Abstract
Background
People with mitochondrial disease (MtD) are susceptible to metabolic decompensation and neurological symptom progression in response to an infection. Increasing evidence suggests that mitochondrial dysfunction may cause chronic inflammation, which may promote hyper-responsiveness to pathogens and neurodegeneration. We sought to examine transcriptional changes between MtD patients and healthy controls to identify common gene signatures of immune dysregulation in MtD.
Methods
We collected whole blood from a cohort of MtD patients and healthy controls and performed RNAseq to examine transcriptomic differences. We performed GSEA analyses to compare our findings against existing studies to identify commonly dysregulated pathways.
Results
Gene sets involved in inflammatory signaling, including type I interferons, interleukin-1β and antiviral responses, are enriched in MtD patients compared to controls. Monocyte and dendritic cell gene clusters are also enriched in MtD patients, while T cell and B cell gene sets are negatively enriched. The enrichment of antiviral response corresponds with an independent set of MELAS patients, and two mouse models of mtDNA dysfunction.
Conclusions
Through the convergence of our results, we demonstrate translational evidence of systemic peripheral inflammation arising from MtD, predominantly through antiviral response gene sets. This provides key evidence linking mitochondrial dysfunction to inflammation, which may contribute to the pathogenesis of primary MtD and other chronic inflammatory disorders associated with mitochondrial dysfunction.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-023-04180-w.
Keywords: Anti-viral signaling, Inflammation, Interferon, Mitochondrial disease, PBMCs
Background
In mitochondrial disease (MtD), a bidirectional relationship between MtD and systemic inflammation emerges, wherein mitochondrial dysfunction may trigger inflammatory cascades, which may then reciprocally contribute to the pathogenesis of MtD. Mouse models have linked primary mitochondrial dysfunction and systemic inflammation. PolgD257A/D257A mutator mice (hereafter: Polg mice), which accumulate mtDNA mutations causing a gradual reduction in mitochondrial respiration [1], display aberrant type I interferon (IFN-I) responses in the innate immune axis leading to immunometabolic dysfunction, accelerated aging, and reduced lifespan [2]. The Ndufs4−/− mouse, a model of neurodegenerative MtD, is also marked by widespread inflammation [3], including increases in serum levels of inflammatory cytokines (IFN-ɣ and IL-6), inflammatory markers in the skin and liver, and numbers of activated microglia [4]. These responses may be initiated in part through mitochondrial components acting as damage-associated molecular patterns (DAMPs) to activate pattern recognition receptor (PRR) signaling, for example mtDNA activation of the cGAS/STING antiviral response or the NLRP3 inflammasome [5–7], which can trigger the production and release of IFN-I and interleukin-1β (IL-1β). In primary MtD, mitochondrial dysfunction may cause these pathways may be continuously activated, leading to chronic inflammation.
Chronic inflammation contributes to numerous disorders, including cardiovascular and metabolic disease, cancer, and neurodegeneration [8, 9]. Neurodegenerative diseases, such as many forms of MtD, may present a uniquely damaging intersection between inflammation and mitochondrial dysfunction. Pro-inflammatory cytokines, released during systemic inflammation, reach the central nervous system (CNS) via multiple pathways including the blood brain barrier, the choroid plexus, and the vagus nerve [10], and may modulate region-specific immune cell activation in the brain [11, 12], leading to microglial activation, cytotoxicity, and immune dysregulation [10, 13]. Microglial activation releases cytokines, such as TNFɑ, which impair neuronal mitochondria, causing both oxidative stress and activating additional inflammatory signaling [14]. Recent studies have demonstrated that depletion of leukocytes, including microglia, abrogates neuronal death in the Ndufs4−/− mouse [15, 16]. Consequently, primary mitochondrial defects may initiate systemic and CNS inflammation, which may contribute to neuronal damage observed in patients with MtD, which manifests clinically as seizures, developmental regression, and degeneration.
To date, most studies on systemic inflammation in MtD have been performed in model organisms. In this study, we aimed to determine if increased inflammation or inflammatory signaling was also observable in human MtD. We performed RNAseq on peripheral blood mononuclear cells (PBMCs) in a heterogeneous group of patients with MtD and controls, using gene set enrichment analysis (GSEA) to identify positively and negatively enriched transcriptional signatures in MtD. GSEA has been extensively validated as a method to identify patterns of gene expression with robust biological relevance [17]. We compared those RNA signatures with those from transcriptomic studies of mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episode (MELAS) patients and two mouse models of mitochondrial dysfunction. Across all four studies we observed enrichment of immune activation and inflammatory gene sets, particularly in antiviral pathways.
Methods
Participants
All 81 participants were consented and enrolled in an IRB approved longitudinal natural history study of viral infection and immunity in children with MtD (NIH MINI Study, NCT01780168, www.clinicaltrials.gov) and evaluated at the NIH Clinical Center. Participant characteristics are shown in Table 1. The diagnosis of MtD was made by the referring provider (i.e., neurologist, clinical geneticist), and Modified Walker criteria score of “probable” (P) or “definite” (D) was assigned. The mean age of the control and MtD cohorts were 14.2 (Std dev = 10.6) and 18.4 (Std dev = 16.1) years of age (P = 0.15), respectively. Molecular testing was available for 30 out of 32 patients (94%) with MtD. The MtD cohort was divided into “treated” (n = 22) or “untreated” (n = 9), based on concurrent medications at the time of sampling. For this study, treatment includes any one or more of the following mitochondria-targeted compounds: Coenzyme Q10 (CoQ10), Sodium bicarbonate, EPI-743, B vitamins (B complex, or B2, B6, B9, or B12), and L-carnitine, collectively termed as “MitoCocktail”. Importantly, MitoCocktail treatment does not exclude the presence of other supplements or medications. “Untreated” indicates the absence of any known MitoCocktail treatments, or other medications or supplements with known direct mitochondrial action.
Table 1.
