Abstract
CcdA is known to be required for the synthesis of c-type cytochromes in Bacillus subtilis, but the exact function of this membrane protein is not known. We show that CcdA also plays a role in spore synthesis. The expression of ccdA and the two downstream genes yneI and yneJ was analyzed. There is a promoter for each gene, but there is only one transcription terminator, located after the yneJ gene. The promoter for ccdA was found to be weak and was active mainly during the transition from exponential growth to stationary phase. The promoters for yneI and yneJ were both active in the exponential growth phase. The levels of the CcdA and YneJ proteins in the membrane were consistent with the observed promoter activities. The ccdA promoter activity was independent of whether the ccdA-yneI-yneJ gene products were absent or overproduced in the cell. It is shown that the four known cytochromes c in B. subtilis and the YneI and YneJ proteins are not required for sporulation. The combined data from analysis of sporulation-specific sigma factor activity, resistance properties of spores, and spore morphology indicate that CcdA deficiency affects stage V in sporulation. We conclude that CcdA, YneI, and YneJ are functionally unrelated proteins and that the role of CcdA in cytochrome c and spore synthesis probably relates to sulfhydryl redox chemistry on the outer surface of the cytoplasmic membrane.
The gram-positive, endospore-forming, bacterium Bacillus subtilis contains four different c-type cytochromes, which are all membrane anchored (2, 49, 52). The heme domain of these cytochromes is located on the outer surface of the cytoplasmic membrane. B. subtilis does not require cytochrome c for aerobic or anaerobic growth under laboratory conditions, and the physiological role of cytochromes of this type in the bacterium is not well understood.
The trademark of c-type cytochromes is that they contain protoheme IX covalently bound to the protein via thioether linkages (1, 29). Cytochrome c synthesis, i.e., the formation of the covalently bound heme, occurs on the outer (periplasmic) side of the cytoplasmic membrane in bacteria. In gram-negative bacteria, this biosynthetic process is assisted by several membrane-bound and periplasmic proteins (see reference 19, 27, and 44 for reviews). B. subtilis ccdA, resB, and resC are hitherto the only genes that have been shown experimentally to be required for cytochrome c synthesis in a gram-positive bacterium (21, 36). Genes encoding B. subtilis CcdA orthologues are present in members of the domains Bacteria (such as Mycobacterium tuberculosis, Helicobacter pylori, Haemophilus influenzae, Treponema pallidum, and cyanobacteria) and Archaea and also in plants (encoded by chloroplast genomes), but experimental data on the gene product in these organisms is not available. Very recently, CcdA was found in Rhodobacter capsulatus and shown to be involved in cytochrome c synthesis (8).
B. subtilis CcdA is an integral membrane protein of 228 or 235 amino acid residues (36). The exact function of this protein in the cell is not known, but it is required for a late step in the cytochrome c maturation pathway, after heme and apocytochrome have been transported across the cytoplasmic membrane (35). The amino acid sequence of CcdA is similar to that of the central part of DsbD (also named DipZ) of Escherichia coli, a protein of 489 residues which is thought to transfer reducing equivalents to disulfide isomerase(s) in the periplasm (6, 25, 31). The C-terminal part of DsbD, which has no similarity to CcdA, contains a thioredoxin-like sequence motif, -CysXaaYaaCys-. Cysteine residues in this motif and those that are invariant in DsbD and CcdA have been shown by site-specific mutagenesis to be functionally important (8, 39). It is notable that the genomes of some bacteria, for example H. influenzae and M. tuberculosis, contain genes for both CcdA and DsbD proteins whereas others, such as E. coli and B. subtilis, contain genes for only one of the two proteins (36). The ccdA gene, positioned at 164° on the B. subtilis chromosomal map, is cotranscribed with two downstream genes, yneI and yneJ (20) (originally named orf120 and orf160, respectively [36]) (Fig. 1). YneI is most probably a single-domain response regulator. The sequence of this 120-residue protein is very similar to that of CheY and Spo0F, for which the three-dimensional structures are known (22, 40). YneJ is a predicted 160-residue integral membrane protein without clear similarity to any protein sequence available in the databases. YneI and YneJ are not required for cytochrome c biogenesis, and no clear difference in phenotype compared to the wild type has been observed with yneI or yneJ insertion mutants (36).
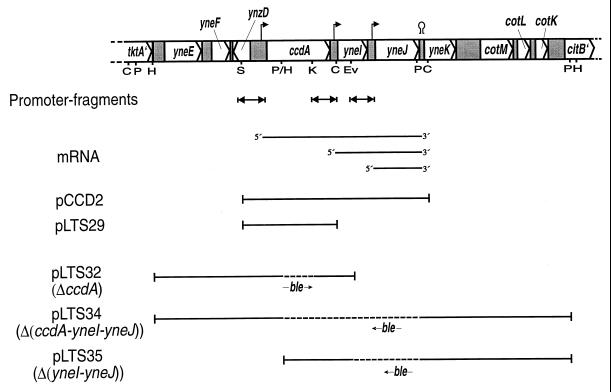
FIG. 1.
Map of the ccdA-yneI-yneJ region in the wild-type B. subtilis chromosome. Transcription initiation sites, as indicated from primer extension analysis, and a termination site are indicated by hooked arrows and a hairpin symbol, respectively. DNA fragments cloned in pDG1728 and analyzed for promoter activity (double arrows) and mRNA species detected previously (36) by Northern blot analysis of strains containing plasmid pCCD2 are also shown. DNAs contained in plasmids pCCD2 and pLTS29, used in complementation studies, and plasmids pLTS32, pLTS34, and pLTS35, used in the construction of strains with deletions, are indicated by bars. The dashed lines in the latter DNAs indicate an approximately 1-kb fragment (not drawn to scale) harboring the ble resistance gene (an arrow indicates the relative orientation of the gene). Restriction enzyme cleavage sites: C, ClaI; E, EcoRI; Ev, EcoRV; H, HindIII; K, KpnI; P, PstI; S, SacI.
Sporulation in B. subtilis, i.e., the conversion of the vegetative cell into a spore, is a process characterized by ordered gene expression and complex morphological changes (10, 41). After the formation of an asymmetrically positioned septum between the mother cell and the forespore, the transition from one developmental stage to the next is governed by four sigma factors, ςF and ςG (forespore specific) and ςE and ςK (mother cell specific). Gene expression in the forespore and the mother cell is coordinated by intercompartment communication, where the appearance of an active sigma factor in one compartment is dependent on the activity of an earlier sigma factor in the other compartment. The end result is a spore much more resistant to heat and chemicals than is the vegetative cell.
To better understand the function of CcdA in B. subtilis and possibly find a role for the YneI and YneJ proteins in the cell, in this study we have analyzed the transcriptional organization of the ccdA-yneI-yneJ gene cluster and the expression of the three genes during growth. We demonstrate that CcdA is an integral membrane protein whose cellular concentration increases at the transition from exponential growth to stationary phase. Strains with ccdA deleted were found to be deficient, but not completely blocked, in the synthesis of spores with normal properties. This defect in sporulation was investigated.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis strains and plasmids used in this work are presented in Table 1. E. coli strains JM83 [ara Δ(lac-proAB) strA thi-1 (φ80 lacZΔM15)] (50), MM294 (supE44 hsdR endA1 thi) (30), and XL1-Blue [supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac F′ proAB+ lacIq lacZΔM15 Tn10 (Tetr)] (4) were used for the propagation of plasmids.
