Abstract
A complete tricarboxylic acid (TCA) cycle is generally considered necessary for energy production from the dicarboxylic acid substrates malate, succinate, and fumarate. However, a Bradyrhizobium japonicum sucA mutant that is missing α-ketoglutarate dehydrogenase is able to grow on malate as its sole source of carbon. This mutant also fixes nitrogen in symbiosis with soybean, where dicarboxylic acids are its principal carbon substrate. Using a flow chamber system to make direct measurements of oxygen consumption and ammonium excretion, we confirmed that bacteroids formed by the sucA mutant displayed wild-type rates of respiration and nitrogen fixation. Despite the absence of α-ketoglutarate dehydrogenase activity, whole cells of the mutant were able to decarboxylate α-[U-14C]ketoglutarate and [U-14C]glutamate at rates similar to those of wild-type B. japonicum, indicating that there was an alternative route for α-ketoglutarate catabolism. Because cell extracts from B. japonicum decarboxylated [U-14C]glutamate very slowly, the γ-aminobutyrate shunt is unlikely to be the pathway responsible for α-ketoglutarate catabolism in the mutant. In contrast, cell extracts from both the wild type and mutant showed a coenzyme A (CoA)-independent α-ketoglutarate decarboxylation activity. This activity was independent of pyridine nucleotides and was stimulated by thiamine PPi. Thin-layer chromatography showed that the product of α-ketoglutarate decarboxylation was succinic semialdehyde. The CoA-independent α-ketoglutarate decarboxylase, along with succinate semialdehyde dehydrogenase, may form an alternative pathway for α-ketoglutarate catabolism, and this pathway may enhance TCA cycle function during symbiotic nitrogen fixation.
Rhizobia are soil bacteria with the unique ability to fix nitrogen in symbiotic association with legumes. Differentiation of rhizobia into the nitrogen-fixing (bacteroid) state involves not only the expression of the genes encoding nitrogenase but also the acclimation of metabolism to the demands of symbiotic nitrogen fixation (6, 7, 17). Specifically, bacteroids must adjust so that they can maintain aerobic metabolism under O2 concentrations 4 orders of magnitude lower than in air. During symbiosis, bacteroids express a terminal oxidase with a very high affinity for oxygen (14). Nevertheless, additional adjustments to central metabolism are presumably required to optimize the flow of ATP and reductant to nitrogenase (6, 7, 17). Of particular concern is the potential for excess reduced pyridine nucleotides to inhibit several enzymes in the tricarboxylic acid (TCA) cycle. Since the main carbon sources for bacteroids are the dicarboxylic acids malate and succinate (22), extensive inhibition of the TCA cycle may, in turn, compromise the supply of ATP and reductant to nitrogenase.
One TCA cycle enzyme that is particularly sensitive to inhibition by a high ratio of NADH to NAD is α-ketoglutarate dehydrogenase (23). Because of this sensitivity, several groups have suggested that bacteroids may use a bypass pathway around this step of the TCA cycle (6, 7, 17). The γ-aminobutyrate (GABA) shunt has received particular attention as a potential bypass, because bacteroids express some of the necessary enzymes. However, the first enzyme of this pathway, glutamate decarboxylase, is usually present only at very low levels in rhizobia (15, 16, 19, 23), raising the question of whether the GABA shunt would be active enough to compensate for inhibition of α-ketoglutarate dehydrogenase. On the other hand, no other potential bypass around α-ketoglutarate dehydrogenase has been demonstrated for rhizobia.
To investigate the presence of pathways in rhizobia that may bypass α-ketoglutarate dehydrogenase, we have been using a sucA mutant of Bradyrhizobium japonicum that lacks this enzyme (11, 12). To construct the sucA mutant, the sucA region from B. japonicum was cloned into a plasmid and subjected to transposon mutagenesis in Escherichia coli. One mutagenized plasmid, containing an insertion in the sucA gene, was then transferred into B. japonicum, and a sucA mutant resulting from double recombination with the suicide plasmid was isolated. The sucA mutation eliminates α-ketoglutarate dehydrogenase activity but has no effect on other TCA cycle enzymes and can be fully complemented by a wild-type copy of the sucA operon (11, 12).
Unlike most bacterial sucA mutants, the B. japonicum mutant is able to grow on malate as its sole carbon source (11). Furthermore, despite having a delayed-nodulation phenotype, the sucA mutant eventually forms bacteroids with apparent nitrogen fixation activity (12, 13). One explanation of these findings is that B. japonicum has an alternative pathway that can bypass the α-ketoglutarate dehydrogenase step of the TCA cycle. In this study we confirm that the sucA mutant forms fully functional bacteroids and demonstrate the presence of a novel α-ketoglutarate-decarboxylating activity in B. japonicum. This decarboxylase activity, along with succinic semialdehyde dehydrogenase, may form a functional bypass around α-ketoglutarate dehydrogenase and may allow the TCA cycle to function under conditions that would otherwise inhibit its operation.
MATERIALS AND METHODS
Plant material and bacterial strains.
Soybean plants (Glycine max L., cv. Stevens) were grown in modified Leonard jars (12). Soybean seed was surface sterilized prior to being planted and inoculated with either wild-type B. japonicum USDA110 or a sucA mutant derivative (LSG184, sucA::Tn10-miniKan [11]). Plants were grown in a growth chamber with a 16-h photoperiod and a day temperature of 27°C and a night temperature of 24°C.
B. japonicum and Mesorhizobium loti R7A (obtained from Clive Ronson, University of Otago) were cultured at 28°C on a defined medium (11) using 20 mM l-arabinose or l-malate as the carbon source and ammonium as the nitrogen source. Rhizobium leguminosarum 3841 and its sucA mutant derivative RU156 (obtained from Philip Poole, University of Reading) were cultured on AMS defined medium (21) with 20 mM l-malate as the carbon source and ammonium as the nitrogen source. Where appropriate, kanamycin was added to 50 μg/ml for R. leguminosarum or 100 μg/ml for B. japonicum. In all B. japonicum experiments, the purity of the wild-type and sucA mutant strains was confirmed by plating them on defined medium with arabinose, defined medium with arabinose and kanamycin (on which the wild type does not grow), defined medium with acetate (on which the sucA mutant does not grow), and Luria-Bertani medium (on which neither B. japonicum strain grows).
