Abstract
Pseudomonas putida S-313 can utilize a broad range of aromatic sulfonates as sulfur sources for growth in sulfate-free minimal medium. The sulfonates are cleaved monooxygenolytically to yield the corresponding phenols. miniTn5 mutants of strain S-313 which were no longer able to desulfurize arylsulfonates were isolated and were found to carry transposon insertions in the ssuEADCBF operon, which contained genes for an ATP-binding cassette-type transporter (ssuABC), a two-component reduced flavin mononucleotide-dependent monooxygenase (ssuED) closely related to the Escherichia coli alkanesulfonatase, and a protein related to clostridial molybdopterin-binding proteins (ssuF). These mutants were also deficient in growth with a variety of other organosulfur sources, including aromatic and aliphatic sulfate esters, methionine, and aliphatic sulfonates other than the natural sulfonates taurine and cysteate. This pleiotropic phenotype was complemented by the ssu operon, confirming its key role in organosulfur metabolism in this species. Further complementation analysis revealed that the ssuF gene product was required for growth with all of the tested substrates except methionine and that the oxygenase encoded by ssuD was required for growth with sulfonates or methionine. The flavin reductase SsuE was not required for growth with aliphatic sulfonates or methionine but was needed for growth with arylsulfonates, suggesting that an alternative isozyme exists for the former compounds that is not active in transformation of the latter substrates. Aryl sulfate ester utilization was catalyzed by an arylsulfotransferase, and not by an arylsulfatase as in the related species Pseudomonas aeruginosa.
The sulfur content of aerobic soils is made up almost entirely of sulfonates and sulfate esters of undefined structure. Inorganic sulfate, by contrast, is comparatively poorly represented and constitutes less than 5% of the sulfur in these environments (2). In order to meet their sulfur requirements, soil bacteria must therefore be able to mobilize this organically bound sulfur and assimilate it into cell material. Utilization of the naturally occurring alkanesulfonates taurine, isethionate, and cysteate as sulfur sources is widespread in soil isolates (24, 38), although complete degradation of these compounds as carbon and energy sources appears to be limited to a few species. The ability to hydrolyze alkyl or aromatic sulfate esters is also common in bacteria from soil and water environments. Arylsulfatase is a stable soil enzyme and has been used as a marker for biological activity in soils (37). Alkyl sulfatase enzymes are also very common and were found in 15% of isolates obtained nonselectively from a noncontaminated environment (52).
In contrast to the naturally occurring alkanesulfonates and aliphatic and aromatic sulfate esters, aromatic sulfonates are regarded as xenobiotic compounds (20) and are produced industrially as surfactants, dyestuffs, and cement additives. These compounds are mineralized as a carbon source by a number of bacterial isolates, but the dioxygenase systems that catalyze the desulfonation step are limited in the range of substrates that they will accept. Species that accept arylsulfonates as a sulfur source, by contrast, tolerate a broad range of different substituents on the aromatic ring (12, 21, 54). The best-characterized such isolate is Pseudomonas putida S-313 (54). Desulfonation of arylsulfonates in this strain is catalyzed by a putative monooxygenase system, which converts the substrate quantitatively to the corresponding phenol (21, 54). Although the sulfonatase has not yet been stabilized in vitro, whole-cell experiments have shown that its expression is repressed during growth with preferred sulfur sources such as sulfate, cysteine, or thiocyanate (5). The arylsulfonatase, and/or an arylsulfonate transport system, is therefore a member of a substantial group of proteins whose synthesis is downregulated in the presence of sulfate or cysteine and derepressed during growth with organosulfur sources or during starvation for sulfur (19, 22, 47).
In a previous study (49), we isolated a number of classes of transposon mutants of P. putida S-313 which had lost the ability to grow with aromatic sulfonates as the sulfur source. Mutants from two of these classes appeared to show a pleiotropic phenotype and were deficient not only in growth with arylsulfonates but also in growth with a variety of other organosulfur compounds. We report here the further characterization of one of these mutant classes and the cloning and molecular characterization of the P. putida ssu gene cluster. Our results show that the ssu gene products, and especially SsuD and SsuF, play a role in desulfurization of a variety of organosulfur compounds in P. putida.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All P. putida strains were grown aerobically at 30°C in a succinate-salts minimal medium (22). Sulfur sources were added as described in Results, to a final concentration of 250 to 500 μM. Escherichia coli was grown aerobically in Luria-Bertani medium (36) at 37°C. Growth was monitored as turbidity at 600 or 650 nm. Antibiotics were added at the following concentrations: for E. coli ampicillin at 100 μg/ml, tetracycline at 25 μg/ml, gentamicin at 15 μg/ml, and kanamycin at 25 μg/ml; for P. putida, tetracycline at 25 μg/ml, gentamicin at 25 μg/ml, and kanamycin at 25 μg/ml. When required in sulfate-free medium, kanamycin chloride (25 μg/ml) was prepared and used as previously described (47). Sulfur-limited solid media were prepared by addition of 1.5% molecular biology grade agarose (Eurobio).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| RK4353 | araD139 Δ(argF-lac)U169 deoC1 flhD5301 fruA25 gyrA219 non-9 relA1 rpsL150 | 39 |
| VJS1778 | RK4353 moaA-250::Tn10d(Tc) | V. Stewart |
| BL21(DE3) | F−ompT [lon] hsdSB (rB− mB−; B strain); λDE3 lysogen with T7 RNA polymerase | Novagen |
| P. putida | ||
| S-313 | Prototroph, arylsulfonate desulfonation | 54 |
| SN34 | ssuE::miniTn5Km | 49 |
| PW2 | ssuB::miniTn5Km | This study |
| PW7 | ssuB::miniTn5Km | This study |
| PW10 | ssuC::miniTn5Km | This study |
| Plasmids | ||
| pBluescript II KS | Apr, cloning vector | Stratagene |
| pBBR1MCS-3 | Tcr, broad-host-range cloning vector | 27 |
| pET-24b | Overexpression vector | Novagen |
| pUCP24 | Gmr, broad-host-range cloning vector | 51 |
| pKLAS-25 | atsBA genes of K. pneumoniae in pBluescript | T. Dierks |
| pME4071 | 7.5-kb ClaI fragment from SN34 in pBluescript | This study |
| pME4368 | ssuE gene of P. putida in pET-24b | This study |
| pME4404 | 1.4-kb KpnI-EcoRI fragment from pME4071 in pBluescript | This study |
| pME4420 | 5.2-kb XhoI-HindIII chromosomal DNA fragment from P. putida S-313 in pBluescript | This study |
| pME4421 | 6.1-kb KpnI-HindIII fragment in pBluescript, lsfA ssuEADCBF | This study |
| pME4423 | 5.0-kb XbaI fragment from pME4421 in pBBR1MCS-3, ssuEADCBF parallel to lac promoter | This study |
| pME4424 | 5.0-kb XbaI fragment from pME4421 in pBBR1MCS-3, ssuEADCBF antiparallel to lac promoter | This study |
| pME4426 | atsA gene of P. aeruginosa in pBBR1MCS-3 | This study |
| pME4441 | atsBA genes of K. pneumoniae in pUCP24 | This study |
| pME4431 | pME4423, ΔssuE | This study |
| pME4432 | pME4423, ΔssuA | This study |
| pME4433 | pME4423, ΔssuD | This study |
| pME4443 | pME4424 with 293 bp upstream of ssuE | This study |
| pME4578 | XhoI-MunI deletion of pME4423, ssuF in pBBR1MCS-3 | This study |
Measurement of growth characteristics.