Patient demographic information
| Subject ID | Age at sample | Sex | Age category | Group | Walker score | Gene | MitoCocktail |
|---|---|---|---|---|---|---|---|
| MINI_01 | 22.9 | F | Adult | Control | |||
| MINI_02 | 12 | M | Ped | Control | |||
| MINI_03 | 51.49 | F | Adult | Control | |||
| MINI_04 | 7.1 | F | Ped | Control | |||
| MINI_05 | 10.38 | F | Ped | Control | |||
| MINI_06 | 12.82 | F | Ped | Control | |||
| MINI_07 | 3.37 | F | Ped | Control | |||
| MINI_08 | 5.52 | M | Ped | Control | |||
| MINI_09 | 5.62 | F | Ped | Control | |||
| MINI_10 | 8.44 | M | Ped | Control | |||
| MINI_11 | 14.72 | M | Ped | Control | |||
| MINI_12 | 3.99 | F | Ped | Control | |||
| MINI_13 | 6.12 | F | Ped | Control | |||
| MINI_14 | 6.88 | F | Ped | Case | D | ATP6 | N |
| MINI_15 | 14.96 | M | Ped | Case | D | PMPCB | Y |
| MINI_16 | 14.55 | F | Ped | Control | |||
| MINI_17 | 10.72 | F | Ped | Case | D | ATP6 | Y |
| MINI_18 | 58.38 | F | Adult | Case | D | TYMP | Y |
| MINI_19 | 53.68 | F | Adult | Case | D | mtDNA deletion | N |
| MINI_20 | 4.58 | M | Ped | Case | D | MT-ND3, MT-ND2 | Y |
| MINI_21 | 8.71 | F | Ped | Case | D | MT-RNR1 | Y |
| MINI_22 | 20.71 | M | Adult | Case | D | NUBPL | Y |
| MINI_23 | 35.2 | M | Adult | Case | D | ATP6 | Y |
| MINI_24 | 11.19 | M | Ped | Case | D | NDUFS2 | Y |
| MINI_25 | 6.64 | F | Ped | Case | D | HIBCH | Y |
| MINI_26 | 34.42 | F | Adult | Case | P | COXIII; MT-CYB | Y |
| MINI_27 | 10.52 | M | Ped | Case | P | MT-CYB | Y |
| MINI_28 | 32.82 | F | Adult | Control | |||
| MINI_29 | 35.81 | F | Adult | Control | |||
| MINI_30 | 32.92 | M | Adult | Control | |||
| MINI_31 | 5.87 | F | Ped | Control | |||
| MINI_32 | 9.23 | M | Ped | Control | |||
| MINI_33 | 12.95 | M | Ped | Control | |||
| MINI_34 | 10.88 | M | Ped | Control | |||
| MINI_35 | 4.99 | M | Ped | Control | |||
| MINI_36 | 3.43 | M | Ped | Control | |||
| MINI_37 | 29.18 | F | Adult | Case | P | MT-ND4 | Y |
| MINI_38 | 13.62 | F | Ped | Control | |||
| MINI_39 | 16.27 | M | Ped | Control | |||
| MINI_40 | 15.21 | M | Ped | Control | |||
| MINI_41 | 4.76 | F | Ped | Case | D | NUBPL | N |
| MINI_42 | 8.38 | M | Ped | Case | P | WARS2, RRM2B | Y |
| MINI_43 | 11.91 | M | Ped | Case | P | MT-RNR2; MT-ND5 | Y |
| MINI_44 | 2.51 | F | Ped | Case | P | unknown | Y |
| MINI_45 | 38.39 | F | Adult | Case | D | MT-TL1; MT-TS1 | Y |
| MINI_46 | 7.97 | F | Ped | Case | D | MT-TL1; MT-TS1 | Y |
| MINI_47 | 11.19 | M | Ped | Case | P | TK2; COX10; SCO1 | N |
| MINI_48 | 7.07 | M | Ped | Case | D | SDHA | N |
| MINI_49 | 14.94 | M | Ped | Case | P | MGME1 | Y |
| MINI_50 | 14.5 | F | Ped | Case | P | MT-ND4; MT-TV | Y |
| MINI_51 | 6.75 | M | Ped | Case | P | NDUFA9; NDUFS2 | N |
| MINI_52 | 21.85 | F | Adult | Case | D | MFN2; MT-ND6 | N |
| MINI_53 | 47.26 | F | Adult | Case | D | MFN2 | N |
| MINI_54 | 12.04 | F | Ped | Case | P | MT-ND1; MT-ND5 | Y |
| MINI_55 | 51.62 | M | Adult | Case | D | MT-ATP6 | Y |
| MINI_56 | 4.06 | M | Ped | Case | D | SURF1 | Y |
| MINI_57 | 4.82 | M | Ped | Case | P | unknown | Y |
| MINI_58 | 35.94 | F | Adult | Control | |||
| MINI_59 | 7.05 | F | Ped | Control | |||
| MINI_60 | 3.82 | M | Ped | Control | |||
| MINI_61 | 24.31 | F | Adult | Control | |||
| MINI_62 | 22.61 | F | Adult | Control | |||
| MINI_63 | 15.15 | F | Ped | Control | |||
| MINI_64 | 35.52 | M | Adult | Control | |||
| MINI_65 | 21.97 | M | Adult | Control | |||
| MINI_66 | 21.93 | M | Adult | Control | |||
| MINI_67 | 19.88 | M | Adult | Control | |||
| MINI_68 | 8.5 | F | Ped | Control | |||
| MINI_69 | 3.81 | M | Ped | Control | |||
| MINI_70 | 9.26 | M | Ped | Control | |||
| MINI_71 | 15.45 | F | Ped | Control | |||
| MINI_72 | 10.99 | F | Ped | Control | |||
| MINI_73 | 6.56 | F | Ped | Control | |||
| MINI_74 | 10.79 | F | Ped | Control | |||
| MINI_75 | 4.95 | F | Ped | Control | |||
| MINI_76 | 9.05 | M | Ped | Control | |||
| MINI_77 | 13.86 | M | Ped | Case | D | mtDNA deletion | N |
| MINI_78 | 8.81 | F | Ped | Control | |||
| MINI_79 | 10.66 | F | Ped | Control | |||
| MINI_80 | 4.95 | M | Ped | Control | |||
| MINI_81 | 8.91 | M | Ped | Control |
Comparative study selection
Transcriptomic datasets were identified and selected from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database. GEO was queried using search terms for mitochondrial disease, human or mouse organism, and excluding iPSC and cell line studies. Search terms for mitochondrial disease included “mito*”, “mtDNA”, “MELAS”, “MERFF”, “PEO”, “Leigh”, “Alpers”, “Barth”, “CPEO”, “KSS”, “LHON”, “MIRAS”, “MNGIE”, “NARP”, “Polg”, “SANDO”, “SCAD”, and “TK2D”.
Transcriptomic analysis
PBMC were isolated from 5 mL whole blood using LeucoSep tubes (Greiner Bio-one) and Ficoll-Paque Plus (GE Healthcare) for density gradient centrifugation, before lysis in TRIzol (Thermo Fisher, Waltham, MA). For RNA extraction samples were batched according to their age, gender and case/control status, and two reference samples were simultaneously processed with each batch. Total RNA was isolated and purified with miRNeasy kit (Qiagen, Hilden, Germany), with RNA quality and quantity estimated using Nanodrop (Thermo Scientific, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Stranded cDNA sequencing libraries were generated with TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA) following the manufacturer’s instructions. Briefly, 500 ng of total RNA was used for mRNA selection. After the reverse transcription to 1st strand cDNA, strand info was reserved with dUTPs during 2nd strand synthesis. The dsDNA fragments then had the addition of a single 'A' base and subsequent ligation of the adapter. The products were then purified and enriched with PCR to create the final cDNA library. The library was qualified with Agilent Bioanalyzer and quantified with Qubit 2.0 fluorometer. The cluster generation and paired-end (2 × 75 bp) sequencing was run on Illumina HiSeq 3000 at NHLBI Sequencing Core. All 81 barcoded samples were pooled for one single run, which yielded at least 25 M passed filter paired reads per sample.