Flow chamber experiments.
Nodules were harvested 5 weeks after inoculation, and bacteroids were purified anaerobically by differential centrifugation (3). Bacteroids (20 to 60 mg [dry weight]), retained above a microporous membrane filter (pore size, 0.45 μm), were incubated in a flow chamber (3) and perfused with buffer containing 40 to 65 μM soybean oxyleghemoglobin as an oxygen carrier and 0.5 mM dl-malate as a carbon source. Analytical methods and calculations were performed essentially as described by Bergersen and Turner (3). In brief, oxygenation of effluent leghemoglobin was monitored spectrophotometrically and used to calculate the oxygen concentration in the flow chamber and the respiration rate of the bacteroids. Fractions of the flow chamber effluent were collected and assayed for the presence of ammonia using a colorimetric method (1), and the results were used to calculate the rate of nitrogen fixation.
Analysis of culture supernatants.
B. japonicum was cultured on defined medium with l-arabinose as the sole carbon source. After the cultures had reached late log phase, the cells were harvested by centrifugation (5 min, 6,000 × g) and washed twice with defined medium with the carbon source omitted. The washed cells were inoculated to an optical density at 630 nm of 0.1 into fresh medium containing 20 mM l-malate for the carbon source. At 0, 24, and 48 h after inoculation, 5-ml samples were removed from the cultures, centrifuged (10 min, 6,000 × g) to remove cells, filtered through 0.2-μm-pore-size filters, and stored at −20°C. Initially, culture supernatants were analyzed by high-performance HPLC liquid chromatography (HPLC) on an HPX-87H column (Bio-Rad). Subsequently, enzymatic methods were used to assay for glutamate and α-ketoglutarate. NAD reduction by glutamate dehydrogenase was used to determine the glutamate concentrations in the culture supernatants (4, 28). Each assay contained 60 μmol of hydrazine, 75 μmol of glycine (pH 9.0), 4 μmol of NAD, and 3 U of glutamate dehydrogenase in a 1.5-ml final volume. α-Ketoglutarate was assayed using aspartate aminotransferase and malate dehydrogenase (28).
Whole-cell CO2 evolution assays.
Cells were harvested by centrifugation (5 min, 6,000 × g), washed, and resuspended in assay buffer (50 mM MOPS [morpholinepropanesulfonic acid; pH 6.8], 1 mM MgCl2, 1 mM K2HPO4). Assay reactions were carried out in stoppered 30-ml Corex tubes in a shaking water bath (27°C), and reaction mixtures contained 0.1 to 1.3 mg (dry weight) of cells in 1 ml of assay buffer. 14C-labeled substrate (0.5 mM final concentration, 0.13 μCi per assay) was added, and the CO2 evolved was trapped on Whatman number 1 filter paper (7 by 20 mm) wetted with 75 μl of 2 M KOH and suspended above the assay mixture. Filters were removed at 15, 30, 45, or 60 min after substrate addition, the 14CO2 was counted in a scintillation counter, and the values were used to calculate the rate of CO2 evolution. Rates were converted to nanomoles per minute per milligram (dry weight) of cells under the assumption that the specific activity of the substrate was not affected by internal metabolites.
Extract preparation.
Cells were harvested by centrifugation (5 min, 6,000 × g), washed, and resuspended in breaking buffer {20 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid; pH 7.0], 100 mM NaCl, 5 mM MgCl2, 0.4 mM EDTA, 1.5 mM dithiothreitol, 4% glycerol} and disrupted in a French pressure cell. For glutamate decarboxylase assays the extract was used directly after centrifugation (20 min, 10,000 × g) to remove unbroken cells and membranes. For all other enzyme assays, the supernatant was desalted by dialysis against breaking buffer with the NaCl omitted.
Enzyme assays.
Glutamate decarboxylase activity was assayed by monitoring 14CO2 evolution from [U-14C]glutamate (Amersham). The assay medium contained 50 mM potassium phosphate (pH 7.0), 30 μM pyridoxal phosphate, 10 mM [U-14C]glutamate (0.05 μCi), and 100 to 400 μg of protein in a final volume of 1 ml. α-Ketoglutarate decarboxylase activity was assayed by monitoring 14CO2 evolution from α-[U-14C]ketoglutarate (New England Nuclear). The assay mixture contained 50 mM TES (pH 6.8), 0.2 mM thiamine pyrophosphate (cocarboxylase), 2 mM MgCl2, 0.01 to 5 mM α-ketoglutarate (0.05 to 0.5 μCi), and 50 to 700 μg of protein in a final volume of 1 ml. For some experiments, 60 μM coenzyme A (CoA), 2.5 mM NAD, or 2.5 mM NADP was also included. Glutamate and α-ketoglutarate decarboxylation reaction mixtures were incubated in stoppered 30-ml Corex tubes at 27°C, and the CO2 evolved was captured on filter paper saturated with 2 N KOH. After 10 to 60 min, the filter paper was removed and the 14CO2 was counted in a scintillation counter. For stoichiometry experiments, the reaction was stopped by the addition of 17 μl of 72% trichloroacetic acid and incubated for a further 2 h before the filter was removed and the 14CO2 was counted.
Succinic semialdehyde dehydrogenase was assayed according to the method of Kouchi et al. (16). The assay mixture contained 50 mM Tris (pH 8.0), 1.4 mM β-mercaptoethanol, 1 mM NAD(P), 2.5 mM succinic semialdehyde, and 50 to 400 μg of protein in a final volume of 1 ml. α-Ketoglutarate dehydrogenase was assayed by monitoring α-ketoglutarate-dependent NAD reduction as described previously (11).
