Highlights
-
•
Hypermutation of the Omicron spike protein resulted in enhanced affinity for ACE2 and great potential for immune evasion.
-
•
Neutralizing antibodies are produced in the serum of people who have been infected with the original SRAS-COV-2 strain, and have a certain protective effect on the virus strain.
-
•
The striking immune evasion ability of the Omicron variant resulted in significantly reduced protection from convalescent sera.
-
•
Pseudovirus neutralization test results obtained results similar to those obtained in the live virus assay, but with better discrimination.
Abstract
The emergence and rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant (BA.1.1) has attracted global attention. The numerous mutations in the spike protein suggest that it may have altered susceptibility to immune protection elicited by the existing coronavirus disease 2019 (COVID-19) infection. We used a live virus neutralization test and SARS-CoV-2 pseudotype vesicular stomatitis virus vector-based neutralization assay to assess the degree of immune escape efficiency of the original, Delta (B1.617.2), and Omicron strains against the serum antibodies from 64 unvaccinated patients who had recovered from COVID-19 and the results were strongly correlated. The convalescent serum neutralization was more markedly reduced against the Omicron variant (9.4–57.9-fold) than the Delta variant (2.0–4.5-fold) as compared with the original strain. Our results demonstrate the reduced fusion and notable immune evasion capabilities of the Omicron variants, highlighting the importance of accelerating the development of vaccines targeting them.
1. Introduction
Since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first discovered in Wuhan, China, in 2019, the coronavirus disease 2019 (COVID-19) pandemic has had a tremendous effect on countries and people worldwide. Containing a single-stranded positive-sense RNA genome, SARS-CoV-2 has a specific proofreading ability, but its replication process nevertheless inevitably has a high error rate (or low viral fidelity) (Robson et al., 2020). Alpha, Beta, Gamma, Delta, and Omicron are the five major variants of concerns (VOCs). Discovered at the end of 2021, Omicron (B.1.1.529) is gradually dominating the pandemic after Delta (B.1.617.2), as forecast by mathematical prediction models (Kumar et al., 2022).
The main structure that determines viral infectivity and antigenicity, the S1 subunit of the spike protein consists of the N-terminal domain (NTD) containing the antigenic supersite and the receptor-binding domain (RBD) that binds to angiotensin-converting enzyme 2 (ACE2), which are the two main antibody-binding and mutagenesis sites (Harvey et al., 2021; McCallum et al., 2021). The Omicron spike protein contains 32 mutations, including 15 and 7 mutation sites in the RBD and NTD regions, respectively, and includes deletions, substitutions, and amino acid insertions (Planas et al., 2022). Omicron covers almost all of the important mutation sites of previously identified mutant strains, such as N501Y, which increases RBD–ACE2 affinity and disrupts neutralizing antibody binding (Hoffmann et al., 2022; Supasa et al., 2021), and D614G, which enhances viral replication and transmission (Tao et al., 2021; Zhou et al., 2021). The human body can produce neutralizing antibodies to viruses and gain immunity through natural infection, vaccination, and mixed immunity. However, different immune levels have different immune effects against different variants. Although some studies have demonstrated the existence of cross-protection between different variants, it is difficult to predict the occurrence of mutation sites, which affects antibody neutralization, and breakthrough infections occur, such as infection with variant Beta (B.1.351), and patients with Gamma (P.1) are at risk of re-infection with the Delta variant (Liu et al., 2021). Unvaccinated people in recovery and those who acquired immunity through two doses of the mRNA COVID-19 vaccine have the weakest neutralizing activity against Omicron compared to the wild-type (WT, origin strain) SARS-CoV-2. Nevertheless, a combination of immunization and booster vaccination involving the vaccination of patients in recovery preserves relatively strong neutralizing activity against Omicron (Carreño et al., 2022; Garcia-Beltran et al., 2022; Li et al., 2022).