Computational analysis
Sequence alignment, quality control filtering, and count matrix generation were performed using STAR [18], QoRTs [19], and RSEM [20] on the NIH HPC Biowulf cluster. All subsequent statistical analysis and graphical presentations were performed in R (https://cran.r-project.org/). RSEM-corrected transcript counts were imported using tximport [21], and differentially expressed genes were identified using DESeq2 [22]. Preliminary gene set enrichment analysis (GSEA) was performed using clusterProfiler [23] and visualized using GOplot [24]. Subsequent GSEA analysis used the fgsea [25] package and blood transcription module (BTM) gene sets [26]. Each BTM is a set of genes, which has been shown to show coherent expression across many biological samples [27, 28]. Gene set variation analysis (GSVA) was used to quantify participant level variation in signatures and test correlation with participant age [29] (Additional file 1: Figure S1C). GEO2R and GEOquery [30] were used to perform differential expression analysis from gene expression microarray data in LIMMA [31], whereas additional RNAseq data was analyzed with DESeq2.
Results
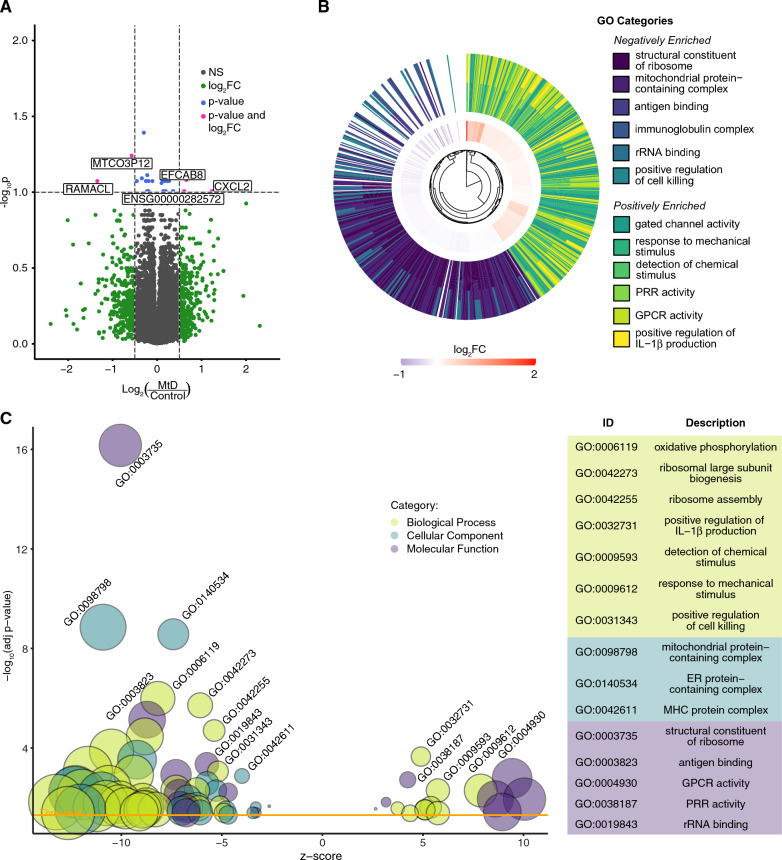
Mouse models of mitochondrial dysfunction have revealed immune signatures marked by elevated inflammation and interferon responses [2, 3, 7, 15]. To understand whether similar transcriptomic signatures occur in people with MtD (Table 1), we performed bulk RNAseq on PBMCs. All participants were in their usual state of health at the time of collection and did not display any symptoms or signs of infection. We examined sample variance using a sample distance correlation matrix of all control participants and MtD patients (Additional file 1: Fig. S1A). Through this analysis, we identified two females (one control, one MtD) that segregated from other samples, and excluded them from subsequent analysis. Using PCA, we observed that our samples do not cluster into diagnosis group (Additional file 1: Fig. S1B). Differential expression analysis identified differentially expressed genes (DEGs) between control and MtD groups (Additional file 2: Table S1). With a threshold of p < 0.1 and log2 fold change (log2FC) of <|0.5|), we detected few DEGs including CXCL2 (Fig. 1A). By ranking the top 50 DEGs by t statistic, we found that the differences in relative expression between these groups was able to improve clustering of control and MtD groups (Additional file 1: Fig. S1C).
Fig. 1.
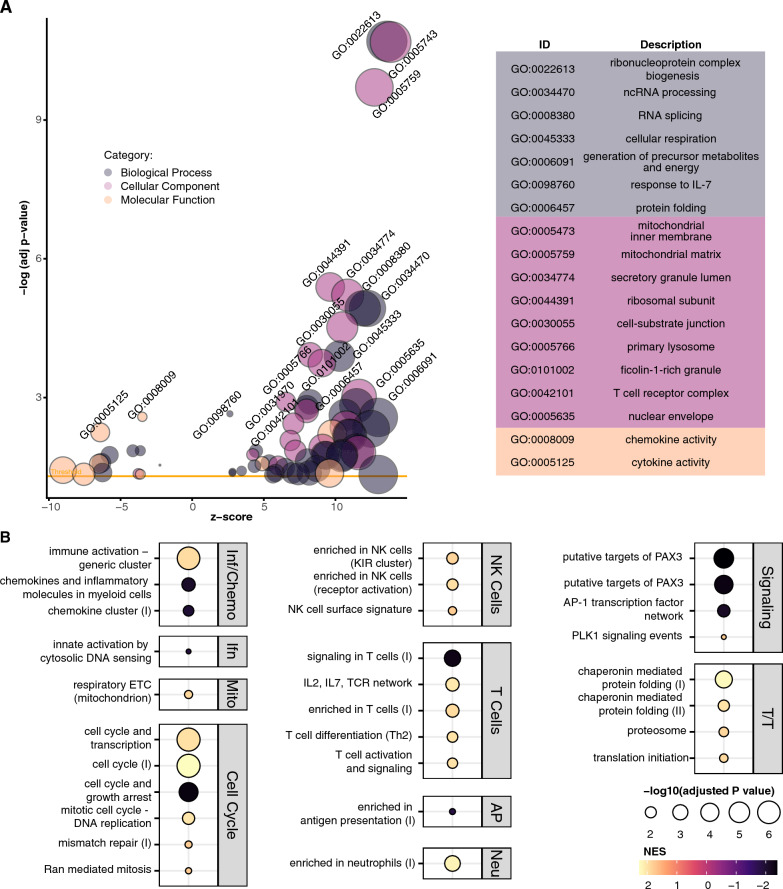
GSEA demonstrates that MtD patients have a positive enrichment in immune activation-associated pathways. A Volcano plot of differential expression analysis between MtD (n = 31) and control participants (n = 48). Log2FC threshold shown >|0.5|, adjusted p value threshold shown at 0.1. B Circular hierarchical cluster plot of top six most significant positive and negative GO categories and associated genes. Inner ring represents log2FC of genes belonging to all twelve categories. Outer ring illustrates the GO functional annotation of each gene. C Bubble plot of significant GO terms following GSEA analysis and reduction to consolidate redundant terms. Bubble size reflects the size of the gene set, while bubble color reflects the GO category of each set. Z score is derived from the comparison of upregulated and downregulated genes in the set. Yellow line indicates the significance threshold of adjusted p < 0.05. For clarity, only a subset of positive and negative enrichment sets is labeled (see Additional file 3: Table S2 for a full list)
To identify patterns of differential gene expression, we performed GSEA using Gene Ontology (GO) categories. This analysis revealed significant positive and negative enrichment of 172 gene sets (adjusted p < 0.05) (Additional file 3: Table S2). In the six most significant positive and negatively enriched categories, we found decreased expression of genes in ribosomal, mitochondrial, B cell and natural killer (NK cell) categories, and increased expression of genes related to stimulus response, ion channel and G protein-coupled receptor (GPCR) activation, pattern recognition receptors (PRR), and interleukin-1β (IL-1β) production (Fig. 1B). Positively enriched gene sets included many involved in immune activation and signaling, including IL-1 and IL-1β production (n = 6), PRR and Toll-like receptor (TLR) binding (n = 2), IFN-β production (n = 2) and viral response regulation (n = 2). Negatively enriched gene sets included mitochondrial proteins and complexes (n = 28), ribosomes and translation (n = 30), natural killer (NK) cell (n = 9), B cell (n = 3), and major histocompatibility complex (MHC) activity (n = 8) (Fig. 1C).
