Summary
The retina is a notable tissue with high metabolic needs which relies on specialized vascular networks to protect the neural retina while maintaining constant supplies of oxygen, nutrients, and dietary essential fatty acids. Here we analyzed the lipidome of the mouse retina under healthy and pathological angiogenesis using the oxygen-induced retinopathy model. By matching lipid profiles to changes in mRNA transcriptome, we identified a lipid signature showing that pathological angiogenesis leads to intense lipid remodeling favoring pathways for neutral lipid synthesis, cholesterol import/export, and lipid droplet formation. Noteworthy, it also shows profound changes in pathways for long-chain fatty acid production, vital for retina homeostasis. The net result is accumulation of large quantities of mead acid, a marker of essential fatty acid deficiency, and a potential marker for retinopathy severity. Thus, our lipid signature might contribute to better understand diseases of the retina that lead to vision impairment or blindness.
Subject areas: Molecular mechanism of behavior, Metabolomics
Graphical abstract

Highlights
-
•
Integrated lipidomic & transcriptomic study of the oxygen-induced retinopathy (OIR)
-
•
OIR increases triglyceride/cholesteryl ester synthesis for lipid droplet formation
-
•
Cholesterol import/export pathways are increased in retinopathy
-
•
OIR increases retinal n-9 lipids (mead acid), a possible marker of disease severity
Molecular mechanism of behavior; Metabolomics
Introduction
Lipid metabolism plays a central role in retina health and disease. For proper biological functioning, the retina depends upon a series of specialized dietary lipids, such as retinol and highly unsaturated fatty acids (HUFA), including docosahexaenoic 22:6 (DHA) and arachidonic 20:4 (ARA) acids. And, similarly to tissues with high metabolic demands such as the heart and skeletal muscle, the retina also relies on glucose and fatty acid β-oxidation in order to supply the energetic needs of the highly demanding photoreceptor cells.1 The vascular network is another notable feature of the retina which, as part of the central nervous system, is protected by specialized barriers: the outer and the inner blood-retina barriers.2 The latter provides nutrients and oxygen to the neural inner retina, while the former feeds the retinal-pigmented epithelial (RPE) layer, which protects the inner retina and nourishes the photoreceptor cells. Thus, it is not surprising that changes that affect retinal lipid metabolism and/or its delicate vascular network, have profound implications in tissue homeostasis and have been implicated as underlying mechanisms for many eye diseases.1,3,4,5,6 Together, these diseases lead to different degrees of vision impairment and blindness, and affect people of all ages, from premature babies to adults with important impact in our society and the health system.
To better understand how changes in the vasculature of the eye affect retinal lipid metabolism and therefore, retinal health, we studied a well-accepted animal model of retinal pathological angiogenesis using state-of-the-art lipidomic methods. The oxygen-induced retinopathy (OIR) mouse model reproduces many of the clinical features observed in premature babies suffering from retinopathy of prematurity (ROP) and to a lesser extent, diabetic retinopathy and other eye diseases.7,8 For instance, the alterations in the vasculature induced by varying retinal oxygen levels in the OIR model cause the loss of astrocytes and microglia,9 reduce neuronal function of the photoreceptor cells (rods and cones),10 and increase neuroinflammation.11,12,13 Here we show that these changes are coupled with intense lipid remodeling in the whole retina. Not surprising, these changes affect cholesterol metabolism, which is essential for neuronal growth and survival since they need cholesterol to repair and build more membranes,14,15 as wells as the metabolism of very long-chain fatty acids, whose oxygenated derivatives (elovanoids) modulate the viability and function of photoreceptor cells and RPE undergoing uncompensated oxidative stress.16,17
Angiogenesis participates in several diseases, collectively known as angiogenesis-dependent diseases.18 Indeed, inhibitors of angiogenesis are already in the clinic and have been effective to treat patients with illnesses as disparate as cancers and ophthalmic disorders, because they share in common the abnormal growth of new blood vessels (pathological angiogenesis). Thus, the OIR model is ideal to study not only retinopathy of prematurity but human angiogenesis-dependent diseases as a whole.7,19 Furthermore, the OIR model has many advantages to study pathological angiogenesis. It relays on isogenic animals (reducing genetic variability) and neovascularization is induced without direct manipulation of the retinas, which allows for good reproducibility while minimizing artifactual and undesired side effects (i.e., caused by the addition of exogenous recombinant growth factors or scaffolds necessary to induce angiogenesis in other mouse models). Additionally, in previous work, we performed a time course comparative transcriptomic analysis of the developmental (healthy) and diseased retinas (OIR model) to show that genes differentially expressed between the physiological and pathological conditions could be used as a new gene signature with high prognostic power for breast cancer, another angiogenesis-dependent disease.19 These data reinforce the notion that the retina and the OIR model are excellent tools to study human angiogenesis-dependent diseases.
Although the OIR model has been the subject of broad molecular characterization by transcriptomic,19,20,21,22,23 proteomic,24,25,26 and metabolomic27 analyses, to the best of our knowledge, to date no detailed lipidomic study has been performed using this mouse model. Here, we performed a global time course lipidomic analysis of pathological (OIR) and healthy retinas. The lipidomic profile of the retinopathic and healthy retinas was then compared with their respective transcriptome to unveil molecular pathways and lipid metabolites that were affected by pathological angiogenesis. Given that angiogenesis is also a hallmark for two of the major eye diseases that lead to blindness, diabetic retinopathy, and age-related macular degeneration, we trust this integrated analysis may provide valuable insights on lipid pathways that could be used as targets for future therapeutic approaches for retinopathy and other retinal diseases dependent on pathological angiogenesis.
Results
OIR promotes intense lipid remodeling in angiogenic retinas
To induce pathological angiogenesis in mice, we used the OIR model as previously described.19,28 In brief, seven-day-old mice (postnatal day 7, P7) with their nursing mothers were submitted to 75% oxygen for 5 days. The high oxygen levels prevent physiological angiogenesis by inhibiting vascular endothelial growth factor (VEGF) production during a crucial time in retinal development (Figure 1A). When mice return to atmospheric oxygen levels, now on P12, the poorly vascularized retina experiences a strong hypoxic condition, which dramatically increases Vegf expression beyond physiological levels. The net result is abnormal growth of blood vessels with the formation of neovascular tufts at the border between the avascular and vascular retina, along with a central area of vaso-obliteration (Figure 1B). To compare the global lipid expression and remodeling in the retina under physiological and the OIR pathological angiogenesis, we performed advanced non-targeted high-resolution lipidomic analysis using retinas from normally developing and OIR mice.
Figure 1.
OIR promotes intense lipid remodeling in angiogenic retinas
(A) Cartoon schematic of the OIR model.
(B) Healthy and OIR mice retinas showing central area vaso-obliteration and vascular tufts.
(C) Number of individual lipid molecular species and their respective lipid classes.
(D) Principal component analysis using the most abundant 300 lipid species.
(E) Heatmap of the top 50 significantly altered lipid molecular species (one-way ANOVA followed by Tukey’s post-hoc analysis, FDR-adjusted p value ≤0.05). Each horizontal row represents a molecular lipid; each vertical column represents a sample. Euclidean distance and Ward cluster algorithm were applied to build the heatmap. The color code bar indicates the log of the fold change of the mean concentration for a given lipid. See also Figures S1 and S2. Abbreviations for lipid categories. SP, sphingolipids; GP, glycerophospholipids; NL, neutral lipids; FFA, free fatty acids; CoQ, coenzyme Q. Lipid classes: Cer, ceramide; SM, sphingomyelin; CL, cardiolipin; LysoPC, lysophosphatidylcholine; PC, phosphatidylcholine; LysoPE, lysophosphatidylethanolamine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; Ch, free cholesterol; CE, cholesteryl ester; DG, diacylglycerol; TG, triacylglycerol; FFA, free fatty acid; o- and p-before lipid class mean plasmanyl- and plasmenyl-phospholipids, respectively.
We identified and quantified a total of 300 lipid species in both physiological (P) and pathological (R) conditions, which were distributed in 20 lipid classes belonging to five different categories: sphingolipids (SP), glycerophospholipids (GP), neutral lipids (NL), free fatty acids (FFA), and coenzyme Q (CoQ) (Figure 1C). Significant lipid remodeling was observed in physiological and pathological retinas with changes in 277 lipids (92% of total) (one-way ANOVA followed by Tukey’s test, FDR-adjusted p value <0.05). Principal component analysis (PCA) showed that there was a good agreement in the lipid profile of our replicas and that changes in retinal lipidome occurs mainly according to time (principal component 1, PC1) and the condition of the retina (physiological or pathological) (PC2) (Figure 1D). In sum, the physiological development of newborn mice retinas promotes, by itself, changes in their lipid profile, which is further affected by retinopathy.
A more detailed analysis was carried out to highlight the main changes induced by retinopathy, or pathological angiogenesis. Lipid alterations are clearly visible during the peak of neovascularization, at postnatal days 15 and 17 (independent of the condition, OIR or physiological retinas). Considering the 50 most significantly altered lipid species, all samples could be correctly grouped according to their lipid composition in time and condition of the retina (Figure 1E). The notable exceptions were the OIR samples at postnatal days 12 and 12.5 (R12 and R12.5), which were all grouped together. This suggests that the first 12 h of relative hypoxia do not induce major changes in retinal lipidome.
Interestingly, the most significantly altered lipid species correspond to neutral lipids, such as cholesteryl esters (CE) and triacylglycerols (TG), in addition to glycerophospholipids esterified to HUFA, for instance DHA (22:6) and ARA (20:4), and very long-chain fatty acids (VLCFA) with more than 30 carbons (Figure 1E). Other main differences: (i) P12, R12, and R12.5 are specially enriched in phosphatidylcholine species linked to saturated and monounsaturated fatty acids (SFA and MUFA, respectively), besides CE molecular species. Concentrations of these species then decrease as time progress, being especially low in the P17 group (physiological condition); (ii) R12 and/or R12.5 also present significant lower concentrations of total lipids, diacylglycerol (DG), phosphatidylglycerol (PG), and sphingomyelin (SM) (Figure S1A), in addition to precursors for the n-3 and n-6 series of fatty acids (Figure S1B); (iii) in general, higher concentrations of TG are observed at days 15 and 17 (P15, R15, P17 and R17), and these lipid species are specially enriched in retinopathy (R17), particularly those linked to DHA and ARA. Finally, (iv) glycerophospholipids esterified to HUFA and VLCFA are enriched at postnatal days 15 and 17, the majority of them in the physiological condition (P15 and P17).