Therefore, at present, exploring the affinity and titer changes of serum antibodies in recovered patients is of great importance for controlling and treating new mutant strains such as Omicron, and booster vaccination increases the neutralizing effect against Omicron. Moreover, the development of variant vaccines with more extensive cross-protection and monoclonal antibodies with stronger affinity is gradually becoming a research focus, which is of great importance for preventing and treating post-COVID-19 syndrome.
2. Material and methods
2.1. Cell culture
Vero cells (ATCC CCL-81, Sinovac Biotech) were cultured in minimum essential medium (MEM, Gibco). Vero E6 (ATCC CRL-1586) and BHK-21-hACE2 cells stably expressing human ACE2 supplied by Prof. F Xiao-Feng Qin were cultured in high-glucose Dulbecco's medium (DMEM, Gibco). All media were supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 25 mM HEPES. The Vero cell medium was supplemented with 2 mM l-glutamine and all cells were passaged every 2–3 days using trypsin-EDTA (0.25%, Gibco).
2.2. Virus stocks
All experiments were performed using three SARS-CoV-2 strains isolated at our Biosafety Level 3 virology laboratory (Zhejiang Provincial Center of Disease Control and Prevention, Hangzhou, China) as previously described(Gao et al., 2020). SARS-CoV-2/Vero/WGF/2020/WZ122 (WT strain/EPI_ISL_12,040,150) and SARS-CoV-2/Vero/LXG/2021/ZJ28 (Delta/B.1.617.2/EPI_ISL_1,911,196) were isolated from a throat swab and cultured in Vero cells, and SARS-CoV-2/VeroE6/DSh/2021ZJ25 (Omicron/B.1.1/EPI_ISL_12,040,149) was grown in Vero E6 cells. WT passage 3, Delta passage 5, and Omicron passage 3 virus-containing supernatants were harvested at 80% cytopathogenic efficiency (CPE) and viral titers were determined by microdose CPE assay. All virus stocks were sequenced with Illumina NextSeq to verify that they contained the expected spike protein sequence and no changes to the furin cleavage sites.
2.3. Blood samples
Zhejiang Provincial Center of Disease Control and Prevention recruited 64 eligible Wuhan COVID-19 convalescents and collected a total of 102 serum samples, collected 1 month after recovery from 38 of them and 12 months after recovery from all volunteers (see Table 1). Briefly, 64 unvaccinated persons with symptom onset between January 11, 2020, and March 7, 2020, were recruited and their blood samples were collected from February 17, 2020, to March 13, 2021. The participants’ clinical information, including disease severity (normal, mild, asymptomatic), the time between symptom onset and sampling, and age, was captured at sampling. The serum samples obtained after 15-min centrifugation at 2000 rpm were stored in a deep freezer at −80 °C and inactivated for 30 min at 56 °C before usage.
Table 1.
Demographics of all serum donors (n = 64).
| Convalescent serum specimens | ||
|---|---|---|
| Sex, no. (%) | ||
| Female | 38 (59.40) | |
| Male | 26 (40.60) | |
| Age (year), no. (%) | ||
| ≤ 30 | 5 (7.81) | |
| 31–40 | 11 (17.19) | |
| 41–50 | 11 (17.19) | |
| > 50 | 37 (57.81) | |
| Severity, no. (%) | ||
| Severe | 0(00.00) | |
| Moderate | 32(50.00) | |
| Mild | 21(32.81) | |
| Asymptomatic | 11(17.19) | |
| Sampling days after PCR confirmation (mean ± SD) | ||
| Approximately 1 month | 38(29.47±10.39)* | |
| Approximately 1 year | 64(390.50±9.56)† |
†Thirty-eight convalescent serum specimens were sampled for approximately 1 month. †Sixty-four convalescent serum specimens were sampled for approximately 1 year.
2.4. Ethical approval
The study protocol was approved by the Ethics Committee of the Zhejiang Provincial Center of Disease Control and Prevention.