TABLE 1.
B. subtilis strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Sourceb or reference |
|---|---|---|
| Strains | ||
| 3G18 | ade met trpC2 | G. Venema |
| 1A1 | trpC2 | BGSCc |
| 1A1-J7 | trpC2 ΔamyE::(ccdA′-lacZ spc) Spr | This work |
| 1A1-J8 | trpC2 ΔamyE::(yneI′-lacZ spc) Spr | This work |
| 1A1-J9 | trpC2 ΔamyE::(yneJ′-lacZ spc) Spr | This work |
| 1A1-DG | trpC2 ΔamyE::(lacZ spc) Spr | This work |
| AD59 | ΩspoIID-lacZ Cmr | 42 |
| AD341 | ΩcotC-lacZ Cmr | 53 |
| AD454 | spoIIIG::neo Nmr | A. Driks |
| JO1 | ade met trpC2 ΔctaCD::ble Pmr | 46 |
| L16205 | trpC2 ΔcccB::tet Tcr | 2 |
| LU60A1 | trpC2 ΔccdA::ble Pmr | LU6018→1A1 |
| LU6018 | ade met trpC2 ΔccdA::ble Pmr | 36 |
| LU62A1 | trpC2 Δ(ccdA-yneI-yneJ)::ble Pmr | pLTS34→1A1 |
| LU62A1R#3 | trpC2 Δ(ccdA-yneI-yneJ)::ble (ccdA suppressor mutation-containing strain derived from LU62A1) Pmr | This work |
| LU63A1 | trpC2 Δ(yneI-yneJ)::ble Pmr | pLTS35→1A1 |
| LU60A1-J7 | trpC2 ΔccdA::ble ΔamyE::(ccdA′-lacZ spc) Spr | This work |
| LU62A1-J7 | trpC2 Δ(ccdA-yneI-yneJ)::ble ΔamyE::(ccdA′-lacZ spc) Spr | This work |
| LU63A1-J7 | trpC2 Δ(yneI-yneJ)::ble ΔamyE::(ccdA′-lacZ spc) Spr | This work |
| LU621 | trpC2 ΩspoIID-lacZ (Cmr) Δ(ccdA-yneI-yneJ)::ble Pmr | AD59→LU62A1 |
| LU623 | trpC2 ΩcotC-lacZ Δ(ccdA-yneI-yneJ)::ble Cmr Pmr | AD341→LU62A1 |
| LU624 | trpC2 spoIIIG::neo Δ(ccdA-yneI-yneJ)::ble Nmr Pmr | AD454→LU62A1 |
| LU625 | trpC2 ΩsspB-lacZ Δ(ccdA-yneI-yneJ)::ble Cmr Pmr | PM73→LU62A1 |
| LU626 | trpC2 ςFspoIIIG::neo Δ(ccdA-yneI-yneJ)::ble Emr Nmr Pmr | PM73→LU624 |
| LU627 | trpC2 ςFspoIIIG::neo sspB-lacZ Δ(ccdA-yneI-yneJ)::ble Emr Nmr Cmr Pmr | PM73→LU626 |
| LUA11 | trpC2 ΩspoIID-lacZ Cmr | AD59→1A1 |
| LUA13 | trpC2 ΩcotC-lacZ Cmr | AD341→1A1 |
| LUA14 | trpC2 spoIIIG::neo Nmr | AD454→1A1 |
| LUA15 | trpC2 ΩsspB-lacZ Cmr | PM73→1A1 |
| LUA16 | trpC2 “ςF” spoIIIG::neo, Emr Nmr | PM73→LUA14 |
| LUA17 | trpC2 “ςF” spoIIIG::neo sspB-lacZ Emr Nmr Cmr | PM73→LUA16 |
| LUT36 | trpC2 ΔcccA::cat ΔcccB::tet ΔctaCD::ble ΔqcrC::neo Cmr Tcr Pmr Nmr | This work |
| PM73 | spoIIACΩspoIIAC (“ςF”) spoIIIGΔ1 sspB-lacZ Emr Cmr | 23 |
| SL6820 | trpC2 lys-3 metB10 ΔqcrC::neo Nmr | 52 |
| Plasmids | ||
| pBLE-1 | Apr Pmr | 12 |
| pBluescript II KS(−), SK(−) | Apr | Stratagene |
| pCCD2 | Cmr Emr; ccdA-yneI-YneJ on 2.1-kb fragment in pHP13 | 36 (Fig. 1) |
| pDGV1 | Cmr | 3 |
| pDG1728 | Apr Spr; B. subtilis amyE deletion/integration vector with promoterless and modified E. coli lacZ | 14 |
| pHP13 | Cmr Emr; B. subtilis/E. coli shuttle vector | 15 |
| pHV32 | Apr Cmr Tcr | 26 |
| pLJJ7 | Apr Spr; 316-bp ccdA promoter fragment in pDG1728 | This work |
| pLJJ8 | Apr Spr; 228-bp yneI promoter fragment in pDG1728 | This work |
| pLJJ9 | Apr Spr; 227-bp yneJ promoter fragment in pDG1728 | This work |
| pLTS1 | Cmr Emr; ccdA on 2.7-kb fragment in pHP13 | 36 |
| pLTS17 | Apr; ccdA on a 2.7-kb fragment in pBluescript SK | 36 |
| pLTS29 | Cmr Emr; ccdA on 1.1-kb fragment in pHP13 | This work (Fig. 1) |
| pLTS32 | Apr Pmr; ccdA on 1.1-kb fragment in pHP13 | 36 (Fig. 1) |
| pLTS33 | Apr Pmr | This work |
| pLTS34 | Apr Pmr | This work (Fig. 1) |
| pLTS35 | Apr Pmr | This work (Fig. 1) |
| pLTS100 | Cmr; ccdA in pDGV1 | This work |
| pMR22 | Apr Tcr | 32 |
| pUC18 | Apr | 50 |
| pΔcccA1 | Apr | 48 |
Apr, Cmr, Emr, Nmr, Pmr, Spr, and Tcr indicate resistance to ampicillin, chloramphenicol, erythromycin, neomycin, phleomycin, spectinomycin, and tetracycline, respectively. “ςF” indicates a mutant variant of the spoIIAC gene encoding a ςF with a Val-233–to–Ala substitution.
Arrows indicate transformation of the indicated strain with chromosomal or plasmid DNA.
Bacillus Genetic Stock Center.
Media and general growth of bacteria.