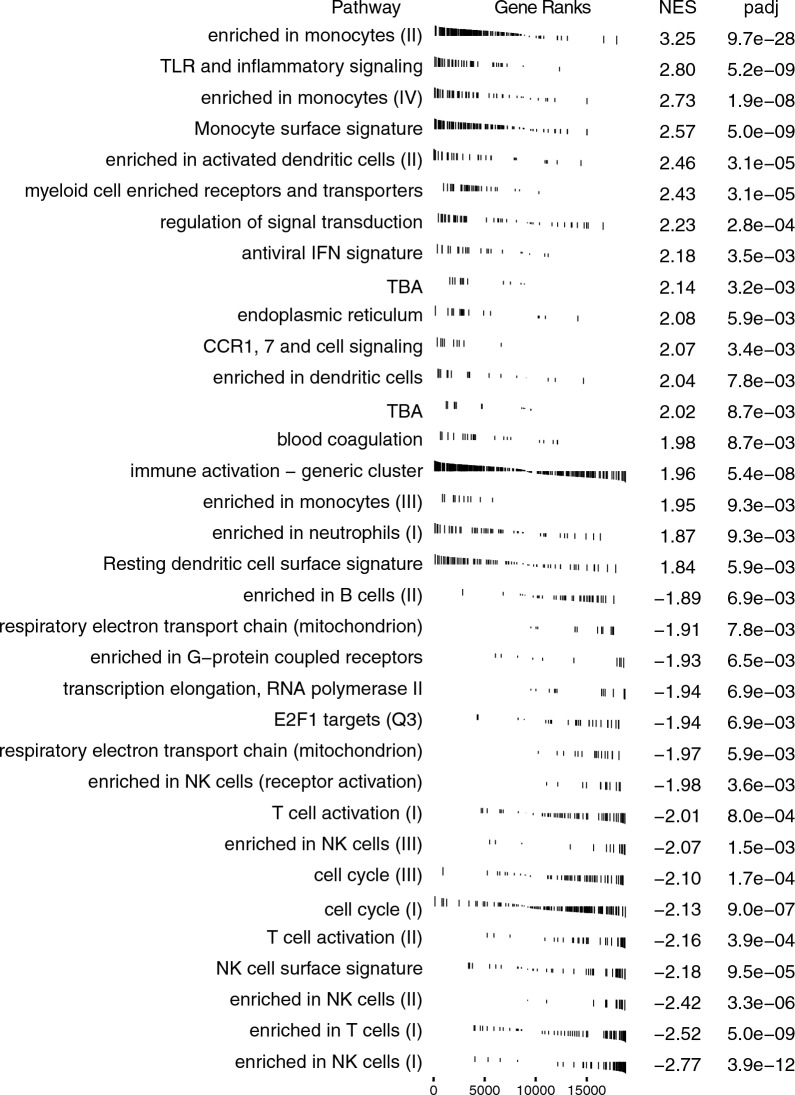
As these results suggested immune and translational dysregulation in our MtD patients, we performed GSEA using blood transcriptional modules (BTMs) to identify targeted pathway enrichment [26]. We identified 62 significant gene sets (Additional file 4: Table S3). Examining the consensus pathways from these modules, we observed a positive enrichment in monocyte, TLR, IFN, and immune activation clusters, and negative enrichment in mitochondria, transcription and translation, and NK and T cell clusters (Fig. 2). We also found positive enrichment of inflammatory response, type I IFN response, and innate antiviral response modules, and negative enrichment of plasma cell/immunoglobulins and B cell modules trending toward significance (p ≤ 0.076).
Fig. 2.
