Abstract
Protein synthesis in eukaryotic organelles such as mitochondria and chloroplasts is widely believed to require a formylated initiator methionyl tRNA (fMet-tRNAfMet) for initiation. Here we show that initiation of protein synthesis in yeast mitochondria can occur without formylation of the initiator methionyl-tRNA (Met-tRNAfMet). The formylation reaction is catalyzed by methionyl-tRNA formyltransferase (MTF) located in mitochondria and uses N10-formyltetrahydrofolate (10-formyl-THF) as the formyl donor. We have studied yeast mutants carrying chromosomal disruptions of the genes encoding the mitochondrial C1-tetrahydrofolate (C1-THF) synthase (MIS1), necessary for synthesis of 10-formyl-THF, and the methionyl-tRNA formyltransferase (open reading frame YBL013W; designated FMT1). A direct analysis of mitochondrial tRNAs using gel electrophoresis systems that can separate fMet-tRNAfMet, Met-tRNAfMet, and tRNAfMet shows that there is no formylation in vivo of the mitochondrial initiator Met-tRNA in these strains. In contrast, the initiator Met-tRNA is formylated in the respective “wild-type” parental strains. In spite of the absence of fMet-tRNAfMet, the mutant strains exhibited normal mitochondrial protein synthesis and function, as evidenced by normal growth on nonfermentable carbon sources in rich media and normal frequencies of generation of petite colonies. The only growth phenotype observed was a longer lag time during growth on nonfermentable carbon sources in minimal media for the mis1 deletion strain but not for the fmt1 deletion strain.
Protein synthesis is initiated with methionine or formylmethionine in all organisms studied to date (23, 33). Of the two species of methionine tRNAs found in all organisms, the initiator is used for initiation of protein synthesis whereas the elongator is used for insertion of methionine into internal peptide linkages. In eubacteria such as Escherichia coli, following aminoacylation of the initiator methionine tRNA (tRNAfMet), the methionyl-tRNA (Met-tRNAfMet) is formylated to formylmethionyl-tRNA (fMet-tRNAfMet). As a consequence, protein synthesis in eubacteria is initiated with formylmethionine (29). The discovery of fMet-tRNA in eukaryotic organelles such as chloroplasts and in the mitochondria of the yeast Saccharomyces cerevisiae, Neurospora crassa, rat liver, and HeLa cells suggested that protein synthesis in these organelles is also initiated with formylmethionine (13, 17, 19, 39, 44). Early evidence for this came from the identification of formylmethionyl-puromycin in several eukaryotic mitochondria and chloroplasts treated with puromycin (5, 6, 14, 16, 28, 35). These results, along with the identification of formylmethionine at the N terminus of several mitochondrially synthesized proteins in S. cerevisiae, N. crassa, and beef heart mitochondria, led to the widespread belief that protein synthesis in all mitochondria is initiated, as in eubacteria, with formylmethionine and that formylation of the initiator Met-tRNA in mitochondria is a prerequisite for its activity in initiation of protein synthesis (8, 40, 46, 50, 55). The finding that the initiation factor IF2 from bovine mitochondria promotes the binding of fMet-tRNAfMet but not of Met-tRNAfMet to mitochondrial ribosomes provides further evidence for this notion (26). However, none of these studies show a strict requirement for fMet-tRNA in vivo.
The formylation of initiator Met-tRNA is catalyzed by the enzyme methionyl-tRNA formyltransferase (MTF). This enzyme is highly specific for the initiator Met-tRNA species (25, 37) and has been found exclusively in the mitochondria of S. cerevisiae, N. crassa and HeLa cells but not in the cytoplasm (13, 17, 19). Bovine mitochondrial MTF has been purified, and the cDNA encoding it has been cloned and sequenced (47). The S. cerevisiae genome contains an open reading frame (ORF YBL03.11 in reference 43) that has 24 to 29% amino acid sequence identity to the eubacterial and bovine mitochondrial MTF. This ORF (Saccharomyces Genome Database YBL013W) encodes a protein of 393 amino acids, including a potential mitochondrial presequence and a highly conserved motif proposed to be the binding site for the N10-formyltetrahydrofolate (10-formyl-THF) substrate. We will refer to this ORF henceforth as the FMT1 gene, encoding the S. cerevisiae MTF.
The formyl group donor in the formylation reaction is 10-formyl-THF (11). S. cerevisiae contains two C1-THF synthase enzymes for the synthesis of 10-formyl-THF, one in the mitochondria and the other in the cytoplasm, encoded by MIS1 and ADE3, respectively (see reference 1 for a review). Both the cytoplasmic and mitochondrial enzymes are trifunctional polypeptides with three enzyme activities: 10-formyl-THF synthetase, 5,10-methenyl-THF cyclohydrolase, and NADP-dependent 5,10-methylene-THF dehydrogenase. S. cerevisiae also expresses a monofunctional NAD-dependent 5,10-methylene-THF dehydrogenase in the cytoplasm, encoded by the MTD1 gene (57). The ADE3 and MTD1 gene products are responsible for cytoplasmic one-carbon interconversions, whereas the MIS1 gene product is responsible for mitochondrial one-carbon interconversions (3, 58).
Shannon and Rabinowitz (41) showed that disruption of the MIS1 gene had no dramatic effects on the growth of S. cerevisiae, suggesting that the MIS1 gene is dispensable in yeast. Also, disruption of the nuclear gene encoding the putative mitochondrial MTF had no effect on viability (43), suggesting that the FMT1 gene is also dispensable in S. cerevisiae, although the growth conditions tested were not specified. These findings are rather surprising. If formylated Met-tRNAfMet is required for initiation of mitochondrial protein synthesis, loss of the enzyme that produces the formyl donor or loss of the enzyme that synthesizes fMet-tRNA would be expected to affect protein synthesis and, thereby, mitochondrial function. Mitochondrial protein synthesis is required for respiratory function in mitochondria, and mutation of genes encoding mitochondrial translation components invariably leads to a respiration-deficient (petite) phenotype (49). Thus, the lack of a dramatic effect on cell growth or respiration upon disruption of the genes coding for these two enzymes would suggest that protein synthesis can be initiated in S. cerevisiae mitochondria without formylation of the initiator tRNA. There are, however, several other possible explanations that need to be ruled out: (i) transport of the cytoplasmically made 10-formyl-THF into mitochondria, (ii) alternate forms of MTF which do not use 10-formyl-THF as a formyl donor (analogous to the formate-dependent glycinamide ribonucleotide transformylase [56]), or (iii) alternate genes for mitochondrial MTF with no homology to MTFs identified thus far.