E. coli strains were grown on Luria agar plates or in Luria broth medium (34). B. subtilis strains were grown on tryptose blood agar base (TBAB) plates (Difco), Difco sporulation (DS) medium (16) [0.8% (wt/vol) Bacto nutrient broth (Difco), 0.1% (wt/vol) KCl, 0.012% (wt/vol) MgSO4 · 7H2O, 0.5 mM NaOH, 1 mM Ca(NO3)2, 10 μM MnCl2, 1 μM FeSO4) plates, or Spizizen minimal medium (38) plates supplemented with required growth factors (10 mg/liter) and with 0.5% (wt/vol) glucose, succinate, or lactate as the carbon source. Nutrient sporulation medium with phosphate (NSMP) (11) or DS medium was used for liquid cultures, which were grown at 37°C in Erlenmeyer glass flasks (culture volume, ≤1/10 the volume of the flask) with indentations, on a rotary shaker at 200 rpm. For detection of β-galactosidase activity on TBAB plates, 80 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-Gal) per liter was included in the medium. The following antibiotics were used when appropriate: ampicillin, 75 mg/liter; chloramphenicol, 12.5 mg/liter (E. coli) or 3 to 5 mg/liter (B. subtilis); erythromycin, 5 mg/liter; neomycin, 5 mg/liter; phleomycin, 1.2 mg/liter; spectinomycin, 150 mg/liter; and tetracycline, 25 mg/liter (B. subtilis).
Construction of plasmids and deletion mutants.
Plasmid pLTS29, containing the ccdA gene, was constructed by first moving the 1.35-kbp SacI-EcoRI fragment from pLTS17 to pUC18. The 1.12-kbp SalI-ClaI fragment from the resulting plasmid, pLTS28, was isolated, and the ends of the fragment were made blunt by treatment with Klenow enzyme in the presence of deoxynucleoside triphosphates. Finally, the fragment was ligated to pHP13 cleaved with SmaI.
pLTS100, a high-copy-number plasmid carrying the ccdA gene, was constructed by moving the 1.2-kb EcoRI-BamHI fragment of pLTS29 to pDGV1.
Plasmid pLTS33 was constructed by cloning the 1.7-kbp PstI fragment of pMR22, which contains the region downstream of the ccdA-yneI-yneJ gene cluster and includes only the last 26 nucleotides of yneJ, into pBLE-1. A plasmid with the PstI fragment in an orientation such that the yneJ end is close to the ble gene (encoding phleomycin resistance) was selected. Plasmids pLTS34 and pLTS35, used to delete the ccdA-yneI-yneJ and yneI-yneJ genes, respectively, were then constructed as follows. (i) The 1.52-kbp HindIII fragment of pLTS1 was moved to pBluescript SK(−) in an orientation such that the ccdA′ part of the fragment is close to the KpnI site in the cloning cassette. The 1.56-kbp EcoRI-KpnI fragment from was then moved to pLTS33, resulting in plasmid pLTS34. (ii) The 844-bp HindIII-EcoRV fragment of pLTS17 was isolated and treated with Klenow enzyme in the presence of deoxynucleoside triphosphates. Subsequently, it was ligated with pBluescript SK(−) cleaved with SmaI (the insert was oriented such that the yneI′ part of the fragment is close to the SacI site in the cloning cassette). The 902-bp EcoRI-SacI fragment from pLTS26 was then moved to pLTS33, resulting in plasmid pLTS35.
Plasmids pLTS34 and pLTS35 were linearized by cleavage with ScaI (which has a unique site in the bla gene) and used to transform B. subtilis 1A1 to phleomycin resistance. The resulting deletions and replacement with the ble gene were confirmed by Southern blot analysis. Note that the ble gene in previously reported deletion mutants obtained using pLTS32 (36) is in the opposite orientation to that in strains obtained from transformation with pLTS34 or pLTS35 (Fig. 1).
Strain LUT36 was constructed by subsequent transformations using linearized pΔcccA1 (to delete the cccA gene) (48) and chromosomal DNA from SL6820 (ΔqcrC::neo), JO1 (ΔctaCD::ble), and L16205 (ΔcccB::tet) and selecting for the respective antibiotic resistance.
Construction of ccdA-lacZ, yneI-lacZ, and yneJ-lacZ transcriptional fusions.
The regions upstream of ccdA (bp 1367 to 1682), yneI (bp 2257 to 2484), and yneJ (bp 2694 to 2920) were amplified by PCR using pCCD2 as template (Fig. 1). The ccdA region was amplified using Taq DNA polymerase (Roche Molecular Biochemicals) and the primer pair 5′ CGGAATTCCTGACTGAGCTCTATCG plus 5′ CGGGATCCATGATTTGACATTCCTTCAAG. The yneI and yneJ regions were amplified using Pwo DNA polymerase (Roche Molecular Biochemicals), and the primer pairs 5′-CGGAATTCTGAAGTGGATAAGGAAGAAC plus 5′-CGGGATCCAACAATCGATTTCCACAG and 5′-CGGAATTCTTAACCTTTGATCCTAAAGC plus 5′-CGGGATCCGGAGTGTTGATACTATATAC, respectively. EcoRI and BamHI restriction sites were added via the primers (underlined). The amplified fragments were cut with EcoRI and BamHI and ligated into pBluescript SK(−) or KS(−). The complete sequence of each cloned DNA fragment was determined to exclude errors introduced by the PCR. The EcoRI and BamHI fragments were then moved to plasmid pDG1728, resulting in plasmids pLJJ7, pLJJ8, and pLJJ9 (Table 1). The lacZ gene of pDG1728 contains the Shine-Dalgarno sequence of the B. subtilis spoVG gene to provide efficient translation of the lacZ gene in B. subtilis. The obtained plasmids and pDG1728, as a negative control, were linearized by digestion with ScaI and integrated, by means of transformation and a double-crossover recombination event, into the amyE locus of B. subtilis strains 1A1, LU60A1, LU62A1, and LU63A1. The desired spectinomycin-resistant transformants obtained were confirmed by erythromycin sensitivity and the lack of α-amylase activity and, in appropriate cases, also for defective cytochrome c synthesis by N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) oxidation staining.
Antisera and immunoblot analysis.
Polyclonal antisera against CcdA and YneJ were obtained by immunizing New Zealand White rabbits with synthetic peptides conjugated to keyhole limpet hemocyanin. The peptides used, YITGVSMDDVKTEK and YRKLHNELQSSNIQMN, correspond to residues 32 to 45 of CcdA and 145 to 160 of YneJ, respectively. Custom peptide synthesis and production of antisera were carried out by Neosystems.
For immunoblot analysis, cell extracts were incubated for 30 min at 40°C in the presence of 0.4% (wt/vol) sodium dodecyl sulfate (SDS), 0.2% (wt/vol) 2-mercaptoethanol, 1.2% (vol/vol) glycerol, and 5 mM Tris buffer (pH 6.8), and the proteins were then fractionated by SDS-polyacrylamide gel electrophoresis on 14% polyacrylamide gels by using the Schägger and von Jagow system (37). The proteins were transferred to Immobilon P membranes (Millipore) using 48 mM Tris–39 mM glycine buffer containing 20% (vol/vol) methanol and a semidry electroblot apparatus. Immunodetection was carried out by chemiluminescence using the ECL system (Pharmacia Amersham). Primary antisera were used at a 500-fold dilution and secondary antibodies, donkey anti-rabbit immunoglobulin G conjugated to horseradish peroxidase, were used at a 3,000-fold dilution.
β-Galactosidase activity measurements.
To determine β-galactosidase activity in cells, growth medium was inoculated with cells from an exponentially growing culture. Samples were withdrawn at intervals during the growth of the batch culture, immediately frozen in liquid nitrogen, and then stored at −20°C. The samples were thawed in cold water, and the cells were collected by centrifugation for 1 min at 20,000 × g in an Eppendorf centrifuge. The cell pellets were suspended in AB buffer (60 mM K2HPO4, 40 mM KH2PO4, 100 mM NaCl). A 175-μl volume of a suspension of an appropriate cell concentration was mixed with 35 μl of 4-methylumbelliferyl-β-d-galactoside (MUG), 0.4 g/liter in dimethyl sulfoxide, and incubated at 23°C on a water bath. At intervals, 50-μl aliquots were withdrawn and diluted in 2.45 ml of AB buffer, and the fluorescence was immediately determined using a Shimadzu RF-5301PC spectrofluorometer with an excitation wavelength of 366 nm (3-nm slit) and an emission wavelength of 445 nm (10-nm slit). The fluorescence of solutions of 4-metylumbelliferone of known concentrations in AB buffer were used to determine the amount of MUG hydrolyzed in the samples. Rates of MUG hydrolysis were obtained from linear plots of product concentration versus time. Specific β-galactosidase activities were calculated for the cell concentration estimated from the optical density at 600 nm (OD600) of the original sample taken from the culture.
Spore assays.
Cultures were grown in 25 ml of NSMP (pH 6.5) at 30°C in a 250-ml Erlenmeyer flask for 2 days. The spore titer was determined by subjecting the cultures to heat, chloroform, or lysozyme treatment. Routinely, 5-ml samples were heated at 80°C for 10 min (unless stated otherwise) or vigorously mixed with 0.6 ml of chloroform for 10 s, or a small volume was diluted 100-fold in minimal salts solution [80.4 mM K2HPO4, 44.1 mM KH2PO4, 0.8 mM MgSO4, 15.1 mM (NH4)2SO4, 3 mM sodium citrate] and incubated with or without lysozyme (0.5 g/liter) at 30°C for 30 min. Serial dilutions were plated on TBAB plates and incubated overnight at 37°C. Dilutions of nontreated samples were plated in parallel.
Electron microscopy.
Cells were grown in NSMP medium (pH 6.5) at 30°C in a 250-ml Erlenmeyer flask for 3 days. Samples (10 ml) were harvested and washed in fresh medium once. The cell pellet was treated with 2.5% glutaraldehyde, fixed in OsO4, and dehydrated in ethyl alcohol, and the sample was finally impregnated in plastic and sections were analyzed by electron microscopy.
Other methods.
B. subtilis chromosomal DNA was isolated by the method of Marmur (24). Plasmid DNA was isolated by using a kit (Maxi Prep [Qiagen] or Jet Prep Plasmid Miniprep [Genomed]). Total RNA from B. subtilis was purified using the RNeasy kit (Qiagen). Primer extension analysis was done as described previously (36) using oligonucleotides complementary to the proximal part of yneI (bp 2609 to 2593, 5′-CAGCTGCTTGTTCACCG) and yneJ (bp 3028 to 3012, 5′-AACAGGCTTGTCAGAGG) (base pair numbering refers to the published sequence [36], accession no. X87845). E. coli was transformed by electroporation and B. subtilis was grown to competence essentially as previously described (18). Oxidation of the artificial electron donor TMPD by colonies on TBAB plates was assayed by the soft-agar overlay method (13). α-Amylase production was assayed by growth on TBAB plates containing 1.5% (wt/vol) starch and then staining colonies with KI-I2 solution. Membranes from B. subtilis strains were isolated from osmotically lysed cells essentially as described by Hederstedt (17) and stored at −80°C. Light absorption spectroscopy was carried out as described previously (36). The protein concentration of membrane fractions was determined using the bicinchoninic acid protein assay (Pierce Chemical Co.) with bovine serum albumin as the standard.
RESULTS
Promoters for the yneI and yneJ genes.
By using Northern blot analysis, ccdA-yneI-yneJ, yneI-yneJ, and yneJ mRNAs have previously (36) been detected (Fig. 1). The ccdA gene is preceded by an apparent ςA-type promoter, and the initiation site for transcription is positioned about 100 bp from the translational start codon (36). Plasmid pCCD2 is a low-copy-number plasmid containing the ccdA-yneI-yneJ gene cluster and flanking DNA (Fig. 1 and Table 1). Primer extension analysis, with total RNA extracted from B. subtilis strain 3G18/pCCD2 grown in NSMP, identified the 5′ end of the yneI-yneJ and yneJ mRNAs (data not shown and Fig. 2). The indicated transcription initiation sites are located in intergenic regions and are both positioned 33 bp from the translation start codon (ATG) for the respective gene. An apparent −10 sequence of a ςA-type promoter, but no corresponding −35 sequence, is present in the promoter regions of yneI and yneJ (Fig. 2).
FIG. 2.
Nucleotide sequence of promoter regions with transcription initiation sites (+1), as determined by primer extension analysis. Apparent ςA-type -35 and -10 promoter sequences, translation start codons, and stop codons are indicated in bold type. There are two possible translation start codons for ccdA, as discussed previously (36). Putative ribosome binding sequences are underlined. The numbering of nucleotides is that used in reference 36.
Promoter activities during growth.
DNA fragments predicted to contain the three promoter regions were amplified by PCR and cloned in front of the promoterless lacZ gene in plasmid pDG1728. The ccdA, yneI, and yneJ fragments contain 224, 202 and 215 bp, respectively, of sequence upstream of +1 and the leader sequence up to (but not including) the ribosome binding sequence (Fig. 1 and 2). Each lacZ transcriptional fusion was integrated in single copy into the amyE locus on the chromosome of B. subtilis 1A1 (parental strain), LU60A1 (ΔccdA), LU62A1 [Δ(ccdA-yneI-yneJ)], and LU63A1 [Δ(yneI-yneJ)] (Table 1; see Materials and Methods for details).
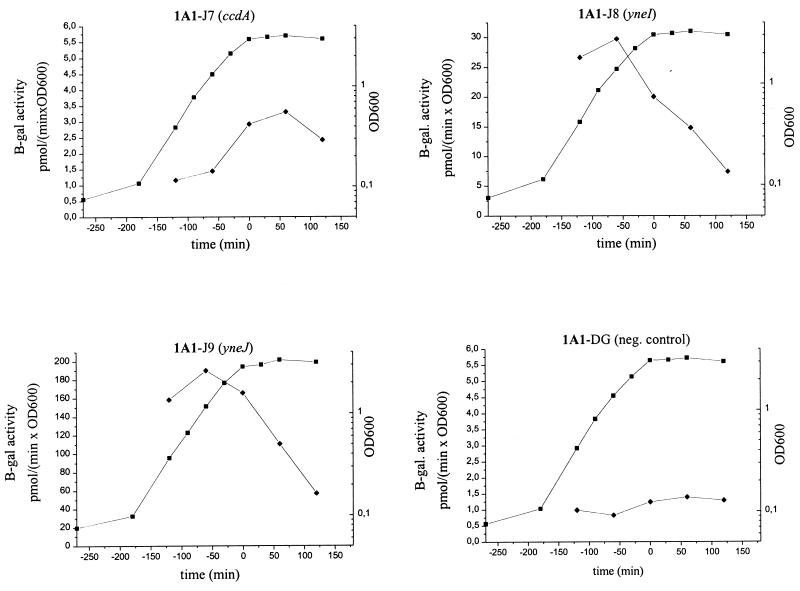
To analyze the relative strength and temporal activity of each promoter during growth, strain 1A1 containing the three different lacZ fusions, i.e., strains 1A1-J7, 1A1-J8, and 1A1-J9, and the negative control, 1A1-DG, were grown in NSMP and the cells were assayed for β-galactosidase enzyme activity (Fig. 3). The results indicate that the ccdA promoter is weak (the maximal β-galactosidase activity was 3 pmol/min × OD600) and active during the transition from exponential growth to stationary phase. In contrast, the yneI and yneJ promoters are active during exponential growth phase and gradually decrease in activity as the culture progresses into stationary phase. The maximal β-galactosidase activity obtained with the yneJ-lacZ gene fusion (190 pmol/min × OD600) was about 9- and 60-fold higher than those obtained with the yneI-lacZ and ccdA-lacZ fusions, respectively.
FIG. 3.
β-Galactosidase activity of B. subtilis strains grown in NSMP. ■, OD600; ⧫, specific enzyme activity. Note the different scales for enzyme activity in the panels. The strains contain the ccdA, yneI, or yneJ promoters fused to a promoterless lacZ gene and integrated into the chromosome in one copy. See Table 1 for a full description of strains.
The β-galactosidase activity profiles of strains with the entire ccdA-yneI-yneJ cluster deleted and containing the respective transcriptional fusion at the amyE locus were essentially the same as those obtained with the different lacZ fusions in the parental genetic background (data not shown). The β-galactosidase activity during growth was also analyzed for strains 1A1-J7, LU60A1-J7, LU62A1-J7, and LU63A1-J7, each harboring one copy of the ccdA promoter-lacZ fusion in the chromosome and containing pLTS1 (ccdA), pCCD2 (ccdA-yneI-yneJ), or pHP13 (plasmid vector). No significant differences in β-galactosidase activity or temporal pattern were observed with these 12 strains compared to strain 1A1-7J (Fig. 3 and data not shown). The combined data strongly suggest that gene products of the ccdA-yneI-yneJ cluster do not regulate the transcription of ccdA. This is based on the assumption that the 316-bp fragment used for promoter activity analysis comprises the entire promoter region with potential regulatory elements.
CcdA and YneJ protein profiles.
The subcellular localization and the steady-state levels of CcdA and YneJ in B. subtilis were analyzed using immunoblot with antisera against oligopeptides corresponding to amino acid residues 32 to 45 of CcdA (assuming ATG to be the start codon [Fig. 2]) and the C-terminal 16 residues of YneJ, respectively. Extracts were prepared from cells grown in NSMP and harvested 1 h before the end of the exponential growth phase (T1), at the end of the exponential growth phase (T0), and 1 h into the stationary phase (T+1), respectively. Typical growth curves are shown in Fig. 3.
A CcdA antigen of 19 kDa and a YneJ antigen of 15 kDa were found in isolated membranes from strain LU62A1/pCCD2 (Fig. 4). As expected, these antigens were not present in membranes of strain LU62A1/pHP13, which lacks the ccdA and yneJ genes (immunoblot not shown). The molecular sizes of the polypeptide antigens as deduced from the SDS-polyacrylamide gels were smaller than those calculated from the DNA sequence, 25 or 26 kDa for CcdA (depending on the translational initiation codon used [Fig. 2]) and 18.3 kDa for YneJ. Such deviations in apparent size are often seen for integral membrane proteins. The CcdA protein could be extracted from the membrane with nonionic detergent, consistent with it being an integral membrane protein.
FIG. 4.
Immunoblot analysis of membrane proteins from different B. subtilis strains, using CcdA antiserum (upper panel) and YneJ antiserum (lower panel). The blots are overexposed to demonstrate the presence of small amounts of CcdA and YneJ antigen. Strain LU62A1 has the ccdA-yneI-yneJ gene cluster deleted, LU60A1 has ccdA deleted, and 1A1 is the parental strain. Plasmids pLTS29 and pCCD2 carry the ccdA gene and the ccdA-yneI-yneJ gene cluster, respectively. The designations −1, 0, and +1 indicate that membranes were isolated from cells harvested 1 h before, at the time, and 1 h after exponential growth ended, respectively. The bands in the top panel seen just above CcdA and in the lower part of the gel are due to cross-reactivity with unrelated antigens and were apparent only on overexposed films.
Even after overexposure of the immunoblot, CcdA antigen was not detected in membranes from strain 1A1, which contains a single copy of the ccdA gene in the chromosome. The protein was found only in strains containing ccdA on a plasmid, e.g., pCCD2 and pLTS29 (Fig. 4, upper panel). YneJ antigen was found in membranes of both strain 1A1 and the ccdA deletion mutant LU60A1 (lower panel). The amount of CcdA polypeptide was small in membranes from exponentially growing cells compared to that in membranes from cells in a later growth stage. YneJ showed a different pattern, with larger amounts present in membranes from exponentially growing cells compared to stationary-phase cells. Thus, the observed variations in cellular amounts of CcdA and YneJ protein are consistent with those in mRNA levels as determined by Northern blot analysis (36) and in activity patterns observed with the ccdA, yneI, and yneJ promoter regions fused to lacZ (Fig. 3).
Phenotype of strains deficient in ccdA, yneI, and yneJ.
B. subtilis strains derived from 1A1 and containing deletions in the ccdA, yneI, and yneJ gene cluster (Fig. 1) were tested for growth on TBAB and minimal plates with glucose, succinate, or lactate as the carbon source. No major differences in growth were seen between the various mutants and the parental strain after incubation at 37°C overnight. Incubation of colonies on TBAB or minimal-succinate plates at room temperature for 2 days, however, resulted in lysis of LU60A1 (ΔccdA) and LU62AI [Δ(ccdA-yneI-yneJ)]. Colonies of strains 1A1 and LU63A1 [Δ(yneI-yneJ)] did not lyse on the plates even after >1 week of incubation at room temperature.
Growth of 1A1 and the derivatives with various deletions in the ccdA-yneI-yneJ region was also tested in NSMP and DS liquid medium. No significant differences in exponential growth rate were observed between the strains, and they reached the same OD at the end of exponential growth phase. A few hours into the stationary phase, the optical density of cultures of strains LU60A1 and LU62A1, both with the ccdA gene deleted, progressively decreased, and this was correlated with a decrease in the number of CFU (data not shown and Table 2). These growth properties of the mutant strains indicated that CcdA deficiency affects sporulation.
TABLE 2.
Sporulation efficiency of B. subtilis strains carrying deletions in the ccdA-yneI-yneJ region
| Strain | Relevant genotype | Total-cell titera (CFU/ml) | Spore titerb (CFU/ml) | Sporulation efficiencyc (%) |
|---|---|---|---|---|
| 1A1 | Wild type | 8.7 × 108 | 7.8 × 108 | 90 |
| LU60A1 | ΔccdA | 1.4 × 108 | 5.2 × 106 | 3.7 |
| LU62A1 | Δ(ccdA-yneI-yneJ) | 2.0 × 108 | 2.7 × 106 | 1.4 |
| LU63A1 | Δ(yneI-yneJ) | 7.6 × 108 | 6.6 × 108 | 87 |
Titer is CFU per milliliter of culture after growth for 2 days at 30°C in NSMP.
Titer after incubation of the sample at 75°C for 10 min.
Sporulation efficiency is calculated as spore titer divided by total cell titer, multiplied by 100.
CcdA-deficient mutants show reduced sporulation efficiency.
The ability of ccdA mutant strains to form spores was analyzed by growing cultures in NSMP for 2 days at 30°C and then determining the fraction of heat- and chloroform-resistant cells in the cultures (Tables 2 and 3). For the parental strain, 1A1, and the yneI-yneJ deletion mutant, LU63A1, the sporulation efficiency was >85%. Strains LU60A1 and LU62A1, with ccdA and ccdA-yneI-yneJ, respectively, deleted, showed a sporulation efficiency of 1 to 6%. This number is much greater than that obtained with a strain completely blocked in an early step in sporulation such as the SpoIIIG-deficient strain LUA14 (Table 3). The spore yields per volume of culture were very low for the CcdA-deficient strains compared to the parental strain because of the cell lysis of the former strains that occurs in the stationary phase. Incubation of spore-containing cultures at 80°C for 10, 15, and 30 min did not reveal any difference in heat sensitivity between spores of strain 1A1 and LU62A1 (data not shown). The results indicate that CcdA-deficient mutants can synthesize spores with normal properties at only a low efficiency.
TABLE 3.
Heat, chloroform, and lysozyme resistance of cells grown for sporulation in NSMP
| Strain | Relevant genotype | Treatmenta | Cell titer before treatment (CFU/ml) | Survivalb (%) |
|---|---|---|---|---|
| 1A1 | Wild type | Heat | 4.1 × 108 | 92 |
| Chloroform | 4.1 × 108 | 89 | ||
| Lysozyme | 4.1 × 108 | 85 | ||
| LU62A1 | Δ(ccdA-yneI-yneJ) | Heat | 0.62 × 108 | 2.6 |
| Chloroform | 0.62 × 108 | 5.6 | ||
| Lysozyme | 0.62 × 108 | 18 | ||
| LUA14 | spoIIIG | Heat | 2.4 × 108 | <0.005 |
| Chloroform | 2.4 × 108 | <0.005 | ||
| Lysozyme | 2.4 × 108 | 2.8 |
See Materials and Methods for details.
Survival is CFU per milliliter after treatment divided by that before treatment, multiplied by 100.
Clones from germinated spores of strains LU60A1 and LU62A1, i.e., cells from colonies obtained on TBAB plates after heat treatment of spore-containing cultures, showed the same reduced sporulation efficiency as LU60A1 and LU62A1. This demonstrated that the heat-resistant spores obtained from cultures of CcdA-defective strains are not due to revertants in the cultures.
B. subtilis wild-type spores are also resistant to lysozyme treatment. Strains with the ccdA gene deleted showed an intermediate sensitivity to lysozyme compared to the parental strain and strain LUA14 blocked at stage III in sporulation (Table 3).
Phase-contrast light microscopy of cultures of LU60A1 and LU62A1, grown for sporulation, showed only a few light-diffracting cells in contrast to corresponding cultures of 1A1, which contained such cells predominantly. Cells in cultures of the CcdA-deficient strains were mostly nonmotile. Thus, it seems as if many cells in a culture of a CcdA-deficient strain can progress in spore synthesis up to, but not beyond, a relatively lysozyme-resistant state where dehydration occurs in the wild-type case, yielding light-diffracting spore structures. A small percentage of the sporulating CcdA-deficient cells apparently are able to complete the differentiation process and mature into strongly light-diffracting heat-resistant spores.
Sporulation-specific sigma factor activity in a CcdA-deficient mutant.
To determine if CcdA is important for the sporulation-specific sigma factor cascade, we analyzed the expression of different promoter-lacZ fusions integrated as a single copy into the amyE locus of B. subtilis strains with and without an intact ccdA gene. The spoIID, sspB, and cotC promoters in the transcriptional fusions we used are dependent on ςE, ςG, and ςK, respectively, for activity. ςF-dependent gene expression was analyzed by the use of strains LUA17 and LU627, which do not synthesize ςG and produce a variant of ςF that acts like ςG, mediating transcription of the sspB promoter-lacZ fusion.
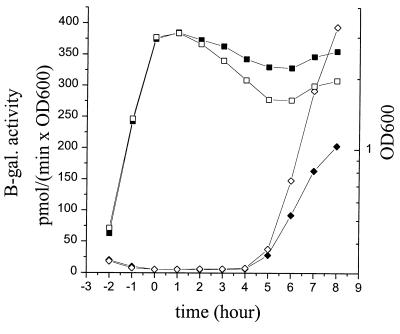
All sporulation-specific sigma factors were found to be active in both the parental genetic background (strains LUA11, LUA13, LUA15, and LUA17) and the strains (LU621, LU623, LU625, and LU627) lacking the ccdA gene, as assayed by the blue color of colonies on TBAB plates supplemented with X-Gal. In liquid NSMP cultures, the expression of the ςE-, ςF-, and ςG-dependent lacZ gene fusions, as determined by β-galactosidase activity, was about the same and appeared at the same time point in the growth curve in the CcdA-defective strains as in the control strains (data not shown). The ςK-dependent gene fusion was expressed to a somewhat higher level in the CcdA-deficient strain LU623 than in strain LUA13 (Fig. 5). It is uncertain whether this observed reproducible minor difference is relevant for a better understanding of the function of CcdA in the cell. Since the expression of the cotC-lacZ fusion is dependent on both ςK and GerE activity (the latest-acting sporulation-related transcription factor) GerE must also be active in a CcdA-defective mutant (41).
FIG. 5.
ςK-dependent expression of a cotC-lacZ gene fusion in the chromosome of the wild-type control strain LUA13 (solid diamonds) and CcdA-deficient strain LU623 (open diamonds) during growth in DS medium. The square symbols show OD600 (solid, LUA13; open, LU623). Time zero is the end of the exponential growth phase.
Morphology of mutant spores.
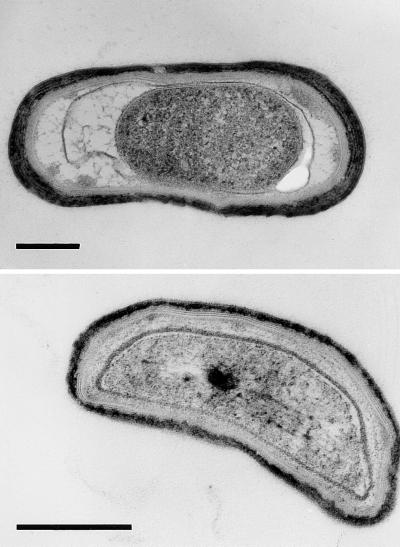
Electron microscopy of samples from sporulating cultures of strain 1A1 and LU62A1 showed more cell lysis in the latter strain. Spore-like structures, often enclosed by the mother cell, were seen in samples of both strains (Fig. 6 and electron micrographs not shown). Those of LU62A1, corresponding to the nonmotile cells seen by light microscopy, showed the main features of B. subtilis spores, i.e., a cortex, a lamellar inner coat, and an electron-dense outer coat. We could not identify a consistent structural difference in the morphology of the spores of 1A1 and LU62A1, except that the cytoplasm of the latter strain appeared more granular. From the sporulation efficiencies of the strains (Tables 2 and 3), less than 10% of the LU62A1 spores observed by electron microscopy would be heat-resistant cells. Judging from the relative lysozyme resistance, the poor light diffraction properties, and the heat and chloroform sensitivity of most spores of CcdA-deficient mutants, we consider it likely that synthesis of the cortex and/or coat and dehydration of the spore are defective.
FIG. 6.
Electron micrographs of two representative B. subtilis LU62A1 cells from a culture grown for sporulation. Bars, 0.4 μm.
Complementation of sporulation defects by the ccdA gene on plasmids.
The deficiency in spore synthesis of strains LU60A1 and LU62A1 was complemented by the ccdA gene or the ccdA-yneI-yneJ gene cluster on plasmids, i.e., by pLTS29 and pCCD2 (data not shown). These plasmids also restored cytochrome c synthesis, as determined by the ability of colonies on TBAB plates to oxidize TMPD. Plasmid pLTS100, a high-copy-number plasmid carrying the ccdA gene, complemented the defect in cytochrome c synthesis in strain LU60A1 and caused a ca. fivefold overproduction of CcdA protein compared to that in LU62A1/pCCD2 (immunoblot not shown). In contrast to strains containing the low-copy-number plasmids pLTS29 or pCCD2 or the vector pGDV1, those containing pLTS100 gave rise to small colonies and were unstable. We do not know if this apparent toxicity of pLTS100 is due to functional activity of CcdA or physical disturbance of the membrane caused by the increased amounts of this integral membrane protein.
A B. subtilis strain with a nonsense mutation in the ccdA gene has recently been isolated and characterized (21). The phenotype of this strain is identical in all aspects to that of LU60A1, with ccdA deleted. This, together with the result of the complementation experiments and the properties of a strain deficient in translation of ccdA (36), demonstrates that the CcdA protein is required for both efficient sporulation and cytochrome c synthesis in B. subtilis.
Isolation of strains with suppressor mutations.
Microcolonies (papilla) appeared within lysed colonies of strain LU62A1 on TBAB plates incubated at room temperature for >1 week. Cells from these microcolonies formed normal-sized colonies when streaked on TBAB plates and oxidized TMPD, albeit more poorly than did 1A1 colonies. These clones were also phleomycin resistant and contained the same ccdA-yneI-yneJ::ble deletion-substitution as in strain LU62A1, as determined by Southern blot analysis of chromosomal DNA. Thus, the clones contain suppressor mutations. The suppressor-containing strains showed wild-type sporulation efficiency (80 to 90%), and light absorption spectroscopy of membranes isolated from one clone, LU62A1R#3, confirmed the presence of c-type cytochromes (spectra not shown). Strain LU62A1R#3 and other suppressor-containing strains differ from LU62A1 in that they do not develop competence. The suppressor mutations can be moved to other strains by transformation and are not linked to the ccdA locus on the chromosome. The results show that the suppressor mutation(s) at one or several loci can restore all known cell defects caused by CcdA deficiency. Despite several attempts, using different strategies and isolates, we have so far not been able to identify any of the suppressor mutations.
c-type cytochromes are not required for sporulation.
The four c-type cytochromes in B. subtilis, cytochrome c550, cytochrome c551, the cytochrome c subunit of the bc complex, and subunit II of the cytochrome caa3 oxidase, are encoded by the cccA, cccB, qcrC, and ctaC genes, respectively (2, 49, 52). Respiration-defective B. subtilis mutants are generally sporulation deficient (43). Defective sporulation of CcdA-deficient strains could therefore be a result of the total lack of cytochrome c. To determine this, we constructed strain LUT36, which has the structural genes for all four cytochromes c deleted. LUT36 was found to have close to normal sporulation efficiency (>60%), demonstrating that cytochrome c is not important for sporulation to occur in B. subtilis.
DISCUSSION
In this work we demonstrate that sporulation in B. subtilis can occur in the absence of the four known c-type cytochromes and that the ccdA gene is required for efficient sporulation. Our available experimental data, taken together, strongly suggest that CcdA deficiency affects a late step in spore synthesis, probably synthesis of the cortex and/or coat and dehydration of the spore.
The ccdA gene is cotranscribed with the yneI and yneJ genes from a promoter upstream of ccdA (36), and in this work we demonstrate that there are also promoters for the synthesis of yneI-yneJ and yneJ mRNAs. These last two promoters show a different temporal pattern of activity during growth of batch cultures compared to the promoter for ccdA. The expression from the ccdA promoter is not regulated by CcdA, YneI, or YneJ, since the absence or overproduction of these three proteins did not affect ccdA promoter activity. Genes for proteins that physically interact are in bacteria often clustered and present in a conserved order on the chromosome (7). B. subtilis genes for functionally closely related proteins, for example enzymes of a biosynthetic pathway, are generally arranged in operons (20). There is, however, no evidence suggesting that the function of the YneI or YneJ protein in the cell is related to that of CcdA. Strains lacking or overexpressing yneI and yneJ are proficient in cytochrome c and spore synthesis. Moreover, the ccdA gene in other gram-positive bacteria of known genome sequence (5) is not flanked by genes corresponding to yneI or yneJ. The B. subtilis cheY and spo0F genes encode single-domain response regulators very similar to YneI, but neither of these genes are flanked by a gene for a membrane protein like YneJ. Although transcription of ccdA, yneI, and yneJ rely on a common terminator located after the yneJ gene, we conclude that the three genes probably encode functionally unrelated proteins. The transcriptional organization of the gene cluster does not cause a problem in the cell, because the promoters in front of the genes progressively increase in strength in steps of approximately 1 order of magnitude.
Cytochromes of the c type are present in exponentially growing B. subtilis cells and increase in concentration together with other cytochromes at the end of exponential growth (45). Transcription of structural genes for c-type cytochromes and ccdA also increases at this growth phase or at the entry into stationary phase in NSMP medium (2, 51). Exponentially growing cells are expected to contain some CcdA protein, since cytochrome c is present. It might be of functional importance to maximize the expression of ccdA when the cell approaches stationary phase, since CcdA is needed for efficient spore synthesis.
The hydrophobicity profile, combined with sequence comparisons and application of the positive-inside rule (47) and topology studies using protein fusions (8, 39), indicates that the CcdA protein has six α-helical transmembrane segments and the C-terminal end exposed on the outer side of the cytoplasmic membrane. Two cysteine residues, located far apart in the primary sequence, are conserved in CcdA sequences (references 8 and 36 and our unpublished data) and are functionally important (8). The amino acid sequence similarity between B. subtilis CcdA and E. coli DsbD and the importance of both proteins for a late step in cytochrome c synthesis (6, 33, 35) suggest that CcdA, together with a thioredoxin-like protein, has a function similar to that of DsbD in the transfer of reducing equivalents across the cytoplasmic membrane (28, 31). If so, the defects observed in CcdA-deficient strains might be explained by inefficient disulfide bond isomerization in proteins localized on the outer surface of the membrane.
Some cortex or coat proteins and extracellular protein factors with a function in spore coat or cortex synthesis possibly contain one or more essential disulfide bonds (reference 9 and references therein). Efficient formation of these bonds and cross-linking of proteins in the coat could be mediated by the activity of CcdA. In the absence of CcdA, proteins with several cysteine residues and located on the outer surface of the membrane would be more commonly misfolded or only slowly folded into a functional state. The resulting small amounts of functional protein in the intermembrane space between the mother cell and the forespore might limit the synthesis of heat-resistant spores, which would be observed as a reduced sporulation efficiency compared to that of wild-type strains.
ACKNOWLEDGMENTS
We thank Ingrid Stål and Hanna Falk-Nilsson for technical assistance, Claes von Wachenfeldt for help in obtaining antisera, Jens Jacobsson for his contribution to the construction of pDG1728 derivatives, Erik Carlemalm for help with electron microscopy of spores, and Alan Driks for strains and helpful discussions.
This work was supported by grants from the Swedish Natural Science Research Council and Maja och Erik Lindqvists Forskningsstiftelse.
REFERENCES
- 1.Barker P D, Ferguson S J. Still a puzzle: why is haem covalently bound in c-type cytochromes? Structure. 1999;7:R281–R290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 2.Bengtsson J, Rivolta C, Hederstedt L, Karamata D. Bacillus subtilis contains two small c-type cytochromes with homologous heme-domains but different types of membrane-anchors. J Biol Chem. 1999;274:26179–26184. doi: 10.1074/jbc.274.37.26179. [DOI] [PubMed] [Google Scholar]
- 3.Bron S. Plasmids. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 75–174. [Google Scholar]
- 4.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 5.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Crooke H, Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase domain. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 7.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 8.Deshmukh M, Brasseur G, Daldal F. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol Microbiol. 2000;35:123–138. doi: 10.1046/j.1365-2958.2000.01683.x. [DOI] [PubMed] [Google Scholar]
- 9.Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortnagel P, Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968;95:1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridén H, Hederstedt L. Role of His residues in Bacillus subtilis cytochrome b558 for haem binding and assembly of succinate:quinone oxidoreductase (complex II) Mol Microbiol. 1990;4:1045–1056. doi: 10.1111/j.1365-2958.1990.tb00677.x. [DOI] [PubMed] [Google Scholar]
- 13.Green G N, Gennis R B. Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J Bacteriol. 1983;154:1269–1275. doi: 10.1128/jb.154.3.1269-1275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guérot-Fleury A M, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 15.Haima P, Bron S, Venema G. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol Gen Genet. 1987;209:335–342. doi: 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- 16.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 17.Hederstedt L. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 1986;126:399–414. doi: 10.1016/s0076-6879(86)26040-1. [DOI] [PubMed] [Google Scholar]
- 18.Hoch J A. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- 19.Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Le Brun, N. E., J. Bengtson, and L. Hederstedt. Genes for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol., in press. [DOI] [PubMed]
- 22.Madhusudan, Zapf J, Whiteley J M, Hoch J A, Xuong N H, Varughese K I. Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis. Structure. 1996;4:679–690. doi: 10.1016/s0969-2126(96)00074-3. [DOI] [PubMed] [Google Scholar]
- 23.Margolis P, Driks A, Losick R. Establishment of a cell type by compartmentalized activation of a transcription factor. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 24.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 25.Missiakas D, Schwager F, Raina S. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 1995;14:3415–3424. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niaudet B, Goze A, Ehrlich S D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982;19:277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- 27.Page D M, Sambongi Y, Ferguson S J. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem Sci. 1998;23:103–108. doi: 10.1016/s0968-0004(98)01173-6. [DOI] [PubMed] [Google Scholar]
- 28.Page D M, Saunders N F W, Ferguson S J. Disruption of the Pseudomonas aeruginosa dipZ gene, encoding a putative protein-disulfide reductase, leads to partial pleiotropic deficiency in c-type cytochrome biogenesis. Microbiology. 1997;143:3111–3122. doi: 10.1099/00221287-143-10-3111. [DOI] [PubMed] [Google Scholar]
- 29.Pettigrew G W, Moore G R. Cytochromes c: biological aspects. Berlin, Germany: Springer Verlag KG; 1987. [Google Scholar]
- 30.Raleigh E, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rietsch A, Berlin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenkranz M S, Dingman D W, Sonenshein A L. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J Bacteriol. 1985;164:155–164. doi: 10.1128/jb.164.1.155-164.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambongi Y, Ferguson S J. Specific thiol compounds complements the deficiency in c-type cytochrome biogenesis in Escherichia coli carrying a mutation in a membrane-bound disulphide isomerase-like protein. FEBS Lett. 1994;353:235–238. doi: 10.1016/0014-5793(94)01053-6. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schiött T, Trone-Holst M, Hederstedt L. Bacillus subtilis CcdA-defective mutants are blocked in a late step of cytochrome c biogenesis. J Bacteriol. 1997;179:4523–4529. doi: 10.1128/jb.179.14.4523-4529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiött T, von Wachenfeldt C, Hederstedt L. Identification and characterization of the ccdA gene required for cytochrome c synthesis in Bacillus subtilis. J Bacteriol. 1997;179:1962–1973. doi: 10.1128/jb.179.6.1962-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 38.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart E J, Katzen F, Beckwith J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 1999;18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stock A M, Mottonen J M, Stock J B, Schutt C E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 41.Stragier P. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 42.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis. How morphological structure could control gene expression. Cell. 1988;532:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 43.Taber H W. Respiratory chains. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 199–212. [Google Scholar]
- 44.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tochikubo K. Changes in terminal respiratory pathways of Bacillus subtilis during germination, outgrowth and vegetative growth. J Bacteriol. 1971;108:652–661. doi: 10.1128/jb.108.2.652-661.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Oost J, von Wachenfeldt C, Hederstedt L, Saraste M. Bacillus subtilis cytochrome oxidase mutants: biochemical analysis and genetic evidence for two aa3-type oxidases. Mol Microbiol. 1991;5:2063–2072. doi: 10.1111/j.1365-2958.1991.tb00829.x. [DOI] [PubMed] [Google Scholar]
- 47.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 48.von Wachenfeldt C, Hederstedt L. Bacillus subtilis 13 kDa cytochrome c-550 encoded by cccA, consists of a membrane-anchor and a heme domain. J Biol Chem. 1990;265:13939–13948. [PubMed] [Google Scholar]
- 49.von Wachenfeldt C, Hederstedt L. Molecular biology of Bacillus subtilis cytochromes. FEMS Microbiol Lett. 1992;100:91–100. doi: 10.1111/j.1574-6968.1992.tb14025.x. [DOI] [PubMed] [Google Scholar]
- 50.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 51.Yu J, Hederstedt L, Piggot P. The cytochrome bc complex (menaquinol:cytochrome c reductase) in Bacillus subtilis has a non-traditional subunit organization. J Bacteriol. 1995;177:6751–6760. doi: 10.1128/jb.177.23.6751-6760.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J, Le Brun N E. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. J Biol Chem. 1998;273:8860–8866. doi: 10.1074/jbc.273.15.8860. [DOI] [PubMed] [Google Scholar]
- 53.Zheng L, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]