Growth experiments were done at 30°C in microtiter plates with 150 μl of culture using a SPECTRAmax Plus microtiter plate reader with SOFTmax PRO software (Molecular Devices), as previously described (48). The turbidity at 650 nm was measured every 5 min, and the plate was shaken for a period of 30 s before each measurement.
For growth under molybdenum starvation conditions, all buffers were made up in plastic ware instead of glassware, and the cells were precultured overnight in molybdenum-free medium. All glassware used for this experiment was washed consecutively with 3 M HCl (three times), 0.4 M NaOH (three times), and double-distilled water (three times).
DNA manipulations.
Plasmid isolation, restriction enzyme digestion, and transformation of E. coli were carried out using published procedures (1). P. putida was transformed by electroporation in 0.1-cm cuvettes (12.5 kV/cm), using a GenePulser apparatus (Bio-Rad, Hercules, Calif.). Southern analysis was carried out by standard methods (1), using digoxigenin-labeled probes labeled by PCR.
Cloning of the ssuEADCBF genes.
The interrupted genes in mutant strains SN34, PW2, PW7, and PW10 were identified by the transposon rescue techniques previously described (19). Suitably sized transposon-containing DNA fragments were identified by Southern analysis using the digoxigenin-labeled kan gene as a probe. For sequencing and complementation studies, a 7.5-kb ClaI fragment from strain SN34 was cloned into linearized, dephosphorylated pBluescript vector, to give plasmid pME4071. A 1.4-kb KpnI-EcoRI fragment from pME4071 was cloned into pBluescript vector to give pME4404, containing ssuE′. A 5.2-kb XhoI-HindIII chromosomal DNA fragment containing ssu′EADCBF was cloned into pBluescript to give pME4420, and this 5.2-kb fragment was subsequently cut out of pME4420 and transferred into pME4404 linearized with XhoI-BamHI, to give pME4421. The entire operon was recloned into pBBR1MCS-3 as an XbaI fragment to give plasmids pME4423 and pME4424, and deletion plasmids were constructed from pME4423 by removal of the following fragments and blunting where required: pME4431 (ΔssuE), XhoI-SapI deletion; pME4432 (ΔssuA), BsrG1-Bpn 11021 deletion; pME4433 (ΔssuD), StuI-Eam 1105I deletion; and pME4578 (ΔssuEADCB), XhoI-MunI deletion. pME4443 was constructed by inserting a 796-bp PCR fragment generated with primers ssuprofor2 (5′-AAGAGCTCCCCAAAGGTTATCGCG-3′) and ssuprorev1 (5′-TCGTGCAAGCGCTCTTCC-3′) into SacI-SapI-digested pME4424. For overexpression, the ssuE gene was cloned into pET-24b (Novagen) as a 592-bp PCR fragment generated with primers ssuEfor (5′-AAGCGCTCATATGCTGGTCGTC-3′) and ssuErev (5′-AAACTCGAGGATGCTCCAGCGGGCGC-3′) to yield plasmid pME4368. Overexpression was carried out in E. coli BL21(DE3), at 25°C.
Enzyme assays.
Flavin reductase activity was assayed as NAD(P)H-dependent flavin mononucleotide (FMN) reduction at 430 nm as previously described (23). Arylsulfatase activity was measured as conversion of nitrocatecholsulfate to nitrocatechol (λ = 515 nm; ɛ = 13,000 M−1) (4). Arylsulfotransferase assays (500-μl mixtures) contained nitrocatecholsulfate (10 mM), 50 μl of cell lysate (prepared with washed, exponential-phase cells), and acceptor (10 mM) (see Table 3) in 70 mM Tris acetate, pH 9.0. The assay mixture was incubated at 30°C for 60 min, during which period enzyme activity was linear with time (data not shown). Release of 4-nitrocatechol was measured spectrophotometrically at 515 nm. When phenols were used as acceptors, the reading was corrected for the absorbance at 515 nm of a blank assay mixture containing no nitrocatecholsulfate.
TABLE 3.
Arylsulfotransferase activity in P. putida S-313
| Acceptor
compound (10 mM) |
Sulfotransferase activity (nmol/min/mg of protein)a |
|---|---|
| Phenol | 13.7 |
| Tyrosine | 0.2 |
| Dopamine | 12.9 |
| Tyramine | 0.85 |
| Catechol | 18.0 |
| 3,4-Dihydroxybenzoate | 1.5 |
| 4-Hydroxybenzoate | 0.3 |
| ATP | 0.1 |
| Ala-Tyr-Ala | 0.7 |
| Tyr-Ala | 0.4 |
| Serine | 0.0 |
| p-Nitrophenol | 1.5 |
| p-Chlorophenol | 9.2 |
| None | 0.1 |
Sulfotransferase activity was measured as nitrocatechol release from nitrocatecholsulfate, as described in Materials and Methods. The results are averages from three independent experiments.
DNA sequence analysis.
DNA sequences were determined on both strands using an ABI Prism 310 sequencer. Analysis of DNA and protein sequences was done with the Genetics Computer Group package. Signal peptide analysis was carried out by the method of Nielsen et al. (33).
Other methods.
Two-dimensional gel electrophoresis was carried out as previously described (19), and proteins were excised from the gel and concentrated by standard methods (10). N-terminal amino acid sequences were determined by Edman degradation using an Applied Biosystems 120A PTH Sequenator. Protein was measured with the Bio-Rad protein reagent (6), using bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in the GenBank database under accession number AF075709.
RESULTS
Mutants of P. putida S-313 with a pleiotropic defect in organosulfur metabolism.
P. putida S-313 is able to grow with a variety of different organosulfur compounds as the sulfur source for growth. To investigate the genetic basis for growth with sulfonates and sulfate esters, we carried out transposon mutagenesis with miniTn5Km and screened the mutants obtained for loss of the ability to grow either with benzenesulfonate or with 5-bromo-4-chloro-indoxylsulfate (X-sulfate) as the sulfur source. Growth of these strains was then further tested in a sulfur-free succinate-salts medium with a variety of organosulfur compounds as the sole sulfur sources. Four of the mutants identified, strains SN34, PW2, PW7, and PW10, were found not only to be defective in desulfurization of benzenesulfonate or X-sulfate but also to have lost the ability to grow with a variety of other organosulfur compounds, including a range of aromatic and aliphatic sulfonates or aromatic and aliphatic sulfate esters. Growth of strain SN34 was studied in more detail (Table 2), and this strain was also found to be unable to grow with methionine or methionine biosynthetic intermediates as the sulfur source, although the other three mutants could desulfurize these compounds. All of the mutant strains grew as fast as the wild-type strain when sulfate, cysteine, or thiocyanate was supplied as the sulfur source, demonstrating that the defects were not located in the sulfate assimilation pathway. Interestingly, in the presence of taurine (2-aminoethanesulfonate) or cysteate, strain SN34 grew at the same rate as the wild type, though it did not grow with isethionate (2-hydroxyethanesulfonate). This suggests that taurine and cysteate are desulfurized by a pathway separate from that for other alkanesulfonates in this species, as previously reported for E. coli with taurine (47), and that the sulfite released by the taurine desulfonation reaction can be utilized normally by strain SN34. Together, these results showed that the transposon insertions in the mutant strains were in a locus (or loci) which was specifically involved in organosulfur utilization and not in the well-characterized sulfate assimilation pathway (28).
TABLE 2.