Thin-layer chromatography and succinic semialdehyde quantification.
Reaction products of the α-ketoglutarate decarboxylation assay were derivatized with 2,4 dinitrophenylhydrazine as follows: 500 μl of the reaction mixture was added to 84 μl of 6.3 mM 2,4 dinitrophenylhydrazine in 3 N HCl and incubated for 45 min at 50°C. The derivatized products were extracted three times with 500 μl of ethyl acetate, and the combined extracts were evaporated to dryness. The residue was redissolved in 20 μl of ethyl acetate, 10 μl of which was spotted onto a Kieselgel 60 F254 thin-layer chromatography plate (Merck) and developed with n-butanol saturated with 3% (vol/vol) NH4OH. The derivatized α-ketoglutarate and succinic semialdehyde could be seen as bright yellow spots on the chromatography plate. Autoradiography was performed using conventional X-ray film and exposure times of 3 to 12 days. Authentic α-ketoglutarate and succinic semialdehyde (Sigma, St. Louis, Mo.) were used as standards.
Succinic semialdehyde was determined colorimetrically after reaction with o-aminobenzaldehyde (26). α-Ketoglutarate was determined by the same enzymatic method used for analyzing culture supernatants.
RESULTS
Nitrogen fixation by isolated mutant bacteroids.
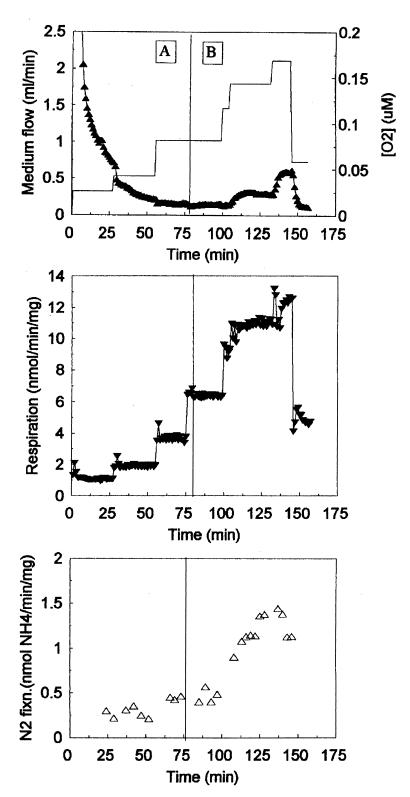
Our previous work showed that nodules formed by the sucA mutant, although containing a reduced number of bacteroids, had some acetylene reduction activity (12). To measure the nitrogen fixation ability of the mutant more directly, we assayed isolated mutant bacteroids in a flow chamber system (3). The reaction buffer contained 40 to 65 μM leghemoglobin to maintain and monitor oxygen levels and 0.5 mM dl-malate as a carbon source. We performed a total of four experiments with the sucA mutant and two experiments with the wild type. A typical experiment using the mutant bacteroids (Fig. 1) showed that they were able to maintain low oxygen levels in the flow chamber and achieve nitrogen fixation and respiration rates similar to those of the wild type. The top and middle panels of Fig. 1 show that when the flow rate of fresh, oxygenated medium into the chamber was increased, the respiration rate of the mutant bacteroids rose in response to the increased supply of oxygen. The bacteroids were able to maintain a low concentration of oxygen in the chamber until, between 100 and 150 min, the rate of oxygen delivery exceeded the respiratory capacity of the bacteroids and the oxygen concentration began to rise. At the end of the experiment, the flow rate was decreased and there was a concomitant decline in respiratory rate and oxygen concentration. Nitrogen fixation (Fig. 1, bottom panel) responded in parallel with respiration.
FIG. 1.
Performance of sucA mutant bacteroids in the flow chamber. Bacteroids were harvested from 5-week-old nodules and placed in a flow chamber with 0.5 mM malate for a carbon source. Two different reaction buffers were used in this experiment. Reaction buffer A was equilibrated with 50% air-50% N2 (10% O2), and buffer B was equilibrated with air (20% O2). Inflowing buffer was changed from A to B at 75 min. The top panel shows the buffer flow rate (solid line) and free oxygen concentration in the flow chamber (▴). The middle and bottom panels show the rates of respiration (▾) and of nitrogen fixation (▵), respectively. Results from one of three similar experiments are shown.
Wild-type bacteroids of B. japonicum strain CB1809 have carbon reserves capable of sustaining nitrogen fixation for extended periods in the absence of added substrate (3). To test the dependence of the sucA mutant bacteroids on an exogenous carbon source, the malate was omitted from the reaction buffer in one experiment. Under these conditions, the mutant was unable to achieve a respiratory rate sufficient to maintain a low oxygen concentration in the flow chamber and very little nitrogen fixation was observed (data not shown). This result indicated that nitrogen fixation by the sucA mutant depended on catabolism of exogenous malate rather than on internal reserves.
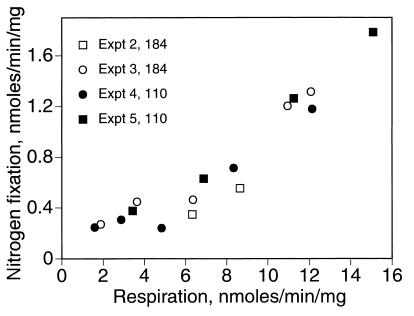
As a further test of the symbiotic function of the mutant bacteroids, we examined the relationship between respiration rate and nitrogen fixation, a measure of metabolic efficiency (2). Steady-state nitrogen fixation rates were plotted against the accompanying respiration rates from four separate experiments, two using wild-type bacteroids and two using sucA mutant bacteroids (Fig. 2). As expected, for both the wild type and mutant, increases in the respiration rate and, consequently, the supply of ATP were accompanied by increases in the rate of nitrogen fixation. Wild-type and mutant values fell along the same curve, providing further evidence that the sucA mutant bacteroids were functioning normally.