2.5. Pseudoviral neutralization assay
The vesicular stomatitis virus (VSV)-based pseudovirus system was used to assess cross-neutralizing activities in 102 convalescent serum specimens. The SARS-CoV-2/WT, SARS-CoV-2/Delta, and SARS-CoV-2/Omicron pseudovirus systems were provided by the Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College. The serum samples were diluted, transferred to 96-well culture plates, and mixed with the pseudoviruses (1 × 105 median tissue culture infectious dose [TCID50]/well).
After 1-h incubation at 37 °C, 2 × 104 per well trypsinized BHK-21-hACE2 cells were added to the 96-well culture plates containing virus–serum. After 48 h, the number of green fluorescent protein–positive cells were collected with a multi-well plate imager (Spark Cyto, Tecan). Then, the reciprocal of the dilution multiple corresponding to a 50% reduction in fluorescence (IC50) as compared to the negative control was designated as the neutralizing antibody titer using nonlinear regression curve fitting (normalized response, variable slope) in GraphPad.
2.6. Live virus neutralization test
The neutralization potential of the convalescent serum samples was measured using a live virus-based microneutralization assay (lVNT). The serum samples were serially diluted 2-fold with cell culture medium and mixed with 100 TCID50/50 µL virus suspension in 96-well plates at a ratio of 1:1. After 2-h incubation, 1–2 × 104 Vero E6 cells were added to the serum–virus mixture and the plates were incubated for 3 days at 37 °C in a 5% CO2 incubator. The cytopathic effect (CPE) of each well was recorded under microscopes and the neutralizing titer was calculated by the dilution number of 50% protective condition.
2.7. Statistical analysis
All data are presented as the mean ± SD. All statistical analyses were conducted using GraphPad Prism 9.4.1. Statistical significance was determined using Kruskal–Wallis test with Dunn's multiple-testing correction. Mann–Whitney two-sided U test were applied to compare two groups. And correlations were assessed by two-sided Spearman rank correlation tests, two important parameters from the correlates analysis shown as R-value and P-value. Values of p < 0.05 are considered statistically significant.
3. Results
3.1. Most patients with COVID-19 developed anti-SARS-CoV-2 antibodies
Between January 11, 2020, and May 7, 2020, we enrolled as a first step 64 convalescent patients who had recovered from COVID-19. They had been diagnosed with SARS-CoV-2 infection by reverse transcription–PCR. The samples from these cases were classified according to the National Institutes of Health COVID-19 clinical categorization criteria into three categories of disease severity: asymptomatic, mild illness, and moderate illness(2021). Individuals who test positive for the SARS-CoV-2 by virologic testing, but show no symptoms of COVID-19, are considered asymptomatic (n = 11). Mild illness (n = 21) is characterized by a combination of symptoms and signs such as fever, cough, sore throat, body aches, headache, muscle pain, nausea, vomiting, diarrhea, or loss of taste or smell. However, individuals with mild illness do not exhibit lower respiratory tract infection. Fifty percent of the cases were moderate COVID-19 (n = 32), defined as having lower respiratory disease, as confirmed by clinical assessment or imaging, and oxygen saturation (SpO2) ≥94% in indoor air at sea level.