Relative growth rates of P. putida strains S-313, SN34, and SN34(pME4423) with different sulfur sources
| Sulfur source | Growth ratea
of strain:
|
||
|---|---|---|---|
| S-313 | SN34 | SN34(pME4423) | |
| Sulfate | 100.0 | 100.0 | 100.0 |
| Cysteine | 90.5 | 97.3 | 91.7 |
| Thiocyanate | 100.6 | 105.3 | 103.6 |
| Benzenesulfonate | 9.8 | 0.0 | 8.6 |
| Toluenesulfonate | 37.7 | 0.0 | 32.2 |
| Pentanesulfonate | 60.9 | 0.0 | 58.6 |
| Methanesulfonate | 79.9 | 3.3 | 101.2 |
| Taurine | 87.2 | 86.4 | 79.9 |
| Cysteate | 60.1 | 54.7 | 58.0 |
| Isethionate | 98.0 | 0.0 | 105.3 |
| Nitrocatecholsulfate | 74.9 | 0.9 | 80.8 |
| Hexylsulfate | 58.9 | 0.0 | 51.5 |
| Homocysteine | 50.3 | 5.0 | 35.5 |
| Methionine | 89.9 | 0.0 | 88.8 |
Growth rates were measured in a succinate-salts growth medium at 30°C in a SPECTRAmax Plus microtiter plate reader, as described in Materials and Methods. Values are relative to the growth rate with sulfate (μ = 0.66 h−1).
Cloning and sequence analysis of the ssuEADCBF genes.
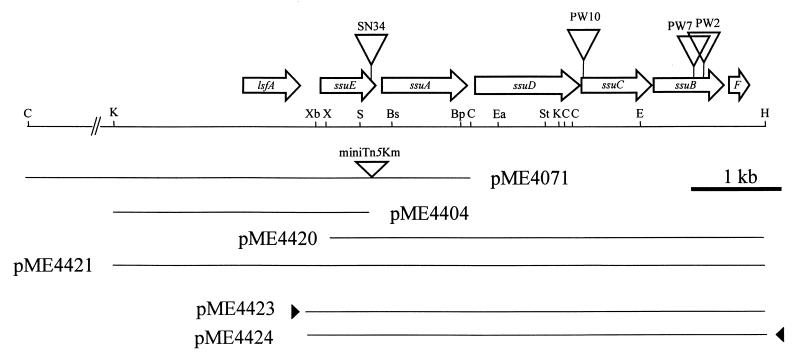
In order to characterize the mutated loci in strain PW2, PW7, PW10, and SN34, the regions flanking the miniTn5 insertions were cloned by transposon rescue techniques and sequenced. Full sequencing of a 5.9-kb fragment (Fig. 1) revealed that the transposon insertions had all occurred in a six-gene operon ssuEADCBF, with SN34 being interrupted 50 bp from the end of the ssuE gene, PW10 being interrupted 100 bp from the start of ssuC, and PW2 and PW7 being interrupted in ssuB (550 and 455 bp from the start of ssuB, respectively). A further open reading frame, lsfA, was identified upstream of ssuE, reading in the same direction as the ssu genes but separated from them by 235 bp. All of the ssu reading frames were preceded by good consensus ribosome binding sites, and the overall GC content of the coding region was 65%. Sequence comparison of the ssu genes revealed that the operon encoded two separate groups of proteins. The first group consists of ssuA, ssuB, and ssuC, which are the genes for a putative ATP-binding cassette (ABC)-type transport system. SsuA is a 200-amino-acid protein showing highest similarity (57% amino acid identity) to the taurine-binding protein (47). This assignment as a periplasmic protein was confirmed by the presence of a putative signal peptide (amino acids 1 to 28). SsuB and SsuC correspond, respectively, to the ATP-binding component (32 to 65% identity to the ATP-binding components of known transporters) and the permease component (25 to 40% identity to known permease components) of such systems. Hydrophobicity analysis of the SsuC protein revealed the presence of six putative transmembrane helices, and SsuB was found to contain the consensus motif expected for the ATP-binding component of a transport system (16).
FIG. 1.
Map of the ssu region of the P. putida chromosome. The positions of the miniTn5Km insertions in strains SN34, PW2, PW7, and PW10 are shown, as are selected restriction sites: Bp, Bpn1102I; Bs, BsrG1; C, ClaI; E, EcoRI; Ea, Eam11051; H, HindIII; K, KpnI; S, SapI; St, StuI. X, XhoI; Xb, XbaI. Several plasmids described in the text are shown, and the location of the lac promoter in the vector is indicated with a solid triangle.
The second group of proteins encoded in the ssu operon are encoded by the ssuD and ssuE genes. These are present in a number of species, and in E. coli they have been shown to encode a two-component desulfonative monooxygenase system. This consists of the NADPH:FMN reductase SsuE and the FMNH2-dependent desulfonative oxygenase SsuD, which catalyzes the desulfonation of unsubstituted alkanesulfonates (13). SsuD displays 22 to 30% identity to other FMNH2-dependent oxygenases, including SnaA (pristinamycin synthase, A subunit, which catalyzes the final step in biosynthesis of the antibiotic pristinamycin [42]), DszA (dibenzothiophene [DBT]) dioxide monooxygenase, which cleaves DBT dioxide to the corresponding hydroxybiphenylsulfinate [15]), and NtaA (nitrilotriacetate monooxygenase [25]). The P. putida ssuD and ssuE gene products are very similar to the MsuE and MsuD proteins of Pseudomonas aeruginosa (30 and 71% amino acid identity, respectively) which are involved in methanesulfonate metabolism in this species (23). Measurements of NADH-dependent FMN reductase activity in crude extracts of strains S-313 and SN34 showed no difference between the two strains, suggesting that more than one FMN reductase may be present in the cell, as has also been observed in P. aeruginosa. From the deletion studies presented below, however, SsuE appears to be required for utilization of aromatic sulfonates, and it may therefore form part of a complex with SsuD.
The distal gene in the ssu gene cluster, ssuF, encodes a small protein (7.6 kDa) with 40 to 43% identity to molybdopterin-binding proteins of Clostridium pasteurianum (18) and which is closely related to molybdopterin-binding proteins of Azotobacter vinelandii (30) and Rhodobacter capsulatus (50).
The gene immediately upstream of the ssu genes, lsfA, encodes a 212-amino-acid protein which belongs to the thiol-specific antioxidant (ahpC-tsaA) family (7), with 44 to 70% identity to the AhpC or TsaA enzymes from the nematode Onchocerca volvulus, yeast, rice, barley, and various bacteria. This large family of related proteins is involved in responses to various types of oxidative stress. The derived N-terminal peptide sequence of LsfA was also 92.9% identical to the N-terminal sequence determined experimentally for protein PA11, one of the sulfate starvation-induced proteins of P. aeruginosa (19, 34). Since expression of protein PA11 is regulated by sulfur supply, it seems likely that lsfA of P. putida is also regulated in the same manner. However, it is probably not cotranscribed with the ssu operon, since sequence analysis revealed a strong terminator signal 40 bp downstream of the lsfA stop codon (see below).
Regulation of SsuF expression by sulfate.