Most of these findings were confirmed by pairwise comparisons (Figure S2). We observed that, at all postnatal days, the pathological condition induced a decrease in the concentration of phospholipids linked to HUFA, including phosphatidylcholines esterified to highly unsaturated VLCFA, with the exception of P15 versus R15, in which no significant change was observed for many of these lipid species (fold change ≥1.5 with FDR-adjusted p value ≤0.05). In OIR, decreased concentrations were also observed for TG species linked to polyunsaturated fatty acids (PUFA, with 2 or 3 unsaturations) and highly unsaturated fatty acids (HUFA, with more than 3 unsaturations) (P12 x R12; P12 x R12.5), in addition to poly- and highly unsaturated CE species (P12 x R12; P12 x R12.5; and P15 x R15). In contrast, a massive increase in the concentration of TG lipid species was detected at postnatal days 15 and mainly 17 in OIR retinas. A similar increase occurred to CE molecular species in OIR retinas at postnatal day 17 (P17 x R17).
Substantial increase in neutral lipid concentration is linked to the formation of lipid droplets and occurrence of reverse cholesterol transport in pathological retinas
OIR clearly drives a substantial increase in total neutral lipid levels, particularly total CE at postnatal day 17, and total TG at postnatal days 15 and 17 (Figure 2A). All CE 15 molecular species were increased in the pathological condition at postnatal day 17 (R17 group), regardless of the degree of unsaturation (Figure 2B). Of these 15 species, 14 were significantly augmented in pathological retinas when compared to healthy retinas (fold change ≥1.5, FDR-adjusted p value ≤0.05) (Figure 2C). The same pattern of increased concentration in retinopathy was observed for all 67 TG species at postnatal day 17 (Figure S3A). In total, 55 TG species had significant increased concentrations due to the disease, regardless of the degree of unsaturation (Figure S3B).
Figure 2.
Increased neutral lipid synthesis and lipid droplets formation in pathological retinas
(A) Normalized mass values (ng of lipids/mg of proteins) for neutral lipid classes in control and OIR retinas. Data are represented as mean ± SEM (Mann-Whitney-Wilcoxon test, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
(B) Heatmap with all CE 15 lipid species at postnatal day 17. The color code bar indicates the log of the fold change of the mean concentration for each given CE.
(C) Pairwise comparisons of CE mass alterations promoted by OIR at postnatal day 17. X axis corresponds to log2(fold change) observed for each CE lipid species. Fold change was set to ≥1.5; FDR-adjusted p value ≤0.05 was used for significance.
(D) Schematic showing the synthesis pathways of TG and CE, neutral lipids stored in lipid droplets.
(E) Schematic showing the formation of CE in the extracellular medium, after efflux of cholesterol and action of LCAT in (apo)lipoproteins. In both diagrams, genes that are differentially expressed (p value ≤0.05) in pathological retinas are highlighted in red (increased expression) or blue (decreased expression) colors. Legend: (+ or -) 0.6≤ log2(fold change) < 1.0; (++ or --) 1.0≤ log2(fold change) < 2.0; (+++ or ---) 2.0≤ log2(fold change) < 3.0; (++++ or ----) 3.0≤ log2(fold change).
(F) Volcano plots with genes whose expression is increased (red) or decreased (blue) in OIR at postnatal days 12, 12.5, 15 and 17. Only genes mentioned in Figures 2D and 2E are shown. See also Figure S3 and Data S1. Abbreviations. G3P, glycerol-3-phosphate; LysoPA, lysophosphatidic acid; PA, phosphatidic acid; PL, phospholipid; MG, monoacylglycerol; LXR, liver X receptor.
Integration of lipidomic with transcriptomic data allowed to better delineate the neutral lipid synthesis pathways that are altered in OIR (Figures 2D and 2E). For the lipidomic-transcriptomic comparison analyses, we employed two criteria: (a) key regulatory genes whose expression levels were significantly changed, considering log2(fold change) > 1.5 or log2(fold change) < 0.5, and p value <0.05; these individual genes are cited in the text and figures. We also considered another situation (b) when the overall expression (regardless of the fold change value) of multiple genes in a given pathway was significantly altered (p value <0.05) in retinopathy (Data S1) (Figures 2D, 2E, and 2F). These altered pathways are indicated in the text and figures as well.
Glycerol-3-phosphate acyltransferase 3 (Gpat3), which converts glycerol-3-phosphate (G3P) to lysophosphatidic acid (LysoPA), is increased in OIR retinas at P15. Increases were also observed for 1-acylglycerol-3-phosphate O-acyltransferase 2 (Agpat2) and phospholipase D1 (Pld1); both lead to the production of phosphatidic acid (PA). Interestingly, phospholipase D family members 4 (Pld4) and 5 (Pld5) are also increased in the disease, although no phospholipase activity has been demonstrated for these isoenzymes29,30 (Figure 2D).
Next, different diacylglycerol-producing pathways were considerably up-regulated in the pathological condition: a) from PA to DG, through the catalysis by lipin 3 (Lpin3). The reverse reaction even seems down-regulated, since a series of diacylglycerol kinases (Dgkb, Dgkg, Dgkq and Dgkz) are moderately reduced in the disease; b) from monoacylglycerol (MG) to DG, with a strong increase in monoacylglycerol O-acyltransferase 2 (Mogat2) gene expression at postnatal days 15 and 17, and a slight decrease in diacylglycerol lipase alpha (Dagla) expression at the same times; c) from phosphatidylcholine (PC), through the action of sphingomyelin synthase 2 (Sgms2), leading to the formation of SM and DG as by-product (Figure 2D).
In the final step, DG is converted to TG through catalysis by diacylglycerol acyltransferase (DGAT) enzymes. Both Dgat1 and Dgat2 mRNAs were present and quantified in normal and OIR retinas, although only the expression of Dgat2 was slightly increased at postnatal day 12.5. Endothelial lipase (Lipg), which can act as a TG lipase, was significantly reduced in retinopathy, corroborating the TG accumulation observed in the lipidomic data. In addition to TG molecular species, the detected increase in CE may be linked to higher expression of the genes sterol O-acyltransferase 1 (Soat1) and especially sterol O-acyltransferase 2 (Soat2), the latter showing high fold change values in the pathological retinas at postnatal days 15 and 17. Indeed, molecular species of TG and CE are stored intracellularly in lipid droplets, and a number of structural markers of lipid droplets presented increased gene expression in retinopathy, including: perilipin 2 (Plin2), perilipin 3 (Plin3), fat storage-inducing transmembrane protein 1 (Fitm1), transmembrane protein 159 (Tmem159/Ldaf1), hypoxia inducible lipid droplet associated (Hilpda), caveolin 1, caveolae protein (Cav1), caveolae associated protein 1 (Cavin1), and caveolae associated protein 3 (Cavin3) (Figure 2D). Collectively, the transcriptomic and lipidomic data suggest that lipid droplet production in the retina is substantially augmented in the pathological retinas.
Transcriptomic analysis also showed that cholesterol import/export through the high-density lipoprotein (HDL) pathway is activated. The expression of the lecithin cholesterol acyltransferase (Lcat) gene is considerably increased in retinopathy at postnatal days 12.5, 15, and 17 (log2 fold change = 1.28, 1.17, and 1.10, respectively), pointing to an increase in the production/maturation of HDL (Figure 2E). The LCAT enzyme esterifies free cholesterol (Ch) on nascent lipoproteins that mediate intraretinal cholesterol transport.31,32 In pre-β-HDL, LCAT catalyses esterification and production of CE, leading to the formation of mature spherical HDL particles.33 Major protein components of HDL34 are also up-regulated in retinopathy according to expression data for the following genes: Lcat, alpha-2-macroglobulin (A2m), amylase 1 (Amy1), complement component 3 (C3), complement component 4B (C4b), clusterin (Clu), glutathione peroxidase 3 (Gpx3), granulin (Grn), hemopexin (Hpx), paraoxonase 1 (Pon1), paraoxonase 3 (Pon3), retinol binding protein 1 (Rbp1) and 4 (Rbp4), transferrin (Trf), transthyretin (Ttr), and wingless-type MMTV integration site family members (Wnt), including member 2 (Wnt2), 5A (Wnt5a), 9A (Wnt9a), 11 (Wnt11), and 16 (Wnt16) (Data S1). These data are also in agreement with the overall increase in the expression of other genes related to cholesterol import/export into cells, such as apolipoprotein B receptor (Apobr), scavenger receptor class B, member 1 (Scarb1/SR-B1), macrophage scavenger receptor 1 (Msr1/SR-A1), and CD36 molecule (Cd36). These indicated receptors, in addition to low-density lipoprotein (LDL) ingestion via phagocytosis and macropinocytosis, are essential for lipoprotein internalization. Lipoproteins are then degraded in lysosomes, releasing cholesterol into the intracellular environment (Figure 2E).
Cholesterol metabolism also relies on a series of esterases to produce more soluble metabolites which are, therefore, easier to be eliminated. So, it is worth noting the unusual high expression of cholesterol 25-hydroxylase (Ch25h) in retinopathy (log2 fold change = 2.78, 2.70, and 3.35 at postnatal days 12.5, 15, and 17, respectively). CH25H catalyses the formation of 25-hydroxycholesterol (25-ChOH), an agonist of nuclear receptors liver X receptors (LXRs).35,36 LXRs regulate gene expression of ATP-binding cassette sub-family A member 1 (Abca1), which is increased in OIR retinas at postnatal days 15 and 17, and promotes cholesterol efflux from cells via lipid-poor apoA-I (pre-β-HDL)37 (Figure 2E). Interestingly, 25-ChOH also restricts activation of SREBP2, a sterol-sensing transcription factor that regulates cholesterol biosynthesis.38 This is in agreement with expression data of 23 genes whose products belong to the cholesterol biosynthesis pathway39 and which are down-regulated in retinopathy (Data S1).