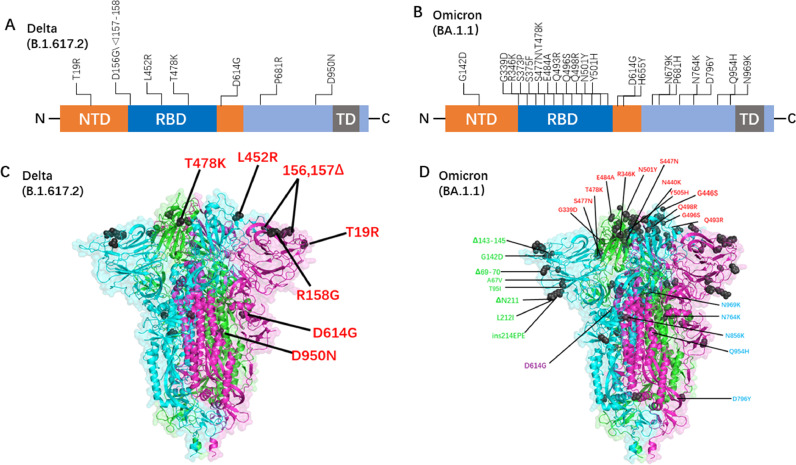
We used live viruses of different mutants and constructed pseudoviruses containing all Delta and Omicron with mutations (Fig. 1), tested the serum-neutralizing antibodies of the recovered patients, and tracked and compared the ability of the SARS-CoV-2 neutralizing antibodies to prevent infection by the mutants. Consistent with previously reported results (Cao et al., 2022), Omicron had a strong immune escape potential and much stronger neutralization escape potential than Delta.
Fig. 1.
(A) Linear mutation diagram of Delta spike proteins (https://outbreak.info). (B) Linear mutation diagram of Omicron spike proteins (https://outbreak.info). (C) Full-length Delta spike proteins (3-dimensional structure) were built to correspond to the relative mutation sites, which were used to construct the pseudotyped Delta virus. (D) Full-length Omicron spike proteins (3-dimensional structure) were built to correspond to the relative mutation sites, which were used to construct the pseudotyped Omicron virus.
3.2. Immune escape of the omicron variant resulted in a significant decrease in the protective effect of convalescent serum
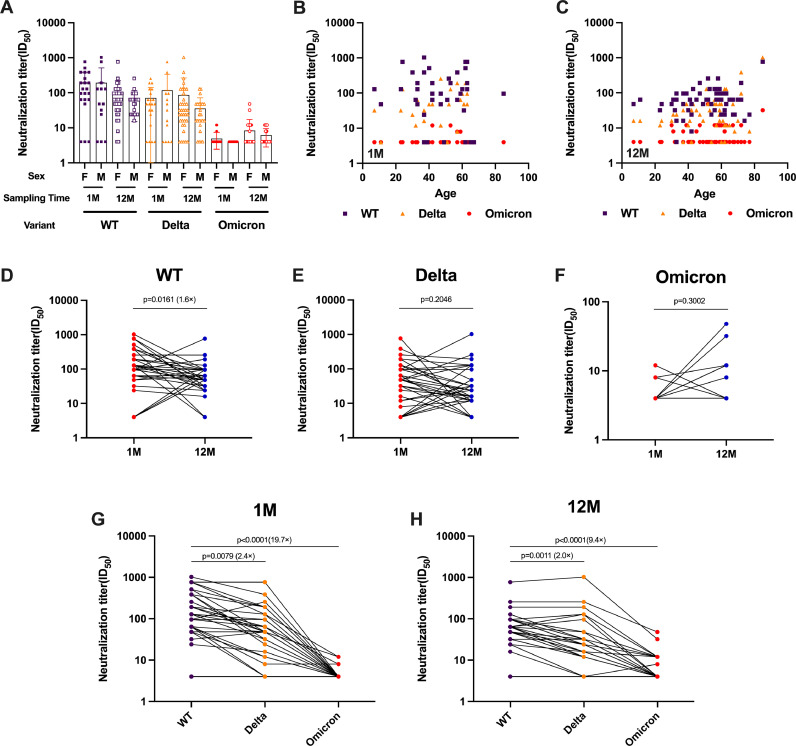
To evaluate the immune evasion of WT, Delta, and Omicron to serum-neutralizing antibodies in recovered WT-infected patients, we obtained longitudinal serum samples from 64 patients who had recovered from COVID-19 and performed a live virus neutralization test (lVNT). The median participant age was 54 years and their serum was sampled prospectively at 1 month and 12 months after recovery. There was no correlation between the level of neutralizing antibodies in convalescent serum and gender or age (Fig. 2A-2C). Over time, serum antibody titers against the WT strain decreased significantly, while neutralizing antibody levels against Delta and Omicron remained relatively stable. The serum neutralizing antibody titer of 1 M against the WT strain (geometric mean titer, GMT 86.5) was 1.6 times higher than 12 M (GMT 57.0) (Fig. 2D). The results of the Delta and Omicron BA.1 variant live virus neutralization assay after two different blood draws showed no statistically significant difference (Fig. 2E-2F). One month after recovery, the neutralizing antibody titer against the wild-type strain was significantly higher than that against the current prevalent mutant strains, Delta (GMT 36) and Omicron BA.1 (GMT 4.4), at 2.4-fold and 19.7-fold, respectively (Fig. 2G). However, the antibody titers of 12 M against the wild-type strain were only 2.0-fold and 9.4-fold higher than those against the Delta (GMT 26.8) and Omicron BA.1 (GMT 5.8) variant strains, respectively, indicating a decrease in the difference (Fig. 2H).