FIG. 2.
Relationship between steady-state rates of respiration and nitrogen fixation. Wild-type (USDA110) and sucA mutant (LSG184) bacteroids were prepared from 5-week-old nodules and assayed in the flow chamber, in experiments similar to that described for Fig. 1. Average respiration and nitrogen fixation rates were calculated for each steady state attained in four separate experiments. Each point represents a single steady state.
Metabolite excretion by the sucA mutant.
To determine whether the sucA mutant excretes metabolites during growth on malate, culture supernatants were assayed for the presence of organic acids. The sucA mutant grows with a doubling time about 30% longer than that of the wild type on malate (11). Nevertheless, by 48 h after inoculation the mutant had consumed all of the malate (as determined by HPLC) in the cultures and attained the same final dry weight as the wild type (data not shown). Preliminary experiments using HPLC indicated that the only detectable metabolite excreted by the mutant was α-ketoglutarate (data not shown). Subsequently, enzymatic methods were used to quantify α-ketoglutarate in the culture supernatants. As expected, wild-type cells did not excrete α-ketoglutarate during growth on malate (Table 1). In contrast, some α-ketoglutarate accumulated in the culture supernatants of the sucA mutant. The amount of α-ketoglutarate was variable and in some cultures actually declined between 24 and 48 h of growth. Plating tests showed that the decline in α-ketoglutarate was not caused by growth of contaminants in the cultures or by reversion of the mutant to a wild-type phenotype. Even at its highest level, the amount of α-ketoglutarate in the culture supernatants never exceeded about 5% of the malate consumed, on a molar basis. Glutamate was never detected in the culture supernatants, either by HPLC or enzymatic determinations. All together, the results indicated that the mutant did not excrete stoichiometric amounts of organic acid metabolites of malate when malate was used as its sole source of carbon.
TABLE 1.
Concentrations of α-ketoglutarate in culture supernatants of B. japonicum culturesa
| No. of h after transfer | Concn (μM) of α-ketoglutarate in culture supernatant of:
|
|
|---|---|---|
| Wild type | sucA mutant | |
| 0 | 24 ± 1 | 20 ± 2 |
| 24 | 11 ± 7 | 90 ± 12 |
| 48 | 6 ± 2 | 159 ± 144 |
Wild-type and sucA mutant cells were grown on minimal medium with arabinose and then washed and transferred to fresh medium containing malate as the carbon source. Culture supernatants were collected at various times after transfer to growth on malate and were analyzed for the presence of α-ketoglutarate using the enzymatic method described in Materials and Methods. Values represent the means ± standard errors of the means of results of three separate experiments.
Decarboxylation of exogenously supplied glutamate and α-ketoglutarate.
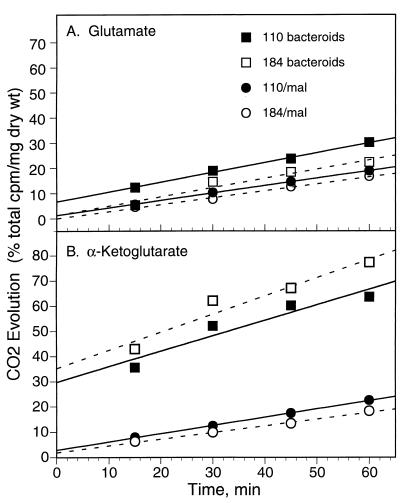
To test for the presence of a bypass pathway around α-ketoglutarate dehydrogenase, we assayed 14CO2 evolution by sucA mutant cells supplied with exogenous α-[U-14C]ketoglutarate or its carbon skeleton equivalent [U-14C]glutamate. Results from a representative subset of these experiments are presented in Fig. 3. CO2 evolution from [U-14C]glutamate was linear for at least an hour for both wild-type and mutant cells that had been cultured on malate and for bacteroids of both strains (Fig. 3A). For cultured cells, rates of CO2 evolution from α-[U-14C]ketoglutarate were also linear for at least an hour (Fig. 3B). For bacteroids of both strains, the kinetics of CO2 evolution from α-[U-14C]ketoglutarate were different, showing an initial rapid rate that declined over the course of the hour-long assay (Fig. 3B).
FIG. 3.
Decarboxylation of exogenous glutamate and α-ketoglutarate by whole cells of B. japonicum. Wild-type and sucA mutant cultured cells (grown with malate [mal]) or bacteroids were harvested, washed, and resuspended in buffer containing either [U-14C]glutamate or α-[U-14C]ketoglutarate. 14CO2 evolution was determined at 15, 30, 45, and 60 min after substrate addition. Results from representative single experiments are shown; decarboxylation rates derived from the complete set of experiments are reported in Results.
The rate of CO2 evolution in each experiment was determined from the slope of the regression line through the four time points. For experiments involving bacteroids and α-ketoglutarate, this method underestimated the probable initial rate of decarboxylation. In two separate experiments, wild-type and sucA mutant cells, cultured on malate, decarboxylated glutamate at 6.3 ± 1.5 and 6.4 ± 0.6 nmol of CO2 per min per mg (dry weight), respectively. The same cells decarboxylated α-ketoglutarate at 8.0 ± 0.2 and 6.9 ± 0.5 nmol of CO2 per min per mg (dry weight), respectively. In single experiments, wild-type and sucA mutant bacteroids decarboxylated glutamate at 9.7 and 8.8 nmol of CO2 per min per mg (dry weight), respectively. α-Ketoglutarate was decarboxylated by wild-type and sucA mutant bacteroids at 15.4 and 17.3 nmol of CO2 per min per mg (dry weight), respectively. In all cases, the rate of CO2 evolution by the sucA mutant was similar to the wild-type rate, providing further evidence that B. japonicum has an alternative to α-ketoglutarate dehydrogenase for catabolizing glutamate and α-ketoglutarate.
Glutamate decarboxylase activity.