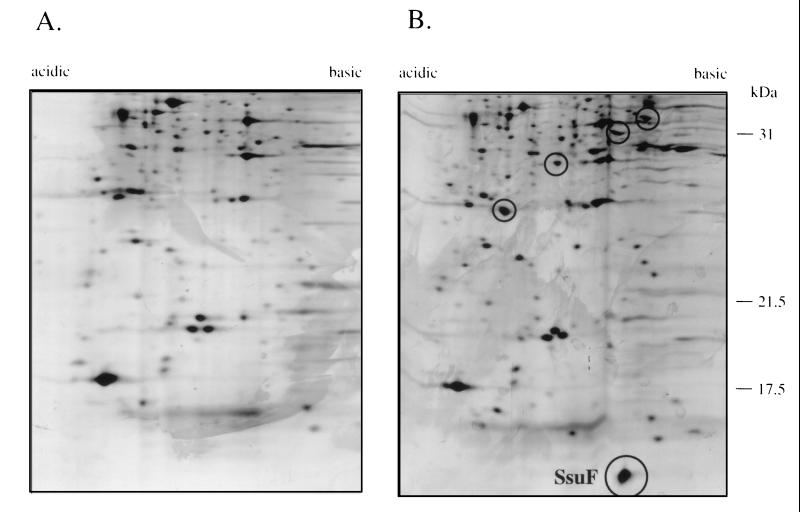
In an earlier study (22) we reported that during growth in the absence of sulfate, cysteine, or thiocyanate, P. putida S-313 responds by synthesis of a set of additional proteins, the sulfate starvation-induced stimulon. The most intense of the proteins detected in the earlier study was a small (7.6-kDa) protein with a pI value of approximately 6.8. This protein was now identified on preparative two-dimensional gels (Fig. 2), excised and concentrated by standard methods (10), and subjected to N-terminal amino acid analysis. The amino acid sequence determined by Edman sequencing was XAINVRNQFKGTVK, which corresponded to the predicted sequence for SsuF. This indicated not only that the ssu operon is expressed only in the absence of sulfate but also that under derepressing conditions the distal gene in this operon is surprisingly strongly expressed.
FIG. 2.
Two-dimensional electropherograms of total cell protein from P. putida S-313. Cells were grown in succinate minimal medium with sulfur for growth provided as inorganic sulfate (A) or toluenesulfonate (B). Proteins that are upregulated during growth in the absence of sulfate are circled, and the SsuF protein is indicated.
SsuF is required for growth with sulfate esters and sulfonates but not with methionine.
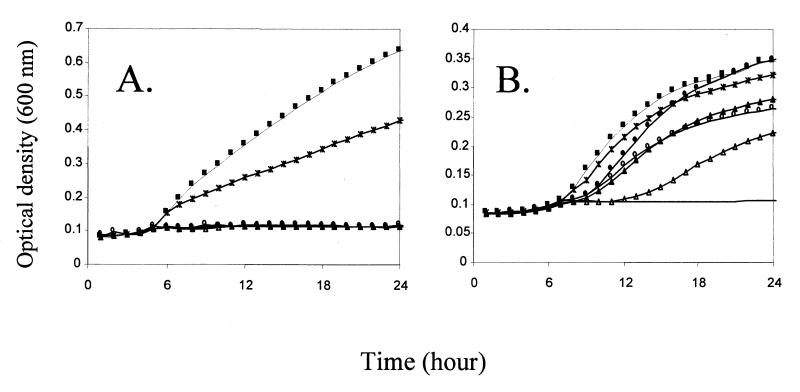
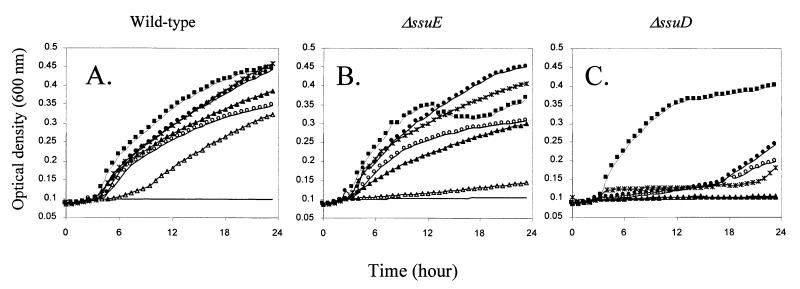
Strains PW2, PW7, and PW10 contained disruptions in the ssuB and ssuC genes (Fig. 1). To test whether the observed phenotype was directly due to the loss of the disrupted genes or to a polar effect on ssuF, mutants PW7 and SN34 were transformed with the ssuF gene on plasmid pME4578. On this construct the ssuF gene is under the control of the lac promoter, which is constitutively expressed in Pseudomonas species. In the presence of ssuF, growth with hexylsulfate or nitrocatecholsulfate was restored in both mutants (Fig. 3). Strain PW7 also regained the ability to utilize sulfonates as the sulfur source when provided with the ssuF gene, but strain SN34 did not, nor did strain SN34 regain the ability to grow with methionine as the sulfur source. This indicated that the SsuF protein is required for growth with sulfonates and sulfate esters but not with methionine. It also provided indirect confirmation that the ssuEADCBF genes are coexpressed as an operon, since the sulfate ester-negative phenotype of strain SN34 was due to the polar effect on ssuF expression of an insertion in ssuE.
FIG. 3.
Growth of P. putida PW7 (A) and P. putida PW7(pME4578) (B) in succinate minimal medium with different sulfur sources. Growth curves were measured in a SPECTRAmax Plus microtiter plate reader, as described in Materials and Methods. Sulfur sources: ■, sulfate; ○, hexylsulfate; ●, nitrocatecholsulfate; ▴, pentanesulfonate; ▵, benzenesulfonate; ∗, methionine. —, no sulfur.
Since the predicted peptide sequence of the 7.6-kDa SsuF protein showed similarity to several molybdopterin-binding proteins, it was tempting to speculate that a molybdoprotein might be involved in sulfate ester and sulfonate metabolism. To test this hypothesis, P. putida S-313 was grown in molybdenum-deficient medium with sulfate as the sulfur source and then transferred to sulfate-free medium with toluenesulfonate as the sole sulfur source. No significant difference in growth pattern was seen between the low-molybdate and high-molybdate media, although it should be noted that complete molybdenum starvation conditions may be difficult to attain (14). However, in a separate experiment we were able to show that an E. coli moaA mutant (strain VJS1778 [39]) which was deficient in molybdopterin biosynthesis was able to grow as well with pentanesulfonate as could the parent strain RK4353. In E. coli, at least, molybdopterin therefore appears not to play a role in the desulfonation process.
The SsuD protein is required for desulfonation of aromatic and aliphatic sulfonates.
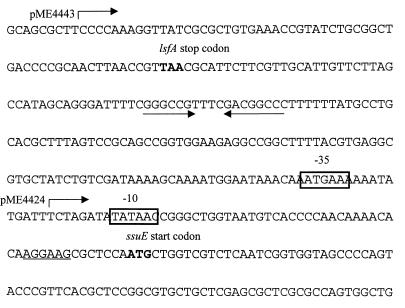
For complementation analysis of strain SN34, the entire ssuEADCBF operon was cloned into the broad-host-range medium-copy-number vector pBBR1MCS-3, to give plasmids pME4423 and pME4424. In pME4423 the ssuEADCBF genes were under the control of the lac promoter of pBBR1MCS-3, while the operon was in the opposite orientation in pME4424 (Fig. 1). These constructs were introduced into strain SN34. Strain SN34(pME4423) displayed the same growth rate as the wild type with all of the sulfur sources tested (Table 2), confirming that the ssuEADCBF operon is sufficient to complement all of the observed growth defects of strain SN34. Introduction of pME4424 into strain SN34 had no effect on the growth characteristics of the mutant strain, suggesting that a part of the ssu promoter region is missing (data not shown). Addition of a further 237-bp fragment to the proximal end of the operon in plasmid pME4424 generated plasmid pME4443 (Fig. 1 and 4). This construct was able to complement the negative growth phenotypes of strain SN34. Sequence analysis revealed that the insert in pME4424 terminated in the middle of a consensus ς70-type promoter about 40 bp upstream of the ssuE translation initiation site but that this promoter was restored in the longer construct, pME4443 (Fig. 4). The presence of a putative rho-independent terminator sequence distal to the lsfA gene also suggests that the lsfA and ssuE genes are transcribed separately.