A knowledge of the state of the initiator tRNA in mitochondria, whether it is in the form of fMet-tRNA or Met-tRNA (52), would allow one to distinguish among the above possibilities. This paper reports on a direct analysis of the state of the initiator tRNA in S. cerevisiae mitochondria in strains carrying the MIS1 and FMT1 gene disruptions. We show that there is no formylation of the initiator Met-tRNA in strains carrying these gene disruptions. Also, these strains grow at nearly wild-type rates in rich medium and on nonfermentable carbon sources requiring full mitochondrial function. There are also no changes in the frequencies of generation of petite colonies, indicating that MIS1 and FMT1 gene disruptions have no effect on mitochondrial protein synthesis. Thus, formylation of the initiator Met-tRNA is not essential for mitochondrial protein synthesis and for mitochondrial function in S. cerevisiae.
MATERIALS AND METHODS
Strains, media, and plasmids.
The S. cerevisiae strains used in this work are summarized in Table 1. Strains 1001, 1049, and 1052 were obtained from B. Purnelle (Universite Catholique de Louvain, Louvain-la-Neuve, Belgium). DAY4Δmis1 was constructed by disruption of the MIS1 gene in DAY4. A 400-bp fragment from the middle of the MIS1 ORF was replaced with a URA3 cassette from plasmid pJR-URA3 (34). A 2-kbp fragment containing the mis1::URA3 disruption construct was used to transform strain DAY4, a haploid ura3− yeast strain, selecting for uracil prototrophy. Yeast transformation was performed using the lithium acetate method (22) to obtain mis1::URA3-disrupted yeast. The URA3 cassette was subsequently evicted from the mis1 locus by using the pHM53-encoded site-specific recombinase (34), resulting in a strain harboring a 400-bp deletion in the middle of the MIS1 locus. The disruption of the MIS1 and FMT1 ORFs was verified by PCR amplification of yeast genomic DNA. Yeast genomic DNA was isolated by the method of Sherman et al. (42). PCR products were separated on a 0.8% agarose gel. A double-disruption strain was constructed by crossing strain 1049 carrying the fmt1 disruption with DAY4Δmis1. Diploids were sporulated, tetrads were dissected, and a haploid spore clone was selected carrying both the fmt1 and mis1 disruptions. This strain was designated WHY2 (Table 1). YEpKS17 contains the MIS1 ORF in the multicopy URA3 yeast vector YEp24 (41). pVT101U is a multicopy URA3 yeast vector lacking an insert (54).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Comments |
|---|---|---|
| 1001 | α ade2 trp1 leu2 his3 ura3 | Wild-type FMT1 |
| 1049 | α ade2 trp1 leu2 his3 ura3 fmt1::URA3 | fmt1 disruptant |
| 1052 | a ade2 trp1 leu2 his3 ura3 fmt1::URA3 | fmt1 disruptant |
| DAY4 | a ser1 ura3 trp1 leu2 his4 | Wild-type MIS1 |
| DAY4Δmis1 | a ser1 ura3 trp1 leu2 his4 Δmis1 | mis1 disruptant |
| WHY2 | a ura3 trp1 leu2 Δmis1 fmt1::URA3 | mis1 fmt1 double disruptant |
Rich medium consisted of 1% yeast extract and 2% Bacto Peptone (Difco) with either 2% glucose (YPED) or 3% glycerol–2% ethanol (YPEG) as the carbon source. Synthetic minimal medium contained 0.7% yeast nitrogen base without amino acids (Difco) and supplemented with the following nutrients when appropriate (final concentration in milligrams per liter): serine, 375; leucine, 30; histidine, 20; tryptophan, 20; and uracil, 20. The synthetic minimal media were supplemented with either 2% glucose (YMD) or 3% glycerol–2% ethanol (YMEG) as the carbon source.
Preparation of yeast mitochondria.
Mitochondria were isolated as described previously (9). Briefly, yeast cells were grown aerobically in 1 liter of medium containing 3 g of yeast extract, 1 g of glucose, 22.5 ml of 85% (wt/vol) lactic acid, 1 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 0.6 g of MgSO4, and 0.3 ml of 1% (wt/vol) FeCl3. The final pH was adjusted to 5.5 with NaOH. Cells were harvested at mid-log phase and converted to spheroplasts using lyticase (Sigma, St. Louis, Mo.) in 1.2 M sorbitol–20 mM KH2PO4 (pH 7.4). Spheroplasts were resuspended in SEM [250 mM sucrose, 1 mM EDTA, 10 mM 3-(N-morpholino)propanesulfonic acid, (pH 7.2)] containing 0.2% (wt/vol) bovine serum albumin and 1 mM phenylmethylsulfonyl fluoride and homogenized using a tight-fitting Teflon homogenizer. Cell debris was pelleted by centrifugation at 1,900 × g. Mitochondria were pelleted at 12,000 × g for 10 min and washed three times by resuspension in 1 ml of SEM and centrifugation at 12,000 × g. An aliquot of the washed mitochondria before the final centrifugation was spread onto yeast extract-tryptone plates to check for bacterial contamination. All preparations contained less than 25 CFU/μl (1 μl represents the extract from ∼108 yeast cells). Mitochondrial pellets were stored at −70°C.
Isolation of RNA from yeast mitochondria.
The yeast mitochondrial RNA was isolated using TRI reagent (Molecular Research Center, Inc., Cincinnati, Ohio). The mitochondrial pellet (∼100 μl in volume) was suspended in TRI reagent (1 ml) and left at room temperature for 5 min. Chloroform (0.2 ml) was added to the suspension, and the mixture was vortexed three times for 15 s each, left for 15 min at room temperature, and centrifuged at 4°C for 30 min. The clear aqueous phase was transferred to a new tube, and isopropanol (0.7 vol) was added. The mixture was left at room temperature for 10 min, and total RNA was collected by centrifugation at 4°C for 15 min. The RNA pellet was washed with 75% ethanol (1 ml), air dried for 5 to 10 min, and dissolved in 10 to 15 μl of 10 mM sodium acetate (pH 4.5). The yield of total mitochondrial RNA from 1 liter of culture was ∼0.5 A260 unit.
Electrophoresis of tRNAs on acid-urea polyacrylamide gels and Northern blot analysis.