Blood transcriptional module gene ranks. GSEA plots of consolidated BTMs illustrating gene ranks in each transcription set. Left-shifted genes have a positive fold change and are found at the beginning of the gene rank list (positive enrichment), while right-shifted genes have a negative fold change and are found at the end of the gene list (negative enrichment). NES and adjusted p value (padj) for each consolidated BTM indicated at right
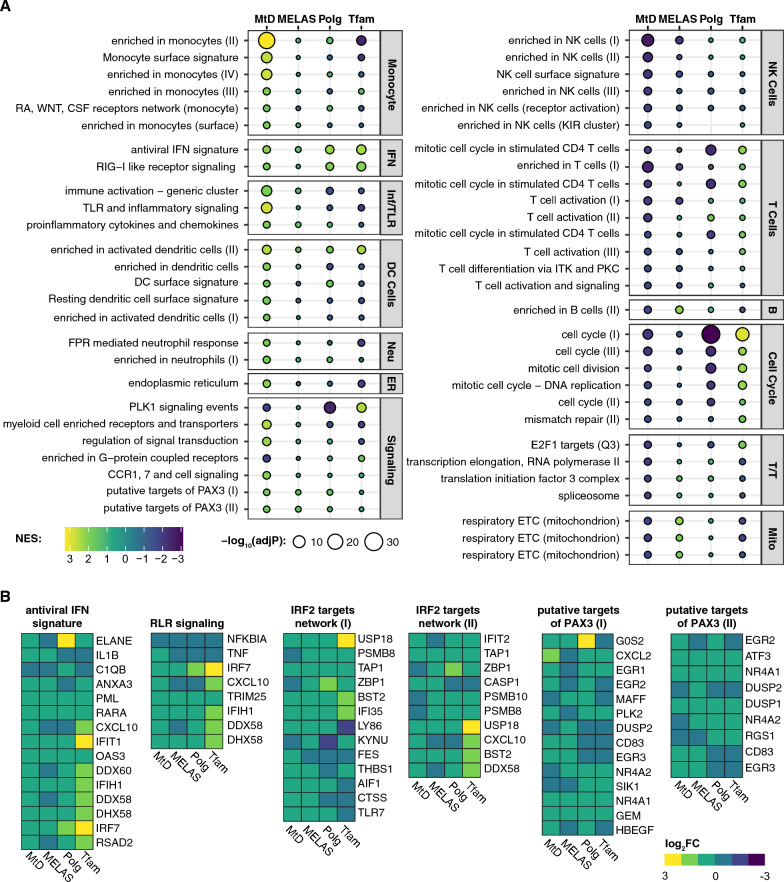
We compared our findings against three other studies: whole blood from MELAS patients [32], bone marrow-derived macrophages (BMDMs) from Polg mutator mice [2], and mouse embryonic fibroblasts from Tfam± mice [7]. We performed BTM GSEA on these sample sets and compared the enrichment of the previously identified MtD-significant modules (Fig. 3A). Examining mitochondrial clusters confirms previously observed trends—MELAS subjects and Polg mice have a positive enrichment, while MtD patients and Tfam± MEFs have a negative enrichment. We discovered key overlaps between MtD patients and the previous MELAS study, including negative enrichment in NK cell and T cell modules. Positive enrichment of monocyte, dendritic cells, and neutrophils are unique to MtD patients, though immune and inflammatory cluster enrichment is shared by MtD and MELAS patients. While these modules were not significant in MELAS patients, related viral sensing and IRF2 targets clusters were (Additional file 4: Table S3). Putative PAX3 target gene sets were positively enriched across all four datasets. We examined the genes that compose antiviral and RIG-I like receptor (RLR) (IFN-related), IRF2 targets, and putative PAX3 target modules (Fig. 3B). In IFN gene sets, we observed increased expression of core cluster genes PML, RARA, IFIT1, OAS3, IFIH1, DHX58, IRF7, and TRIM25 in all four datasets. In IRF2 targets, which were significantly enriched in MELAS subjects, Polg mice, and Tfam± MEFs, USP18, TAP1, BST2, IFI35 were consistently increased. Finally, in putative PAX3 targets, NR4A1, GEM, ATF3, and DUSP1 were increased in all four datasets.
Fig. 3.
BTM enrichment of MtD patients, MELAS patients, and comparable mouse models of MtD. A NES plot of significant BTMs identified in MtD patients. Bubble size represents adjusted p value, bubble color represents normalized enrichment score (NES). Corresponding results from MELAS, Polg, and Tfam GSEA analysis are shown. Modules are grouped into categories shown at right. IFN: interferon; Inf/TLR: Inflammation and Toll-like receptor signaling; Neu: Neutrophils; B: B cells; T/T: Transcription/Translation; Mito: mitochondrion. See Additional file 4: Table S3 for full results. B Log2FC of genes from indicated modules
To find explainable sources of variance within our dataset, we considered the sex and age of our participants, as PCA plots suggested that a proportion of variance could be explained by sex (Additional file 1: Fig. S1B). We assessed the contribution of age by performing gene set variation analysis (GSVA) by evaluating a subset of BTMs across our sample population against age (Additional file 1: Fig. S2). We did not observe a significant trend between age and diagnosis group. We divided our samples based on sex and re-performed differential expression analysis. In male samples, we continued to observe poor within-diagnosis sample correlation (Additional file 1: Fig. S3A) and observed that a significant proportion of variance remained unexplained (Additional file 1: Fig. S3B). We observed similar trends in female samples (Additional file 1: Fig. S3C and S3D). In males, we observed that only four DEGs crossed a log2FC >|0.5| and p < 0.1 (Additional file 1: Fig. S4A and Additional file 2: Table S1). Nonetheless, two downregulated genes, FCRL6 and KLRC4-KLRK1, are linked to cytotoxic T and/or NK cells. Ranking DEGs by t-statistic and examining the relative expression of the top 25 genes improved clustering between male control and MtD (Additional file 1: Fig. S4B). In females, no genes were significantly differentially expressed (Additional file 1: Fig. S4C and Additional file 2: Table S1), but we observed within-diagnosis group clustering when examining the t-statistic ranked top 25 DEGs (Additional file 1: Fig. S4D). Ranking by t-statistic and splitting genes into “upregulated” and “downregulated” categories, we examined potential intersections of control and MtD differential gene expression in the full dataset, males, and females. Using the top 500 genes in each category, both upregulated (Additional file 1: Fig. S5A) and downregulated (Additional file 1: Fig. S5B) intersections demonstrated a majority of uniquely identified genes in the full, male, and female datasets, but with larger intersections between the full and male datasets, and the full and female datasets. At the intersection of all three sets, we found 26 upregulated genes and 27 downregulated genes. The upregulated genes include TMEM106A, TRIM14, and LPAR1, which are associated with positive regulation of NFκB signaling. In contrast, the downregulated genes include RPL4P4, RPS7P1, RPL13AP5, RPS15P4, RPL7P9, and RPL7AP30, which are processed ribosomal pseudogenes. In aggregate, the function of these genes is unclear, however, as new functions have recently been described for several pseudogenes [33], it is possible that these genes are related to dysregulated ribosomal or RNA processing patterns observed in the GSEA (Additional file 3: Table S2).
