice.
-
59.
Wash cells with 2 mL cell culture medium A and spin once at 7 min at 800 g.
-
60.
Use the BD cytofix/ cytoperm kit according to manufactures instructions and thereafter add antibodies for intracellular detection of IFNγ and TNFα. Incubate 20 min on ice.
-
61.
Wash cells with 2 mL cell culture medium A and spin once at 7 min at 800 g.
-
62.
Add 0.2 mL cell culture medium A and acquire cells on 8+ parameter flow cytometer.
-
63.
Analyze cells according to the strategy depicted in Figure 6.
Figure 6.
dNK do not degranulate or produce cytokines in response to HLA-G+ EVT
(A–C) dNK were cultured alone (unstimulated), with PMA/I or with primary HLA-G+ EVT for 16 h in the presence of CD107a antibodies and analyzed for the percentage of (A) CD107a+ cells, (B) IFNɣ+ cells and (C) TNFα+ cells. Co-culture with PMA/I resulted in high CD107a, IFNɣ and TNFα expression whereas HLA-G+ EVT did not elicit responses above the unstimulated control levels.
Expected outcomes
This protocol is useful for the isolation and purification of leukocytes from the human placenta. Expected cell yield are include in Table 1. Furthermore, in Figures 3 and 4 we demonstrate how these cells can be analyzed for functional changes upon co-cultures with HLA-G+ EVT.
Table 1.
Leukocyte yield after FACS sort
| Decidua parietalis | Decidua basalis | Villi | |
|---|---|---|---|
| NK cells >37 week | 0.2–0.8 × 106 | 0.3–1.0 × 106 | 0.1–0.3 × 106 |
| CD4+ T cells >37 week | 0.3–0.8 × 106 | 0.3–1.0 × 106 | 0.4–1.2 × 106 |
| CD8+ T cells >37 week | 0.3–0.8 × 106 | 0.3–1.0 × 106 | 0.4–1.2 × 106 |
| CD14+ macrophages >37 week | 0.1–0.5 × 106 | 0.2–0.6 × 106 | 0.1–0.5 × 106 |
Limitations
The protocol works best when using fresh placental tissues obtained within 2 h after delivery. Tissues up to 10 h after delivery can be used but generally result in lower cell yields.9 The placental tissues should be stored at room temperature at all times. Refrigerated but not frozen tissues can be used but will result in low lymphocyte yields.
Troubleshooting
Problem 1
Cell yields should be within the ranges depicted in (Table 1). Cell yields below the expected range can be improved through the following adjustments.
Potential solutions
-
•Optimize tissue digestion, steps 23–24:
-
○If tissue digestion is ineffective and after 75 min of digestion more than 5 mL of the original 10 mL tissue is left, the Tissue Digestion Enzyme Cocktail A may have lost activity. This will result in low cell yields. Increase Tissue Digestion Enzyme Cocktail A concentration in step 11 or prepare new stocks of Tissue Digestion Enzyme Cocktail A.
-
○If during tissue digestion the fluid viscosity increases significantly and gelatin like tissue blobs appear, the DNase in Tissue Digestion Enzyme Cocktail A may have lost its activity. This will result in low cell viability and yields. Add an additional 0.1–0.2 mg/ mL DNAse to the tissue in step 11 or prepare new Tissue Digestion Enzyme Cocktail A stocks.
-
○If tissue digestion is excessive and tissue volume after 45 min of digestion is less than 2 mL, this may also result in low cell viability and yields. Reduce Tissue Digestion Enzyme Cocktail A concentration by 25%–50% (step 11) to slow down the digestion.
-
○
-
•Optimize Percoll gradient centrifugation, steps 30–36 and Methods Videos S1, S2, and S3.
-
○If Percoll rings look fuzzy without clearly separated bands, this will significantly reduce cell yields.
-
○
-
•Use freshly collected placental tissues (preferably within 2 h after delivery) at room temperature for best cell yields and viability, steps 5–7.
-
○Refrigerated or old (>5 h after delivery) placentas will have significantly lower cell yields and cell viability.
-
○
Resource availability
All of the reagents used and listed in the key resources table can be substituted by similar reagents from other vendors.
Lead contact
Further information should be directed to and will be fulfilled by the lead contact Tamara Tilburgs email: Tamara.Tilburgs@cchmc.org.