Lipid saturation degree is markedly altered by OIR toward the production of MUFA and PUFA with up to 3 unsaturations
Total SFA, MUFA, PUFA, and HUFA (examples in Figure 3A) were quantified considering all detected lipid classes in the lipidomic analysis. In our analysis, we considered PUFA all fatty acids with two or three unsaturations, and HUFA all those with more than three unsaturations, including, for instance, DHA and ARA. In general, OIR retinas showed an increase in MUFA concentrations compared to healthy retinas at postnatal days 12, 12.5, and 17 (Figure 3B). In contrast, PUFA concentration was decreased at P12.5, but subsequently recovered at P15 and increased at P17, while HUFA concentration was generally decreased in OIR retinas at all different time points.
Figure 3.
Lipid saturation is altered in OIR favoring the production of MUFA and PUFA with up to 3 unsaturations
(A) Examples of SFA, MUFA, PUFA, and HUFA found in OIR retinas.
(B) Percentage of SFA, MUFA, PUFA, and HUFA in physiological (controls, N = 6) and pathological (OIR, N = 6) retinas at postnatal days 12, 12.5, 15, and 17. Data are represented as mean ± SEM (Student’s t-test, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
(C) Percentage of MUFA, PUFA, and HUFA in specific lipid classes, including neutral (CE and TG) and membrane (PE, pPE, and PI) lipid species. Data are represented as mean ± SEM (Mann-Whitney-Wilcoxon test; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001). See also Figure S4 and Supplemental Material - Venn diagram.
When we consider the lipid classes that are mostly enriched with HUFA—cholesteryl esters (CE), phosphatidylethanolamines (PE), PE plasmalogens (pPE) and phosphatidylinositols (PI)—in addition to triacylglycerols (TG), this pattern is even more apparent (Figure 3C). All these lipid classes showed higher concentrations of MUFA and lower concentrations of HUFA in OIR. In terms of fatty acids with two or three unsaturations, CE and TG molecular species linked to PUFA showed a tendency to increase in retinopathy, and a similar trend was observed for PE and PI.
Next, we used the Venn diagram to screen for common lipid features associated with OIR (fold change ≥1.5, FDR-adjusted p value ≤0.05) (Figure S4A). Among the five lipid species that were found altered at all postnatal days, four species - FFA 20:3 (n-9), PI 18:0/20:3, PC 16:0/22:3 e CE 20:3 – were increased in OIR, while only one of them, specifically a plasmalogen esterified to HUFA (pPE p18:2/22:6), was decreased. Notably, in agreement with the unsaturation data, all increased lipid species were linked to fatty acyl chains containing three double bonds (20:3 or 22:3) (Figure S4B).
The concentration of phosphatidylcholines (PC) linked to VLCFA is decreased in OIR retinas
Very long-chain highly unsaturated fatty acids (VLC-HUFA) are found mainly in retina, brain, testis, and spermatozoa.40 We identified and quantified 15 PC lipid species esterified to VLC-HUFA with at least 32 carbons of length in all collected samples. Among these phospholipids, PC (34:6/22:6) and PC (36:6/22:6) were the most abundant in retinas, with the VLC-HUFA esterified to the sn-1 position of the glycerol-phosphocholine backbone (Figure 4A). No VLC-HUFA bound to other types of phospholipids was identified; all of them are linked to PC. Retinopathy drove a significant decrease in total mass values of these PC species, which is seen at postnatal days 12, 12.5, and 17 (Figure 4B). The same trend is clearly seen in 11 of the 15 individual species of PC linked to VLC-HUFA that we could identify in lipidomic analysis (Figure 4C).
Figure 4.
Phosphatidylcholines (PC) linked to VLCFA in OIR retinas
(A) Representative structures of the two most abundant PC-VLCFA found in normal and OIR retinas.
(B) Normalized mass values (ng of lipids/mg of proteins) for total PC-VLCFA.
(C) Normalized mass values (ng of lipids/mg of proteins) for individual PC-VLCFA species. PC-VLCFA species were identified and quantified in all physiological (control, N = 6 replicates) and pathological (OIR, N = 6 replicates) retinas collected at postnatal days 12, 12.5, 15, and 17. Only PC species linked to at least one fatty acid with 32 or more carbons were considered. Data are represented as mean ± SEM (Mann-Whitney-Wilcoxon test; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
OIR retinas present decreased synthesis of n-3 and n-6 fatty acids and up-regulation of the n-9 alternative pathway of fatty acid desaturation
The pattern of decreased HUFA, such as DHA (22:6, n-3) and ARA (20:4, n-6), as well as VLC-HUFA (n-3 or n-6), led us to investigate how different fatty acid synthesis pathways behave in retinopathy. Both the n-3 (or ω-3) and n-6 (or ω-6) fatty acid pathways comprise a series of products derived biosynthetically from essential fatty acids, ingested in the diet. In our lipidomic analysis, we were able to differentiate specific FFA isomers through their retention time in the column used in high-performance liquid chromatography (HPLC). These isomers present the same molecular mass but different positions of double bonds, i.e., different “n” values. In general, considering two isomers, the FFA with a higher “n” value had a longer retention time in our method (Figure S5A). This is shown in detail for FFA 20:3, whose m/z value generated two peaks in our analysis, one corresponding to FFA 20:3 n-3 and n-6, and the other to FFA 20:3 n-9, the latter with a longer retention time (Figure S5B).
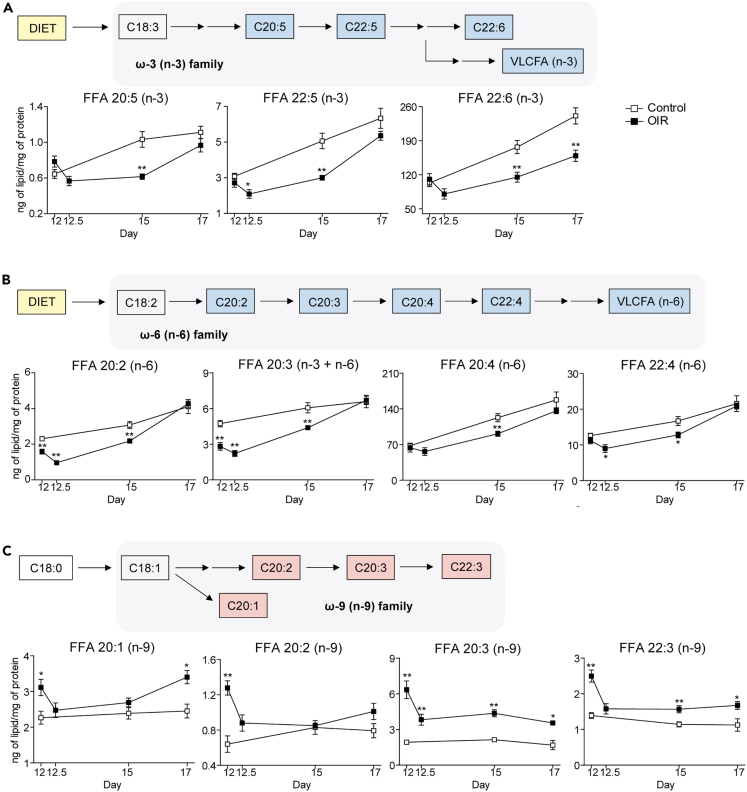
Derived from the α-linolenic acid (18:3, n-3), n-3 family includes fatty acids such as eicosapentaenoic acid 20:5, n-3 (EPA), docosapentaenoic acid 22:5, n-3 (DPA), and DHA (22:6, n-3). All of them are significantly decreased in retinopathy (Figure 5A). Similarly, the n-6 family of fatty acids, derived from linoleic acid (18:2, n-6), also shows up in lower concentrations in OIR retinas. This is clearly seen for several intermediates of the n-6 pathway, including eicosadienoic acid (20:2, n-6), dihomo-γ-linolenic acid (DGLA, 20:3, n-6), ARA (20:4, n-6), and adrenic acid (AdA, 22:4, n-6) (Figure 5B). This indicates a chronic deficiency of n-3 and n-6 precursors from the early stages of the retinopathic process affecting the synthesis of all subsequent derived long-chain fatty acids. Indeed, the n-3 and n-6 synthesis pathways are the precursors of VLC-HUFA. Their down-regulation helps to explain, for example, the decreased concentration of phosphatidylcholine species linked to VLC-HUFA in the pathological condition (Figure 4). This also explains the overall decrease in the number of unsaturations in most of the lipid classes analyzed in this study (Figure 3).
Figure 5.
OIR retinas decreased synthesis of n-3 and n-6 fatty acids and up-regulation of the n-9 alternative pathway of fatty acid desaturation
(A–C) Schemes showing intermediates of the n-3 (A), n-6 (B), and n-9 (C) fatty acid synthesis pathways which are significantly altered in the retinas of OIR mice. Normalized mass values (ng of lipids/mg of proteins) are presented for each of these intermediates in physiological (control, N = 6 replicates) and pathological (OIR, N = 6 replicates) retinas at postnatal days 12, 12.5, 15 and 17. Data are represented as mean ± SEM (Mann-Whitney-Wilcoxon test; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001). See also Figure S5.
On the other hand, as seen earlier, retinopathy induces increased concentration of MUFA and PUFA with up to 3 unsaturations. This finding correlates with up-regulation of de novo fatty acid synthesis, generating products with 1, 2, or 3 unsaturations. In a situation of essential fatty acid deficiency (EFAD), there is possibly an accumulation of MUFA and PUFA belonging to families other than n-3 and n-6. This includes, for example, the n-9 (or ω-9) family. We were able to detect and quantify four products from the n-9 fatty acid family: eicosenoic acid (20:1, n-9), eicosadienoic acid (20:2, n-9), mead acid (20:3, n-9), and docosatrienoic acid (22:3, n-9), all of them showing an increasing trend in the OIR retinas (Figure 5C).
We also looked at expression data for a number of genes (elongases and desaturases) linked to the n-3, n-6, and n-9 pathways and none of them showed significant expression difference in OIR, including those described for mead acid synthesis, namely Elovl5, Fads1, and Fads241 (Data S1). However, it is important to note that mead acid and other n-9 fatty acids are thought to be produced by the same enzymes that synthesize n-3 and n-6 fatty acids. This is true for the enzymes Elovl5, Fads1, and Fads2. Therefore, expression data alone would not be sufficient to explain why n-3 and n-6 fatty acids are decreased while n-9 fatty acids are increased in OIR.