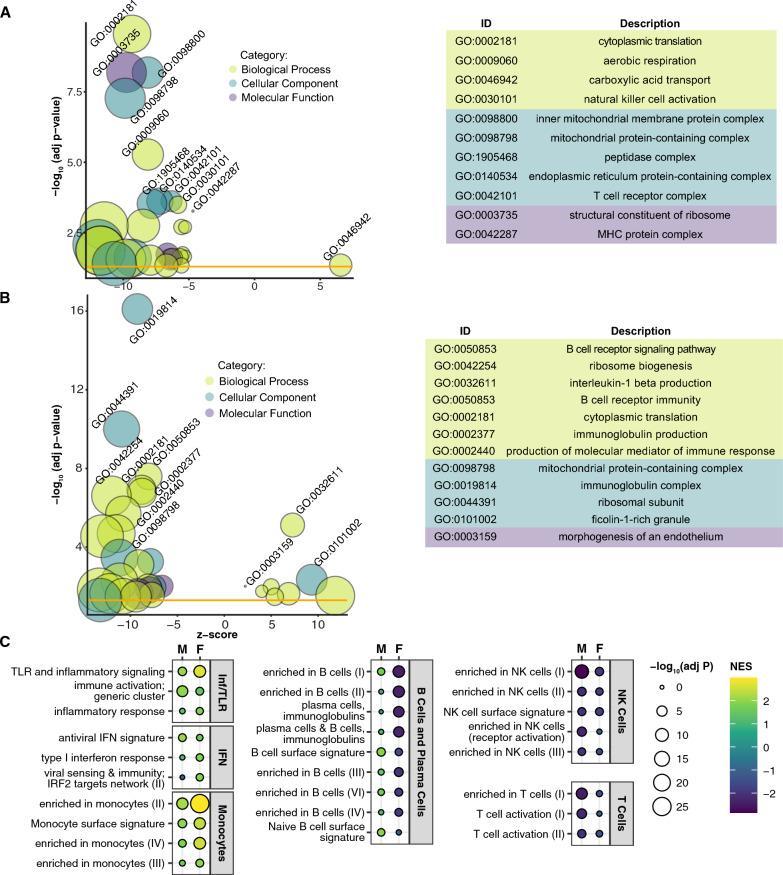
In males, GSEA of GO terms identified 87 significant gene sets, including negative enrichment of gene sets relating to ribosomes (n = 12), mitochondria (n = 23), MHCs (n = 2), NK cells (n = 9) and T cells (n = 3) (Fig. 4A and Additional file 3: Table S2). In females, significant GO GSEA results (n = 78) revealed similar negatively enriched gene sets, including ribosomes (n = 21), mitochondria (n = 3), and B cells (n = 5), but also positively enriched gene sets including IL-1β production (n = 6) and endothelial morphogenesis (n = 2) (Fig. 4B and Additional file 3: Table S2). Based on these disparate results, we compared male and female GSEA using BTMs. While there were many overlapping significantly enriched categories (Additional file 4: Table S3), we observed intriguing differences between the two groups (Fig. 4C). TLR and inflammatory signaling and monocyte gene sets are more positively enriched in female MtD samples than males, and IRF2 targets are significantly enriched in females. In contrast, several B cell gene sets are significantly negatively enriched in females, whereas these sets are either nonsignificant or positively enriched in males. Finally, we observed that while NK and T cell gene sets are negatively enriched in males and females, these sets were more significant in males.
Fig. 4.
Male and female MtD patients show differential enrichment of immune and inflammatory gene sets. A Bubble plot of male controls (n = 22) and MtD patients (n = 16), B of female controls (n = 27) and MtD patients (n = 16) shows significant GO terms following GSEA analysis and consolidation of redundant terms. A, B Bubble size reflects the size of the gene set, while bubble color reflects the GO category of each set. Z score is derived from the comparison of upregulated and downregulated genes in the set. Yellow line indicates the significance threshold of adjusted p < 0.05. See Additional file 3: Table S2 for a full list of significant GO terms. C NES plots from selected modules from males (M) and females (F). Bubble size reflects adjusted p value, bubble color reflects NES. Modules are grouped into categories shown at right. IFN; interferon, Inf/TLR; Inflammation and Toll-like receptor signaling. See Additional file 4: Table S3 for full results from all datasets
We further grouped MtD patients into “treated” (n = 22) and “untreated” (n = 9) conditions, on the basis of receiving any class of mitochondria-related treatment, or "MitoCocktail" (see Methods for treatment details). While differential gene expression analysis again did not detect any single significant genes (Additional file 2: Table S1), GSEA of GO terms detected significant enrichment of 151 categories (Fig. 5A and Additional file 3: Table S2). MtD patients treated with MitoCocktail had significant positive enrichment of mitochondrial gene sets, including oxidative phosphorylation, mitochondrial translation, and purine metabolism (n = 43), ribonucleoproteins and rRNA (n = 16), RNA and RNA splicing (n = 12), cell division (n = 8), and protein folding and localization (n = 14) related gene sets. We also observed positive enrichment of defense response and viral process (n = 4), T cells (n = 3) and response to IL-7 (n = 2) gene sets. Negatively enriched categories included chemokine and cytokine (n = 3), receptor activity (n = 6) and response to IL-1, negative regulation of NFκB, and MHC protein (n = 1, respectively) gene sets. Pursuing these findings in GSEA of BTM terms (Fig. 5B and Additional file 4: Table S3), we found that treated patients had a majority positive enrichment of NK and T cell gene sets, and positive enrichment of protein folding genes. In contrast, chemokine, cytosolic DNA sensing, and PAX3 target gene sets were negatively enriched.
Fig. 5.
Positive enrichment of mitochondrial, NK cell, and T cell gene sets in MtD patients treated with MitoCocktail. GSEA analysis of MtD patients treated with MitoCocktail (n = 22) against MtD patients not undergoing treatment (n = 9). A Bubble plot of significant GO terms in treated MtD patients following GSEA analysis and reduction to consolidate redundant terms. Bubble size reflects the size of the gene set, while bubble color reflects the GO category of each set. Z score is derived from the comparison of upregulated and downregulated genes in the set. Yellow line indicates the significance threshold of adjusted p < 0.05. For clarity, only a subset of positive and negative enrichment sets is labeled (see Additional file 3: Table S2 for a full list). B NES plot of a subset of significant BTMs identified in treated MtD patients. Bubble size represents adjusted p value, bubble color represents normalized enrichment score (NES). Modules are grouped into categories shown at right. Inf/Chemo: Inflammation and Chemokines; Ifn: Interferon/Antiviral Sensing; Neu: Neutrophils; Mito: mitochondrion; T/T: Transcription/Translation. See Additional file 4: Table S3 for full results
Discussion
Our study is among the first to examine the transcriptomic profile of PBMCs from a cohort of MtD patients. We used GSEA to identify key pathway perturbations, and, using both GO terms and validated BTMs, we observed consistent set enrichments. We observed positive enrichment of IL-1β, IFN, and TLR/PRR GO terms, and monocyte, TLR, and IFN signaling BTMs, suggesting elevated basal levels of inflammation in MtD patients. Conversely, we observed negative enrichment of ribosomal proteins, cell killing, and mitochondrial GO terms, and T cell, NK cell, B cell, and transcription and translation BTMs, suggesting suppression of immune system activity coupled with deficits in translation. These data are a valuable resource to inform future study of the mechanisms of peripheral inflammation in patients with MtD.