The various forms of tRNAs were separated and detected as described previously (52), except that 0.16 A260 unit of mitochondrial RNA was applied to the 6.5% polyacrylamide gel. The mitochondrial tRNAs migrate slower on the gel than the corresponding Escherichia coli tRNAs. Therefore, the nucleic acids in an 11-cm segment of gel, starting with the xylene cyanol dye and going toward the bromphenol blue dye, were transferred by electroblotting to Nytran Plus membrane (Schleicher & Schuell, Keene, N.H.) in 1× TAE (50× TAE is 242 g of Tris base, 57.1 ml of glacial acetic acid, and 100 mM EDTA [pH 8.0]) at 40 V for 2 h. The tRNAs in the membrane were detected by hybridization with sequence-specific oligonucleotide probes. Prehybridization and hybridization were carried out for 4 and 16 h, respectively, in 4× SET (20× SET contains 87 g of NaCl, 46.5 g of Tris base, and 40 ml EDTA in 500 ml) containing 100 μg of salmon sperm DNA per ml, 1% sodium dodecyl sulfate, and 10× Denhardt's solution. The oligonucleotide 5′TAGCAATAATACGATTTG3′, which is complementary to nucleotides 56 to 73 of the S. cerevisiae mitochondrial tRNAfMet, was used to detect the mitochondrial tRNAfMet.
Deacylation of aminoacyl-tRNAs.
Aminoacyl-tRNA was deacylated in 0.1 M Tris-HCl (pH 9.4) at 37°C for 1 h. Alternatively, the aminoacyl-tRNA was incubated with 10 mM CuSO4 in 0.1 M Tris-HCl (pH 8.0) at room temperature for 15 min. Copper sulfate treatment hydrolyzes aminoacyl-tRNAs but not formylaminoacyl-tRNAs (38).
Rates of chemical deacylation of Tyr-tRNATyr and Met-tRNAMet.
Total E. coli tRNA (0.5 A260 unit) was aminoacylated at 37°C for 30 min in 20 mM imidazole (pH 7.5)–150 mM NH4Cl–10 mM MgCl2–0.1 mM EDTA–10 μg of bovine serum albumin per ml–2 mM ATP, with either methionine (100 μM) plus a saturating amount of purified E. coli Met-tRNA synthetase or tyrosine (25 μM) plus a saturating amount of purified E. coli Tyr-tRNA synthetase. The tRNAs were quantitatively aminoacylated under these conditions. The aminoacyl-tRNAs were isolated by phenol-chloroform extraction followed by ethanol precipitation (52). For measurement of rates of deacylation, the aminoacyl-tRNAs were incubated with 0.1 M Tris-HCl (pH 9.4) at 37°C and aliquots were taken out at various times and frozen. At the end of the incubation period, the samples were thawed and loaded onto a 6.5% acid–urea gel for separation of tRNA and aminoacyl-tRNA (12). The tRNAs in the gel were transferred to Nytran Plus membrane and detected by hybridization with oligonucleotides complementary to either nucleotides 10 to 25 of E. coli tRNATyr or nucleotides 40 to 56 of E. coli tRNAfMet. The amount of radioactivity in the tRNA bands was determined by quantification using a Molecular Dynamics PhosphorImager.
RESULTS
S. cerevisiae strains carrying disruptions in the MIS1 and FMT1 genes.
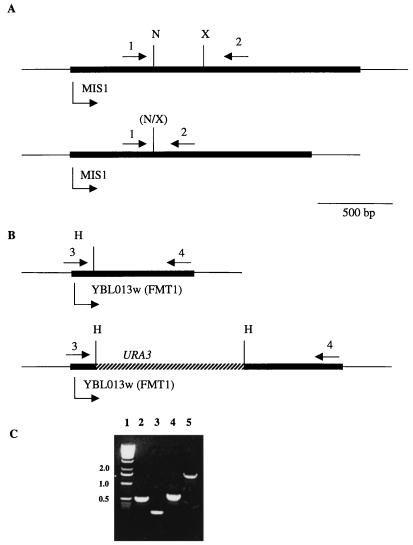
The MIS1 gene was disrupted as described in Materials and Methods in the haploid strain DAY4 to yield DAY4Δmis1. Haploid yeast strain 1052 harboring a disruption of the putative MTF ORF (FMT1; YBL013W) was obtained from B. Purnelle. The genotypes of these mutants and the parent strains are summarized in Table 1. The disruptions were verified by PCR analysis of genomic DNA isolated from each strain. Disruption of MIS1 gave the expected 400-bp difference when genomic DNA from DAY4 and DAY4Δmis1 was amplified with primers 1 and 2 (Fig. 1, compare lanes 2 and 3). Disruption of FMT1 gave the expected 1,100-bp difference when genomic DNA from 1001 (wild type) and 1052 (disruptant) was amplified with primers 3 and 4 (compare lanes 4 and 5).
FIG. 1.
Disruption of MIS1 and FMT1 (YBL013W). (A) Intact MIS1 locus (top) and disrupted locus (bottom). (B) Intact FMT1 locus (top) and URA3-disrupted locus (bottom). Arrows above the constructs indicate oligonucleotide primers 1, 2, 3, and 4. Arrows below the constructs indicate the direction of transcription of the MIS1, FMT1, or URA3 genes. N, NarI; X, XbaI; H, HindIII (C) Agarose gel showing PCR products of yeast genomic DNA from DAY4 (MIS1 wild type, lane 2), DAY4Δmis1 (mis1 disruptant, lane 3), 1001 (FMT1 wild type, lane 4), and 1052 (fmt1 disruptant, lane 5). The MIS1 locus was amplified with primers 1 plus 2; the FMT1 locus was amplified with primers 3 plus 4. Lane 1 contains size standards. The numbers on the left indicate the sizes in kilobase pairs of the standard.
Lack of formylation in vivo of the mitochondrial initiator Met-tRNA in strains disrupted at the MIS1 or FMT1 locus.