Discussion
By carefully identifying and quantifying the lipid composition of the retina and mapping them to their respective metabolic pathways (for which we had information regarding mRNA expression for their enzymes and proteins), we have identified a lipid signature for the pathological angiogenesis of the OIR model. This signature points to a prominent role for cholesterol esterification, lipid droplet formation, reverse cholesterol transport, and increased concentration of n-9 fatty acids (for example, mead acid) in the retina.
At first glance, the most striking result is the increase in the concentration of neutral lipids (mostly CE and TG species) in the retinopathic retinas. In agreement with the lipidomics, transcriptomic data indicate TG formation, since the pathways leading to its synthesis are in general significantly augmented. The same is observed for the expression of genes leading to CE synthesis (Soat1 and, particularly, Soat2), as well as structural proteins for lipid droplet production (Plin2, Plin3, Fitm1, Tmem159/Ldaf1, Hilpda, Cav1, Cavin1, Cavin3). Lipid droplets are important organelles for fat storage, which are composed mainly of CE and TG within their lipid core, enclosed by a monolayer of phospholipids and proteins. This is in agreement with the recent observation that the retina relies on a steady supply of fatty acids in order to provide the energetic needs of the photoreceptor cells. These cells express high levels of the very low-density lipoprotein (VLDL) receptor, enabling the uptake of TG to be metabolized by β-oxidation.1
But the role of lipid droplets in the retina is still not fully understood. For instance, reactive oxygen species (ROS) production is increased in the OIR model and in retinopathies in general, as well as in many other eye diseases.42,43 So, besides the role of lipid droplets in energy storage, they may also act to buffer the concentration of toxic lipid species44 and have a prominent role in limiting the levels of ROS in cells by capturing PUFA from cellular membranes and thus preventing lipid peroxidation.45 Another interesting observation is that RPE cells contain organelles named retinosomes, which are retinyl ester-containing lipid droplets important to keep a proper supply of the visual chromophore 11-cis-retinal to the photoreceptor cells.46 Changes in the formation of these lipid droplets (retinosomes) have been associated with pathological conditions in the retina of drosophila and rat,47,48 in addition to patients suffering from age-related macular degeneration (AMD).49 Although in our lipidomic analysis we did not look specifically at retinyl esters, it is noteworthy the increase in expression of lecithin-retinol acyltransferase (Lrat), which encodes the enzyme responsible for the esterification of trans-retinal, producing trans-retinyl esters. Lrat is substantially augmented in retinopathic retinas (log2 fold change = 2.24 at postnatal day 17, compared to the physiological condition).
Alongside Lrat, Soat1, and Soat2, which are responsible for the intracellular esterification of lipids stored in lipid droplets, we also observed in OIR a significant increase in gene expression of Lcat (log2 fold change = 1.28, 1.17, and 1.11 at postnatal days 12.5, 15, and 17, respectively). The LCAT enzyme is responsible for the production of extracellular CE, converting cholesterol and phosphatidylcholines to CE and lysophosphatidylcholine (LysoPC). LCAT has a defined role in the growth and maturation of HDL particles33 and cholesterol efflux from macrophages via ABCA1. LCAT deficiency is characterized by two different phenotypes with distinct levels of LCAT activity: familial LCAT deficiency (FLD) and fish eye disease (FED), both leading to low HDL cholesterol levels and corneal opacification.50 This is in agreement with our lipid signature, which shows a prominent role of the cholesterol import/export pathway in retinopathy.
The retina differs from the central nervous system with regard to cholesterol metabolism, a key factor in retinal pathological conditions as mentioned above. While the brain synthesizes all cholesterol it uses, the retina can both synthesize it and uptake cholesterol from circulation.14,15 Thus, it is interesting that under retinopathic condition, several genes associated with cholesterol esterification and transport (Soat1, Soat2, Lcat, Abca1, besides lipoprotein receptors such as Apobr, Scarb1, Msr1, and Cd36) are up-regulated, indicating an increase in the import/export of cholesterol (i.e., from circulation to the RPE, then, from the RPE to the photoreceptor cells and the neural retina). At the same time, 23 genes related to cholesterol biosynthesis are reasonably decreased in OIR (Data S1), suggesting that intraretinal cholesterol synthesis is not the main factor behind the increase in CE levels.
One limitation of our study is that we cannot discriminate which cell is responsible for CE synthesis and export, such as glial or tissue macrophages. However, if we look at evidence from literature, we observe that different animal models of eye diseases (i.e., NaIO3-induced model of dry AMD and Bietti crystalline dystrophy zebrafish model)51,52 showed accumulation of lipid droplets in the RPE. There has also been report of lipid droplet formation in astrocytes.53,54 Considering that large epidemiologic studies point to a strong correlation between circulating lipoprotein species, such as HDL, and the severity of retinopathy and other eye diseases, one cannot rule out the contribution of infiltrating macrophages as the source of accumulation of cholesterol in the retina. In this context, lipid-lowering treatments with statin and fibrate have been shown to improve diabetic retinopathy-related conditions.55,56,57 Similarly to what occurs in atherosclerosis, we observed that key genes for reverse cholesterol transport showed significant increased expression in OIR retinas, as well as multiple markers for tissue macrophages (Itgam/Cd11b, Cd14, Fcgr3/Cd16, Fcgr1/Cd64, Cd68, Ccr5, Adgre1) and glial macrophages (Trem2, Tyrobp, Cd33, Tlr1, Tlr2, Tlr3, Tlr4, Tlr7, Tlr12, Tlr13), in addition to markers for HDL particles (A2m, Amy1, C3, C4b, Clu, Gpx3, Grn, Hpx, Lcat, Pon1, Pon3, Rbp1, Rbp4, Trf, Ttr, Wnt2, Wnt5a, Wnt9a, Wnt11, Wnt16).34 Therefore, potential targets for drugs are provided, in particular those directed to neutral lipid (CE and TG) synthesis pathways, reverse cholesterol transport and the inflammatory status of pathological retinas.
On the other hand, it is important to note that part of these markers have been associated with intraretinal lipid transport, which is dependent on HDL-like particles and class B scavenger receptors31 that are up-regulated in the OIR transcriptome analysis. Distinct from the blood-brain barrier, the blood-retina barrier allows for the uptake of cholesterol rich lipoproteins from circulation, mostly mediated by the RPE layer. These HDL-like particles are then formed via ABCA1 transporters and taken up by photoreceptor and ganglion cells in the retina using scavenger receptors.15 Indeed, photoreceptor cells shed approximately 10% of their outer segment membranes every day. Because of the decrease in dietary n-3 and n-6 lipids, which are essential for proper photoreceptor transduction pathways (in particular, DHA),58,59,60 the increase in HDL-like particle formation during OIR may reflect the concomitant necessary increase in lipid transport in order to supply enough of these lipids to the photoreceptor cells. In aggregate, our data suggest that intraretinal lipoprotein/cholesterol transport pathways are substantially increased in retinopathy. In agreement with this is the observation that Ch25h expression is also substantially increased by OIR.19 CH25H catalyzes the conversion of cholesterol to 25-hydroxycholesterol (25-ChOH), a product which can act as an activator of LXR receptors and then promote cholesterol efflux to (apo)lipoproteins via ABCA1. CH25H was previously shown to contribute to a pathological angiogenesis gene signature able to predict breast cancer patient survival.19 Besides being an LXR activator, its product, 25-ChOH, presents anticancer61 and antiviral62 activities in mammalian cells, working against viruses such as HIV, EboV, Influenza, and others by affecting their entry and replication in the cell. The role of CH25H and 25-ChOH in retinopathy and other eye diseases is still unknown.
In addition to neutral lipid increase and aspects related to the “inflammatory environment” of OIR retinas, we detected changes in lipid desaturation pathways as another hallmark of pathological angiogenesis. OIR retinas present higher concentrations of MUFA than healthy retinas at all postnatal days we analyzed. They also contain higher levels of PUFA with up to three unsaturations at postnatal day 17. Nevertheless, n-3 and n-6 HUFA species (with more than three unsaturations) were considerably decreased in the pathological condition. The same happened to PC molecular species linked to VLC-HUFA, unusual lipid species found in retina and a few other vertebrate tissues, such as brain and testis. In retinas, they seem to play a crucial role in maintaining membrane fluidity, which is essential to the long-term survival and function of photoreceptor outer segments.63,64 They may also modulate other membrane biophysical properties, including permeability, compression, fusion, and flipping.64 Loss of VLC-HUFA, for example, was detected in AMD and diabetic donor eyes40,65,66 and linked to a reduction in rod b-wave amplitudes and to death of rod cells over time.63,67
The sharp decrease in highly unsaturated species led us to further investigate different pathways of fatty acid synthesis in healthy and OIR retinas. The decrease in n-3 and n-6 FFA, for example EPA, DPA, and DHA (n-3 family) and eicosadienoic acid, DGLA, ARA e AdA (n-6 family), are in agreement with previously published data showing that premature infants have lower blood concentrations of DHA and ARA, a condition of essential fatty acid deficiency (EFAD) that correlates with ROP progression.68,69,70,71 EFAD is tightly associated with changes in lipid desaturation profiles and the promotion of alternative fatty acid desaturation pathways, which in turn affects, for instance, the fatty acid profile of cells in culture, augmenting the concentration of MUFA instead of PUFA/HUFA.72 Lipid metabolism plasticity is also observed in some cancer cell lines. Some of them exploit an unusual pathway that desaturates palmitate to sapienate, a monounsaturated fatty acid of the n-10 family that supports membrane formation and proliferation.73
It is noteworthy that deficiency in n-3 and n-6 essential fatty acids in OIR promotes a significant increase in mead acid in the retina. Mead acid is an indicator of EFAD41 and, interestingly, it is substantially increased at all postnatal days in the pathological condition but not in the physiological retinas. To the best of our knowledge, this has not been reported before and it is a clear indication that EFAD might be an early event in ROP. Whether retinal mead acid comes from the alternative lipid synthesis pathway that leads to the production of n-9 fatty acids in the retina or from extraretinal tissues due to an increase in uptake from plasma, it is still an open question. DHA, ARA, and mead acid are all synthesized by similar pathways that share the enzymes encoded by Elovl5, Fads1, and Fads2 genes; thus, the retina has all enzymes necessary to produce mead acid, including Elovl5. Interestingly, a report in bioRxiv suggests that ELOVL5 substrate preference could be modulated by GSK3: GSK3 phosphorylation of ELOVL5 leads to the production of DHA and ARA. However, hypoxia pathways and other tyrosine kinases (i.e., VEGFR) inhibit GSK3 (via Akt/PKB), preventing the phosphorylation of ELOVL5 and promoting mead acid production.74 Of course, it is critical to further validate this study and to better understand Akt, GSK3, and ELOVL5 phosphorylation status in retina cells under physiological and OIR condition to confirm this hypothesis.