FIG. 4.
Nucleotide sequence of the intergenic region between the P. putida lsfA and ssuE genes. The stop and start codons of lsfA and ssuE, respectively, are shown; the ribosome binding site of ssuE is underlined; and a putative rho-independent terminator structure downstream of lsfA is indicated with arrows. The −10 and −35 sequences of a putative ς70-dependent promoter are boxed. Plasmids pME4424 and pME4443 begin at the points indicated.
Deletions were then constructed in the ssuE, ssuA, and ssuD genes on pME4423 as described in Materials and Methods, and the resultant plasmids were tested for their ability to complement the growth phenotype of strain SN34. Deletion of ssuA had no effect on growth with any sulfur source tested (not shown). If ssuABC encodes a sulfonate transport system, as suggested above, it must therefore be duplicated on the chromosome. The FMN reductase encoded by ssuE was not required for growth with aliphatic sulfonates or with sulfate esters (Fig. 5B). This agrees with the fact that no decrease in FMN reductase activity was observed in strain SN34, compared with the wild-type strain, and is presumably due to the presence of other FMN reductases in the cell. Surprisingly, growth with aromatic sulfonates required SsuE, and in this more complex enzyme system (49) the SsuE protein may be directly involved in a larger enzyme complex. To confirm that the SsuE protein indeed catalyzed FMN reduction, as expected by comparison to E. coli SsuE and P. aeruginosa MsuE, it was overexpressed in E. coli BL21(DE3) using plasmid pME4368, and FMN reductase activities in cell extracts were measured. SsuE revealed NADPH-dependent FMN reductase activity and lower levels of FAD reductase activity (32% of the value with FMN). No activity was observed with NADH.
FIG. 5.
Growth of derivatives of P. putida SN34 in succinate minimal medium with different sulfur sources. Growth curves were measured in a SPECTRAmax Plus microtiter plate reader, as described in Materials and Methods. (A) SN34(pME4423); (B) SN34(pME4431); (C) SN34(pME4433). Sulfur sources: ■, sulfate; ○, hexylsulfate; ●, nitrocatecholsulfate; ▴, pentanesulfonate; ▵, benzenesulfonate; ∗, methionine. —, no sulfur.
No growth was seen with pentanesulfonate or benzenesulfonate in the absence of SsuD (Fig. 5C). With sulfate esters as the sulfur source, growth was subject to an extremely long lag phase, although the final growth yield was not reduced. We conclude that the SsuD protein is the monooxygenase responsible for cleavage both of alkanesulfonates (as in E. coli [13]; P. putida ssuD restored growth with alkanesulfonates to an in-frame ssuD deletion mutant of E. coli [C. Wietek, unpublished data]) and of arylsulfonates. Growth with methionine or homocysteine was also not observed in the absence of ssuD. P. putida lacks the reverse transsulfuration pathway from methionine to cysteine that is present in P. aeruginosa, and methionine-sulfur is therefore cleaved to methanethiol by methionine lyase and probably converted to cysteine via oxidation to methanesulfonate and oxygenolytic cleavage to sulfite (48). It was unclear why the loss of the SsuD oxygenase had an effect on growth with sulfate esters, so this was examined in more detail.
Hydrolysis of arylsulfate esters in P. putida by an arylsulfotransferase.
To further investigate the role of the ssu gene products in sulfonate and sulfate ester utilization, we wished to compare the levels of enzyme activities responsible for desulfurization of these compounds, including alkanesulfonatase and alkyl- and arylsulfatase activities, in the wild-type and mutant strains. However, this was frustrated by the fact that sulfatases and sulfonatases are expressed only in the absence of sulfate or thiocyanate, i.e., during growth with organosulfur compounds as the sulfur source (5, 22, 48). The pleiotropic growth phenotype of the mutant strains meant that they could not be cultivated under any conditions where the ssu genes were expressed.
Although strain SN34's inability to grow with sulfonates was clearly related to the loss of the SsuD sulfonatase, it was not clear how the loss of growth with sulfate esters could be explained for any of the mutants. In the related organism P. aeruginosa, arylsulfate utilization is dependent on the arylsulfatase protein, an enzyme which requires a posttranslational modification of an active-site formylglycine residue from a cysteine residue (11). The mechanism by which this modification occurs has not yet been elucidated. To test whether any of the ssu gene products might play a role in this modification process, we transformed strains S-313 and SN34 with the arylsulfatase gene (atsA) from P. aeruginosa and with the related atsBA genes from Klebsiella pneumoniae (41). However, no significant difference was seen in sulfatase activities between the wild type and the mutant, ruling out a role for the ssu gene products in this process. In fact, we found that P. putida does not contain an arylsulfatase gene of the type found in P. aeruginosa, since Southern analysis of the P. putida S-313 chromosome using the P. aeruginosa atsA gene as a probe gave no signal, even under reduced-stringency conditions (data not shown). This was a surprising result, since arylsulfatase activity has been previously reported for this isolate (22), although attempts to purify it were unsuccessful and enzyme activity was rapidly lost when the cells were lysed (our unpublished results). However, when phenol was added to the arylsulfatase assay, a dramatic increase in desulfation of nitrocatecholsulfate to nitrocatechol was observed. We therefore postulate that P. putida grows with aromatic sulfates by transferring the sulfate moiety onto an uncharacterized acceptor, from which it can be incorporated into the sulfate assimilation pathway. A range of possible sulfate acceptors were tested for their ability to stimulate arylsulfotransferase activity (Table 3). Using nitrocatecholsulfate as a donor substrate, catechol was the best acceptor, followed by phenol, dopamine, and p-chlorophenol in decreasing order. Tyrosine and tyramine were poor acceptors, and serine did not act as an acceptor at all. Tyrosine-containing di- and tripeptides were not significantly better acceptors than tyrosine alone, but the data do not rule out the possibility that a tyrosine-containing protein is the natural acceptor in P. putida. Furthermore, it is noteworthy that with phenol, tyramine, or 4-hydroxybenzoate as an acceptor, the presence of a second hydroxyl group in the ortho position significantly enhanced activity. p-Chlorophenol was a better acceptor than p-nitrophenol, possibly because p-chlorophenol is expected to be present mainly in the protonated form under the assay conditions used, while p-nitrophenol is not.
DISCUSSION
Desulfurization of aromatic sulfonates by P. putida S-313 is carried out by a monooxygenase system, which incorporates molecular oxygen into the phenol product (54), releasing the sulfur moiety as sulfite (49). It has not yet been possible to stabilize this desulfonating oxygenase activity outside the cell, but a genetic approach has led to the identification of three loci which are required for growth of P. putida S-313 with aromatic sulfonates as the sulfur source (49). The first of these loci, the asf gene cluster, comprised genes for a transcriptional regulator, an electron transport system, and a putative sulfonate-binding protein but not for the desulfonating monooxygenase itself (49). In this report we show that the second of these three loci consisted of the ssuEADCBF genes, containing the FMNH2-dependent monooxygenase gene ssuD, which has previously been associated with alkanesulfonate metabolism in E. coli and Bacillus subtilis (13, 45). For P. putida S-313, we have shown that the ssu operon not only is required for growth with alkanesulfonates but is central to much of organosulfur metabolism. SsuD was required for the desulfonation of alkyl- and arylsulfonates, was involved in growth with sulfate esters but was not essential in this process, and also enabled the cell to grow with methionine and methionine biosynthetic intermediates such as homocysteine. The ssuF gene product was found to be involved in the utilization of aromatic and aliphatic sulfate esters as well as aromatic and aliphatic sulfonates.