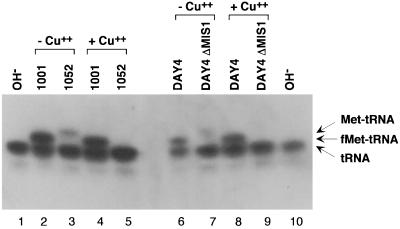
tRNA was isolated under acidic conditions from mitochondria obtained from the four strains. The cells were grown in medium containing lactate as a nonfermentable carbon source. Since yeast must actively respire to grow well in this medium, it gives a good yield of intact, functional mitochondria (59). Acid-urea gel electrophoresis (52) was used to resolve the mitochondrially encoded tRNAfMet into three forms: uncharged tRNAfMet, Met-tRNAfMet, and fMet-tRNAfMet. A labeled oligonucleotide complementary to nucleotides 56 to 73 of the tRNAfMet was used to probe a Northern blot of the gel. Figure 2 shows a typical Northern blot analysis of total yeast mitochondrial tRNA from the wild-type strain (lanes 2, 4, 6, and 8), the fmt1-disrupted strain (lanes 3 and 5), and the mis1-disrupted strain (lanes 7 and 9). Lanes 1 and 10 contain deacylated tRNAfMet as markers. Uncharged tRNAfMet (bottom band) was present in all strains, whereas formylated Met-tRNAfMet (middle band) was detectable only in the wild-type strains (1001 and DAY4). The mutant strains contained instead small amounts of charged but unformylated Met-tRNAfMet (top band, lanes 3 and 7). Treatment of tRNA with copper sulfate prior to electrophoresis resulted in the disappearance of the aminoacyl-tRNA band but not the formylaminoacyl-tRNA band, confirming the identity of the upper bands seen in the mutant strains as unformylated Met-tRNAfMet (compare lane 3 to lane 5 and lane 7 to lane 9). These data show that there is no formylation of Met-tRNAfMet in mitochondria from either the fmt1-disrupted or mis1-disrupted strains and provide further support for the identification of ORF YBL013W as the yeast methionyl-tRNA formyltransferase gene.
FIG. 2.
RNA blot hybridization of mitochondrial tRNAfmet from wild-type (lanes 2, 4, 6, and 8), fmt1-disrupted (lanes 3 and 5), and mis1-disrupted (lanes 7 and 9) strains. The various forms of the tRNA were separated on a 6.5% polyacrylamide gel in 8 M urea and 0.2 M sodium acetate (pH 5.0) and transferred to Nytran Plus membrane. A total of 0.16 A260 unit of mitochondrial RNA was loaded. Lanes 1 and 10 contain the deacylated wild-type control. An oligonucleotide complementary to nucleotides 56 to 73 of the S. cerevisiae mitochondrial initiator tRNA was used as a hybridization probe.
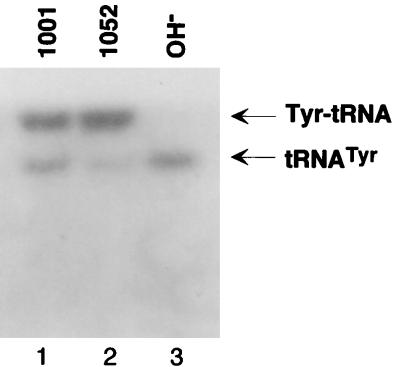
A somewhat surprising result is the limited amount of aminoacylated mitochondrial Met-tRNAfMet (Fig. 2, lanes 3 and 7) found in strains carrying the fmt1 or the mis1 disruptions. This is unlike the situation in E. coli, where a block in formylation of the initiator tRNAfMet leads to an essentially quantitative accumulation of the tRNA as Met-tRNAfMet (24, 53). The ester linkage between the methionine and the tRNA is known to be more labile than that between formylmethionine and tRNA (38). Therefore, one possible explanation for the above result is that methionine is cleaved off the Met-tRNAfMet during the prolonged workup (6 h or longer) necessary for the isolation of yeast mitochondria prior to isolation of the mitochondrial RNA. To test this possibility, we probed another blot of the mitochondrial RNA preparation with an oligonucleotide complementary to yeast mitochondrial tRNATyr. The results (Fig. 3) show that while this tRNA is present mostly in the aminoacylated form as Tyr-tRNATyr, in this case also there is a substantial amount of uncharged tRNATyr.
FIG. 3.
RNA blot hybridization of mitochondrial tRNATyr from wild-type (lane 1) and fmt1-disrupted (lane 2) strains of yeast. Other details are as in the legend to Fig. 2. Lane 3 contains the deacylated wild-type control. The blot was probed with an oligonucleotide complementary to mitochondrial tRNATyr.
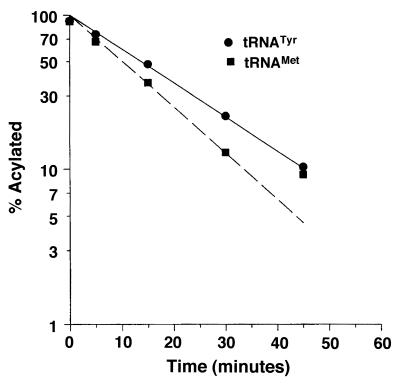
The difference between the extent of accumulation of Met-tRNAfMet (Fig. 2) and Tyr-tRNATyr (Fig. 3) in the mitochondria of strains carrying the fmt1 and mis1 disruptions is probably due to the different stabilities of ester linkage between methionine and tRNAfMet versus tyrosine and tRNATyr. To investigate this possibility, we prepared E. coli Tyr-tRNATyr and Met-tRNAfMet and monitored the rates of base-catalyzed deacylation of these aminoacyl-tRNAs in 0.1 M Tris-HCl (pH 9.4) at 37°C (for details, see Materials and Methods). Radioactivity in the tRNA and aminoacyl-tRNA bands was quantified using a PhosphorImager. A plot of the percentage of residual aminoacyl-tRNA versus time (Fig. 4) showed that the ester linkage between tyrosine and tRNATyr is more stable (half-life of 15 min) than the ester linkage between methionine and tRNAfMet (half-life of 10 min).
FIG. 4.
Time course of deacylation of E. coli Tyr-tRNATyr and Met-tRNAfMet in 0.1 M Tris-HCl (pH 9.4) at 37°C. Total charged tRNA isolated as described in Materials and Methods was incubated for the time indicated, mixed with an equal volume of acid-urea sample loading dye, and placed on dry ice. When the time course measurement was finished, the samples were thawed and loaded onto a 6.5% acid–urea gel. The tRNAs were detected using 5′-32P-labeled oligonucleotides complementary to either E. coli tRNATyr or E. coli tRNAfMet. The amount of radioactivity in the tRNA and aminoacyl-tRNA bands was quantitated using a PhosphorImager and used to calculate the percent residual aminoacyl-tRNA.
Growth rate of strains carrying the MIS1 and FMT1 gene disruptions.