Our data is therefore in agreement with the notion that n-3 and n-6 essential fatty acids supplementation might be beneficial for prevention of ROP, as it has been demonstrated by several studies.75,76,77 Nevertheless, the presence of mead acid in the retina (either endogenously produced or taken up from plasma) may reduce or hinder some effects of lipid supplementation by outcompeting with the dietary n-3 and n-6 essential fatty acids, and may explain some inconsistent observations regarding lipid supplementation.78 Mead acid and other n-9 lipids, may be used as substitutes for other PUFA and HUFA in biological membranes,41 as indicated by the desaturation profile in the lipidomic data which shows that HUFA are being replaced by mono- and polyunsaturated fatty acids (MUFA and PUFA, respectively) with up to three unsaturations in pathological angiogenesis. Finally, considering that mead acid concentration in blood could be used to identify patients with EFAD, conceivably, it could also be a potential biomarker for ROP. Further studies are necessary to answer these important questions regarding mead acid synthesis in the retina, the phosphorylation control of ELOVL5, as well as n-3 and n-6 supplementation in ROP.
In sum, these are important findings, which might have implications for the diagnosis, treatment and prevention of ROP, a condition for which the OIR mouse model recapitulates several hallmarks of the human disease.8 Changes in lipid metabolism have also been described for diabetic retinopathy,55,56,57,79 glaucoma,80 and age age-related macular degeneration.81,82 Therefore, it is possible that our integrated lipidomic/transcriptomic study may reflect changes in lipid metabolism associated with pathological conditions of the eye. A better understanding of lipid metabolism in the retina may translate into improved therapeutic and diagnostic alternatives for the treatment and prevention of human eye diseases.
Limitations of the study
The OIR model is a robust and reliable method to study retinal angiogenesis in vivo, and the use of isogenic animals minimizes genetic background variability and allows for reproducible and quantifiable results. Nevertheless, exposure to high oxygen levels (75%) can be detrimental to the animals, reducing postnatal weight gain and aggravating the retinopathic state. To minimize these effects and reduce vessel loss and variability in neovascularization, we avoided large litter size (8 or less pups) to improve postnatal weight gain, and animals with less than 5 g at P17 were excluded from the study, as recommended.83,84 Another aspect that merits comments is the difficulty in assigning specific lipid synthesis pathways and transcript expression to precise cells in the retina. Therefore, we used bona fide molecular markers along with data from the literature in order to suggest and indicate the most likely cell types candidates responsible for some of the changes in lipid metabolism that we observed in our study. Nevertheless, one should be cautious and understand that other cell types may also be involved in these processes. Further studies are necessary to better delineat these associations. Finally, for the lipidomic analysis, we used the LC-MS untargeted measurement and standards that did not take into consideration more specific types of lipids, such as oxylipins and oxysterols.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Butylated hydroxytoluene; 2,6-Di-tert-butyl-4-methylphenol | Sigma-Aldrich | Cat#B1378 |
| Deferoxamine mesylate salt | Sigma-Aldrich | Cat#D9533 |
| Cholest-5-en-3β-yl (decanoate); cholesteryl decylate | Sigma-Aldrich | Cat#S436739 |
| N-decanoyl-D-erythro-sphingosine | Avanti Polar Lipids | Cat#860510 |
| N-heptadecanoyl-D-erythro-sphingosine | Avanti Polar Lipids | Cat#860517 |
| 1′,3′-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-glycerol | Avanti Polar Lipids | Cat#750332 |
| 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#110686 Cat#855676 |
| 1-(10Z-heptadecenoyl)-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat#856707 |
| 1,2-diheptadecanoyl-sn-glycero-3-phosphate | Avanti Polar Lipids | Cat#830856 |
| 1,2-dimyristoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#850345 |
| 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#850360 |
| 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat#850745 |
| 1,2-diheptadecanoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat#830756 |
| 1,2-diheptadecanoyl-sn-glycero-3-phospho-(1′-rac-glycerol) | Avanti Polar Lipids | Cat#830456 |
| 1,2-diheptadecanoyl-sn-glycero-3-phospho-L-serine | Avanti Polar Lipids | Cat#840028 |
| N-heptadecanoyl-D-erythro-sphingosylphosphorylcholine | Avanti Polar Lipids | Cat# 860585 |
| 1,2,3-tritetradecanoyl-sn-glycerol; Trimyristin | Sigma-Aldrich | Cat#T2500100 |
| Critical commercial assays | ||
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| Deposited data | ||
| Oxygen-Induced Retinopathy RNA-Seq | Guarischi-Sousa et al.19 | Sequence Read Archive (SRA), BioProject PRJNA483866 |
| RNA-Seq of Mus musculus: newborn retina | Guarischi-Sousa et al.19 | Sequence Read Archive (SRA), SRP155931 |
| Lipidomics raw data | This paper | MetaboLights, MTBLS6965, www.ebi.ac.uk/metabolights/MTBLS6965 |
| Lipid quantification data | This paper | Mendeley Data, https://doi.org/10.17632/sn5hz3s992.1 |
| Experimental models: Organisms/strains | ||
| C57BL/6j mice, Strain #:000664 | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Software and algorithms | ||
| Analyst®TF 1.7.1 | Sciex | RRID:SCR_015785 |
| DESeq2 | Bioconductor | RRID:SCR_015687; https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| GraphPad Prism 8.4.3 | GraphPad Software | RRID:SCR_002798 |
| MetaboAnalyst 5.0 | Xia et al.94 | RRID:SCR_015539; https://www.metaboanalyst.ca/ |
| MultiQuant® 3.0.3 | Sciex | N/A |
| PeakView® 2.2 | Sciex | RRID:SCR_015786 |
| Other | ||
| KEGG: Kyoto Encyclopedia of Genes and Genomes | Kanehisa.90 | RRID:SCR_012773; https://www.genome.jp/kegg/ |
| LIPID MAPS | Fahy et al.91 | RRID:SCR_006579; https://www.lipidmaps.org/ |
| Reactome | Fabregat et al.92 | RRID:SCR_003485; https://reactome.org/ |
| WikiPathways | Pico et al.93 | RRID:SCR_002134; https://www.wikipathways.org/index.php/WikiPathways |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ricardo José Giordano (giordano@iq.usp.br).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study approval
All experimental procedures were approved by the Animal Study Ethics Committee from the Chemistry Institute (protocol number 73/2017). C57BL/6j mice (RRID:IMSR_JAX:000664) were maintained at the animal facility of the Chemistry Institute and Pharmacy School of the University of São Paulo.
Murine model of OIR
In the OIR model, pathological angiogenesis is induced in mice through their exposure to variable oxygen levels. Thus, mouse pups with their nursing mothers were kept at 75% O2 from postnatal day 7 (P7) until day 12 (P12). Mice were then returned to ambient air (∼20.8% O2) and retinas (N = 6) were collected at different time points (P12, P12.5, P15 and P17) for lipid extraction. Retinas from mouse pups under physiological development were also collected.
Method details
Lipid extraction
Lipid extraction was performed according to a method adapted from Bligh and Dyer (1959).86 Retinas were initially lysed in 600 μL of 50 mM phosphate buffer (pH 7.4) containing 100 μM deferoxamine mesylate (Cat# D9533, Sigma-Aldrich), with zirconia beads and a Mixer Mill MM 301 homogenizer (Retsch GmbH, Haan, Germany). Lysis was performed with a cycle of 1 min at a frequency of 30 s−1 10 μL aliquots of the prepared homogenates were used to quantify proteins with Pierce BCA Protein Assay Kit (Cat# 23227, Thermo Fisher Scientific). 500 μL aliquots were then used in the lipid extraction procedure. These aliquots were mixed with 400 μL of ice-cold methanol containing 100 μM butylated hydroxytoluene (Cat# W218405, Sigma-Aldrich), and 100 μL of lipid internal standards (Table S1), which are necessary for semi-quantification of lipid molecular species. 2.0 mL of chloroform: ethyl acetate (4:1) were added to each mixture, followed by vortexing during 30 s. After centrifugation at 1,500 x g for 2 min at 4°C, the lower phase containing the total lipid extracts (TLE) was transferred to new tubes and dried under N2 gas. Dried TLE were dissolved in 100 μL of isopropanol and the HPLC injection volume was set at 1 μL.
Lipidomic analysis
Non-targeted lipidomic analysis of major lipids was performed by reverse-phase liquid chromatography coupled to mass spectrometry (RPLC-MS).87,88 In detail, TLE were analyzed by electrospray ionization time-of-flight mass spectrometry (ESI-TOFMS, Triple TOF 6600, Sciex, Concord, USA) interfaced with ultra HPLC (UHPLC Nexera, Shimadzu, Kyoto, Japan). The samples were loaded into a C18 column (1.6 μm, 2.1 mm i.d. x 100 mm, CORTECS, Waters Corporation, Milford, USA) with a flow rate of 0.2 mL/min and an oven temperature maintained at 35°C. For the liquid chromatography, mobile phase A consisted of water: acetonitrile (60:40), while mobile phase B was composed of isopropanol: acetonitrile: water (88:10:2). Mobile phases A and B were supplemented with ammonium acetate or ammonium formate (at a final concentration of 10 mM) for experiments performed in negative or positive ionization modes, respectively. The linear gradient used in RPLC was 40%–100% B over the first 10 min and held at 100% B for additional 2 min, decreased from 100 to 40% B during the next 1 min, and held at 40% B for the remaining 7 min, totalizing a 20 min run. The MS was operated in both positive and negative ionization modes, and the scan range set at a mass-to-charge ratio of 200–2000 Da. Data for lipid molecular species identification and quantification were obtained by Information Dependent Acquisition (IDA). Data acquisition was performed with a period cycle time of 1.05 s with 100 ms acquisition time for MS1 scan and 25 ms acquisition time to obtain the top 36 precursor ions. Data acquisition was performed using AnalystTF 1.7.1 (RRID:SCR_015785) with an ion spray voltage of −4.5 kV and 5.5 kV for negative and positive modes, respectively, and the cone voltage at +/− 80 V. The curtain gas was set at 25 psi, nebulizer and heater gases at 45 psi and interface heater at 450°C. The MS/MS data were analyzed with PeakView 2.2 (RRID:SCR_015786), and lipid molecular species were manually identified with the help of an in-house manufactured Excel-based macro. The lipid quantification was performed with MultiQuant 3.0.3, in which peak areas of precursor ions were normalized to those of the internal standards. Final data were expressed as mass of lipid species per mass of total proteins, and graphs were constructed with GraphPad Prism 8.4.3 (RRID:SCR_002798).