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We would like to thank Sherry Thornton and the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center for all their help with flow cytometry and cell sorting, all nurses and physicians of participating hospitals for their efforts in collecting placental materials, and all Tilburgs Lab members and CCHMC immunology faculty for their helpful discussions. This work was supported by Cincinnati Children’s Research Foundation (CCRF) Tilburgs Lab start-up funds (T.T.), the CCRF Trustee Award (T.T.), Burroughs Wellcome Fund Next Gen Pregnancy Award #NGP10115 (T.T.), and the March of Dimes Ohio Collaborative grant (T.T.). N.I. is supported by a fellowship from the AXA Research Fund, Paris, and the National Institute for Health Research (NIHR) (17/63/26). Part of the work was also funded by the National Institutes of Health R01HD102050 (C.G.).
Author contributions
Designed research, N.I., C.G., T.T.; performed research, N.I., Z.K., S.M., T.T.; analyzed data, N.I., Z.K., S.M., T.T.;, visualized data, N.I., Z.K., T.T.; writing - original draft, N.I. and T.T.; writing - review and editing, Z.K., S.M., C.G.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102277.
Contributor Information
Nadia M. Ikumi, Email: nadia.ikumi@uct.ac.za.
Tamara Tilburgs, Email: tamara.tilburgs@cchmc.org.
Data and code availability
This study did not generate or analyze datasets.
References
- 1.Ikumi N.M., Pillay K., Tilburgs T., Malaba T.R., Dzanibe S., Enninga E.A.L., Chakraborty R., Lamorde M., Myer L., Khoo S., et al. T-cell homeostatic imbalance in placentas from women with human immunodeficiency virus in the absence of vertical transmission. J. Infect. Dis. 2021;224:S670–S682. doi: 10.1093/infdis/jiab192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tilburgs T., Crespo Â.C., van der Zwan A., Rybalov B., Raj T., Stranger B., Gardner L., Moffett A., Strominger J.L. Human HLA-G+ extravillous trophoblasts: immune-activating cells that interact with decidual leukocytes. Proc. Natl. Acad. Sci. USA. 2015;112:7219–7224. doi: 10.1073/pnas.1507977112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvany-Celades M., van der Zwan A., Benner M., Setrajcic-Dragos V., Bougleux Gomes H.A., Iyer V., Norwitz E.R., Strominger J.L., Tilburgs T. Three types of functional regulatory T cells control T cell responses at the human maternal-fetal interface. Cell Rep. 2019;27:2537–2547.e5. doi: 10.1016/j.celrep.2019.04.109. [DOI] [PubMed] [Google Scholar]
- 4.Crespo Â.C., Mulik S., Dotiwala F., Ansara J.A., Sen Santara S., Ingersoll K., Ovies C., Junqueira C., Tilburgs T., Strominger J.L., Lieberman J. Decidual NK cells transfer granulysin to selectively kill bacteria in trophoblasts. Cell. 2020;182:1125–1139.e18. doi: 10.1016/j.cell.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Zwan A., Bi K., Norwitz E.R., Crespo A.C., Claas F.H.J., Strominger J.L., Tilburgs T. Mixed signature of activation and dysfunction allows human decidual CD8+ T cells to provide both tolerance and immunity. Proc. Natl. Acad. Sci. USA. 2018;115:385–390. doi: 10.1073/pnas.1713957115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papuchova H., Kshirsagar S., Xu L., Bougleux Gomes H.A., Li Q., Iyer V., Norwitz E.R., Strominger J.L., Tilburgs T. Three types of HLA-G+ extravillous trophoblasts that have distinct immune regulatory properties. Proc. Natl. Acad. Sci. USA. 2020;117:15772–15777. doi: 10.1073/pnas.2000484117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton I., Ikumi N.M., Kshirsagar S., Goodman W., Tilburgs T. Utilizing primary HLA-G+ extravillous trophoblasts and HLA-G+ EVT-like cell lines to study maternal-fetal interactions. STAR Protocols. 2023;4:102276. doi: 10.1016/j.xpro.2023.102276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crespo Â.C., Strominger J.L., Tilburgs T. Expression of KIR2DS1 by decidual natural killer cells increases their ability to control placental HCMV infection. Proc. Natl. Acad. Sci. USA. 2016;113:15072–15077. doi: 10.1073/pnas.1617927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Egmond A., van der Keur C., Swings G.M.J.S., van Beelen E., van Zijl L., Scherjon S.A., Claas F.H.J. Preservation of human placenta facilitates multicenter studies on the local immune response in normal and aberrant pregnancies. J. Reprod. Immunol. 2013;98:29–38. doi: 10.1016/j.jri.2013.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets.