Abstract
Background & Aims
Noninvasive modalities for assessing active endoscopic and histologic inflammation in Crohn’s disease and ulcerative colitis patients are critically needed. Fecal wash host shed-cell transcriptomics has been shown to be a robust classifier of endoscopic and histologic inflammation in inflammatory bowel disease patients with distal colitis. Whether such fecal washes can inform on inflammatory processes occurring in more proximal intestinal segments is currently unknown.
Methods
Fifty-nine inflammatory bowel disease patients and 50 controls were prospectively enrolled. Biopsy specimens and fecal washes from the distal colon, proximal colon, and terminal ileum were compared. Host transcriptomics were performed on the biopsy specimens and fecal washes obtained during colonoscopy at predefined locations throughout the colon and terminal ileum and results were associated with concurrent clinical, endoscopic, and histologic parameters.
Results
We found that host transcriptomics of distal fecal washes robustly classify histologic inflammation in ileal and proximal colonic Crohn’s disease, even without distal colonic involvement (area under the receiver operating characteristic curve, 0.94 ± 0.09). We further found that fecal washes consist of modules of co-expressed genes of immune, stromal, and epithelial origin that are indicative of endoscopic disease severity. Fecal wash host transcriptomics also captures expression of gene modules previously associated with a lack of response to biological therapies.
Conclusions
Our study establishes the accuracy of distal colonic fecal washes for identifying and scoring inflammatory processes throughout the entire ileal–colonic axis.
Keywords: IBD, Transcriptomics, Histology, Therapy Outcome
Graphical abstract
Summary.
Biopsy specimens and fecal washes (suctioned during colonoscopy) from the distal colon, proximal colon, and terminal ileum of inflammatory bowel disease patients vs controls were compared. Host transcriptomics of distal fecal washes identified the location and severity of inflammation in the ileum and colon.
Inflammatory bowel diseases (IBDs) are autoinflammatory conditions associated with chronic inflammation of the gastrointestinal tract. Ulcerative colitis (UC) invariably involves the colon, whereas Crohn’s disease (CD) can affect any colonic segment as well as the small intestine and even the upper gastrointestinal tract.1 Colonoscopy with biopsies currently is considered the gold standard for IBD diagnosis, stratification, and monitoring. Nevertheless, colonoscopy is an invasive procedure, heralding risks of perforation and infection.2 Development of less-invasive monitoring is an outstanding challenge. Moreover, because only approximately 50% of IBD patients achieve endoscopic/histologic remission with the different currently available biological therapies, identification of noninvasive biomarkers that can assist in diagnosis, predict outcome, and guide therapy selection is needed.3,4
We have recently shown that host shed cell messenger RNAs (mRNAs) can be retrieved successfully from fecal washes and that transcriptomics of the host mRNA is associated significantly with endoscopic and even histologic healing in IBD patients with distal colitis.5 Furthermore, compared with biopsy-obtained transcriptomics, fecal wash transcriptomics had a stronger association with local pathologic findings. Fecal washes of patients with histologic inflammation were enriched in inflammatory monocytes, regulatory T cells, natural killer cells, and innate lymphoid cells. The enrichment of these cell types, critical for the inflammatory process, as well as the ability to capture pathologic processes that might be missed in localized biopsies, may explain the higher statistical power of fecal wash host transcriptomics. Whether similar statistical power exists for IBD that does not involve the distal colon is currently unknown.
In mice, intestinal cells continue to be viable for several hours after shedding,6 strengthening the notion that distal colonic fecal washes may also inform on proximal colonic and terminal ileal mucosal inflammation. Assessing whether distal fecal washes can provide information as to ileal/proximal colonic inflammation is of high clinical importance. This potentially could obviate the need for a full ileocolonoscopy for assessment of endoscopic and histologic inflammation and would enable diagnosing and localizing mucosal inflammation using a noninvasive method. Moreover, it may prove a powerful tool, enabling analytic summation of processes along the gut axis, which may not be feasible even with multiple biopsies along the intestinal tract. To explore this, we performed a comprehensive analysis of fecal washes obtained from several segments throughout the colon and terminal ileum in Crohn's patients with terminal ileitis, naïve to biological therapy. These were compared with colocalized biopsy specimens from IBD patients with distal colitis and of healthy controls.
Results
Host Transcriptomics of Fecal Washes From Different Intestinal Segments Captures Information That Is Distinct From Same-Segment Biopsy Transcriptomics
To assess the information captured by fecal wash host transcriptomics in IBD we performed colonoscopies on 59 IBD patients and 50 controls (with no signs of inflammation observed on colonoscopy) (Figure 1A). Clinical and demographic parameters of the 3 study subgroups (control, CD, and UC) are presented in Table 1. We sampled biopsies and matching fecal washes from multiple segments: the distal (left sided, sigmoid) colon, proximal (right) colon, and terminal ileum (TI). Our cohort included 56 patients and controls from our previous study of distal colitis and 53 new patients and controls aimed specifically at enriching for CD pathology without distal involvement (Figure 2A, Supplementary Table 1).5 We sequenced the RNA from these samples using the mcSCRBseq protocol7 (see the Methods section), a unique molecular identifier (UMI)-based sensitive protocol ideally suited for low-quality mRNA samples. We mapped the reads to the human genome (Supplementary Table 2), resulting in 11,275 ± 2728 genes per biopsy sample and 5973 ± 3134 genes per fecal wash sample. Cluster analysis (Figure 1B) and principal component analysis (Figure 1C) of all fecal washes and biopsy specimens showed clear separation between the biopsy and fecal wash samples. Fecal wash samples were enriched in immune-related genes, such as IL1B, CXCL8, and TNFAIP6 (Figure 2B, Supplementary Table 3). Biopsy specimens were enriched in genes associated with stromal components (COL6A3), plasma cells (JCHAIN), and epithelial cells (PIGR). Notably, as previously shown in distal fecal washes of IBD patients with exclusively left-sided (distal) inflammation,5 pairs of fecal wash samples of patients with histologic inflammation were significantly more correlated compared with mixed pairs, containing 1 sample with and 1 without histologic inflammation (median Spearman correlation, 0.66 vs 0.55, respectively; P = 2.8e-110) (Figure 1D). In contrast, pairs of biopsy specimens with histologic inflammation were as correlated as mixed pairs (median Spearman correlation, 0.68 vs 0.69, respectively; P = 3.3e-01) (Figure 1D). Compared with biopsy specimens, fecal washes of histologically inflamed patients had a significantly higher representation of multiple immune cell populations involved directly in the inflammatory process, including inflammatory monocytes, innate lymphoid cells (ILCs), regulatory T cells (Tregs), and natural killer cells (Figure 3, Supplementary Table 4). Fecal wash host transcriptomics therefore contain information that is distinct from biopsies and that correlates with inflammation status.
Figure 1.
Host transcriptomics of fecal washes from different intestinal segments captures information that is distinct from same-segment biopsy transcriptomics. (A) Experimental layout. (B) Clustergram of transcriptomes of biopsy specimens (purple) and fecal washes (yellow), color-coded by intestinal segment source and inflammation status. Red indicates high expression, blue indicates low expression. Representative gene names are shown on the right. (C) Principal component analysis of biopsy specimens (purple) and fecal washes (yellow). Black circles highlight inflamed samples. Parentheses show the percentage of explained variance by each PC. (D) Spearman correlations between the transcriptomes of pairs of either biopsy specimens or fecal washes that were both annotated as inflamed and mixed pairs (one annotated as inflamed and the other not). (B–D) Biopsy specimens were considered inflamed if they showed histologic inflammation, fecal washes were considered inflamed if at least 1 segmental biopsy specimen from the patient showed inflammation. (A) Created with BioRender.com. D, distal colon; P, proximal colon; TI, terminal ileum; mRNA, messenger RNA; PC1, principal component 1; PC2, principal component 2.
Table 1.
Demographic and Clinical Characteristics of Study Participants
| Control | Crohn's disease | Ulcerative colitis | |
|---|---|---|---|
| N (appeared in Ungar et al5) | 50 (31) | 30 (7) | 29 (18) |
| Female gender, n (%) | 26 (52) | 14 (46.6) | 14 (48.2) |
| Age, y, median (IQR) | 53.5 (34–70.7) | 33.5 (26.2–51) | 55 (43–68) |
| Age at diagnosis, y, median (IQR) | a | 29 (22.2–38.7) | 34 (20–43) |
| Weight, kg, median (IQR) | 72 (63.6–81.1) | 67.5 (56–75) | 69 (53.7–85.7) |
| Smoking at induction, n (%) | 6 (12) | 5 (16.6) | 2 (6.9) |
| Past intestinal resection, n (%) | 0 (0) | 2 (6.6) | 3 (10.3) |
| Concomitant illnesses, n (%) | 28 (56) | 5 (16.6) | 12 (41.3) |
| Disease location, n (%) | a | Ileal, 16 (53.3) | Proctitis, 1 (3.4) |
| Colonic, 6 (20) | Left-sided colitis, 13 (44.8) | ||
| Ileocolonic, 8 (26.6) | Pancolitis, 15 (51.7) | ||
| Concomitant biological therapy, n (%) | 0 (0) | 7 (23.3) | 6 (20.6) |
| Concomitant immunomodulator therapy, n (%) | 0 (0) | 0 (0) | 1 (3.4) |
| Concomitant mesalamine therapy, n (%) | 0 (0) | 4 (13.3) | 13 (44.8) |
| Concomitant steroidal therapy, n (%) | 0 (0) | 1 (3.3) | 3 (10.3) |
| Calprotectin level, mcg/g, median (IQR)b | a | 110 (54.7–274.5) | 682 (84.7–1000) |
| Endoscopic inflammation, n (%)c | a | 23 (76.6) | 16 (55.1) |
| Histologic inflammation, n (%)d | a | 21 (70) | 22 (75.8) |
IQR, interquartile range.
Not applicable for control patients.
Calprotectin levels were not available for all patients included. They were measured for patients with apparent inflammation on colonoscopy.
Endoscopic inflammation was defined as appearance of ulcers/mucosal inflammation on colonoscopy.
Histologic inflammation was defined as active histologic disease activity as per the pathologist’s assessment.
Figure 2.
Illustration of sample classification and differences in gene expression between biopsy specimens and fecal washes. (A) Heatmap showing the source of the patient samples in the study cohort, with reference to the previous data set from Ungar et al.5 (B) Differential gene expression between fecal washes and biopsy specimens. Red indicates significantly different genes (fold change >2 and q value <0.1). Shown are the names of representative differentially expressed genes.
Figure 3.
Computational deconvolution shows differences in cell composition between biopsy specimens and fecal washes. (A) Clustergram of inferred cell proportions. Table was standardized to Z-scores. The red dashed box highlights fecal washes enriched in inflamed samples. Colorbars annotate samples by sample type, disease, and inflammation status. (B) Violin plots of the inferred proportions of distinct cell types in biopsy specimens (top) and fecal washes (bottom). Noninflamed samples include both controls and noninflamed IBD patients. Biopsy specimens were considered inflamed if they showed histologic inflammation, fecal washes were considered inflamed if at least 1 segmental biopsy from the patient showed inflammation. DC2, dendritic cells type 2; GC, germinal center; ILC, innate lymphoid cells; NK, natural killer; TA, transit amplifying; Treg, regulatory T cell.
Host Transcriptomics of Distal Fecal Washes Captures Active Inflammation in Ileal or Proximal Colonic CD
We next turned to assess whether distal fecal washes could identify inflammation in proximal intestinal segments (right colon, TI). Similar to Ungar et al,5 when analyzing the host transcriptomics of distal fecal washes of UC and controls, we observed distinct separation between the patients with and without histologic inflammation (Figure 4A and B). When analyzing CD patients and controls we observed similar separation between patients with and without histologic inflammation (Figure 4C and D). Notably, patients without distal involvement, including patients with TI inflammation and no colonic inflammation, clustered based on the transcriptomics of their distal fecal washes (Figure 4C and D). Distal fecal washes of inflamed CD patients showed increased levels of inflammatory genes such as NFKBIA, IL1RN, CCR1, IFIT2, and S100A9 (Figure 4E). We built a classifier of histologic inflammation based on the summed expression of inflammatory signature genes (see the Methods section) and applied it to either UC or CD patients’ distal fecal washes or distal biopsy specimens of both. Receiver operating characteristic curve analysis showed significantly higher classification power based on the fecal wash transcriptomics compared with biopsies: the area under the curve (AUC) of 0.94 ± 0.07 for UC distal fecal washes and the AUC of 0.94 ± 0.09 for distal fecal washes of CD patients without distal colonic involvement, compared with the AUC of 0.82 ± 0.20 for all distal biopsies (Figure 4F). Host transcriptomics of distal fecal washes therefore serve as powerful classifiers not only of distal colonic inflammation in UC patients, but also of CD inflammation, including when the inflammatory segments are ileal and no colonic involvement is observed.
Figure 4.
Host transcriptomics of distal fecal washes captures active inflammation in ileal or proximal colonic CD. (A) Clustergram of distal fecal washes from UC patients and controls, color-coded by disease and segment of inflammation. (B) Principal component analysis of UC and control distal fecal washes. Percentages in in parentheses explain the variance of each PC. (C) Clustergram of distal fecal washes from CD patients and controls, color-coded by disease and segment of inflammation. (D) Principal component analysis of CD and control distal fecal washes. Parentheses show the variance explained by each PC. Blue indicates no inflammation, green indicates inflammation that includes the distal colon (left), pink indicates inflammation in the proximal colon (right) or proximal colon + TI, light pink indicates inflammation only in the TI. (A and C) Red indicates high expression, blue indicates low expression, representative gene names are shown on the right. Red dashed box highlight fecal washes enriched in inflamed samples. (E) Differential gene expression of inflamed and noninflamed distal fecal washes from CD patients and controls. Red indicates significantly different genes (fold change >2 and q value <0.1). (F) Receiver operating characteristic (ROC) curve of the classification performance of inflammation status based on distal fecal washes of controls and UC patients (orange), distal fecal washes of controls and CD patients that do not involve the distal colon (black), and distal biopsy specimens of controls vs UC and CD patients (purple). Biopsy specimens were considered inflamed if they showed histologic inflammation, fecal washes were considered inflamed if at least 1 segmental biopsy specimen from the patient showed inflammation. CT, control; inf, inflamed; PC1, principal component 1; PC2, principal component 2.
Transcriptomics of Distal Fecal Washes Contain Information Referring to the Inflamed Intestinal Segment
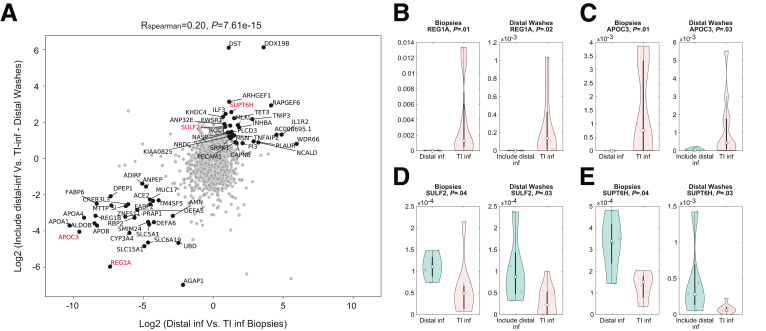
Our finding that distal fecal washes can classify active histologic inflammation, even in proximal intestinal segments, prompted us to ask whether distal fecal washes contain a signature of the site of inflammation. To this end, we focused on patients with active inflammation and performed differential gene expression between distal and ileal inflamed biopsy specimens, as well as between distal fecal washes of patients with TI inflammation only and patients with distal colonic involvement. We found that both biopsy specimens and distal fecal washes contained differentially expressed genes (Supplementary Table 5). Notably, the differences in expression were correlated significantly between the distal fecal washes and the biopsy specimens (R = 0.20; P = 7.61e-15) (Figure 5A). Genes that showed correlated differences in expression between IBD patients with only TI vs distal colonic inflammation included REG1A (Figure 5B) and APOC3 (Figure 5C), which were expressed more highly in patients with TI inflammation, and SULF2 (Figure 5D) and SUPT6H (Figure 5E), which were expressed more highly in patients with distal colonic inflammation, both in biopsy specimens and distal fecal washes. Distal colonic fecal washes therefore contain signatures not only of active inflammation but also of the intestinal segment where inflammation occurs.
Figure 5.
Transcriptomics of distal fecal washes contain information referring to the inflamed intestinal segment. (A) Scatter plot of the expression ratios between inflamed samples of CD patients with distal colon inflammation and ones with only TI inflammation. X-axis shows the ratios in the segmental biopsy specimens, the Y-axis shows the ratios in distal fecal washes. (B–E) Violin plots of the expression of representative genes shown in red in panel A. Biopsy specimens were considered inflamed if they showed histologic inflammation, fecal washes were considered inflamed if at least 1 segmental biopsy from the patient showed inflammation. inf, inflamed.
Distal Fecal Washes Contain Modules of Co-expressed Immune, Stromal, and Epithelial Genes Associated With Inflammation Severity
Our computational deconvolution showed enriched representation of distinct immune cell types in fecal washes of patients with histologic inflammation. Importantly, other cellular components also were represented in fecal washes, including epithelial and stromal cell types (Figure 3, Supplementary Table 4). To explore the transcriptomic landscape of immune, stromal, and epithelial genes in inflamed and noninflamed patients, we used a single-cell atlas of the human colon8 to extract genes that are expressed solely by these coarse-grained cell types (Methods section, Supplementary Table 6). We clustered the distal fecal wash samples based on these distinct gene sets (Figure 6). Unbiased clustering showed several modules of co-expressed genes for each of these cellular subsets. We assigned each sample a module score based on the summed expression of the respective epithelial (Epi), stromal (Str) and Immune (Imm) module genes. Epithelial genes clustered into 4 modules, including a module enriched in inflamed samples (Epi1, median module score of 0.2 in inflamed fecal washes vs 0.087 in noninflamed fecal washes; P = 3.3e-03). The Epi1 module was enriched for interleukin (IL)2/signal transducer and activator of transcription (STAT)5, transforming growth factor β, and P53 signaling pathways (Supplementary Table 7) and contained genes previously associated with IBD, including DUOXA2,9 SAA1,10 and IL13RA111 (Figure 6A). Stromal genes clustered into 3 modules, including a module enriched in inflamed samples (Str3, median module score of 0.5 in inflamed fecal washes vs 0.08 in noninflamed fecal washes; P = 2.8e-03). Str3 included genes previously shown to be expressed in inflammatory fibroblasts, such as CHI3L1,12 CCL2,13 MMP3,14 and IFITM315 (Figure 6B). Immune genes clustered into 3 modules, including a module enriched in inflamed samples (Imm3, median module score of 0.8 in inflamed fecal washes vs 0.32 in noninflamed fecal washes; P = 1.6e-03), which contained genes previously associated with intestinal inflammation, such as CXCR4,16 CSF3R,17 IL1B,18 and OSM19 (Figure 6C). Both Str3 and Imm3 modules were enriched in inflammation-associated pathways such as interferon γ, tumor necrosis factor α (TNF-α), IL6/Janus kinase/STAT3, and IL2/STAT5 signaling (Supplementary Table 7).
Figure 6.
Distal fecal washes contain modules of co-expressed immune, stromal, and epithelial genes associated with inflammation severity. Clustergram of epithelial genes (A), stromal genes (B), and immune genes (C). Red indicates high expression, blue indicates low expression. Modules are based on the gene hierarchical clustering, with relevant branches colored accordingly. Right: Representative module genes are shown. Color bars annotate disease and inflammation status. Red dashed boxes highlight a branch of fecal washes enriched in inflamed samples. (D) Clustergram of the module scores for distal fecal wash samples show co-expression of the Str3, Imm3, and Epi1 modules. Red indicates high expression, blue indicates low expression. Colorbars annotate disease and inflammation severity score. Right: Heatmap shows the median of module scores over the discrete inflammation severity score classes (see the Methods section). Each row was normalized to the maximum across severity classes. Fecal washes were considered inflamed if at least 1 segmental biopsy from the patient showed inflammation. Epi, epithelial; Imm, immune; inf, inflamed; Non-inf, noninflamed; Str, stromal.
We found that patients clustered according to their module scores, with increased combined expression of Str3, Imm3, and Epi1 in the distal fecal washes of patients with histologic inflammation (Figure 6D). Notably, the module scores showed trends that were correlated with the severity of inflammation, as computed by clinical, inflammatory, and endoscopic parameters (Methods section, Figures 6D and 7, Supplementary Table 8). For example, stromal modules Str3 and Str1 increased or decreased, respectively, with inflammation severity in a gradual manner, Epi3 began to decrease in patients with moderate inflammation, and Str2 peaked in patients with mild inflammation (Figures 6D and 7). When considering only endoscopic score as a proxy for severity, we further found that module scores such as Imm3 and Str3 increased monotonically (R = 0.83, P = 3.0e-07 and R = 0.79, P = 3.4e-06, respectively) (Figure 8A). In contrast, calprotectin levels changed nonmonotonically and showed substantially decreased correlation with endoscopic severity (R = 0.41; P = 5.0e-02) (Figure 8B). Our analysis therefore highlights modules of epithelial, stromal, and immune genes co-expressed in shed cells that are enriched in patients with histologic inflammation and are highly associated with endoscopic inflammation severity.
Figure 7.
Module scores of co-expressed immune, stromal, and epithelial genes show trends that correlate with inflammation severity. Shown are the module scores of each of the 10 modules (Figure 6D) binned according to classes of inflammation severity (Supplementary Table 8). The classes were binned by the percentile of the predefined inflammation severity score (see the Methods section, controls were assigned a severity score of 0). Shown are Spearman correlation values between the module scores and severity scores of distal fecal washes from patients with inflammation and controls. Fecal washes were considered inflamed if at least 1 segmental biopsy from the patient showed inflammation. Epi, epithelial; Imm, immune; inf, inflamed; Non-inf, noninflamed; Str, stromal.
Figure 8.
Fecal wash host transcriptomics correlates with endoscopic severity score significantly more than calprotectin levels. (A) Module scores of co-expressed immune, stromal, and epithelial gene modules (Figure 6D) are in correlation with endoscopic severity score. Module scores are binned according to classes of endoscopic severity scores (controls were assigned an endoscopic score of 0) (Methods section, Supplementary Table 8). Shown are Spearman correlation values between the module scores and endoscopic severity scores of distal fecal washes from patients with inflammation and controls. Blue frames and red frames highlight gene modules that significantly decrease and increase with endoscopic inflammation severity, respectively (P < .05). (B) Correlation between calprotectin levels and endoscopic scores of distal fecal washes from patients with inflammation (Methods section, Supplementary Table 8). Fecal washes were considered inflamed if at least 1 segmental biopsy from the patient showed inflammation. Epi, epithelial; Imm, immune; Str, stromal.
Modules of Co-expressed Genes That Correlate With Response to Biological Therapy Carry Information on Inflammation Severity in Fecal Washes
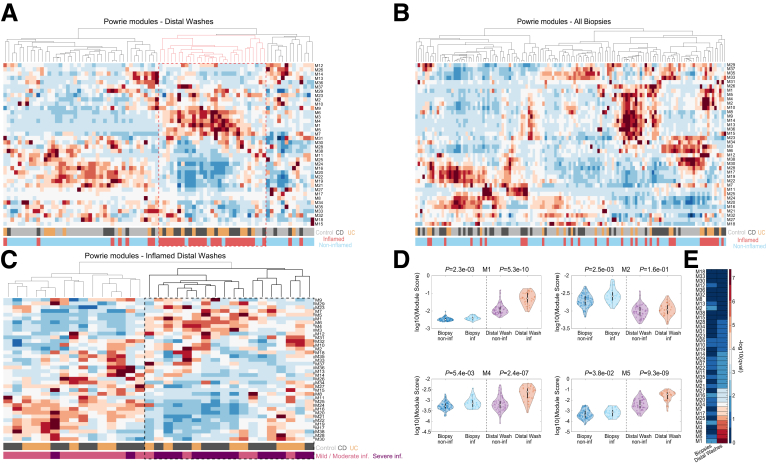
A recent study of biopsy transcriptomics in IBD patients by Powrie and colleagues20 defined gene modules that were shown to correlate with either response or lack of response to biological therapies (anti-TNF and anti-integrin) and to corticosteroids. We sought to explore the expression of these modules in the distal fecal washes (Figure 9A) and biopsy specimens (Figure 9B) of our cohort. We generated a module score for each sample based on the sum of the respective module genes and clustered the data. We found that distal fecal washes of inflamed patients clustered based on this module sum signature (Figure 9A and C). Patients with histologic inflammation had higher expression of modules M1, M2, M4, and M5 (Figure 9D), which previously were shown to correlate with IBD severity.20 Notably, the difference in M1, M4, and M5 module scores was significantly higher in the distal fecal washes compared with the biopsy specimens (Figure 9D and E, Supplementary Table 9). When clustering only the inflamed distal fecal washes, based on the module scores, we identified 2 clusters of patients, one of which strongly enriched in modules that corresponded to nonresponse to the aforementioned therapies (M4, M5) (Figure 9C). Notably, this cluster was enriched in patients with more severe disease status, as determined by clinical, inflammatory, and endoscopic parameters (13 of 15 samples, hypergeometric P value of 4.5e-05) (Methods section, Supplementary Table 8). Our analysis indicates that distal fecal washes, including from CD patients with no distal colitis, contain a strong transcriptomic signature of gene modules associated with response to biological therapy that correlate with disease severity.
Figure 9.
Modules of co-expressed genes that correlate with response to biological therapy carry information on inflammation severity in fecal washes. (A–C) Summed expression of previously identified modules that showed correlation with response to anti-TNF/anti-integrin therapies.20 (A) Distal fecal washes, red dashed box highlights a branch of fecal washes enriched in inflamed samples, increased in modules M1, M3, M4, M5, and M6. (B) All biopsies. (A and B) Color bars annotate disease and inflammation status. (C) Only inflamed distal fecal washes. Color bars annotate disease and inflammation severity (see the Methods section). Bold dashed box highlights fecal washes of patients with severe inflammation. (D) Scores of representative modules correlating with nonresponse to anti-TNF/anti-integrin therapies, showing more significant differences in expression between inflamed and noninflamed distal fecal washes compared with biopsies for M1, M4, and M5, but not M2. A pseudonumber (2.4163e-05) was added to the module scores before applying the log10 transform. (E) Higher statistical significance in the differences in module expression between inflamed and noninflamed samples in distal fecal washes compared with biopsies. Shown are –log10 (q values). Biopsy specimens were considered inflamed if they showed histologic inflammation, fecal washes were considered inflamed if at least 1 segmental biopsy specimen from the patient showed inflammation. inf, inflamed; non-inf, noninflamed.
Discussion
Fecal wash host transcriptomics has been shown to be a powerful predictor of active endoscopic and histologic inflammation in IBD patients with distal (left-sided) colonic inflammation.5 Here, we sought to extend these results and assess the power of distal fecal wash host transcriptomics in identifying the existence, severity, and location of inflammation in CD, which often involves inflammation in more proximal segments of the bowel, such as the right colon or the terminal ileum. We showed that fecal wash host transcriptomics provides information about histologic inflammation even in CD terminal ileitis. Notably, fecal wash transcriptomics showed higher sensitivity and specificity in identifying inflammation when compared with transcriptomics of biopsy specimens (Figure 4F). This higher statistical power may be a result of the fact that fecal washes capture cells that are shed throughout the gastrointestinal tract and therefore are not sensitive to biases related to the precise location from which a biopsy specimen is obtained. Moreover, fecal washes show higher representation of several immune cell types involved directly in the inflammatory process, such as inflammatory monocytes, ILCs, natural killer cells, and Tregs (Figure 3). Furthermore, fecal wash host transcriptomics provides information not only about the location of inflammation, but also the severity of inflammation (Figures 6D, 9C, and 7). In a recent study,6 shed cells in mice showed elongated lifetimes of several hours, and a higher stability in the colon compared with the small intestine. Such stability can explain the detection of cellular signs of ileal inflammation in distal colonic fecal washes. Corroborating studies will show whether distal fecal washes further contain signatures of more proximal intestine pathologies, such as celiac disease.
Our study showed that fecal wash host transcriptomics reveals gene modules such as Str3, Imm2, and Imm3, that are significantly more correlated with endoscopic severity scores compared with calprotectin levels (Figure 8). Because the therapeutic approach in IBD is based on endoscopic disease severity, fecal wash host transcriptomics can offer a noninvasive modality for therapy selection.21 We further found that expression in distal fecal washes of gene modules previously shown to correlate with lack of response to biological therapy20 correlates with a severe disease phenotype, in either CD or UC patients, as per endoscopy, inflammatory and clinical parameters (see the Methods section). The genes we have found to be increased in fecal washes of inflamed patients also showed an overlap with molecular biomarkers measured in biopsy specimens and blood samples of IBD patients with active endoscopic and histologic disease who were more resistant to therapy.22
We showed that distal fecal washes can predict histologic inflammation in the TI, potentially obviating the need for ileocolonoscopy for these indications. The fact that distal fecal washes can be obtained in a less-invasive and more accessible manner compared with colonoscopy pertains to its easy utilization. It remains to be seen whether similar signatures of active inflammation in UC and CD exist in host transcriptomics obtained from stool samples.
To conclude, our study showed that distal fecal washes from IBD patients are associated with endoscopic disease severity and histologic inflammation, regardless of the site of inflammation. Hence, even TI inflammation can be inferred from the distal fecal washes. Furthermore, distal fecal washes show variable signatures of modules of co-expressed genes, previously shown to be associated with response to biological therapy.20 The information contained in distal fecal wash host transcriptomics regarding inflammation location and severity could be used to guide patient-specific therapy. This is critical, given that current clinical remission rates with different biological agents are only approximately 30%–60%.23,24 Future prospective studies of fecal host transcriptomics of patients at diagnosis will establish the ability to predict response and to establish companion diagnostic signatures for specific drugs for CD and UC.
Methods
Patient Population
This study was approved by the Sheba Medical Center Helsinki Committee (reference number 8796). In this cross-sectional study, patients undergoing a lower endoscopy at Sheba Medical Center Gastroenterology Institute were recruited. The study group consisted of 56 patients and controls from our previously published cohort and 53 newly recruited patients.5 The 53 newly recruited patients comprised 19 controls, 11 UC patients, and 23 CD patients. In total, 24 ileal (involving TI)/ileocolonic CD patients were compared with a total of 35 colitis patients (comprising 29 UC and 6 Crohn's colitis patients) and 50 control non-IBD patients. All control patients had a lower endoscopy performed for unrelated reasons (colorectal screening, and so forth), and patients with IBD underwent the procedure because of clinical indications as per the decision of their treating physician irrespective of the present study. Five UC patients from the previous cohort were excluded because they had a sigmoidoscopy performed, as opposed to a full colonoscopy (Figure 2A, Supplementary Table 1), because sigmoidoscopies only provide information about the distal colon. All IBD patients had colonoscopies spanning the TI to the distal colon, enabling annotation of the precise segment of inflammation. Clinical and demographic parameters were obtained from questionnaires at the time of enrollment and from patients’ computerized medical records (Table 1).
Sample Collection
On colonoscopy, biopsy specimens (2 consecutive biopsy specimens per patient, ie, double bite) were obtained from the designated areas: distal colon (left, sigmoid), proximal colon (right, between hepatic flexure and cecum), and TI, and fecal fluid was suctioned from the same areas. Fecal washes were obtained while advancing the scope (before any through-the-scope washing was applied) and biopsy specimens from corresponding areas were obtained during withdrawal. In patients with endoscopic inflammation of the colon/ileum, the biopsy specimens were obtained from the inflamed area, adjacent to ulcers. Samples were snap-frozen in liquid nitrogen and stored at −80°C until further analysis.
Study Outcomes
The primary outcome was to assess whether the transcriptomic profiles of distal (left-sided) fecal washes obtained during the colonoscopy were associated with histologic inflammation in the distal colon/proximal colon/TI in the same patient.
Secondary outcomes included a comparison of transcriptomic profiles of fecal washes with transcriptomics of colonic/ileal biopsy specimens. Furthermore, we aimed to analyze the cellular composition of distal vs proximal colonic and ileal fecal washes using computational deconvolution based on the single-cell RNA sequencing data. Finally, an analysis was performed based on a previous study by Powrie and colleagues,20 to identify gene modules characterizing patients with primary nonresponse to several therapeutic agents targeting IBD.
Exclusion Criteria
The exclusion criteria were as follows: age younger than 18 years, undetermined diagnosis of UC or CD (IBD unclassified), missing clinical/demographic data, and patients undergoing sigmoidoscopy/failing to complete a full colonoscopy in UC/ileocolonoscopy in CD.
Definition of Endoscopic Mucosal Healing and Histologic Remission
Endoscopic and histologic inflammation were graded according to standardized indices and by blinded gastroenterologists and pathologists, respectively. Endoscopic scores were determined prospectively during colonoscopy. Mucosal healing was defined as an absence of ulcers and a lack of inflammation on endoscopic examination, for CD and UC, respectively.25 Histologic inflammation was determined by a certified pathologist at the Sheba Medical Center based on biopsy specimens from the same colon (distal/proximal) or ileal (TI) region used for the biopsy transcriptomics. Assessment of histologic disease activity was performed using the Nancy Score for UC and the Global Histological Disease Activity Score for CD. Because histologic score assessment has not been fully standardized in the previous literature for CD, a dichotomous assessment only was performed (active histologic disease activity/lack of histologic disease activity).
Calprotectin Measurement
Calprotectin was assessed using a previously established Quantum-blue fCAL extended point-of-care testing assay (Bülmann, Schönenbuch, Switzerland).26 Calprotectin levels were measured prospectively from fecal washes at the beginning of each colonoscopy. Calprotectin was measured only in patients with apparent inflammation on colonoscopy.
Disease Severity Assessment
The disease severity analysis was based on several parameters. The first parameter was the endoscopic score (Mayo endoscopic score of 0 to 3 for UC27 and the Simple Endoscopic Score for CD,28 with scores stratified per endoscopic remission/mild/moderate/severe, 0–3). The threshold for mucosal healing (lack of endoscopic inflammation) was defined as no ulcers or mucosal inflammation. This annotation was performed by a gastroenterologist blinded to the bioinformatic analysis. The second parameter was the fecal wash calprotectin levels (mcg/g). Calprotectin values were stratified as accepted, normal (<100; score, 0), mild to moderate (100–300; score, 1) and severe (>300; score, 2).29 The third parameter was the previous use of steroids, with a score of 0 or 1. The fourth parameter was previous events of IBD exacerbation leading to hospitalization, with a score of 0 or 1. An inflammation severity score was calculated as the mean of these 4 values for each patient in the severity analysis (Supplementary Table 8). In Figures 6D and 7, patients were binned into 4 groups based on equal percentile thresholds of the severity scores of patients with inflammation. In Figure 9C, patients were classified into 2 groups according to their values less than/greater than the median severity score (0.67).
RNA Extraction
For colonic biopsy specimens, snap-frozen tissues (2 × 2 mm) were thawed in 300 μL Tri-reagent (Sigma Aldrich) and homogenized mechanically with bead beating, followed by a short centrifugation step to pull-down beads and any tissue leftovers. For fecal washes, Tri-reagent was added at a ratio of 3:1, samples were allowed to thaw on ice, followed by thorough mixing. A first centrifugation step was used (1 min, 18,000 rpm) to eliminate fecal solids. After this, ethanol was added at a ratio of 1:1 to the supernatant from the previous step and continued according to the manufacturer’s instructions of the Direct-zol Mini and Micro Prep Kit (R2052; ZYMO Research).
Bulk RNA Sequencing of Samples
RNA was processed by the mcSCRBseq protocol7 with minor modifications. For biopsy specimen RNA, a reverse-transcription (RT) reaction was applied to 10 ng total RNA. For fecal wash RNA, the RT reaction was started with one third of the total eluted volume with a final reaction volume of 20 μL (1× Maxima H Buffer, 1 mmol/L deoxynucleoside triphosphate, 2 μmol/L TSO∗ E5V6NEXT, 7.5% PEG8000, 20 U Maxima H enzyme, and 2 μL barcoded RT primer). Subsequent steps were applied as mentioned in the protocol. The library preparation was performed using the Nextera XT kit (Illumina) on 0.6 ng amplified complementary DNA. The library final concentration of 2 nmol/L was loaded on a NovaSeq 6000 (Illumina) sequencing machine that aimed for 10–20 million reads per sample with the following setting: Read1, 16 bp; Index1, 8 bp; and Read2, 66 bp.
Bioinformatics and Computational Analysis
Illumina output sequencing raw files were demultiplexed to FASTQ files using the bcl2fastq package. To obtain the UMI counts, FASTQ files were aligned to the human reference genome (GRCh38.91) using the zUMI package.30 Statistical analyses were performed with MATLAB R2022a. Mitochondrial genes and non–protein-coding genes were removed from the analysis. Protein-coding genes were extracted using the annotation in the Ensembl database (BioMart) for reference genome GRch38 version 91, using the R package biomaRt (version 2.44.4). Gene expression for each sample consequently was normalized by the sum of the UMIs of the remaining genes that individually took up less than 10% of the total UMI sum. Clustering and principal component analysis were performed in MATLAB using the Z-score–transformed expression matrix. Clustering was performed with the MATLAB function clustergram, using Spearman correlation distances. Differential gene expression was performed using Kruskal–Wallis tests and Benjamini–Hochberg false discovery rate (FDR) corrections. Computational deconvolution was performed using CIBERSORTx31 using signature tables obtained from a single-cell atlas of the human colon8 (Supplementary Table 4). Original cell-type annotations were used, but subsequently coarse-grained into a small number of cell types. M cells were removed from the analysis because of their low abundance.
Classifier of Inflammation
Samples used to build the classifier were fecal washes or biopsy specimens taken from the distal segment of the colon, with total UMI counts higher than 10,000. Classification was based on a parameter that represented the summed expression of inflammation-related genes, extracted as follows. For 40 iterations, the data set was split into a training set (70%) and a test set (30%). Differential gene expression between the inflamed and noninflamed training set samples was calculated. Genes with a maximal normalized expression higher than 10-4 over all the included samples that were expressed in more than 5% of the samples were further considered. The Kruskal–Wallis test followed by the Benjamini–Hochberg FDR correction was performed. Genes whose mean expression in inflamed samples was more than 2-fold higher than the mean in noninflamed samples, with FDR q-value less than 0.1 were selected as inflammatory markers. Inversely, genes whose mean expression in inflamed samples was less than 2-fold lower than the mean in noninflamed samples, with FDR q-value less than 0.1 were chosen as noninflammatory markers. Those genes were identified based on the training set only.
Next, for the test set samples, the sums of inflammatory markers and noninflammatory markers were calculated, after the gene expression levels for each gene were normalized internally by their maximal values across the test set. An inflammation score was calculated for each test set sample, as follows: (sum of normalized inflammation markers)/(sum of normalized inflammatory markers + sum of normalized noninflammatory markers). The receiver operating characteristic curve32 and subsequent false-positive rate, true-positive rate, and AUC were recorded for each test set to examine the classification of samples of inflamed patients and noninflamed patients based on the inflammation score.
The division of the data set into training and test sets was performed randomly 40 times. False-positive rates over all 40 iterations were binned into 20 equal intervals (0–0.05, 0.05–0.1…0.95–1), and the means and standard errors (SEs) of the false-positive rates and the corresponding true-positive rates over each bin were calculated (Figure 4F).
This analysis was performed 3 times to classify different types of groups: the first classification was performed on distal fecal washes of control and UC patients, and the classification criterion was either histologic inflammation in the distal and/or proximal segment of the colon or no inflammation at all. The second classification was performed on distal fecal washes taken from control and CD patients, with the classification criterion of either histologic inflammation in the proximal colon and/or TI, whereas the distal segment was not histologically inflamed or no histologic inflammation at all. The third analysis was performed on distal biopsies. Biopsy specimens taken from the distal colon of controls, CD patients, and UC patients were used to classify histologic inflammation or no inflammation in the distal colon. In case no markers passed the differential gene expression thresholds, the iteration was skipped for the specific training and test set.
Gene Modules
To extract gene modules based on specific intestinal cell types, a single-cell atlas of the human colon8 was used to extract signatures of 3 cell types: immune, stroma, and epithelia. Expression levels of these coarse-grained cell types were determined as the maximum over the average expression of the single-cell RNA sequencing cluster of cell types belonging to the coarse-grained cell type (Supplementary Table 6). We defined a signature for each one of the coarse-grained cell types by including genes with an expression level above 5 × 10-6 of the UMI sum and that were expressed at least 3 times higher than any other coarse-grained cell type.
Next, we found the signature genes of each coarse-grained cell type in our data set, and specifically included here the fecal washes taken from the distal colon (Figure 6). For each cell type, expression of each signature gene was divided by the sum of the cell-type–specific signature genes. This yielded 3 tables, representing the fraction of each signature gene from the total sum of the cell type–specific signature genes. Clustering analysis was performed on the Z-score of the coarse-grained cell type tables, including samples with a UMI count higher than 5000 for epithelial genes, 2000 for stromal genes, and 2000 for immune genes and including genes expressed above 2 × 10-3 of the summed expression of cell type–specific genes in at least 1 sample. Signature genes were classified manually into 4 epithelial (Figure 6A), 3 stromal (Figure 6B), and 3 immune (Figure 6C) modules, based on the clustergram (Supplementary Table 7). The score of each module in each sample was calculated as the sum of the expression of genes belonging to the same module (from the signature tables renormalized over the sum of the respective cell type–specific signatures). The module scores of all samples were standardized (Z-score) and clustered using the MATLAB clustergram function with Spearman correlation distances (Figure 6D). To this end, we included samples that both individually passed the cell type–specific UMI sums of the signature genes and had a total UMI count greater than 10,000. Enrichr was used to identify pathways enriched in each of the modules (MSigDB Hallmark 2020 data set) (Supplementary Table 7).33
CRediT Authorship Contributions
Stav Dan (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Writing – original draft: Equal; Writing – review & editing: Supporting)
Bella Ungar (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Writing – original draft: Equal; Writing – review & editing: Supporting)
Shani Ben-Moshe (Formal analysis: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Keren Bahar Halpern (Methodology: Supporting)
Miri Yavzori (Methodology: Supporting)
Ella Fudim (Methodology: Supporting)
Orit Picard (Methodology: Supporting)
Chaya Mushka Abitbol (Methodology: Supporting; Project administration: Supporting)
Sivan Harnik (Methodology: Supporting)
Iris Barshack (Methodology: Supporting)
Uri Kopylov (Supervision: Supporting)
Shomron Ben-Horin (Conceptualization: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Shalev Itzkovitz (Conceptualization: Lead; Investigation: Equal; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest These authors disclose the following: Shomron Ben-Horin has received consulting and advisory board fees and/or research support from AbbVie, MSD, Janssen, Takeda, and CellTrion; Uri Kopylov has received speaker and advisory fees from AbbVie, Jannsen, Gilead, MSD, Medtronic, and Takeda, and research support from Jannsen, Medtronic, and Takeda; and Bella Ungar has received consultation fees from Neopharm, Takeda, Janssen, and AbbVie. The remaining authors disclose no conflicts.
Funding Supported by the Wolfson Family Charitable Trust, the Edmond de Rothschild Foundations, the Fannie Sherr Fund, the Dr Beth Rom-Rymer Stem Cell Research Fund, the Minerva Stiftung grant, the Weizmann-Sheba joint research program, the Israel Ministry of Innovation, Science & Technology, the Israel Science Foundation grant 1486/16, the European Research Council under the European Union’s Horizon 2020 research and innovation program grant 768956, the Chan Zuckerberg Initiative grant CZF2019-002434, the Bert L. and N. Kuggie Vallee Foundation, and the Howard Hughes Medical Institute international research scholar award (S.I.); and by the Talpiot medical excellence grant and the Weizmann-Sheba joint research program (B.U.).
Data Availability Statement All data relevant to the study are included in the article or uploaded as supplemental information.