One possible alternative route for decarboxylation of α-ketoglutarate is via glutamate decarboxylase and the GABA shunt. Therefore, we assayed for glutamate decarboxylase activity in crude extracts from wild-type and sucA mutant cells. In agreement with previous reports (11, 16, 23), we found only very low glutamate decarboxylase activity in both cultured cells and bacteroids of either strain (Table 2).
TABLE 2.
Various enzyme activities of crude extracts from wild-type and sucA mutant B. japonicuma
| Enzyme | Activity (nmol min−1 mg of protein−1) of:
|
|||
|---|---|---|---|---|
| Wild type
|
sucA mutant
|
|||
| Malate grown | Bacteroids | Malate grown | Bacteroids | |
| Glutamate decarboxylase | 0.42 ± 0.08 | 0.42 | 0.30 ± 0.30 | 0.42 |
| α-Ketoglutarate dehydrogenase | 46.0 ± 7.6 | 23.0 ± 2.4 | −0.2 ± 3.0 | 2.8 ± 0.4 |
| α-Ketoglutarate decarboxylase | 7.0 ± 2.0 | 17.0 ± 1.2 | 6.2 ± 2.0 | 18.0 ± 1.0 |
| Succinic semialdehyde dehydrogenase (NAD) | 12.8 ± 0.4 | 11.1 ± 0.8 | 23.2 ± 0.4 | 18.2 ± 2.0 |
| Succinic semialdehyde dehydrogenase (NADP) | 7.9 ± 0.9 | 9.4 ± 1.3 | 17.7 ± 2.1 | 14.0 ± 2.5 |
Cells were either cultured on minimal medium with malate (malate grown) or isolated from 5-week-old soybean nodules (bacteroids). Cells were harvested, extracts were prepared, and enzymes were assayed as described in Materials and Methods. Values are single determinations (for bacteroid glutamate decarboxylase assays) or the means ± standard errors of the means of results from either two (malate grown) or three (bacteroid) separate experiments.
α-Ketoglutarate decarboxylase activity.
In contrast to the low glutamate decarboxylase activity, cell extracts from both wild-type and sucA mutant cells showed an α-ketoglutarate-decarboxylating activity that, unlike α-ketoglutarate dehydrogenase, did not require CoA (Tables 2 and 3). The sucA mutant had the same amount of activity as the wild type, whether as cultured cells or bacteroids. Decarboxylation was linear for at least 1 h and was proportional to the amount of protein added (data not shown). Boiled extracts were inactive.
TABLE 3.
Effect of CoA addition on the rate of α-ketoglutarate decarboxylationa
| Crude extracts | α-KG decarboxylation (nmol min−1 mg of protein−1)
|
|
|---|---|---|
| Without CoA | With CoA | |
| wt, cultured | 1.6 | 5.7 |
| sucA, cultured | 1.5 | 1.6 |
| wt, bacteroids | 3.3 | 6.4 |
| sucA, bacteroids | 3.2 | 3.4 |
Crude extracts were prepared from wild type (wt) or sucA mutant (sucA) cells cultured on malate or isolated from nodules (bacteroids). Extracts were assayed for α-ketoglutarate (α-KG) decarboxylation with or without CoA. Assay mixtures contained 1 mM α-ketoglutarate (0.1 μCi), 0.12 to 0.3 mg of protein, thiamine pyrophosphate, and NAD. Assays were terminated 10 min after substrate addition. All values are from single determinations.
Identification of the product of α-ketoglutarate decarboxylation.
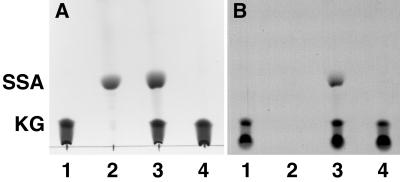
A likely product of α-ketoglutarate decarboxylation, in the absence of CoA and pyridine nucleotides, is succinic semialdehyde. To determine whether succinic semialdehyde was formed by the activity in B. japonicum extracts, reaction mixtures were derivatized with 2,4 dinitrophenylhydrazine and analyzed by thin-layer chromatography. A radioactive product that cochromatographed with authentic succinic semialdehyde was formed in reaction mixtures containing B. japonicum crude extract but not in those where extract was omitted (Fig. 4).
FIG. 4.
Thin-layer chromatography of α-ketoglutarate decarboxylation reaction products. α-Ketoglutarate decarboxylase activity was assayed using extract from wild-type bacteroids, and the reaction products were treated with 2,4 dinitrophenylhydrazine. The derivatized reaction products were analyzed by thin-layer chromatography and autoradiography as described in Materials and Methods. α-Ketoglutarate and succinic semialdehyde were derivatized in the same way and used as standards. (A) Chromatogram of the following samples: α-[U-14C]ketoglutarate substrate (lane 1), unlabeled succinic semialdehyde (lane 2), products of an α-ketoglutarate decarboxylation assay containing wild-type bacteroid extract (0.35 mg of protein) (lane 3), and products of an α-ketoglutarate decarboxylation assay with the extract omitted (lane 4). (B) Autoradiograph of the chromatogram shown in panel A. This figure was composed in Adobe Photoshop 3.04, using scanned images. Abbreviations: KG, α-ketoglutarate; SSA, succinic semialdehyde.
An attempt was made to determine the stoichiometry of the α-ketoglutarate decarboxylation reaction. For example, in one experiment 420 nmol of α-ketoglutarate was consumed and 318 nmol of succinic semialdehyde was formed, approximating the expected 1:1 ratio and confirming that succinic semialdehyde is a product of CoA-independent α-ketoglutarate decarboxylation. However, the amount of CO2 evolved was consistently lower than expected. In four separate experiments, the ratio of succinic semialdehyde formed to CO2 evolved was 1.85 ± 0.06. The reason for this discrepancy is unclear but may arise from interfering activities in the crude extracts used in these experiments or from incomplete recovery of evolved CO2. CO2 evolution assays must be acidified at the end of the incubation period to allow quantitative recovery of dissolved CO2 (27). Our experimental apparatus required the assay tubes to be opened briefly for the addition of acid, leading to some loss of the gas-phase CO2 in the stoichiometry experiments.