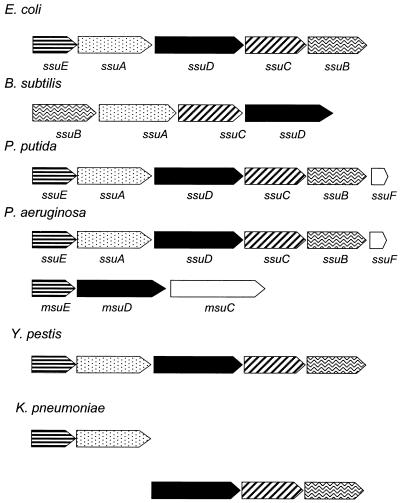
The ssu operon has been identified in several bacterial species, including both gram-negative and gram-positive species and enteric and soil bacteria (45, 46; Pseudomonas Genome Project website [http://www.pseudomonas.com/]) (Fig. 6). The exact ordering of the genes within the operon varies between species, but the oxygenase gene ssuD is always present, together with genes encoding an ABC-type transporter. Except in B. subtilis, an NAD(P)H-dependent FMN reductase gene is also always present. These species can all utilize aliphatic sulfonates as sulfur sources, and in B. subtilis, P. putida, and E. coli mutation of the ssu operon leads to loss of this ability (references 45 and 46 and this report). However, of the species studied, only P. putida S-313 is able to utilize aromatic sulfonates as the sulfur source. We hypothesized that slight differences in the primary structure of the SsuD protein might lead to a relaxation of the substrate specificity of the oxygenase and hence the acceptance of aromatic substrates (the E. coli and P. putida SsuD proteins show only 77% amino acid identity), but as transformation of the P. putida ssu operon into E. coli did not allow the latter organism to desulfonate benzenesulfonate, this was clearly not the case. In fact, growth of P. putida with aromatic sulfonates requires two further gene clusters, consisting of the asfABC genes (encoding a putative reductase, a ferredoxin, and a periplasmic binding protein) and the atsRBC genes (encoding an ABC-type transport system which is also present in P. aeruginosa [accession no. Z48540]) (49). Transformation of the asf operon into P. aeruginosa allowed this organism to grow with benzenesulfonate as the sulfur source (52). Deletion studies (reference 49 and this study) showed that whereas aliphatic desulfonation required only the SsuD and SsuF proteins (SsuE is also presumably involved, but the flavin reductase function is probably duplicated within the cell), the minimum set of proteins required for the aromatic desulfonation reaction was SsuE, SsuD, SsuF, AsfA, and AsfB. Work to reconstitute the enzyme complex with these proteins and characterize the desulfonative enzyme activity in vitro is continuing in our laboratory.
FIG. 6.
Genetic organization of related ssu and
msu operons. The enzymes encoded in each gene cluster are
putative oxygenases (■), NADH-dependent FMN reductases
( ), and
the components of ABC-type transporters: periplasmic solute-binding
proteins (
), ATP-binding proteins
(
), and
the components of ABC-type transporters: periplasmic solute-binding
proteins (
), ATP-binding proteins
( ), and permease proteins
(
). No definite function is known for the products of
the ssuF and msuC genes (reference
23 and this paper). The genes are from E.
coli (46, 47), B. subtilis (45),
P. putida (reference 49 and this paper),
P. aeruginosa (23; Pseudomonas Genome
Project [http://www.pseudomonas.com/), and the unfinished chromosomes
of Yersinia pestis and K. pneumoniae
(Yersinia pestis Sequencing Group, Sanger Centre
[ftp://ftp.Sanger.ac.uk/pub/pathogens/yp/]; Klebsiella
pneumoniae Sequencing Group, University of Washington
Genome Center
[http://genome.wustl.edu/gsc/Projects/bacterial/klebsiella/klebsiella.shtml]).
), and permease proteins
(
). No definite function is known for the products of
the ssuF and msuC genes (reference
23 and this paper). The genes are from E.
coli (46, 47), B. subtilis (45),
P. putida (reference 49 and this paper),
P. aeruginosa (23; Pseudomonas Genome
Project [http://www.pseudomonas.com/), and the unfinished chromosomes
of Yersinia pestis and K. pneumoniae
(Yersinia pestis Sequencing Group, Sanger Centre
[ftp://ftp.Sanger.ac.uk/pub/pathogens/yp/]; Klebsiella
pneumoniae Sequencing Group, University of Washington
Genome Center
[http://genome.wustl.edu/gsc/Projects/bacterial/klebsiella/klebsiella.shtml]).
From a mechanistic standpoint, the difference in biochemistry observed between the aromatic and aliphatic desulfonation processes is not surprising. Desulfonation of aliphatic sulfonates involves oxygenation at the α-position to the sulfonate group to generate a hydroxy sulfonate which spontaneously decomposes to the corresponding aldehyde and sulfite (13, 43). Oxygenation of benzenesulfonate, by contrast, requires the temporary disruption of the aromatic ring, to an unknown intermediate which decomposes to yield the corresponding phenol, releasing the sulfonate moiety as sulfite. The additional enzyme components required for aromatic desulfonation include a reductase-ferredoxin couple, and we speculate that this complex, perhaps in connection with the FMN reductase SsuE, provides reducing equivalents at the correct potential. Interestingly, in several of the FMNH2-dependent monooxygenases investigated, the FMN reductase has been shown to be involved with the oxygenase component at best as part of a loose complex, and the reduced FMN is a cosubstrate of the monooxygenase rather than an enzyme-bound prosthetic group as is the case for other flavin-dependent di- and monooxygenases. The enzymes of this family that have been studied include the nitrilotriacetate (NTA) and EDTA monooxygenase enzyme systems (44, 53), the DBT monooxygenase DszC (15), and the methanesulfonate desulfonatase from P. aeruginosa, MsuED (23). For the first two it was also shown that the reductase could be replaced by the unrelated Vibrio fischeri flavin reductase without loss of function. Aliphatic desulfonation in P. putida occurred in the absence of SsuE (Fig. 5), demonstrating that here too, SsuE can be replaced by other cellular FMN reductases. In contrast, aromatic desulfonation required the presence of SsuE. The reductase protein therefore not only may be required to supply diffusible FMNH2 but may also play a structural role in the enzyme complex catalyzing aromatic (but not aliphatic) desulfonation, although there is no direct evidence for this yet.
The family of bacterial FMNH2-dependent monooxygenases that use FMNH2 as a cosubstrate rather than as a prosthetic group is relatively small and includes the sulfonate oxygenases SsuD and MsuD; the NTA, EDTA, and DBT oxygenases mentioned above; an oxygenase involved in synthesis of the antibiotic pristinamycin IIB (42); and bacterial luciferase (31). Whereas the NTA system is quite substrate specific (NTA was the only substrate found for the NTA oxygenase [44]), the EDTA monooxygenase accepts a broad range of aminopolycarboxylate substrates, and mutant analysis showed that the SsuD protein is also involved in metabolism of a variety of sulfonates. Interestingly, SsuD is not required for desulfurization of the natural sulfonates taurine and cysteate (Table 2), an effect which may be mediated by charge and lipophilicity differences between xenobiotic substrates and natural sulfonates. It should be noted, however, that the natural substrates of the bacterial enzymes might not yet have been discovered. For the EDTA oxygenase, these could include natural aminocarboxylate metallophores (25), whereas for SsuD we anticipate that many uncharacterized sulfonates may also be present in nature.