Supplementary Material
References
- 1.Guan Q. A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res. 2019;2019 doi: 10.1155/2019/7247238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spiceland C.M., Lodhia N. Endoscopy in inflammatory bowel disease: role in diagnosis, management, and treatment. World J Gastroenterol. 2018;24:4014–4020. doi: 10.3748/wjg.v24.i35.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paramsothy S., Rosenstein A.K., Mehandru S., Colombel J.-F. The current state of the art for biological therapies and new small molecules in inflammatory bowel disease. Mucosal Immunol. 2018;11:1558–1570. doi: 10.1038/s41385-018-0050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh S., George J., Boland B.S., et al. Primary non-response to tumor necrosis factor antagonists is associated with inferior response to second-line biologics in patients with inflammatory bowel diseases: a systematic review and meta-analysis. J Crohns Colitis. 2018;12:635–643. doi: 10.1093/ecco-jcc/jjy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ungar B., Yavzori M., Fudim E., et al. Host transcriptome signatures in human faecal-washes predict histological remission in patients with IBD. Gut. 2022;71:1988–1997. doi: 10.1136/gutjnl-2021-325516. [DOI] [PubMed] [Google Scholar]
- 6.Halpern K.B., Kohanim Y.K., Biram A., et al. The cellular states and fates of shed intestinal cells. [DOI] [PubMed]
- 7.Bagnoli J.W., Ziegenhain C., Janjic A., et al. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat Commun. 2018;9:2937. doi: 10.1038/s41467-018-05347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smillie C.S., Biton M., Ordovas-Montanes J., et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacFie T.S., Poulsom R., Parker A., et al. DUOX2 and DUOXA2 form the predominant enzyme system capable of producing the reactive oxygen species H2O2 in active ulcerative colitis and are modulated by 5-aminosalicylic acid. Inflamm Bowel Dis. 2014;20:514–524. doi: 10.1097/01.MIB.0000442012.45038.0e. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.-Y., Hall J.A., Kroehling L., et al. Serum amyloid A proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell. 2020;180:79–91.e16. doi: 10.1016/j.cell.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magyari L., Kovesdi E., Sarlos P., et al. Interleukin and interleukin receptor gene polymorphisms in inflammatory bowel diseases susceptibility. World J Gastroenterol. 2014;20:3208–3222. doi: 10.3748/wjg.v20.i12.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutschmann C., Roggenbuck D., Schierack P. The loss of tolerance to CHI3L1 – a putative role in inflammatory bowel disease? Clin Immunol. 2019;199:12–17. doi: 10.1016/j.clim.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Martin J.C., Chang C., Boschetti G., et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penrose H.M., Iftikhar R., Collins M.E., et al. Ulcerative colitis immune cell landscapes and differentially expressed gene signatures determine novel regulators and predict clinical response to biologic therapy. Sci Rep. 2021;11:9010. doi: 10.1038/s41598-021-88489-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., Dassopoulos T., Cope L., et al. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 16.Werner L., Guzner-Gur H., Dotan I. Involvement of CXCR4/CXCR7/CXCL12 interactions in inflammatory bowel disease. Theranostics. 2013;3:40–46. doi: 10.7150/thno.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashton J.J., Boukas K., Davies J., et al. Ileal transcriptomic analysis in paediatric Crohn’s disease reveals il17- and NOD-signalling expression signatures in treatment-naïve patients and identifies epithelial cells driving differentially expressed genes. J Crohns Colitis. 2020;15:774–786. doi: 10.1093/ecco-jcc/jjaa236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aschenbrenner D., Quaranta M., Banerjee S., et al. Deconvolution of monocyte responses in inflammatory bowel disease reveals an IL-1 cytokine network that regulates IL-23 in genetic and acquired IL-10 resistance. Gut. 2021;70:1023–1036. doi: 10.1136/gutjnl-2020-321731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West N.R., Hegazy A.N., Owens B.M.J., et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich M., Pohin M., Jackson M.A., et al. IL-1-driven stromal–neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat Med. 2021;27:1970–1981. doi: 10.1038/s41591-021-01520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peyrin-Biroulet L., Sandborn W., Sands B.E., et al. Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE): determining therapeutic goals for treat-to-target. Am Coll Gastroenterol. 2015;110:1324. doi: 10.1038/ajg.2015.233. [DOI] [PubMed] [Google Scholar]
- 22.Argmann C, Hou R, Ungaro RC, et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut. Published online September 15, 2022. https://doi.org/gutjnl-2021-326451. [DOI] [PMC free article] [PubMed]
- 23.Gisbert J.P., Chaparro M. Predictors of primary response to biologic treatment [anti-TNF, vedolizumab, and ustekinumab] in patients with inflammatory bowel disease: from basic science to clinical practice. J Crohns Colitis. 2020;14:694–709. doi: 10.1093/ecco-jcc/jjz195. [DOI] [PubMed] [Google Scholar]
- 24.Greuter T., Rieder F., Kucharzik T., et al. Emerging treatment options for extraintestinal manifestations in IBD. Gut. 2021;70:796–802. doi: 10.1136/gutjnl-2020-322129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungar B., Levy I., Yavne Y., et al. Optimizing anti-TNF-α therapy: serum levels of infliximab and adalimumab are associated with mucosal healing in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2016;14:550–557.e2. doi: 10.1016/j.cgh.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 26.Coorevits L., Baert F.J., Vanpoucke H.J.M. Faecal calprotectin: comparative study of the Quantum Blue rapid test and an established ELISA method. Clin Chem Lab Med. 2013;51:825–831. doi: 10.1515/cclm-2012-0386. [DOI] [PubMed] [Google Scholar]
- 27.Mohammed Vashist N., Samaan M., et al. Endoscopic scoring indices for evaluation of disease activity in ulcerative colitis. Cochrane Database Syst Rev. 2018;1:CD011450. doi: 10.1002/14651858.CD011450.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daperno M., D’Haens G., Van Assche G., et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc. 2004;60:505–512. doi: 10.1016/s0016-5107(04)01878-4. [DOI] [PubMed] [Google Scholar]
- 29.De Vos M., Louis E.J., Jahnsen J., et al. Consecutive fecal calprotectin measurements to predict relapse in patients with ulcerative colitis receiving infliximab maintenance therapy. Inflamm Bowel Dis. 2013;19:2111–2117. doi: 10.1097/MIB.0b013e31829b2a37. [DOI] [PubMed] [Google Scholar]
- 30.Parekh S., Ziegenhain C., Vieth B., et al. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience. 2018;7:giy059. doi: 10.1093/gigascience/giy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman A.M., Steen C.B., Liu C.L., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan J., Upadhye S., Worster A. Understanding receiver operating characteristic (ROC) curves. Can J Emerg Med. 2006;8:19–20. doi: 10.1017/s1481803500013336. [DOI] [PubMed] [Google Scholar]
- 33.Kuleshov M.V., Jones M.R., Rouillard A.D., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.