Fig. 2.
Shown are live virus neutralization test results from 128 serum samples obtained from 64 volunteers 1 or 12 months after recovery from infection with the original strain (WT). Neutralization of authentic viruses was performed by cytopathic effect (CPE)-based assay using a viral titer of 100 TCID50. The neutralization titer of the serum sample was calculated as the reciprocal of the highest dilution that protected more than 50% of cells from CPE. Sera with different neutralization titer against WT, Delta, and Omicron at different times are connected by lines. The numbers on the graph represent the ratio of geometric mean titers (GMT). (A-C) The 50% neutralization titer (NT50) by sex (A) and age (B, C) after 1 month or 12 months. (D–F) Neutralization analysis of convalescent serum against the origin strain (WT) (D), Delta (E), and Omicron (F) live viruses at 1 month and 12 months. The neutralization activity of serum from 64 patients who had recovered from coronavirus disease 2019 (COVID-19) was tested. Data represent the median effective dose (ED50) of three independent experiments. (G, H) Convalescent serum neutralization sensitivity against Delta and Omicron at 1 month (G) and 12 months (H) after recovery. Data represent the ED50 of six independent experiments. Statistics were calculated using Kruskal–Wallis test with Dunn's multiple-testing correction (A-C, G-H), Mann–Whitney U test (D–F). Correlations were assessed by two-sided Spearman rank correlation tests (B, C).
3.3. SARS-CoV-2 pseudoviruses neutralized by convalescent serum also demonstrated lower neutralization titers than infectious variants
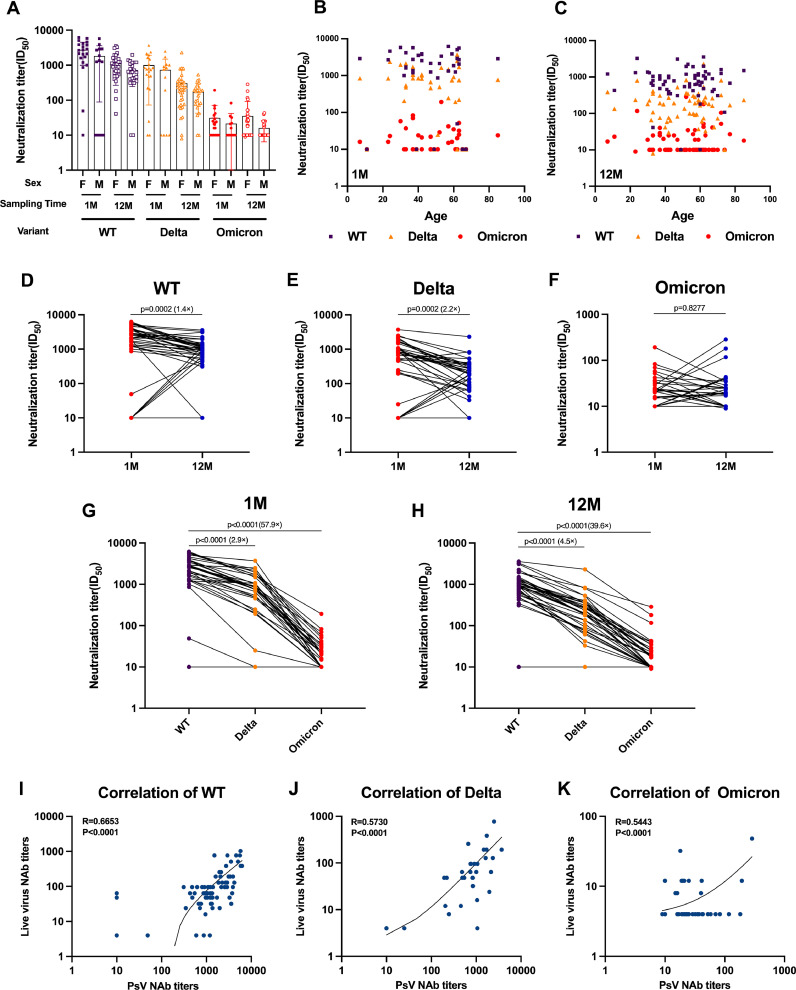
We used a SARS-CoV-2 spike protein-pseudotyped VSV vector-based neutralization assay (pVNT) (Bošnjak et al., 2021; Hu et al., 2021; Ma et al., 2022; Yuan et al., 2021) and obtained results similar to those obtained in the live virus assay (Fig. 3I-3K), but with better discrimination. Gender and age were found to have no impact on serum-neutralizing antibody levels (Fig. 3A-3C). Over time, the decline in neutralizing antibody titers against the WT was similar to that of live virus, with a 1.4-fold decrease in antibody titers (GMT 672.8) at 12 M compared to 1 M (GMT 1116.0) (Fig. 3D). The decline in serum-neutralizing antibody levels was more obvious for the Delta variant, with geometric mean titers decreasing from 390.2 at 1 M post-recovery to 148.4 at 12 M post-recovery (Fig. 3E). In contrast, neutralizing antibody levels against Omicron remained relatively stable (Fig. 3F). At 1 M post-recovery, neutralizing antibody titers against the WT strain was 2.9-fold and 57.9-fold higher than those against the Delta and Omicron BA.1 (GMT 19.3), respectively (Fig. 3G). Similarly, at 12 months post-recovery, antibody titers against the wild-type strain were 4.5-fold higher and 39.6-fold higher than those against the Delta and Omicron BA.1 variant strains (GMT 17.0), respectively (Fig. 3H).
Fig. 3.
Pseudovirus neutralization test results. (A-C) NT50 by sex (A) and age (B,C) after 1 month or 12 months. (D–F) Neutralization analysis of convalescent serum against the WT (D), Delta (E), and Omicron (F) pseudoviruses at 1 month and 12 months after recovery. The neutralization activity of serum from 64 patients who had recovered from COVID-19 was tested. Data represent the ED50 of three independent experiments. (G, H) Convalescent serum neutralization sensitivity against Delta and Omicron at 1 month (G) and 12 months (H) after recovery. The neutralization ED50 and ratio compared to the origin strain WT is indicated. Data represent the ED50 of six independent experiments. (I-K) Correlation between live virus-neutralizing antibody titers and pseudovirus-neutralizing antibody titers of WT, Delta and Omicron strains. Statistics were calculated using Kruskal–Wallis test with Dunn's multiple-testing correction (A-C, G-H), Mann–Whitney U test (D–F). Correlations were assessed by two-sided Spearman rank correlation tests (B-C,I-K).