Upstream of the ssu operon in P. putida S-313 a further open reading frame was located, which we have designated lsfA. The N-terminal sequence of the encoded protein is >90% identical to that of the sulfate-repressed protein PA11 from P. aeruginosa (19). The pa11 gene was identified in the preliminary release of the P. aeruginosa genome (http://www.pseudomonas.com/), and the PA11 and LsfA proteins were found to be 88% identical, confirming that lsfA encodes the P. putida homologue of PA11. By sequence comparison, LsfA is a putative member of the 1-Cys family of thiol-specific antioxidants (TSA), which are important in protecting bacterial cells from peroxides (7, 32), and the P. aeruginosa LsfA protein indeed showed the expected TSA activity (our unpublished results). The upregulation of antioxidant proteins (LsfA in P. putida, LsfA and the alkylhydroperoxide reductase AhpC in P. aeruginosa [34], and AhpC in E. coli [35]) under sulfate limitation conditions may be a response to increased levels of desulfonative FMNH2-dependent monooxygenase systems such as SsuED or MsuED. When the cells are grown under aerobic conditions in the absence of sulfate, expression of the ssuE gene will lead to the generation of excess FMNH2 in the cell, and if suitable substrates (e.g., sulfonates) are not present to use up these reducing equivalents, they will rapidly react with molecular oxygen, generating superoxide radicals that can lead to considerable cellular damage. The cell responds to this kind of oxidative challenge by inducing a number of antioxidant genes, under the control of the OxyR and SoxRS systems (40), including genes encoding protector proteins of the TSA-AhpC family (7). It will be useful to see whether expression of the lsfA gene (which is separate from that of ssu [Fig. 4]) is also under the control of these oxidative stress regulators and if the cells perceive sulfate starvation stress primarily as an oxidative challenge as well as a nutrient stress. Interestingly, the B. subtilis ssu operon is located directly downstream of the catalase gene katA, suggesting that this genetic arrangement may be a more general phenomenon.
The SsuD monooxygenase was also involved in growth of P. putida S-313 with sulfate esters (Fig. 5). This was unexpected, since sulfatases are hydrolytic enzymes, and desulfonative oxygenation of these compounds has previously been demonstrated only for monomethylsulfate (17). Closer examination revealed that P. putida S-313 does not in fact synthesize an arylsulfatase enzyme. Instead, during growth with nitrocatecholsulfate, the sulfate moiety is transferred onto an unidentified acceptor by an arylsulfotransferase and then presumably transferred from the acceptor into the cysteine biosynthesis pathway. Sulfotransferases are well-characterized enzymes in mammalian systems, where they are involved in detoxification processes and catalyze the transfer of activated sulfate from 3′-phosphoadenosine-5′-phosphosulfate (PAPS) onto phenolic acceptor substrates (9). A different type of sulfotransferase is found in bacteria, which accepts phenyl sulfate esters instead of PAPS as donors of the sulfate moiety (3, 8, 29) and is probably involved in sulfation of peptidyl tyrosine residues (26). The arylsulfotransferase of P. putida S-313 may catalyze the reverse reaction to that seen in eukaryotes, thereby generating PAPS for cysteine biosynthesis, or alternatively, it may transfer the sulfate onto an unidentified intermediate molecule from which it can be oxygenatively cleaved by the SsuD monooxygenase to yield sulfite. The details of this reaction are currently under investigation in our laboratory.
ACKNOWLEDGMENTS
We are grateful to A. M. Cook for helpful discussions. Thomas Dierks and Valley Stewart kindly provided strains and plasmids.
This work was supported in part by the Swiss National Science Foundation (grant no. 31-49435.96), the Netherlands Organization for Scientific Research, and the Swiss Federal Office for Education and Sciences (grant no. 97.0190, as part of the EC program SUITE, contract no. ENV4-CT98-0723).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1987. [Google Scholar]
- 2.Autry A R, Fitzgerald J W. Sulfonate S—a major form of forest soil organic sulfur. Biol Fertil Soils. 1990;10:50–56. [Google Scholar]
- 3.Baek M C, Kim S K, Kim D H, Kim B K, Choi E C. Cloning and sequencing of the Klebsiella K-36 astAgene, encoding an arylsulfate sulfotransferase. Microbiol Immunol. 1996;40:531–537. doi: 10.1111/j.1348-0421.1996.tb01105.x. [DOI] [PubMed] [Google Scholar]
- 4.Beil S, Kehrli H, James P, Staudenmann W, Cook A M, Leisinger T, Kertesz M A. Purification and characterization of the arylsulfatase synthesized by Pseudomonas aeruginosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA) Eur J Biochem. 1995;229:385–394. doi: 10.1111/j.1432-1033.1995.0385k.x. [DOI] [PubMed] [Google Scholar]
- 5.Beil S, Kertesz M A, Leisinger T, Cook A M. The assimilation of sulfur from multiple sources and its correlation with expression of the sulfate-starvation-induced stimulon in Pseudomonas putidaS-313. Microbiology. 1996;142:1989–1995. doi: 10.1099/13500872-142-8-1989. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Chae H Z, Robison K, Poole L B, Church G, Storz G, Rhee S G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai C L L, Lowe G. The mechanism and stereochemical course of sulfuryl transfer catalyzed by the aryl sulfotransferase from EubacteriumA-44. Bioorg Chem. 1992;20:181–188. [Google Scholar]
- 9.Coughtrie M W H, Sharp S, Maxwell K, Innes N P. Biology and function of the reversible sulfation pathway catalysed by human sulfotransferases and sulfatases. Chem Biol Interact. 1998;109:3–27. doi: 10.1016/s0009-2797(97)00117-8. [DOI] [PubMed] [Google Scholar]
- 10.Dainese-Hatt P, Quadroni M, Staudenmann W, James P. Concentration of and SDS removal from proteins isolated from multiple two dimensional electrophoresis gels. Eur J Biochem. 1997;246:336–343. doi: 10.1111/j.1432-1033.1997.00336.x. [DOI] [PubMed] [Google Scholar]
- 11.Dierks T, Miech C, Hummerjohann J, Schmidt B, Kertesz M A, von Figura K. Posttranslational formation of formylglycine in prokaryotic sulfatases by modification of either cysteine or serine. J Biol Chem. 1998;273:25560–25564. doi: 10.1074/jbc.273.40.25560. [DOI] [PubMed] [Google Scholar]
- 12.Dudley M W, Frost J W. Biocatalytic desulfurization of arylsulfonates. Bioorg Med Chem. 1994;2:681–690. doi: 10.1016/0968-0896(94)85018-6. [DOI] [PubMed] [Google Scholar]
- 13.Eichhorn E, van der Ploeg J R, Leisinger T. Characterization of a two component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 14.Gibson J, Dispensa M, Fogg G C, Evans D T, Harwood C S. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J Bacteriol. 1994;176:634–641. doi: 10.1128/jb.176.3.634-641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray K A, Pogrebinsky O S, Mrachko G T, Xi L, Monticello D J, Squires C H. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat Biotechnol. 1996;14:1705–1709. doi: 10.1038/nbt1296-1705. [DOI] [PubMed] [Google Scholar]
- 16.Higgins C F. ABC transporters—from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 17.Higgins T P, Hewlins M J E, White G F. A C-13-NMR study of the mechanism of bacterial metabolism of monomethyl sulfate. Eur J Biochem. 1996;236:620–625. doi: 10.1111/j.1432-1033.1996.00620.x. [DOI] [PubMed] [Google Scholar]