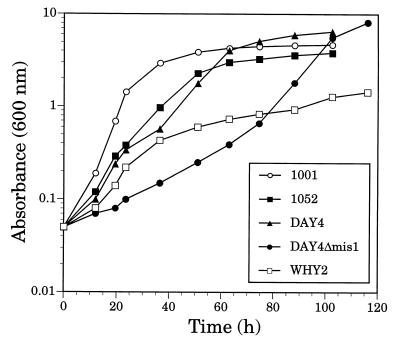
The complete absence of fMet-tRNAfMet in strains carrying the disruptions led us to investigate more closely whether these strains had any growth defects. A classic diagnostic test of mitochondrial function in yeast is to grow cells on nonfermentable carbon sources such as lactate or glycerol plus ethanol. Cells with defective mitochondria will grow poorly or not at all under these conditions (59). The growth rates of the wild type and the single and double disruptants were determined in both rich and minimal media using 3% glycerol plus 2% ethanol as the nonfermentable carbon source. The mutant strains grew at nearly the same rates as the corresponding wild-type strains in rich medium (YPEG) (Table 2). When the strains were grown in a synthetic minimal medium on the glycerol-ethanol carbon source (YMEG), the mis1-disrupted strain (DAY4Δmis1) had a significantly longer lag time, although after about 80 h it achieved a growth rate approaching that of its wild-type parent, DAY4 (9.1- and 6.4-h doubling times, respectively [Fig. 5 and Table 2]). The fmt1-disrupted strain (strain 1052) grew similarly to its wild-type parent (strain 1001) (8.3- and 6.4-h doubling times, respectively). To confirm that the longer lag observed for DAY4Δmis1 on YMEG was due to loss of mitochondrial C1-THF synthetase, a plasmid carrying the wild-type MIS1 gene (YEpKS17) was introduced into the mutant strain. This plasmid also carries the URA3 gene, complementing the ura3-52 mutation in DAY4Δmis1. As a control, DAY4Δmis1 was transformed with another URA3 plasmid (pVT101U) that lacks the MIS1 gene. The plasmid-borne MIS1 gene completely rescued the long lag of DAY4Δmis1, whereas pVT101U had no effect on lag time (data not shown). One explanation for the lag observed in DAY4Δmis1 is that the mutation does limit growth on nonfermentable carbon sources and the eventual attainment of a normal growth rate is due to the appearance of cells harboring a second mutation that suppresses the growth defect of the mis1 disruption. This possibility was tested by harvesting cells from the DAY4Δmis1 culture at the end of the experiment in Fig. 5, and repeating the growth curve determination with cells grown in fresh YMEG. The same lag was observed (data not shown), ruling out the selection of a revertant or a second-site mutation. Thus, it seems likely that the pronounced lag time in growth seen with the mis1-disrupted strain is more a reflection of a nutritional limitation than an effect on initiation of protein synthesis.
TABLE 2.
Growth rates of various yeast strains
| Strain | Doubling time (h) for growth on:
|
|
|---|---|---|
| YPEG | YMEG | |
| 1001 | 3.0 | 6.4 |
| 1052 | 3.0 | 8.3 |
| DAY4 | 3.7 | 6.4 |
| DAY4Δmis1 | 4.5 | 9.1a |
This is the growth rate achieved after ∼80 h of growth.
FIG. 5.
Growth of wild-type and mutant yeast strains in minimal medium with a nonfermentable carbon source (YMEG). Cultures were inoculated at an initial density of 0.05 A600 unit in synthetic minimal medium supplemented with 3% glycerol and 2% ethanol as the carbon sources. The genotypes of the strains are as follows: 1001, MIS1+ FMT1+; 1052, MIS1+ fmt1−; DAY4, MIS1+ FMT1+; DAY4Δmis1, mis1− FMT1+; and WHY2, mis1− fmt1−.
The mis1 fmt1 double-disruption strain (WHY2) also grew at nearly wild-type rates on glycerol plus ethanol on both rich (5-h doubling time) and minimal (8-h doubling time) media. Interestingly, on YMEG, WHY2 did not exhibit the long lag phase observed for DAY4Δmis1 and reached stationary phase at a lower cell density than did either of the single disruptants (Fig. 5). Disruption of the FMT1 gene in the Δmis1 background apparently suppressed the effect of the mis1 disruption, but the mechanism of suppression is not known. In any event, the double disruptant was able to grow on nonfermentable carbon sources.
These growth results confirm and extend earlier reports. Shannon and Rabinowitz (41) reported that disruption of MIS1 had no effect on growth on fermentable (glucose) or nonfermentable (glycerol) carbon sources. However, those experiments used only rich media; synthetic minimal medium was not tested. Skala et al. (43) did not describe the conditions under which they examined the fmt1 disruptants; they reported only that the disruptants were viable.
Frequency of generation of petite colonies in strains carrying disruptions in the MIS1 and the FMT1 gene.
One of the hallmarks of S. cerevisiae mutants impaired in mitochondrial protein synthesis is extreme instability of the mitochondrial DNA, giving rise to cytoplasmic petite derivatives at frequencies approaching 100% (30). These petite derivatives represent ρ0 (no mitochondrial DNA) or ρ− (partially deleted mitochondrial DNA) mutants. We used two different assays to test whether disruption of the FMT1 or MIS1 genes increased the frequency of petite derivatives. Respiration-competent cells can be distinguished from respiration-incompetent cells on plates containing ethanol plus glycerol supplemented with 0.1% glucose. Cells that cannot respire yielded small (petite) colonies on these plates, whereas respiration-competent yeasts gave normal-size colonies. Table 3 shows that although the two parental strains (DAY4 and 1001) have different inherent frequencies of petite derivatives, neither disruption increases that frequency. A second assay relies on the ability of actively respiring yeast to reduce a tetrazolium salt to a colored precipitate (32). Colonies grown on rich plates are overlaid with agar containing 0.1% 2,3,5-triphenyltetrazolium chloride. Within 3 h, respiration-competent colonies will be deep red whereas respiration-incompetent colonies will remain white. This assay gave slightly higher overall frequencies of petite derivatives, but again, no significant differences between the mutants and their wild type parents were observed (Table 3). These results further confirm that neither disruption significantly impaired mitochondrial protein synthesis.
TABLE 3.
Frequency of petite derivatives in various yeast strains
| Strain | % of petite derivatives in:
|
|
|---|---|---|
| 0.1% glucose assay | Tetrazolium assay | |
| 1001 | 1 | 3.5 |
| 1052 | 0.5 | 1.4 |
| DAY4 | 2.5 | 4.5 |
| DAY4Δmis1 | 4.3 | 4.1 |
DISCUSSION
We have shown that protein synthesis in yeast mitochondria can be initiated without formylation of the initiator Met-tRNAfMet. This conclusion is based on the finding that in cells carrying disruptions in the MIS1 or FMT1 gene, there is no formylation in vivo of the initiator Met-tRNAfMet. However, cells carrying these gene disruptions grow quite well on nonfermentable carbon sources requiring mitochondrial protein synthesis and function.
The MIS1-encoded C1-THF synthase is just one of three isozymes in S. cerevisiae capable of producing 10-formyl-THF. A mis1-disrupted strain such as DAY4Δmis1 is still capable of synthesizing 10-formyl-THF in the cytoplasm via the enzymes encoded by the ADE3 and MTD1 genes. The absence of any fMet-tRNAfMet in the mitochondria of the mis1-disruptant indicates that cytoplasmic 10-formyl-THF does not enter the mitochondria to any significant extent. Similarly, disruption of the FMT1 gene resulted in a total lack of formylation of the Met-tRNAfMet in vivo. Besides providing strong support to the assumption that the S. cerevisiae YBL013W ORF codes for the mitochondrial methionyl-tRNA formyltransferase, this result also rules out the possibility of the existence of any redundant and/or alternate forms of MTF for formylation of the initiator Met-tRNAfMet.
The fmt gene encoding methionyl-tRNA formyltransferase has also been disrupted in eubacteria such as E. coli and Pseudomonas aeruginosa. In E. coli, this mutation causes a severe growth defect but the cells remain viable (18). In P. aeruginosa, the disruption causes a less severe but still significant effect on the rate of cell growth, a 3-fold increase in doubling time for P. aeruginosa compared to a 10-fold increase for E. coli (31). In contrast, the growth rate of the fmt1-disrupted yeast strain is essentially the same as that of the parental wild-type strain in both rich and minimal media containing glycerol and ethanol as the nonfermentable carbon sources. Furthermore, there was essentially no difference between the parental and fmt1-disrupted strain in the frequency of formation of petite colonies. These results suggest that the overall rates of protein synthesis in yeast mitochondria are not very different when initiated with fMet-tRNAfMet versus Met-tRNAfMet.
How is mitochondrial protein synthesis initiated in yeast lacking fMet-tRNAfMet? Genetic studies with eubacteria may provide some clues. Strains of Streptococcus faecalis and mutant strains of E. coli are known that can grow in media free of folic acid and its coenzymes, initiating protein synthesis with unformylated Met-tRNAfMet (4, 36). These strains contain a tRNAfMet that is lacking in one of the base modifications, with uridine instead of ribothymidine (T) found in loop IV (also called the T loop) of all tRNAs (10). The absence of T in tRNAs from S. faecalis grown in folate-free medium occurs because the source of the methyl group for the enzymatic methylation of U to T in tRNA is 5,10-methylene-THF in S. faecalis, Bacillus subtilis, and presumably other gram-positive eubacteria (45). In contrast, in E. coli, as in many other organisms studied to date, the methyl group donor for this reaction is S-adenosylmethionine. The mutant E. coli strain that can grow in the absence of folate is also partially lacking in T, but this is due to reduced activity of the tRNA uracil 5-methylase in the mutant strain (4). Preliminary results indicated that this mutant strain also overproduces initiation factor IF2 by about three- to fourfold. The absence of T in tRNAfMet from two different organisms, S. faecalis and E. coli, that grow without requiring formylation of the initiator Met-tRNAfMet suggests that replacement of T with U in the tRNAfMet somehow enables the initiator Met-tRNAfMet to initiate protein synthesis without formylation.
It is unlikely that a similar “undermodification” of U to T in yeast mitochondrial initiator tRNA is responsible for initiation with Met-tRNAfMet. While N. crassa mitochondrial initiator tRNA normally contains U in place of T in loop IV (20), the S. cerevisiae mitochondrial initiator tRNA is known to contain T (45). S. cerevisiae has a single gene (TRM2) for tRNA uracil-5 methylase, which encodes both the cytoplasmic and mitochondrial forms of the enzyme (21). However, this enzyme is known to use S-adenosylmethionine for methylation of U to T (21). Therefore, both the mitochondrial mis1- and fmt1-disrupted strains of S. cerevisiae would be expected to contain a full complement of the base modifications in their tRNAs, including the T in loop IV.
It is possible that disruption of MIS1 or FMT1 genes results in overproduction of the yeast mitochondrial IF2 (IF-2mt) and that this compensates for the lack of formylation of the initiator Met-tRNAfMet (4). This would mean that the yeast IF-2mt is capable of interacting with unformylated Met-tRNAfMet in vivo. Yeast IF-2mt has not been identified biochemically; however, the IFM1 gene (51) encodes a protein with significant homology to the human IF-2mt (27) and disruption of the IFM1 gene causes a defect in mitochondrial protein synthesis, resulting in the petite phenotype (51), suggesting that the IFM1 gene product is important for mitochondrial protein synthesis. In contrast to yeast IF-2mt, bovine IF-2mt has been purified and shown to promote the binding of fMet-tRNA to mitochondrial ribosomes in a GTP- and AUG-dependent manner (26). Bovine IF-2mt is reported to be inactive with unformylated Met-tRNAfMet in vitro (26).
Protein synthesis is initiated with formylmethionine in the mitochondria of a wide range of organisms from fungi to mammals. The finding that protein synthesis in S. cerevisiae can be initiated with methionine using an unformylated Met-tRNAfMet and without any significant effect on the overall growth rate in nonfermentable media requiring mitochondrial protein synthesis suggests that at least in S. cerevisiae, the role of formylation of the initiator Met-tRNAfMet is quite subtle. Given the strong conservation of initiator Met-tRNA formylation in mitochondria from such a wide range of organisms, it is likely that formylation of the mitochondrial initiator Met-tRNA provides at least an incremental advantage to the cell. The retention of initiator tRNA formylation in mitochondria of S. cerevisiae would thus be an example of what has been called “the ruthless delicacy of the selection” (48), which ensures the strict conservation, across a wide phylogenetic spectrum, of a feature that provides even the slightest advantage to the organism.