Succinic semialdehyde dehydrogenase activity.
Succinic semialdehyde dehydrogenase was found in all of the B. japonicum extracts, using either NAD or NADP as the electron acceptor (Table 2). Activity with NAD was always higher than with NADP (Table 2), and the activities were not additive (data not shown). Extracts from both bacteroids and cultured cells of the sucA mutant showed higher succinic semialdehyde dehydrogenase activities than those of comparable extracts from the wild type (Table 2).
Properties of the α-ketoglutarate decarboxylase activity.
Thiamine pyrophosphate (cocarboxylase) was required for full activity of the CoA-independent α-ketoglutarate decarboxylase (Table 4). In contrast, the decarboxylation activity did not require pyridine nucleotides. Although there was α-ketoglutarate-dependent, CoA-independent NAD and NADP reduction by crude extracts from both the wild type and the mutant, the addition of pyridine nucleotides resulted in no stimulation of CO2 evolution activity (Table 4). The NAD(P) reduction may have resulted from the activity of a dehydrogenase in the crude extracts, probably succinic semialdehyde dehydrogenase, that is able to oxidize the product of α-ketoglutarate decarboxylation.
TABLE 4.
Effects of various additions on CoA-independent α-ketoglutarate decarboxylationa
| Addition(s) | Rate of α-ketoglutarate decarboxylation (nmol min−1 mg of protein−1) |
|---|---|
| None | 1.9 |
| TPP | 5.7 |
| NAD | 1.8 |
| NADP | 1.9 |
| TPP and NAD | 5.8 |
| TPP and NADP | 5.7 |
Assay mixtures contained 1 mM α-ketoglutarate (0.416 μCi/assay) and extract from sucA mutant bacteroids amounting to 0.086 mg of protein. All values are from single determinations. Abbreviation: TPP, thiamine pyrophosphate.
In some cases, pyruvate decarboxylase (EC 4.1.1.1) shows activity with α-ketoglutarate (24). To test whether α-ketoglutarate was being decarboxylated by an enzyme for which pyruvate was the preferred substrate, we tested for competitive inhibition of α-ketoglutarate decarboxylation by pyruvate. Excess cold pyruvate did not inhibit α-ketoglutarate decarboxylation, suggesting that the CO2 evolution activity that we observed did not result from a side reaction of pyruvate decarboxylase. Likewise, excess cold α-ketobutyrate did not inhibit CoA-independent α-ketoglutarate decarboxylation. In contrast, succinic semialdehyde, added in 2.5- to 5-fold molar excess relative to the concentration of α-ketoglutarate, inhibited α-ketoglutarate decarboxylation by 25 to 30%, indicating that the activity may be subject to product inhibition.
α-Ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase activity in other rhizobia.
To see whether CoA-independent α-ketoglutarate decarboxylation was confined to B. japonicum, we assayed crude extracts prepared from three other rhizobial strains (Table 5). M. loti showed a level of α-ketoglutarate decarboxylase activity even higher than that of B. japonicum. R. leguminosarum, both a wild-type strain and a sucA mutant (28), also showed small amounts of activity. All three strains had substantial succinic semialdehyde dehydrogenase activity.
TABLE 5.
Activities of α-ketoglutarate-catabolizing enzymes in crude extracts from various rhizobial strainsa
| Strain | Sp act (nmol min−1 mg of protein−1) of:
|
|||
|---|---|---|---|---|
| KGDH | KDC | SSDH (NAD) | SSDH (NADP) | |
| M. loti R7A | 22.7 | 86.9 | 45.2 | 109.8 |
| R. leguminosarum 3841 (wild type) | 91.6 | 2.4 | 12.4 | 6.0 |
| R. leguminosarum RU156 (sucA mutant) | 0 | 5.6 | 10.8 | 4.4 |
Cells were grown on minimal medium with malate for a carbon source. Crude extracts were prepared and assayed as described in Materials and Methods. All values are from single determinations. Abbreviations: KGDH, α-ketoglutarate dehydrogenase; KDC, α-ketoglutarate decarboxylase; SSDH, succinic semialdehyde dehydrogenase.
DISCUSSION
The results of this study provide further evidence that B. japonicum possesses a functional bypass around the α-ketoglutarate dehydrogenase step of the TCA cycle. Flow chamber experiments showed that sucA mutant bacteroids are not impaired by the loss of α-ketoglutarate dehydrogenase, even when relying on exogenous malate to support nitrogen fixation. The mutant is able to grow on malate as its sole carbon source, without excreting stoichiometric amounts of organic acid metabolites of malate. Furthermore, the mutant can decarboxylate exogenously supplied α-ketoglutarate and glutamate at rates similar to those of the wild type. Together, these results strongly suggest that B. japonicum has an alternative to the conventional TCA cycle for the catabolism of dicarboxylic acids.
The GABA shunt is the usual α-ketoglutarate dehydrogenase bypass pathway proposed for rhizobia. In this pathway, α-ketoglutarate is first converted to glutamate, which is then decarboxylated to GABA by glutamate decarboxylase. GABA is then transaminated to form succinic semialdehyde, which is oxidized via succinic semialdehyde dehydrogenase to succinate, and thereby rejoins the TCA cycle. All bacteroids examined so far have had succinic semialdehyde dehydrogenase activity comparable to those of other TCA cycle enzymes (25 to 34 nmol min−1 mg of protein−1 [8, 15, 16, 19]). In contrast, the amount of glutamate decarboxylase activity found in bacteroids has generally been much lower (0 to 1.4 nmol min−1 mg of protein−1 [15, 16, 19]). An exception is one study that found high levels of glutamate decarboxylase activity in Sinorhizobium meliloti (8). This group also isolated a mutant strain in which reduced levels of succinic semialdehyde dehydrogenase were correlated with an inability to grow on glutamate and reduced rates of nitrogen fixation (9). This finding, although not yet corroborated by genetic evidence from other rhizobia, has sustained interest in the GABA shunt as a potentially important bypass pathway during symbiosis.