Transcriptomic analysis
Transcriptomic data described by Guarischi-Sousa et al.19 were re-analyzed in order to allow their integration with lipidomic data presented here. Gene expression was analyzed in pairs covering the pathological and physiological conditions at a same time point, that is, R12 x P12, R15 x P15 and R17 x P17, in addition to R12.5 x P12. Log2(fold change) values were obtained for the expression of 32,569 genes, using DESeq2 R package (RRID:SCR_015687),89 from which differentially expressed lipid metabolism-related genes (p value <0.05 or FDR-adjusted p value <0.05) were annotated manually and with the aid of KEGG (RRID:SCR_012773),90 LIPID MAPS (RRID:SCR_006579),91 Reactome (RRID:SCR_003485)92 and WikiPathways (RRID:SCR_002134)93 databases for Mus musculus.
Quantification and statistical analysis
Statistical analysis for lipidomic data was performed with MetaboAnalyst 5.0 (RRID:SCR_015539)94 and R. We used MetaboAnalyst to build heatmaps and PCA graphs, and also to obtain ‘fold change’ values, following Xia and Wishart’s protocols.95 Data were log transformed (base 10) prior to statistical analysis. All groups were compared by one-way ANOVA followed by Tukey’s HSD post-hoc test (FDR-adjusted p value ≤0.05). For pairwise comparisons, bar graphs were constructed with log2(fold change) values of lipid species whose variation was significant (Student’s t-test, FDR-adjusted p value ≤0.05, and fold change set to ≥1.5). In the case of Figures 2A, 3B, 3C, 4B, 4C, 5A–5C, and S1 (graphs: log2 of normalized mass values), Shapiro-Wilk normality test96 was conducted to check if the data followed a normal distribution. Also, Levene’s test97 was performed to verify the homoscedasticity of the data. Both tests demonstrated that the data did not meet the assumption for parametric tests. Thus, Mann-Whitney-Wilcoxon test (also known as Mann-Whitney U test or Wilcoxon Rank-Sum Test) was used to verify if the differences observed between normal and OIR retinas were significant (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001). Only data presented in Figures 3B and S1 followed a normal distribution and were homoscedastic. In these cases, Student’s t-tests were performed (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001). In lipidomics, data for sample R12 (#3) was of poor quality (total ion chromatogram with very low intensity), probably due to problems with lipid extraction or sample injection. Thus, data from this sample was excluded from the final analysis. More statistical details can be found in the figure legends. For transcriptomics, the distribution and homoscedasticity of data were also verified before statistical analysis. All data were obtained from independent samples. Data with normal distribution and homoscedastic were tested using Student’s t-test (two group comparisons) or ANOVA followed by Tukey’s post-hoc test with Benjamin-Hochberg p value adjustment (three or more group comparisons). Data with nonnormal distribution and/or heteroscedastic were tested using the Mann-Whitney-Wilcoxon Test (two group comparisons), or Kruskal-Wallis test followed by Dunn’s post-hoc test with Benjamini-Hochberg p value adjustment98 (three or more group comparisons).
Acknowledgments
This work was supported by research grants from São Paulo Research Foundation (FAPESP; www.fapesp.br) (grants 2019/25828-8 to RJG, CEPID-Redoxoma 2013/07937-8 to SM and fellowship 2017/13804-1 to AI), the National Council for Scientific and Technological Development (CNPq; www.cnpq.br) (grants 310485/2021-5 to RJG and 313926/2021-2 to SM) and research fellowships. This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; www.capes.gov.br), Finance Code 001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization, R.J.G. and S.M. with support of all co-authors; Methodology, R.J.G., S.M., A.I., and L.C.A.; Formal Analysis, A.I. and J.S.M.; Investigation, A.I. and L.C.A.; Resources, R.J.G., S.M., and J.C.S.; Data Curation, A.I., L.C.A., and M.Y.Y.; Writing – Original Draft, R.J.G., S.M., A.I., and L.C.A.; Writing – Review & Editing, all authors and co-authors; Visualization, R.J.G., S.M., A.I., and L.C.A.; Supervision, R.J.G., S.M., and J.C.S.; Funding Acquisition, R.J.G. and S.M.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: May 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106777.
Contributor Information
Sayuri Miyamoto, Email: miyamoto@iq.usp.br.
Ricardo José Giordano, Email: giordano@iq.usp.br.
Supplemental information
Data regarding the transcripts (mRNA) cited in the manuscript (Figures 2D and 2E) and/or linked to HDL formation and maturation, cholesterol and fatty acid biosynthesis, and specific retinal cells (glia and macrophages)
Data and code availability
-
•
As previously reported,19 sequencing metadata for the RNA-seq data used in the manuscript have been deposited at the Sequence Read Archive (SRA) under accession number BioProject PRJNA483866. RNA-seq data is available under accession number SRP155931. Lipidomics raw files have been deposited at MetaboLights85 under the unique identifier MTBLS6965 (https://www.ebi.ac.uk/metabolights/MTBLS6965). Lipid quantification data are available at Mendeley Data (https://doi.org/10.17632/sn5hz3s992.1). They are publicly available as of the date of publication. Accession numbers are also listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Joyal J.S., Sun Y., Gantner M.L., Shao Z., Evans L.P., Saba N., Fredrick T., Burnim S., Kim J.S., Patel G., et al. Retinal lipid and glucose metabolism dictates angiogenesis through the lipid sensor Ffar1. Nat. Med. 2016;22:439–445. doi: 10.1038/nm.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Díaz-Coránguez M., Ramos C., Antonetti D.A. The inner blood-retinal barrier: cellular basis and development. Vis. Res. 2017;139:123–137. doi: 10.1016/j.visres.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campochiaro P.A. Molecular pathogenesis of retinal and choroidal vascular diseases. Prog. Retin. Eye Res. 2015;49:67–81. doi: 10.1016/j.preteyeres.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fliesler S.J. Introduction to the thematic review series: seeing 2020: lipids and lipid-soluble molecules in the eye. J. Lipid Res. 2021;62:100007. doi: 10.1016/j.jlr.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahdenranta J., Pasqualini R., Schlingemann R.O., Hagedorn M., Stallcup W.B., Bucana C.D., Sidman R.L., Arap W. An anti-angiogenic state in mice and humans with retinal photoreceptor cell degeneration. Proc. Natl. Acad. Sci. USA. 2001;98:10368–10373. doi: 10.1073/pnas.181329198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Z., Kern T.S., Hellström A., Smith L.E.H. Fatty acid oxidation and photoreceptor metabolic needs. J. Lipid Res. 2021;62:100035. doi: 10.1194/jlr.TR120000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith L.E., Wesolowski E., McLellan A., Kostyk S.K., D'Amato R., Sullivan R., D'Amore P.A. Oxygen-induced retinopathy in the mouse. Invest. Ophthalmol. Vis. Sci. 1994;35:101–111. [PubMed] [Google Scholar]
- 8.Kim C.B., D'Amore P.A., Connor K.M. Revisiting the mouse model of oxygen-induced retinopathy. Eye Brain. 2016;8:67–79. doi: 10.2147/EB.S94447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell M.I., Aguilar E., Jacobson R., Trauger S.A., Friedlander J., Siuzdak G., Friedlander M. Maintaining retinal astrocytes normalizes revascularization and prevents vascular pathology associated with oxygen-induced retinopathy. Glia. 2010;58:43–54. doi: 10.1002/glia.20900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vessey K.A., Wilkinson-Berka J.L., Fletcher E.L. Characterization of retinal function and glial cell response in a mouse model of oxygen-induced retinopathy. J. Comp. Neurol. 2011;519:506–527. doi: 10.1002/cne.22530. [DOI] [PubMed] [Google Scholar]
- 11.Saito Y., Geisen P., Uppal A., Hartnett M.E. Inhibition of NAD(P)H oxidase reduces apoptosis and avascular retina in an animal model of retinopathy of prematurity. Mol. Vis. 2007;13:840–853. [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Wang D., Liu Y., Luo Y., Ma W., Xiao W., Yu Q. Neuronal-driven angiogenesis: role of NGF in retinal neovascularization in an oxygen-induced retinopathy model. Invest. Ophthalmol. Vis. Sci. 2010;51:3749–3757. doi: 10.1167/iovs.09-4226. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson-Berka J.L., Deliyanti D., Rana I., Miller A.G., Agrotis A., Armani R., Szyndralewiez C., Wingler K., Touyz R.M., Cooper M.E., et al. NADPH oxidase, NOX1, mediates vascular injury in ischemic retinopathy. Antioxidants Redox Signal. 2014;20:2726–2740. doi: 10.1089/ars.2013.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietschy J.M., Turley S.D. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J. Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Fliesler S.J., Bretillon L. The ins and outs of cholesterol in the vertebrate retina. J. Lipid Res. 2010;51:3399–3413. doi: 10.1194/jlr.R010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jun B., Mukherjee P.K., Asatryan A., Kautzmann M.A., Heap J., Gordon W.C., Bhattacharjee S., Yang R., Petasis N.A., Bazan N.G. Elovanoids are novel cell-specific lipid mediators necessary for neuroprotective signaling for photoreceptor cell integrity. Sci. Rep. 2017;7:5279. doi: 10.1038/s41598-017-05433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazan N.G. Overview of how N32 and N34 elovanoids sustain sight by protecting retinal pigment epithelial cells and photoreceptors. J. Lipid Res. 2021;62:100058. doi: 10.1194/jlr.TR120001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat. Rev. Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 19.Guarischi-Sousa R., Monteiro J.S., Alecrim L.C., Michaloski J.S., Cardeal L.B., Ferreira E.N., Carraro D.M., Nunes D.N., Dias-Neto E., Reimand J., et al. A transcriptome-based signature of pathological angiogenesis predicts breast cancer patient survival. PLoS Genet. 2019;15:e1008482. doi: 10.1371/journal.pgen.1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa K., Yoshida S., Kadota K., Nakamura T., Niiro H., Arakawa S., Yoshida A., Akashi K., Ishibashi T. Gene expression profile of hyperoxic and hypoxic retinas in a mouse model of oxygen-induced retinopathy. Invest. Ophthalmol. Vis. Sci. 2010;51:4307–4319. doi: 10.1167/iovs.09-4605. [DOI] [PubMed] [Google Scholar]