Finally, our finding that S. cerevisiae can grow quite well without formylation of the mitochondrial initiator Met-tRNA raises the question whether other eukaryotic cells will behave similarly. The yeast S. cerevisiae is, in many respects, an exception among eukaryotes in terms of mitochondrial function. First, S. cerevisiae, unlike most other eukaryotes, is a facultative anaerobe and can grow without mitochondrial function. Second, S. cerevisiae mitochondrial DNA encodes fewer species of mitochondrial membrane proteins than in other eukaryotes (15). For example, it does not encode any of the components of the multisubunit enzyme NADH-ubiquinone oxidoreductase (NADH dehydrogenase [ND]) whereas N. crassa and animal cell mitochondrial DNAs encode at least six or seven of the ND subunits (2, 7). In bovine heart mitochondria, besides cytochrome oxidase subunits I and II and the mitochondrially made subunits of ATPase (8, 46, 55), all of the mitochondrially made ND subunits are thought to retain the formylmethionine residue at the N terminus (J. E. Walker, personal communication). Therefore, formylation of the initiator Met-tRNA could well be important in beef heart mitochondria, although the retention of formylmethionine could also be simply due to the lack of a peptide deformylase activity in mitochondria. It would clearly be interesting to study whether formylation of the mitochondrial initiator Met-tRNA is more important in the mitochondria of Neurospora and animal cells than in S. cerevisiae.
ACKNOWLEDGMENTS
We thank the anonymous reviewers of this paper for their prompt review of the manuscript and for their very thoughtful comments and suggestions. We thank Mike Dyson and Anne Kowal for gifts of purified Met-tRNA synthetase and Tyr-tRNA synthetase, Shannon Reed for technical assistance, and Annmarie McInnis for patience and care in the preparation of the manuscript.
This work was supported by grants R37GM17151 (U.L.R.) and RR09276 (D.R.A.) from the National Institutes of Health.
REFERENCES
- 1.Appling D R. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991;5:2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- 2.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 3.Barlowe C K, Appling D R. Molecular genetic analysis of Saccharomyces cerevisiae C1-tetrahydrofolate synthase mutants reveals a noncatalytic function of the ADE3 gene product and an additional folate-dependent enzyme. Mol Cell Biol. 1990;10:5679–5687. doi: 10.1128/mcb.10.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumstark B R, Spremulli L L, RajBhandary U L, Brown G M. Initiation of protein synthesis without formylation in a mutant of Escherichia coli that grows in the absence of tetrahydrofolate. J Bacteriol. 1977;129:457–471. doi: 10.1128/jb.129.1.457-471.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchetti R, Lucchini G, Crosti P, Tortora P. Dependence of mitochondrial protein synthesis initiation on formylation of the initiator methionyl-tRNAf. J Biol Chem. 1977;252:2519–2523. [PubMed] [Google Scholar]
- 6.Bianchetti R, Lucchini G, Sartirana M L. Endogenous synthesis of formyl-methionine peptides in isolated mitochondria and chloroplasts. Biochem Biophys Res Commun. 1971;42:97–102. doi: 10.1016/0006-291x(71)90367-6. [DOI] [PubMed] [Google Scholar]
- 7.Breitenberger C A, RajBhandary U L. Some highlights of mitochondrial research based on analyses of Neurospora crassa mitochondrial DNA. Trends Biochem Sci. 1985;10:478–483. [Google Scholar]
- 8.Chomyn A, Hunkapiller M W, Attardi G. Alignment of the amino terminal amino acid sequence of human cytochrome c oxidase subunits I and II with the sequence of their putative mRNAs. Nucleic Acids Res. 1981;9:867–877. doi: 10.1093/nar/9.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daum G, Bohni P C, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 10.Delk A S, Rabinowitz J C. Partial nucleotide sequence of a prokaryote initiator tRNA that functions in its non-formylated form. Nature. 1974;252:106–109. doi: 10.1038/252106a0. [DOI] [PubMed] [Google Scholar]
- 11.Dickerman H W, Steers E, Jr, Redfield B G, Weissbach H. Methionyl soluble ribonucleic acid transformylase. I. Purification and partial characterization. J Biol Chem. 1967;242:1522–1525. [PubMed] [Google Scholar]
- 12.Drabkin H J, RajBhandary U L. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol Cell Biol. 1998;18:5140–5147. doi: 10.1128/mcb.18.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epler J L, Shugart L R, Barnett W E. N-formylmethionyl transfer ribonucleic acid in mitochondria from Neurospora. Biochemistry. 1970;9:3575–3579. doi: 10.1021/bi00820a011. [DOI] [PubMed] [Google Scholar]
- 14.Feldman F, Mahler H R. Mitochondrial biogenesis. Retention of terminal formylmethionine in membrane proteins and regulation of their synthesis. J Biol Chem. 1974;249:3702–3709. [PubMed] [Google Scholar]
- 15.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 16.Galper J B, Darnell J E. Mitochondrial protein synthesis in HeLa cells. J Mol Biol. 1971;57:363–367. doi: 10.1016/0022-2836(71)90354-8. [DOI] [PubMed] [Google Scholar]
- 17.Galper J B, Darnell J E. The presence of N-formyl-methionyl-tRNA in HeLa cell mitochondria. Biochem Biophys Res Commun. 1969;34:205–214. doi: 10.1016/0006-291x(69)90633-0. [DOI] [PubMed] [Google Scholar]
- 18.Guillon J-M, Mechulam Y, Schmitter J-M, Blanquet S, Fayat G. Disruption of the gene for Met-tRNAfMet formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halbreich A, Rabinowitz M. Isolation of Saccharomyces cerevisiae mitochondrial formyltetrahydrofolic acid:methionyl-tRNA transformylase and the hybridization of mitochondrial fmet-tRNA with mitochondrial DNA. Proc Natl Acad Sci USA. 1971;68:294–298. doi: 10.1073/pnas.68.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heckman J E, Hecker L I, Schwartzbach S D, Barnett W E, Baumstark B, RajBhandary U L. Structure and function of initiator methionine tRNA from the mitochondria of Neurospora crassa. Cell. 1978;13:83–95. doi: 10.1016/0092-8674(78)90140-x. [DOI] [PubMed] [Google Scholar]
- 21.Hopper A K, Furukawa A H, Pham H D, Martin N C. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell. 1982;28:543–550. doi: 10.1016/0092-8674(82)90209-4. [DOI] [PubMed] [Google Scholar]
- 22.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C P, Dyson M R, Mandal N, Varshney U, Bahramian B, RajBhandary U L. Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc Natl Acad Sci USA. 1992;89:9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C P, Seong B L, RajBhandary U L. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J Biol Chem. 1991;266:18012–18017. [PubMed] [Google Scholar]