Although B. japonicum has the other two enzymes required for operation of the GABA shunt, the amount of glutamate decarboxylase found is very low (references 11, 16, and 23 and this study), suggesting that the pathway is not very active. We have now found a CoA-independent α-ketoglutarate decarboxylase activity in B. japonicum that is capable of forming succinic semialdehyde directly from α-ketoglutarate. The activity requires thiamine pyrophosphate but not CoA or pyridine nucleotides and is thus distinct from α-ketoglutarate dehydrogenase. The results suggest that wild-type B. japonicum can decarboxylate α-ketoglutarate via two different enzymes, one being the canonical α-ketoglutarate dehydrogenase and the other being a CoA- and NAD(P)-independent activity.
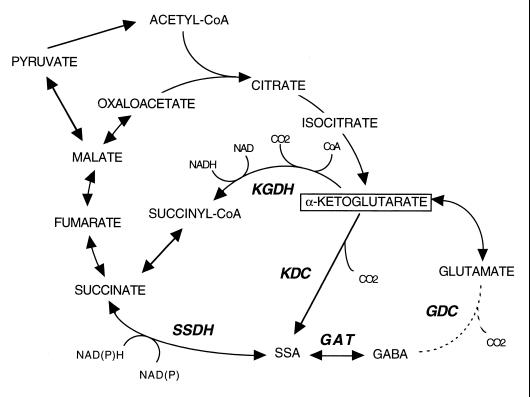
We propose that the CoA-independent α-ketoglutarate decarboxylase activity in B. japonicum, along with succinic semialdehyde dehydrogenase, forms a functional bypass around the α-ketoglutarate dehydrogenase step of the TCA cycle (Fig. 5). The phenotype of the sucA mutant implies that this bypass can support TCA cycle activity sufficient for growth on malate and malate-dependent nitrogen fixation. Such a pathway operates as part of a modified TCA cycle in Euglena gracilis mitochondria (24, 25) and has also been proposed, on the basis of genome sequence analysis, for bacteria that lack the structural genes for α-ketoglutarate dehydrogenase (5).
FIG. 5.
Proposed pathways for α-ketoglutarate catabolism in B. japonicum. KGDH, α-ketoglutarate dehydrogenase; KDC, α-ketoglutarate decarboxylase; SSDH, succinic semialdehyde dehydrogenase; SSA, succinic semialdehyde; GAT, γ-aminobutyrate aminotransferase; GDC glutamate decarboxylase.
Two enzymes that catalyze the conversion of α-ketoglutarate to succinic semialdehyde, E. gracilis α-ketoglutarate decarboxylase (EC 4.1.1.71; 24) and a bifunctional protein encoded by menD, have been described (20). The latter enzyme is part of the menaquinone biosynthetic pathway, and the succinic semialdehyde produced by decarboxylation of α-ketoglutarate would normally be consumed by a second activity on the enzyme to form 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid (SHCHC). However, α-ketoglutarate decarboxylation can occur independently of SHCHC synthesis, and succinic semialdehyde is then released from the enzyme (20). Since the gene encoding E. gracilis α-ketoglutarate decarboxylase has not yet been cloned, it is not known whether it is related to menD. However both proteins are 62,000 Da in size and are homomultimers in their active form (18, 25). Whether the B. japonicum activity is encoded by a homolog of menD or by a unique gene must await further study.
A potential advantage of having an alternative pathway for α-ketoglutarate catabolism is that succinic semialdehyde dehydrogenase, unlike α-ketoglutarate dehydrogenase, can reduce NADP instead of NAD. Theoretically, operation of the bypass may facilitate continued flux of carbon through the TCA cycle under conditions, such as oxygen limitation, that cause a buildup of excess NADH and feedback inhibition of α-ketoglutarate dehydrogenase. Drawbacks of using the bypass instead of α-ketoglutarate dehydrogenase would be the need to synthesize succinyl-CoA from succinate and the loss of the substrate-level phosphorylation catalyzed by succinyl-CoA synthetase. These disadvantages may account for the sucA mutant's slower than normal growth on malate (11) and delayed-nodulation phenotype (12, 13). For wild-type B. japonicum, having two pathways for α-ketoglutarate catabolism may allow greater flexibility in partitioning the reductant generated by the TCA cycle in response to changing metabolic demands.
Two other rhizobia, M. loti and R. leguminosarum, also showed CoA-independent α-ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase activity, suggesting that the bypass we have proposed for B. japonicum is distributed widely among rhizobia. The α-ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase bypass may be a general adaptation for catabolizing dicarboxylic acids under the microaerobic conditions inside legume nodules. Indeed, one study showed that succinic semialdehyde dehydrogenase activity was required for optimal nitrogen fixation in S. meliloti (9). Structural genes for succinic semialdehyde dehydrogenase have been found on the symplasmid of NGR234 (10) and in the symbiosis island of M. loti (C. W. Ronson, personal communication), suggesting a symbiotic function. Whether an α-ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase bypass is required for symbiotic nitrogen fixation must await the analysis of mutants specifically defective in this pathway.
ACKNOWLEDGMENTS
This work was supported in part by USDA-NRICGP grant CSREES 98-353-5-6909 to D.W.E., an Australian Research Council grant to D.A.D., and the Interdisciplinary Plant Group at the University of Missouri.
We thank Clive Ronson and Philip Poole for kindly providing bacterial strains for this work.