While we observed few significant DEGs, our sample population is highly heterogenous. We included male and female, adult and pediatric patients and controls, and patients with multiple MtD disease-causing variants, including both mtDNA and nDNA mutations. Few significant DEGs were detected in the MELAS study, despite restricting to adult patients with the same m.A3243G variant [32]. Further, this distinction in study populations likely underlies the oppositional enrichment between MtD and MELAS groups in mitochondrial gene sets. The MELAS study found a positive enrichment of mitochondrial genes, and our confirmation of this finding with our BTM GSEA approach is an important validation of our technique. The significant negative enrichment of mitochondrial genes among our MtD patients may reflect the differing genetic origins of MtD.
Across all four datasets, we observed a positive enrichment of antiviral IFN or viral sensing signaling modules, supporting previous findings. In Tfam± MEFs, a pathway was identified by which mitochondrial stress promotes the release of mtDNA into the cytosol, activating the cGAS-STING-IRF3 antiviral response [7]. In Polg BMDMs, type I IFN signaling genes are basally elevated [2]. Together, these demonstrated increased IFN signaling and inflammatory signaling in multiple mouse models of mitochondrial dysfunction. We found significant enrichment of similar pathways in our MtD patients and in MELAS, suggesting basal IFN and antiviral pathway hyperactivation may be common mechanisms in MtD. We identified significant enrichment of an “innate activation by cytosolic DNA sensing” module in Polg and Tfam datasets, but this module was not enriched in either MtD or MELAS. Consequently, while we confirm enrichment of IFN and inflammatory pathways in human MtD, we emphasize that additional research will be needed to characterize underlying mechanisms.
Our dataset allowed us to perform a preliminary analysis of male and female patients with MtD. While sex did not drive all variance observed, we found interesting differences between males and females. In males, T cell and NK cell GO terms were significantly negatively enriched. In females, B cell and immunoglobulin GO terms were negatively enriched, while IL-1β and ficolin-1 gene sets were positively enriched. BTM terms furthered these findings, demonstrating oppositional enrichment of B cell gene sets between males and females, stronger negative enrichment of T cell and NK cell sets in males, and stronger positive enrichment of monocyte and TLR sets in females. This intriguing segregation of enriched pathways suggests potential sex-specific manifestations of immune dysfunction in MtD. Further, we compared MtD patients treated with MitoCocktail against untreated MtD patients. This comparison suggested that patients receiving treatment have positive enrichment of mitochondrial, ribosomal, and RNA processing gene sets, and of NK and T cell gene sets. In contrast, we observed negative enrichment of chemokine, cytosolic DNA sensing, and PAX3 target gene sets. These results are also preliminary, particularly as our treated and untreated groups are unbalanced. However, these enrichments further support the link between mitochondrial dysfunction and immune dysregulation, as they suggest that treatments supporting mitochondrial function affect immune related gene set expression.
Importantly, there are key limitations to this study. Our findings are based on GO and GSEA methods, which are valuable approaches to identifying gene networks and patterns of subtle changes in gene expression in heterogeneous sample populations. However, these findings are limited to gene expression in uninfected MtD patients and controls, and it will nonetheless be essential for future studies to validate these findings with other methods. For example, gene expression changes or perturbations in cytokine levels in the presence or absence of infection may yield more information about a potential mitochondrial dysfunction-induced hyperinflammatory response, as previously suggested by animal models. Further, though our study suggests potential cell-type specific changes in gene expression in MtD, single cell analysis of PBMC cell types or other tissue-specific immune cells may provide more insight into the mechanisms underlying these observations. We have also presented preliminary data suggesting that there are differences in gene expression patterns in male and female MtD, as well as in treated and untreated MtD. We would like to stress that these findings especially should be validated with other methodologies, in larger patient cohorts, and in other models of MtD. Finally, clinical observations have suggested the coincidence of infection and exacerbation of MtD symptoms. Further animal model and clinical studies directed at this relationship may be necessary to further understand MtD progression, and to characterize the role of immune dysfunction.
Conclusions
Using multiple GSEA approaches, we demonstrate that MtD patients have positively enriched IFN, inflammatory signaling, and monocyte gene sets, and negatively enriched NK cell, T cell, and ribosomal gene sets. Our approach confirms the enrichment of type I IFN signaling in two mouse models of mitochondrial dysfunction and detects a previously unreported enrichment of viral sensing genes in a study of MELAS patients. MtD and mitochondrial dysfunction may induce basal peripheral inflammation, which likely contributes to the susceptibility to elevated inflammatory responses and sepsis in people with MtD. Further, peripheral inflammation exacerbates neuroinflammation and neurodegeneration in animal models [34, 35]. This may point to an important mechanism underlying the acceleration of neurodegeneration following infection in MtD, and the exacerbation of neurodegeneration in disorders with mitochondrial dysfunction.
Supplementary Information
Additional file 1: Figure S1. Sample variance and differential gene expression analysis. Figure S2. Gene set-based comparison of age and diagnosis group. Figure S3. Male and female sample variance analysis. Figure S4. Male and female differential gene expression analysis. Figure S5. Intersection of differentially expressed genes across all control-MtD comparisons.
Additional file 2: Table S1. Differential gene expression results from DESeq2.
Additional file 3: Table S2. GSEA results using GO terms.
Additional file 4: Table S3. GSEA results using BTM terms.
Acknowledgements
The authors would like to acknowledge the United Mitochondrial Disease Foundation and all the patients and volunteers who participated in the study.
Abbreviations
- MtD
Mitochondrial disease
- MELAS
Mitochondrial encephalomyopathy, lactic acid, and stroke-like episodes syndrome
- IFN
Interferon
- DAMP
Damage-associated molecular pattern
- PRR
Pattern-recognition receptor
- CNS
Central nervous system
- PBMCs
Peripheral blood mononuclear cells
- GSEA
Gene set enrichment analysis
- GSVA
Gene set variant analysis
- NES
Normalized enrichment score
- NK cell
Natural killer cell
- TLR
Toll-like receptor
- BTM
Blood transcriptional module
- BMDM
Bone marrow-derived macrophage
- RLR
RIG-I-like receptor
Author contributions
PJM conceived the project and edited the original and revised paper, APW, JST, and RA contributed to the design of the study and edited the original and revised paper, FC conducted original analysis and data visualization, EBW conducted additional analysis, interpretation, and data visualization, and wrote and edited the original and revised paper, YL performed and managed data from Polg mouse studies, EMG-L and SK recruited patients and managed data under the MINI study and edited the original and revised paper, JJ and MLS managed data under the MINI study and edited the original and revised paper, and JC and AM collected and processed samples through CHI. All authors read and approved the final manuscript.
Funding
This research was supported by the National Institutes of Health, as a part of the Intramural Research Programs of NHGRI, NIAID, as well as other institutes supporting the Trans-NIH Center for Human Immunology, Autoimmunity, and Inflammation. This research was supported by award W81XWH-20-1-0150 to APW from Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Programs. Additional support was provided by NIH Grant R01HL148153 to APW.