3.4. Symptom severity has little effect on protection against reinfection after recovery
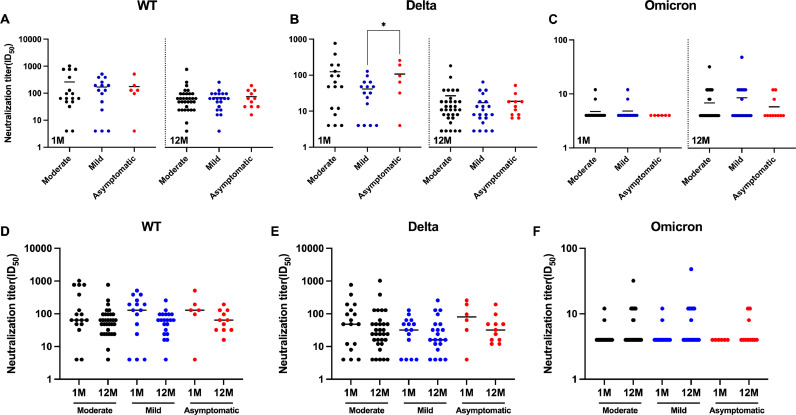
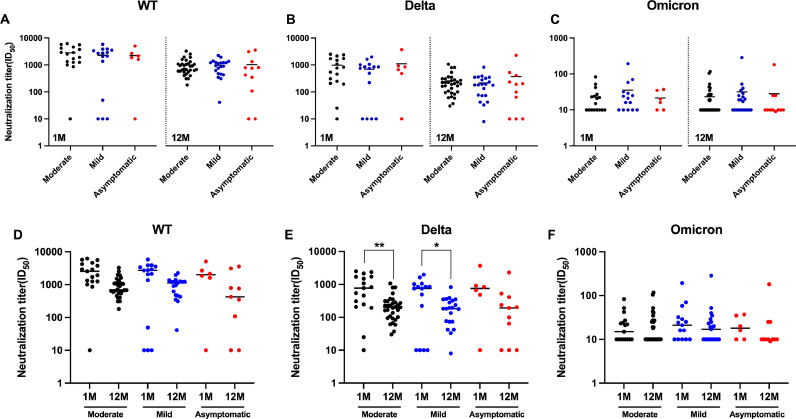
We categorized the volunteers who participated in this study into mild, moderate, and asymptomatic groups based on the severity of their infections. We found that patients with different severity levels generally had no statistically significant differences in serum neutralizing antibody titers against WT, Delta, and Omicron BA.1 at 1 M or 12 M post-recovery, except for mild and asymptomatic patients who showed statistical differences in neutralizing antibody titers against Delta at 1 M post-recovery (Fig. 4A-4C). Moreover, there were no statistically significant differences in serum-neutralizing antibody experiments at different times after recovery for the same severity level, whether against WT, Delta, or Omicron BA.1 (Fig. 4D-4F).
Fig. 4.
Shown are live virus neutralization test results of neutralizing antibodies against three virus strains by serum from recovered patients with different COVID-19 severity. (A–F) Convalescent serum neutralization analysis at 1 month and 12 months post-recovery. (A–C) Comparison of serum neutralizing antibody levels at different severity levels after the same recovery time. (D–E) Comparison of serum neutralizing antibody levels after different recovery times of the same severity. Statistics were calculated using Mann–Whitney two-sided U test and Kruskal–Wallis test with Dunn's multiple-testing correction.
The results of the pVNT were similar to those of the lVNT. Statistically significant differences in neutralizing antibody titers against Delta at different times post-recovery were only observed for the moderate and mild infection groups (Fig. 5E), while no significant differences were observed in other groups (Fig. 5A-5D, 5F).
Fig. 5.
Shown are pseudoviral neutralization test results of neutralizing antibodies against three virus strains by serum from recovered patients with different COVID-19 severity. (A–F) Convalescent serum neutralization analysis at 1 month and 12 months post-recovery. (A–C) Comparison of serum neutralizing antibody levels at different severity levels after the same recovery time. (D–E) Comparison of serum neutralizing antibody levels after different recovery times of the same severity. Statistics were calculated using Mann–Whitney two-sided U test and Kruskal–Wallis test with Dunn's multiple-testing correction.