- 21.Bartakova A., Weiner G., Oldham M., Ferrara N., Daneman R., Nudleman E. Endothelial cell specific gene expression changes in Oxygen Induced Retinopathy (OIR) Investigative Ophthalmology & Visual Science. 2018;59:5470. [Google Scholar]
- 22.Zhou H., Song H., Wu Y., Liu X., Li J., Zhao H., Tang M., Ji X., Zhang L., Su Y., et al. Oxygen-induced circRNA profiles and coregulatory networks in a retinopathy of prematurity mouse model. Exp. Ther. Med. 2019;18:2037–2050. doi: 10.3892/etm.2019.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zasada M., Madetko-Talowska A., Revhaug C., Rognlien A.G.W., Baumbusch L.O., Książek T., Szewczyk K., Grabowska A., Bik-Multanowski M., Józef Pietrzyk J., et al. Short- and long-term impact of hyperoxia on the blood and retinal cells' transcriptome in a mouse model of oxygen-induced retinopathy. Pediatr. Res. 2020;87:485–493. doi: 10.1038/s41390-019-0598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S.J., Jin J., Kim Y.J., Kim Y., Yu H.G. Retinal proteome analysis in a mouse model of oxygen-induced retinopathy. J. Proteome Res. 2012;11:5186–5203. doi: 10.1021/pr300389r. [DOI] [PubMed] [Google Scholar]
- 25.Tu C., Beharry K.D., Shen X., Li J., Wang L., Aranda J.V., Qu J. Proteomic profiling of the retinas in a neonatal rat model of oxygen-induced retinopathy with a reproducible ion-current-based MS1 approach. J. Proteome Res. 2015;14:2109–2120. doi: 10.1021/pr501238m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vähätupa M., Nättinen J., Jylhä A., Aapola U., Kataja M., Kööbi P., Järvinen T.A.H., Uusitalo H., Uusitalo-Järvinen H. SWATH-MS proteomic analysis of oxygen-induced retinopathy reveals novel potential therapeutic targets. Invest. Ophthalmol. Vis. Sci. 2018;59:3294–3306. doi: 10.1167/iovs.18-23831. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y., Tan W., Zou J., Cao J., Huang Q., Jiang B., Yoshida S., Li Y. Metabolomics analyses of mouse retinas in oxygen-induced retinopathy. Invest. Ophthalmol. Vis. Sci. 2021;62:9. doi: 10.1167/iovs.62.10.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaloski J.S., Redondo A.R., Magalhães L.S., Cambui C.C., Giordano R.J. Discovery of pan-VEGF inhibitory peptides directed to the extracellular ligand-binding domains of the VEGF receptors. Sci. Adv. 2016;2:e1600611. doi: 10.1126/sciadv.1600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frohman M.A. The phospholipase D superfamily as therapeutic targets. Trends Pharmacol. Sci. 2015;36:137–144. doi: 10.1016/j.tips.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egea-Jimenez A.L., Zimmermann P. Phospholipase D and phosphatidic acid in the biogenesis and cargo loading of extracellular vesicles. J. Lipid Res. 2018;59:1554–1560. doi: 10.1194/jlr.R083964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tserentsoodol N., Gordiyenko N.V., Pascual I., Lee J.W., Fliesler S.J., Rodriguez I.R. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol. Vis. 2006;12:1319–1333. [PubMed] [Google Scholar]
- 32.Saadane A., Mast N., Dao T., Ahmad B., Pikuleva I.A. Retinal hypercholesterolemia triggers cholesterol accumulation and esterification in photoreceptor cells. J. Biol. Chem. 2016;291:20427–20439. doi: 10.1074/jbc.M116.744656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih A.Y., Sligar S.G., Schulten K. Maturation of high-density lipoproteins. J. R. Soc. Interface. 2009;6:863–871. doi: 10.1098/rsif.2009.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontush A., Chapman M.J. John Wiley & Sons; 2011. High-density Lipoproteins: Structure, Metabolism, Function and Therapeutics. [Google Scholar]
- 35.Komati R., Spadoni D., Zheng S., Sridhar J., Riley K.E., Wang G. Ligands of therapeutic utility for the liver X receptors. Molecules. 2017;22 doi: 10.3390/molecules22010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Wei Z., Ma X., Yang X., Chen Y., Sun L., Ma C., Miao Q.R., Hajjar D.P., Han J., Duan Y. 25-Hydroxycholesterol activates the expression of cholesterol 25-hydroxylase in an LXR-dependent mechanism. J. Lipid Res. 2018;59:439–451. doi: 10.1194/jlr.M080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuchel M., Rader D.J. Macrophage reverse cholesterol transport: key to the regression of atherosclerosis? Circulation. 2006;113:2548–2555. doi: 10.1161/CIRCULATIONAHA.104.475715. [DOI] [PubMed] [Google Scholar]
- 38.Raghow R., Yellaturu C., Deng X., Park E.A., Elam M.B. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol. Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 39.WikiPathways Cholesterol metabolism with bloch and kandutsch-russell pathways (Mus musculus) 2022. https://www.wikipathways.org/pathways/WP4718.html
- 40.Agbaga M.P., Mandal M.N.A., Anderson R.E. Retinal very long-chain PUFAs: new insights from studies on ELOVL4 protein. J. Lipid Res. 2010;51:1624–1642. doi: 10.1194/jlr.R005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichi I., Kono N., Arita Y., Haga S., Arisawa K., Yamano M., Nagase M., Fujiwara Y., Arai H. Identification of genes and pathways involved in the synthesis of Mead acid (20:3n-9), an indicator of essential fatty acid deficiency. Biochim. Biophys. Acta. 2014;1841:204–213. doi: 10.1016/j.bbalip.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Deliyanti D., Lee J.Y., Petratos S., Meyer C.J., Ward K.W., Wilkinson-Berka J.L., de Haan J.B. A potent Nrf2 activator, dh404, bolsters antioxidant capacity in glial cells and attenuates ischaemic retinopathy. Clin. Sci. 2016;130:1375–1387. doi: 10.1042/CS20160068. [DOI] [PubMed] [Google Scholar]
- 43.Shi X., Li P., Liu H., Prokosch V. Oxidative stress, vascular endothelium, and the pathology of neurodegeneration in retina. Antioxidants. 2022;11:543. doi: 10.3390/antiox11030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey A.P., Koster G., Guillermier C., Hirst E.M.A., MacRae J.I., Lechene C.P., Postle A.D., Gould A.P. Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell. 2015;163:340–353. doi: 10.1016/j.cell.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orban T., Palczewska G., Palczewski K. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem. 2011;286:17248–17258. doi: 10.1074/jbc.M110.195198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Den Brink D.M., Cubizolle A., Chatelain G., Davoust N., Girard V., Johansen S., Napoletano F., Dourlen P., Guillou L., Angebault-Prouteau C., et al. Physiological and pathological roles of FATP-mediated lipid droplets in Drosophila and mice retina. PLoS Genet. 2018;14:e1007627. doi: 10.1371/journal.pgen.1007627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yako T., Otsu W., Nakamura S., Shimazawa M., Hara H. Lipid droplet accumulation promotes RPE dysfunction. Int. J. Mol. Sci. 2022;23:1790. doi: 10.3390/ijms23031790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apte R.S. Targeting tissue lipids in age-related macular degeneration. EBioMedicine. 2016;5:26–27. doi: 10.1016/j.ebiom.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehta R., Elías-López D., Martagón A.J., Pérez-Méndez O.A., Sánchez M.L.O., Segura Y., Tusié M.T., Aguilar-Salinas C.A. LCAT deficiency: a systematic review with the clinical and genetic description of Mexican kindred. Lipids Health Dis. 2021;20:70. doi: 10.1186/s12944-021-01498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sreekumar P.G., Su F., Spee C., Araujo E., Nusinowitz S., Reddy S.T., Kannan R. Oxidative stress and lipid accumulation augments cell death in LDLR-deficient RPE cells and Ldlr(-/-) mice. Cells. 2022;12:43. doi: 10.3390/cells12010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao P., Jia D., Li P., Huang Y., Hu H., Sun K., Lv Y., Chen X., Han Y., Zhang Z., et al. Accumulation of lipid droplets in a novel Bietti crystalline dystrophy zebrafish model with impaired PPARalpha pathway. Invest. Ophthalmol. Vis. Sci. 2022;63:32. doi: 10.1167/iovs.63.5.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smolič T., Tavčar P., Horvat A., Černe U., Halužan Vasle A., Tratnjek L., Kreft M.E., Scholz N., Matis M., Petan T., et al. Astrocytes in stress accumulate lipid droplets. Glia. 2021;69:1540–1562. doi: 10.1002/glia.23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cashikar A., Rios D., Timm D., Romero J., Strickland M., Long J., Han X., Holtzman D., Paul S. Regulation of astrocyte lipid metabolism and ApoE secretion by the microglial oxysterol, 25-hydroxycholesterol. J. Lipid Res. 2023:100350. doi: 10.1016/j.jlr.2023.100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Modjtahedi B.S., Bose N., Papakostas T.D., Morse L., Vavvas D.G., Kishan A.U. Lipids and diabetic retinopathy. Semin. Ophthalmol. 2016;31:10–18. doi: 10.3109/08820538.2015.1114869. [DOI] [PubMed] [Google Scholar]
- 56.Chou Y., Ma J., Su X., Zhong Y. Emerging insights into the relationship between hyperlipidemia and the risk of diabetic retinopathy. Lipids Health Dis. 2020;19:241. doi: 10.1186/s12944-020-01415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Busik J.V. Lipid metabolism dysregulation in diabetic retinopathy. J. Lipid Res. 2021;62:100017. doi: 10.1194/jlr.TR120000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bush R.A., Malnoë A., Remé C.E., Williams T.P. Dietary deficiency of N-3 fatty acids alters rhodopsin content and function in the rat retina. Invest. Ophthalmol. Vis. Sci. 1994;35:91–100. [PubMed] [Google Scholar]