REFERENCES
- 1.Bergersen F J. Measurement of nitrogen fixation by direct means. In: Bergersen F J, editor. Methods for evaluating biological nitrogen fixation. Chichester, United Kingdom: John Wiley; 1980. pp. 65–110. [Google Scholar]
- 2.Bergersen F J. Physiological and biochemical aspects of nitrogen fixation by bacteroids in soybean nodule cells. Soil Biol Biochem. 1997;29:875–880. [Google Scholar]
- 3.Bergersen F J, Turner G L. Bacteroids from soybean root nodules: respiration and N2-fixation in flow-chamber reactions with oxyleghaemoglobin. Proc R Soc Lond B. 1990;238:295–320. [Google Scholar]
- 4.Bernt E, Bergmeyer H U. l-Glutamate UV-assay with glutamate dehydrogenase and NAD. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 2nd ed. New York, N.Y: Academic Press; 1974. pp. 1704–1708. [Google Scholar]
- 5.Cordwell S. Microbial genomes and “missing” enzymes: redefining biochemical pathways. Arch Microbiol. 1999;172:269–279. doi: 10.1007/s002030050780. [DOI] [PubMed] [Google Scholar]
- 6.Day D A, Copeland L. Carbon metabolism and compartmentation in nitrogen-fixing legume nodules. Plant Physiol Biochem. 1991;29:185–201. [Google Scholar]
- 7.Dunn M F. Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia. FEMS Microbiol Rev. 1998;22:105–123. doi: 10.1111/j.1574-6976.1998.tb00363.x. [DOI] [PubMed] [Google Scholar]
- 8.Fitzmaurice A M, O'Gara F. Glutamate catabolism in Rhizobium meliloti. Arch Microbiol. 1991;155:422–427. [Google Scholar]
- 9.Fitzmaurice A M, O'Gara F. A Rhizobium meliloti mutant, lacking a functional γ-aminobutyrate (GABA) bypass, is defective in glutamate catabolism and symbiotic nitrogen fixation. FEMS Microbiol Lett. 1993;109:195–202. [Google Scholar]
- 10.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:392–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 11.Green L S, Emerich D W. Bradyrhizobium japonicum does not require α-ketoglutarate dehydrogenase for growth on succinate or malate. J Bacteriol. 1997;179:194–201. doi: 10.1128/jb.179.1.194-201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green L S, Emerich D W. The formation of nitrogen-fixing bacteroids is delayed but not abolished in soybean infected by an α-ketoglutarate dehydrogenase-deficient mutant of Bradyrhizobium japonicum. Plant Physiol. 1997;114:1359–1368. doi: 10.1104/pp.114.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green L S, Emerich D W. Light microscopy of early stages in the symbiosis of soybean with a delayed-nodulation mutant of Bradyrhizobium japonicum. J Exp Bot. 1999;50:1577–1585. [Google Scholar]
- 14.Hennecke H. Rhizobium respiration to support symbiotic nitrogen fixation. In: Elmerich C, Kondorosi A, Newton W E, editors. Biological nitrogen fixation for the 21st century. Dordrecht, The Netherlands: Kluwer Academic; 1998. pp. 429–434. [Google Scholar]
- 15.Jin H N, Dilworth M J, Glenn A R. 4-Aminobutyrate is not available to bacteroids of cowpea Rhizobium MNF2030 in snake bean nodules. Arch Microbiol. 1990;153:455–462. [Google Scholar]
- 16.Kouchi H, Fukai K, Kihara A. Metabolism of glutamate and aspartate in bacteroids isolated from soybean root nodules. J Gen Microbiol. 1991;137:2901–2910. [Google Scholar]
- 17.McDermott T R, Griffith S M, Vance C P, Graham P H. Carbon metabolism in Bradyrhizobium japonicum bacteroids. FEMS Microbiol Rev. 1989;63:327–340. [Google Scholar]
- 18.Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 642–656. [Google Scholar]
- 19.Miller R W, McRae D G, Joy K. Glutamate and γ-aminobutyrate metabolism in isolated Rhizobium meliloti bacteroids. Mol Plant-Microbe Interact. 1991;4:37–45. [Google Scholar]
- 20.Palaniappan C, Sharma V, Hudspeth M E S, Meganathan R. Menaquinone (vitamin K2) biosynthesis: evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and α-ketoglutarate decarboxylase activities. J Bacteriol. 1992;174:8111–8118. doi: 10.1128/jb.174.24.8111-8118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole P S, Schofield N A, Reid C J, Drew E M, Walshaw D L. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 22.Rosendahl L, Glenn A R, Dilworth M J. Organic and inorganic inputs into legume root nodule nitrogen fixation. In: Dilworth M J, Glenn A R, editors. Biology and biochemistry of nitrogen fixation. Amsterdam, The Netherlands: Elsevier; 1991. pp. 259–291. [Google Scholar]
- 23.Salminen S O, Streeter J G. Factors contributing to the accumulation of glutamate in Bradyrhizobium japonicum bacteroids. J Gen Microbiol. 1990;136:2119–2126. [Google Scholar]
- 24.Shigeoka S, Onishi T, Maeda K, Nakano Y, Kitaoka S. Occurrence of thiamin pyrophosphate-dependent 2-oxoglutarate decarboxylase in mitochondria of Euglena gracilis. FEBS Lett. 1986;195:43–47. [Google Scholar]
- 25.Shigeoka S, Nakano Y. Characterization and molecular properties of 2-oxoglutarate decarboxylase from Euglena gracilis. Arch Biochem Biophys. 1991;288:22–28. doi: 10.1016/0003-9861(91)90160-k. [DOI] [PubMed] [Google Scholar]
- 26.Soda K, Toyama S, Misono H, Hirasawa T, Asada K. Spectrophotometric determination of glyoxylic acid with o-aminobenzaldehyde and glycine, and its application to enzyme assay. Agric Biol Chem. 1973;37:1393–1400. [Google Scholar]
- 27.Steinberg D, Udenfriend S. The measurement of radioisotopes. Methods Enzymol. 1957;4:425–472. [Google Scholar]
- 28.Walshaw D W, Wilkinson A, Mundy M, Smith M, Poole P S. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology. 1997;143:2209–2221. doi: 10.1099/00221287-143-7-2209. [DOI] [PubMed] [Google Scholar]