- 18.Hinton S M, Slaughter C, Eisner W, Fisher T. The molybdenum-pterin binding protein is encoded by a multigene family in Clostridium pasteurianum. Gene. 1987;54:211–220. doi: 10.1016/0378-1119(87)90489-6. [DOI] [PubMed] [Google Scholar]
- 19.Hummerjohann J, Kuttel E, Quadroni M, Ragaller J, Leisinger T, Kertesz M A. Regulation of the sulfate starvation response in Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology. 1998;144:1375–1386. doi: 10.1099/00221287-144-5-1375. [DOI] [PubMed] [Google Scholar]
- 20.Kertesz M A, Cook A M, Leisinger T. Microbial metabolism of sulfur- and phosphorus-containing xenobiotics. FEMS Microbiol Rev. 1994;15:195–215. doi: 10.1111/j.1574-6976.1994.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 21.Kertesz M A, Kölbener P, Stockinger H, Beil S, Cook A M. Desulfonation of linear alkylbenzenesulfonate surfactants and related compounds by bacteria. Appl Environ Microbiol. 1994;60:2296–2303. doi: 10.1128/aem.60.7.2296-2303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kertesz M A, Leisinger T, Cook A M. Proteins induced by sulfate limitation in Escherichia coli, Pseudomonas putida, or Staphylococcus aureus. J Bacteriol. 1993;175:1187–1190. doi: 10.1128/jb.175.4.1187-1190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kertesz M A, Schmidt K, Wüest T. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J Bacteriol. 1999;181:1464–1473. doi: 10.1128/jb.181.5.1464-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King J E, Quinn J P. The utilization of organosulphonates by soil and freshwater bacteria. Lett Appl Microbiol. 1997;24:474–478. [Google Scholar]
- 25.Knobel H R, Egli T, van der Meer J R. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintziiATCC 29600. J Bacteriol. 1996;178:6123–6132. doi: 10.1128/jb.178.21.6123-6132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobashi K, Kim D H. A novel sulfotransferase sulfates tyrosine-containing peptides and proteins. Biochem Biophys Res Commun. 1986;140:38–42. doi: 10.1016/0006-291x(86)91054-5. [DOI] [PubMed] [Google Scholar]
- 27.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 514–527. [Google Scholar]
- 29.Lee N S, Kim B T, Kim D H, Kobashi K. Purification and reaction mechanism of arylsulfate sulfotransferase from HaemophilusK-12, a mouse intestinal bacterium. J Biochem. 1995;118:796–801. doi: 10.1093/oxfordjournals.jbchem.a124982. [DOI] [PubMed] [Google Scholar]
- 30.Luque F, Mitchenall L A, Chapman M, Christine R, Pau R N. Characterization of genes involved in molybdenum transport in Azotobacter vinelandii. Mol Microbiol. 1993;7:447–459. doi: 10.1111/j.1365-2958.1993.tb01136.x. [DOI] [PubMed] [Google Scholar]
- 31.Meighen E A. Genetics of bacterial bioluminescence. Annu Rev Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- 32.Netto L E S, Chae H Z, Kang S W, Rhee S G, Stadtman E R. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties: TSA possesses thiol peroxidase activity. J Biol Chem. 1996;271:15315–15321. doi: 10.1074/jbc.271.26.15315. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Quadroni M, James P, Dainese-Hatt P, Kertesz M A. Proteome mapping, mass spectrometric sequencing and reverse transcriptase-PCR for characterisation of the sulfate starvation-induced response in Pseudomonas aeruginosaPAO1. Eur J Biochem. 1999;266:986–996. doi: 10.1046/j.1432-1327.1999.00941.x. [DOI] [PubMed] [Google Scholar]
- 35.Quadroni M, Staudenmann W, Kertesz M, James P. Analysis of global responses by protein and peptide fingerprinting of proteins isolated by two-dimensional gel electrophoresis: application to the sulfate-starvation response of Escherichia coli. Eur J Biochem. 1996;239:773–781. doi: 10.1111/j.1432-1033.1996.0773u.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sarathchandra S U, Perrott K W. Determination of phosphatase and arylsulfatase activities in soils. Soil Biol Biochem. 1981;13:543–545. [Google Scholar]
- 38.Seitz A P, Leadbetter E R, Godchaux I W. Utilization of sulfonates as sole sulfur source by soil bacteria including Comamonas acidovorans. Arch Microbiol. 1993;159:440–444. [Google Scholar]
- 39.Stewart V, Macgregor C H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlGloci. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 41.Szameit C, Miech C, Balleininger M, Schmidt B, von Figura K, Dierks T. The iron sulfur protein AtsB is required for posttranslational formation of formylglycine in the Klebsiellasulfatase. J Biol Chem. 1999;274:15375–15381. doi: 10.1074/jbc.274.22.15375. [DOI] [PubMed] [Google Scholar]
- 42.Thibaut D, Ratet N, Bisch D, Faucher D, Debussche L, Blanche F. Purification of the two-enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin II-B during the last step of pristinamycin II-A biosynthesis. J Bacteriol. 1995;177:5199–5205. doi: 10.1128/jb.177.18.5199-5205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thysse G J E, Wanders T H. Initial steps in the degradation of n-alkane-1-sulphonates by Pseudomonas. Antonie Leeuwenhoek. 1974;40:25–37. doi: 10.1007/BF00394550. [DOI] [PubMed] [Google Scholar]
- 44.Uetz T, Schneider R, Snozzi M, Egli T. Purification and characterization of a two-component monooxygenase that hydroxylates nitrilotriacetate from “Chelatobacter” strain ATCC 29600. J Bacteriol. 1992;174:1179–1188. doi: 10.1128/jb.174.4.1179-1188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Ploeg J R, Cummings N J, Leisinger T, Connerton I F. Bacillus subtilisgenes for the utilization of sulfur from aliphatic sulfonates. Microbiology. 1998;9:2555–2561. doi: 10.1099/00221287-144-9-2555. [DOI] [PubMed] [Google Scholar]
- 46.van der Ploeg J R, Iwanicka-Nowicka R, Bykowski T, Hryniewicz M, Leisinger T. The Cbl-regulated ssuEADCB gene cluster is required for aliphatic sulfonate-sulfur utilization in Escherichia coli. J Biol Chem. 1999;174:29358–29365. doi: 10.1074/jbc.274.41.29358. [DOI] [PubMed] [Google Scholar]
- 47.van der Ploeg J R, Weiss M A, Saller E, Nashimoto H, Saito N, Kertesz M A, Leisinger T. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol. 1996;178:5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vermeij P, Kertesz M A. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J Bacteriol. 1999;181:5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vermeij P, Wietek C, Kahnert A, Wüest T, Kertesz M A. Genetic organization of sulfur-controlled aryl desulfonation in Pseudomonas putidaS-313. Mol Microbiol. 1999;32:913–926. doi: 10.1046/j.1365-2958.1999.01398.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang G, Angermuller S, Klipp W. Characterization of Rhodobacter capsulatusgenes encoding a molybdenum transport system and putative molybdenum-pterin-binding proteins. J Bacteriol. 1993;175:3031–3042. doi: 10.1128/jb.175.10.3031-3042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West S E H, Schweizer H P, Dall C, Sample A K, Runyenjanecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 52.White G F, Russell N J, Day M J. A survey of sodium dodecyl-sulfate (SDS) resistance and alkylsulfatase production in bacteria from clean and polluted river sites. Environ Poll Ser A. 1985;37:1–11. [Google Scholar]
- 53.Witschel M, Nagel S, Egli T. Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J Bacteriol. 1997;179:6937–6943. doi: 10.1128/jb.179.22.6937-6943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zürrer D, Cook A M, Leisinger T. Microbial desulfonation of substituted naphthalenesulfonic acids and benzenesulfonic acids. Appl Environ Microbiol. 1987;53:1459–1463. doi: 10.1128/aem.53.7.1459-1463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]