- 59.Grossfield A., Feller S.E., Pitman M.C. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pitman M.C., Grossfield A., Suits F., Feller S.E. Role of cholesterol and polyunsaturated chains in lipid-protein interactions: molecular dynamics simulation of rhodopsin in a realistic membrane environment. J. Am. Chem. Soc. 2005;127:4576–4577. doi: 10.1021/ja042715y. [DOI] [PubMed] [Google Scholar]
- 61.Ortiz A., Gui J., Zahedi F., Yu P., Cho C., Bhattacharya S., Carbone C.J., Yu Q., Katlinski K.V., Katlinskaya Y.V., et al. An interferon-driven oxysterol-based defense against tumor-derived extracellular vesicles. Cancer Cell. 2019;35:33–45.e6. doi: 10.1016/j.ccell.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lembo D., Cagno V., Civra A., Poli G. Oxysterols: an emerging class of broad spectrum antiviral effectors. Mol. Aspect. Med. 2016;49:23–30. doi: 10.1016/j.mam.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 63.Bennett L.D., Hopiavuori B.R., Brush R.S., Chan M., Van Hook M.J., Thoreson W.B., Anderson R.E. Examination of VLC-PUFA-deficient photoreceptor terminals. Invest. Ophthalmol. Vis. Sci. 2014;55:4063–4072. doi: 10.1167/iovs.14-13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hopiavuori B.R., Anderson R.E., Agbaga M.P. ELOVL4: very long-chain fatty acids serve an eclectic role in mammalian health and function. Prog. Retin. Eye Res. 2019;69:137–158. doi: 10.1016/j.preteyeres.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorusupudi A., Liu A., Hageman G.S., Bernstein P.S. Associations of human retinal very long-chain polyunsaturated fatty acids with dietary lipid biomarkers. J. Lipid Res. 2016;57:499–508. doi: 10.1194/jlr.P065540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorusupudi A., Rallabandi R., Li B., Arunkumar R., Blount J.D., Rognon G.T., Chang F.Y., Wade A., Lucas S., Conboy J.C., et al. Retinal bioavailability and functional effects of a synthetic very-long-chain polyunsaturated fatty acid in mice. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2017739118. e2017739118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bennett L.D., Brush R.S., Chan M., Lydic T.A., Reese K., Reid G.E., Busik J.V., Elliott M.H., Anderson R.E. Effect of reduced retinal VLC-PUFA on rod and cone photoreceptors. Invest. Ophthalmol. Vis. Sci. 2014;55:3150–3157. doi: 10.1167/iovs.14-13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lapillonne A., Eleni dit Trolli S., Kermorvant-Duchemin E. Postnatal docosahexaenoic acid deficiency is an inevitable consequence of current recommendations and practice in preterm infants. Neonatology. 2010;98:397–403. doi: 10.1159/000320159. [DOI] [PubMed] [Google Scholar]
- 69.Fu Z., Lofqvist C.A., Shao Z., Sun Y., Joyal J.-S., Hurst C.G., Cui R.Z., Evans L.P., Tian K., SanGiovanni J.P., et al. Dietary ω-3 polyunsaturated fatty acids decrease retinal neovascularization by adipose–endoplasmic reticulum stress reduction to increase adiponectin. Am. J. Clin. Nutr. 2015;101:879–888. doi: 10.3945/ajcn.114.099291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lofqvist C., Najm S., Hellgren G. Association of retinopathy of prematurity with low levels of arachidonic acid: a secondary analysis of a randomized clinical. JAMA OPHTHALMOLOGY. 2018;136:1078. doi: 10.1001/jamaophthalmol.2017.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomita Y., Usui-Ouchi A., Nilsson A.K., Yang J., Ko M., Hellstrom A., Fu Z. Metabolism in retinopathy of prematurity. Life. 2021;11:1119. doi: 10.3390/life11111119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Else P.L. The highly unnatural fatty acid profile of cells in culture. Prog. Lipid Res. 2020;77:101017. doi: 10.1016/j.plipres.2019.101017. [DOI] [PubMed] [Google Scholar]
- 73.Vriens K., Christen S., Parik S., Broekaert D., Yoshinaga K., Talebi A., Dehairs J., Escalona-Noguero C., Schmieder R., Cornfield T., et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayashi Y., Yamano M., Kono N., Arai H., Fujiwara Y., Ichi I. Phosphorylation of Elovl5 changes its substrate preference to synthesize Mead acid in response to essential fatty acid deficiency. bioRxiv. 2020 doi: 10.1101/2020.01.31.929224. Preprint at. [DOI] [Google Scholar]
- 75.Connor K.M., SanGiovanni J.P., Lofqvist C., Aderman C.M., Chen J., Higuchi A., Hong S., Pravda E.A., Majchrzak S., Carper D., et al. Increased dietary intake of omega-3-polyunsaturated fatty acids reduces pathological retinal angiogenesis. Nat. Med. 2007;13:868–873. doi: 10.1038/nm1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sapieha P., Stahl A., Chen J., Seaward M.R., Willett K.L., Krah N.M., Dennison R.J., Connor K.M., Aderman C.M., Liclican E., et al. 5-Lipoxygenase metabolite 4-HDHA is a mediator of the antiangiogenic effect of omega-3 polyunsaturated fatty acids. Sci. Transl. Med. 2011;3:69ra12. doi: 10.1126/scitranslmed.3001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Z., Lofqvist C.A., Shao Z., Sun Y., Joyal J.S., Hurst C.G., Cui R.Z., Evans L.P., Tian K., SanGiovanni J.P., et al. Dietary omega-3 polyunsaturated fatty acids decrease retinal neovascularization by adipose-endoplasmic reticulum stress reduction to increase adiponectin. Am. J. Clin. Nutr. 2015;101:879–888. doi: 10.3945/ajcn.114.099291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu Z., Yan W., Chen C.T., Nilsson A.K., Bull E., Allen W., Yang J., Ko M., SanGiovanni J.P., Akula J.D., et al. Omega-3/Omega-6 long-chain fatty acid imbalance in phase I retinopathy of prematurity. Nutrients. 2022;14:1333. doi: 10.3390/nu14071333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tikhonenko M., Lydic T.A., Wang Y., Chen W., Opreanu M., Sochacki A., McSorley K.M., Renis R.L., Kern T., Jump D.B., et al. Remodeling of retinal Fatty acids in an animal model of diabetes: a decrease in long-chain polyunsaturated fatty acids is associated with a decrease in fatty acid elongases Elovl2 and Elovl4. Diabetes. 2010;59:219–227. doi: 10.2337/db09-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Léger-Charnay E., Gambert S., Martine L., Dubus E., Maire M.A., Buteau B., Morala T., Gigot V., Bron A.M., Bretillon L., Masson E.A.Y. Retinal cholesterol metabolism is perturbated in response to experimental glaucoma in the rat. PLoS One. 2022;17:e0264787. doi: 10.1371/journal.pone.0264787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Leeuwen E.M., Emri E., Merle B.M.J., Colijn J.M., Kersten E., Cougnard-Gregoire A., Dammeier S., Meester-Smoor M., Pool F.M., de Jong E.K., et al. A new perspective on lipid research in age-related macular degeneration. Prog. Retin. Eye Res. 2018;67:56–86. doi: 10.1016/j.preteyeres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 82.Park S.J., Park D.H. REvisiting lipids in REtinal diseases: a focused review on age-related macular degeneration and diabetic retinopathy. J. Lipid Atheroscler. 2020;9:406–418. doi: 10.12997/jla.2020.9.3.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashton N., Ward B., Serpell G. Effect of oxygen on developing retinal vessels with particular reference to the problem of retrolental fibroplasia. Br. J. Ophthalmol. 1954;38:397–432. doi: 10.1136/bjo.38.7.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu C.H., Wang Z., Sun Y., Chen J. Animal models of ocular angiogenesis: from development to pathologies. Faseb. J. 2017;31:4665–4681. doi: 10.1096/fj.201700336R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., O'Donovan C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48:D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 87.Chaves-Filho A.B., Pinto I.F.D., Dantas L.S., Xavier A.M., Inague A., Faria R.L., Medeiros M.H.G., Glezer I., Yoshinaga M.Y., Miyamoto S. Alterations in lipid metabolism of spinal cord linked to amyotrophic lateral sclerosis. Sci. Rep. 2019;9:11642. doi: 10.1038/s41598-019-48059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Macedo F., Martins G.L., Luévano-Martínez L.A., Viana G.M., Riske K.A., Inague A., Yoshinaga M.Y., Aguilaniu H., Miyamoto S., Glezer I., da Cunha F.M. Lipase-like 5 enzyme controls mitochondrial activity in response to starvation in Caenorhabditis elegans. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158539. doi: 10.1016/j.bbalip.2019.158539. [DOI] [PubMed] [Google Scholar]
- 89.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 91.Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Jr., Murphy R.C., Raetz C.R.H., Russell D.W., Seyama Y., Shaw W., et al. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 92.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pico A.R., Kelder T., van Iersel M.P., Hanspers K., Conklin B.R., Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xia J., Wishart D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 96.SHAPIRO S.S., WILK M.B. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 97.Olkin I. Stanford University Press; 1960. Contributions to Probability and Statistics; Essays in Honor of Harold Hotelling. [Google Scholar]
- 98.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data regarding the transcripts (mRNA) cited in the manuscript (Figures 2D and 2E) and/or linked to HDL formation and maturation, cholesterol and fatty acid biosynthesis, and specific retinal cells (glia and macrophages)
Data Availability Statement
-
•
As previously reported,19 sequencing metadata for the RNA-seq data used in the manuscript have been deposited at the Sequence Read Archive (SRA) under accession number BioProject PRJNA483866. RNA-seq data is available under accession number SRP155931. Lipidomics raw files have been deposited at MetaboLights85 under the unique identifier MTBLS6965 (https://www.ebi.ac.uk/metabolights/MTBLS6965). Lipid quantification data are available at Mendeley Data (https://doi.org/10.17632/sn5hz3s992.1). They are publicly available as of the date of publication. Accession numbers are also listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.