- 26.Liao H-X, Spremulli L L. Initiation of protein synthesis in animal mitochondria. Purification and characterization of translational initiation factor 2. J Biol Chem. 1991;266:20714–20719. [PubMed] [Google Scholar]
- 27.Ma L, Spremulli L L. Cloning and sequence analysis of the human mitochondrial translation initiation factor 2 cDNA. J Biol Chem. 1995;270:1859–1865. doi: 10.1074/jbc.270.4.1859. [DOI] [PubMed] [Google Scholar]
- 28.Mahler H R, Dawidowicz K, Feldman F. Formate as a specific label for mitochondrial translation products. J Biol Chem. 1972;247:7439–7442. [PubMed] [Google Scholar]
- 29.Marcker K, Sanger F. N-Formyl-methionyl-sRNA. J Mol Biol. 1964;8:835–840. doi: 10.1016/s0022-2836(64)80164-9. [DOI] [PubMed] [Google Scholar]
- 30.Myers A M, Pape L K, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton D T, Creuzenet C, Mangroo D. Formylation is not essential for initiation of protein synthesis in all eubacteria. J Biol Chem. 1999;274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 32.Ogur M, St. John R, Nagai S. Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- 33.RajBhandary U L. Initiator transfer RNAs. J Bacteriol. 1994;176:547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roca J, Gartenberg M R, Oshima Y, Wang J C. A hit-and-run system for targeted genetic manipulations in yeast. Nucleic Acids Res. 1992;20:4671–4672. doi: 10.1093/nar/20.17.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sala F, Kuntzel H. Peptide chain initiation in homologous and heterologous systems from mitochondria and bacteria. Eur J Biochem. 1970;15:280–286. doi: 10.1111/j.1432-1033.1970.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 36.Samuel C E, D'Ari L, Rabinowitz J C. Evidence against the folate-mediated formylation of formyl-accepting methionyl transfer ribonucleic acid in Streptococcus faecalis R*. J Biol Chem. 1970;245:5115–5121. [PubMed] [Google Scholar]
- 37.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J. 1998;17:6819–6826. doi: 10.1093/emboj/17.23.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schofield P, Zamecnik P C. Cupric ion catalysis in hydrolysis of aminoacyl-tRNA. Biochim Biophys Acta. 1968;155:410–416. doi: 10.1016/0005-2787(68)90185-8. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz J H, Meyer R, Eisenstadt J M, Brawerman G. Involvement of N-formylmethionine in initiation of protein synthesis in cell-free extracts of Euglena gracilis. J Mol Biol. 1967;25:571–574. doi: 10.1016/0022-2836(67)90210-0. [DOI] [PubMed] [Google Scholar]
- 40.Sebald W, Wachter E, Tzagoloff A. Identification of amino acid substitutions in the dicyclohexylcarbodiimide-binding subunit of the mitochondrial ATPase complex from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur J Biochem. 1979;100:599–607. doi: 10.1111/j.1432-1033.1979.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 41.Shannon K W, Rabinowitz J C. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J Biol Chem. 1988;263:7717–7725. [PubMed] [Google Scholar]
- 42.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 43.Skala J, van Dyck L, Purnelle B, Goffeau A. The sequence of an 8 kb segment on the left arm of chromosome II from Saccharomyces cerevisiae identifies five new open reading frames of unknown functions, two tRNA genes and two transposable elements. Yeast. 1992;8:777–785. doi: 10.1002/yea.320080911. [DOI] [PubMed] [Google Scholar]
- 44.Smith A E, Marcker K A. N-formylmethionyl transfer RNA in mitochondria from yeast and rat liver. J Mol Biol. 1968;38:241–243. doi: 10.1016/0022-2836(68)90409-9. [DOI] [PubMed] [Google Scholar]
- 45.Staben C, Rabinowitz J C. Formation of formylmethionyl-tRNA and initiation of protein synthesis. In: Blakley R L, Benkovic S J, editors. Folates and pterins. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1984. pp. 457–495. [Google Scholar]
- 46.Steffens G J, Buse G. Studies on cytochrome c oxidase IV. Primary structure and function of subunit II. Hoppe Seylers Z Physiol Chem. 1979;360:613–619. [PubMed] [Google Scholar]
- 47.Takeuchi N, Kawakami M, Omori A, Ueda T, Spremulli L L, Watanabe K. Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure. J Biol Chem. 1998;273:15085–15090. doi: 10.1074/jbc.273.24.15085. [DOI] [PubMed] [Google Scholar]
- 48.Thompson R C, Cline S W, Yarus M. Site directed mutagenesis of the anticodon region: the “universal U” is not essential to tRNA synthesis and function. In: Grunberg-Manago M, Safer B, editors. Interaction of translational and transcriptional controls in the regulation of gene expression. Developments in biochemistry series. Vol. 24. New York, N.Y: Elsevier Science Publishing Co.; 1982. pp. 189–202. [Google Scholar]
- 49.Tzagoloff A, Dieckmann C L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzagoloff A, Macino G, Sebald W. Mitochondrial genes and translation products. Annu Rev Biochem. 1979;48:419–441. doi: 10.1146/annurev.bi.48.070179.002223. [DOI] [PubMed] [Google Scholar]
- 51.Vambutas A, Ackerman S H, Tzagoloff A. Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur J Biochem. 1991;201:643–652. doi: 10.1111/j.1432-1033.1991.tb16325.x. [DOI] [PubMed] [Google Scholar]
- 52.Varshney U, Lee C-P, RajBhandary U L. Direct analysis of aminoacylation levels of tRNAs in vivo. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 53.Varshney U, Lee C P, RajBhandary U L. From elongator tRNA to initiator tRNA. Proc Natl Acad Sci USA. 1993;90:2305–2309. doi: 10.1073/pnas.90.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vernet T, Dignard D, Thomas D Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 55.Walker J E, Lutter R, Dupuis A, Runswick M J. Identification of the subunits of F1F0-ATPase from bovine heart mitochondria. Biochemistry. 1991;30:5369–5378. doi: 10.1021/bi00236a007. [DOI] [PubMed] [Google Scholar]
- 56.Warren M S, Mattia K M, Marolewski A E, Benkovic S J. The transformylase enzymes of de novo purine biosynthesis. Pure Appl Chem. 1996;68:2029–2036. [Google Scholar]
- 57.West M G, Barlowe C K, Appling D R. Cloning and characterization of the Saccharomyces cerevisiae gene encoding NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase. J Biol Chem. 1993;268:153–160. [PubMed] [Google Scholar]
- 58.West M G, Horne D W, Appling D R. Metabolic role of cytoplasmic isozymes of 5,10-methylenetetrahydrofolate dehydrogenase in Saccharomyces cerevisiae. Biochemistry. 1996;35:3122–3132. doi: 10.1021/bi952713d. [DOI] [PubMed] [Google Scholar]
- 59.Yaffe M P. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]