Availability of data and materials
All summary level data supporting the findings of this study are available within the paper and its Supplementary Information. These data have also been deposited into the database of Genotypes and Phenotypes (dbGaP) under accession number phs003299.v1. Individual level data are not openly available due to patient privacy and are available from the corresponding authors under reasonable request and institutional approval. This study also used publicly available datasets from accession numbers GSE171960, GSE14882, and GSE63767 (GEO). Code for the analyses in this study are available at https://github.com/foocheung/mini_study_2019.
Declarations
Ethics approval and consent to participate
The NIH Institutional Review Board approved the study protocol with approval number 11-HG-0053. Written informed consent from all participants or participant parents was obtained prior to enrollment in the study. All patient data was de-identified before analysis.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
A. Phillip West, Email: awest@tamu.edu.
Peter J. McGuire, Email: peter.mcguire@nih.gov
References
- 1.Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, et al. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 2009;10(2):131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Lei Y, Guerra Martinez C, Torres-Odio S, Bell SL, Birdwell CE, Bryant JD, et al. Elevated type I interferon responses potentiate metabolic dysfunction, inflammation, and accelerated aging in mtDNA mutator mice. Sci Adv. 2021;7(22):eabe7548. doi: 10.1126/sciadv.abe7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z, Wei W, Yang M, Du Y, Wan Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 2014;20(3):483–498. doi: 10.1016/j.cmet.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu AK, Song L, Murray KD, van der List D, Sun C, Shen Y, et al. Mitochondrial complex I deficiency leads to inflammation and retinal ganglion cell death in the Ndufs4 mouse. Hum Mol Genet. 2015;24(10):2848–2860. doi: 10.1093/hmg/ddv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchi S, Guilbaud E, Tait SWG, Yamazaki T, Galluzzi L. Mitochondrial control of inflammation. Nat Rev Immunol. 2022;23:1–15. doi: 10.1038/s41577-022-00760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dela Cruz CS, Kang MJ. Mitochondrial dysfunction and damage associated molecular patterns (DAMPs) in chronic inflammatory diseases. Mitochondrion. 2018;41:37–44. doi: 10.1016/j.mito.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Missiroli S, Genovese I, Perrone M, Vezzani B, Vitto VAM, Giorgi C. The role of mitochondria in inflammation: from cancer to neurodegenerative disorders. J Clin Med. 2020;9(3):740. doi: 10.3390/jcm9030740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suss P, Hoffmann A, Rothe T, Ouyang Z, Baum W, Staszewski O, et al. Chronic peripheral inflammation causes a region-specific myeloid response in the central nervous system. Cell Rep. 2020;30(12):4082–4095. doi: 10.1016/j.celrep.2020.02.109. [DOI] [PubMed] [Google Scholar]
- 11.Alves de Lima K, Rustenhoven J, Da Mesquita S, Wall M, Salvador AF, Smirnov I, et al. Meningeal gammadelta T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nat Immunol. 2020;21(11):1421–1429. doi: 10.1038/s41590-020-0776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horowitz AM, Fan X, Bieri G, Smith LK, Sanchez-Diaz CI, Schroer AB, et al. Blood factors transfer beneficial effects of exercise on neurogenesis and cognition to the aged brain. Science. 2020;369(6500):167–173. doi: 10.1126/science.aaw2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnusen AF, Hatton SL, Rani R, Pandey MK. Genetic defects and pro-inflammatory cytokines in Parkinson's disease. Front Neurol. 2021;12:636139. doi: 10.3389/fneur.2021.636139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Horssen J, van Schaik P, Witte M. Inflammation and mitochondrial dysfunction: a vicious circle in neurodegenerative disorders? Neurosci Lett. 2019;710:132931. doi: 10.1016/j.neulet.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 15.Stokes JC, Bornstein RL, James K, Park KY, Spencer KA, Vo K, et al. Leukocytes mediate disease pathogenesis in the Ndufs4(KO) mouse model of Leigh syndrome. JCI Insight. 2022;7(5):e156522. doi: 10.1172/jci.insight.156522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilar K, Comes G, Canal C, Quintana A, Sanz E, Hidalgo J. Microglial response promotes neurodegeneration in the Ndufs4 KO mouse model of Leigh syndrome. Glia. 2022;70(11):2032–2044. doi: 10.1002/glia.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartley SW, Mullikin JC. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinform. 2015;16(1):224. doi: 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3):100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 25.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021:060012.
- 26.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15(2):195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Joseph Z, Gerber GK, Lee TI, Rinaldi NJ, Yoo JY, Robert F, et al. Computational discovery of gene modules and regulatory networks. Nat Biotechnol. 2003;21(11):1337–1342. doi: 10.1038/nbt890. [DOI] [PubMed] [Google Scholar]
- 28.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14(4):271–280. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Mende S, Royer L, Herr A, Schmiedel J, Deschauer M, Klopstock T, et al. Whole blood genome-wide expression profiling and network analysis suggest MELAS master regulators. Neurol Res. 2011;33(6):638–655. doi: 10.1179/1743132810Y.0000000016. [DOI] [PubMed] [Google Scholar]
- 33.Cheetham SW, Faulkner GJ, Dinger ME. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat Rev Genet. 2020;21(3):191–201. doi: 10.1038/s41576-019-0196-1. [DOI] [PubMed] [Google Scholar]
- 34.Kitazawa M, Oddo S, Yamasaki TR, Green KN, LaFerla FM. Lipopolysaccharide-induced inflammation exacerbates tau pathology by a cyclin-dependent kinase 5-mediated pathway in a transgenic model of Alzheimer's disease. J Neurosci. 2005;25(39):8843–8853. doi: 10.1523/JNEUROSCI.2868-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krstic D, Madhusudan A, Doehner J, Vogel P, Notter T, Imhof C, et al. Systemic immune challenges trigger and drive Alzheimer-like neuropathology in mice. J Neuroinflamm. 2012;9:151. doi: 10.1186/1742-2094-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Sample variance and differential gene expression analysis. Figure S2. Gene set-based comparison of age and diagnosis group. Figure S3. Male and female sample variance analysis. Figure S4. Male and female differential gene expression analysis. Figure S5. Intersection of differentially expressed genes across all control-MtD comparisons.
Additional file 2: Table S1. Differential gene expression results from DESeq2.
Additional file 3: Table S2. GSEA results using GO terms.
Additional file 4: Table S3. GSEA results using BTM terms.
Data Availability Statement
All summary level data supporting the findings of this study are available within the paper and its Supplementary Information. These data have also been deposited into the database of Genotypes and Phenotypes (dbGaP) under accession number phs003299.v1. Individual level data are not openly available due to patient privacy and are available from the corresponding authors under reasonable request and institutional approval. This study also used publicly available datasets from accession numbers GSE171960, GSE14882, and GSE63767 (GEO). Code for the analyses in this study are available at https://github.com/foocheung/mini_study_2019.