4. Discussion
This study involved a cohort of COVID-19 patients with prospective follow-up over 1 year. It presents essential information on the persistence of SARS-CoV-2 antibodies after mild COVID-19. Our data are in good agreement with preliminary reports on the effect of Omicron on the in vitro neutralizing activity of convalescent serum (Cele et al., 2022; Gallais et al., 2021; Rössler et al., 2022). Previous studies have shown that many mutations in the Omicron variant significantly reduced the neutralizing sensitivity to convalescent serum, and our results also showed that the level of neutralizing antibodies against Omicron BA.1 was significantly lower than that of WT and Delta strains, which also consistent with a related report describing a significantly reduced protection against Omicron reinfection (Pulliam et al., 2022). We used an in vitro assay system that used both live virus and pseudovirus and obtained consistent results. The level of neutralizing antibodies against SARS-CoV-2 mutants in recovered patients was low and tended to decline over time. Therefore, we predict that vaccines targeting SARS-CoV-2 prototype antigens will be less protective against Omicron variants and that protection will decrease over time. Therefore, it is necessary to carry out booster vaccination of existing vaccines, as well as the development of vaccines targeting new mutant strains of SARS-CoV-2.
The sharp increase in Omicron mutation sites resulted in severe inhibition of the convalescent serum neutralization activity. The Omicron variant shares RBD mutations with previously focused variants: K417N (Lys417→Asn), T478K, and N501Y (Asn501→Tyr), where residue Y501 mutated in Omicron to create a π-stacking interaction with Y41 in ACE2. In contrast, the disappearance of the vital salt bridge between residue K417 and ACE2 residue D30 decreases affinity (Cho et al., 2021; Laffeber et al., 2021; Liu et al., 2021; Mannar et al., 2022, 2021; Zhu et al., 2021). The N501Y and K417N mutations confer increased and decreased binding affinity to ACE2, respectively. These mutational effects on ACE2 affinity generally remained unchanged when mutations were present in combination. Nevertheless, the Omicron RBD contains other mutations, most of which reduce receptor binding in high-throughput assays. Substitution of residue R493 was observed at the Q493R (Gln493→Arg), G496S (Gly496→Ser), and Q498R mutation sites, where residue R493 replaced the hydrogen bond of ACE2 residue E35 with a new salt bridge, residue S496 generated a hydrogen bond to form a new cross-link with ACE2 residue K353 at the interface, residue R498 cross-linked with the ACE2 residue D38 formed a new salt bridge while maintaining a hydrogen-bonded cross-link with ACE2 residue Q42 (Mannar et al., 2022). However, mutations at the G339D (Gly339→Asp), N440K (Asn440→Lys), S447N (Ser447→Asn), and Q498R (Gln498→Arg) sites led to decreased affinity (Starr et al., 2020; Zahradník et al., 2021). The Omicron RBD binds ACE2 2.5 times more than the original SARS-CoV-2 strain, and although several RBD mutations reduce ACE2 binding, the Omicron spike protein is generally compensatory for ACE2 binding affinity mutation (Cameroni et al., 2022; Shah and Woo, 2021).
In conclusion, numerous mutations on the surface of the Omicron spike protein, including the immunodominance of RBD, are expected to help the virus escape the serum-neutralizing antibodies caused by previous SARS-CoV-2 infection. As the mutation site of the Omicron variant and previous variants has certain repeatability, designing a next-generation COVID-19 vaccine based on the Omicron spike protein is expected to achieve broad-spectrum protection against all variants. Meanwhile, we still recommend increasing the vaccination rate of the third dose, starting the vaccination program for the fourth dose, and carrying out sequential vaccination to curb the COVID-19 pandemic.
Funding
This study was supported by Key Research and Development Program of Zhejiang Province (program number 2021C03044), Major Health Science and Technology Projects of Zhejiang Province (projects number WKJ-ZJ-2105), Science and Technology Program of Zhejiang Province (grant no. 2022C03017), and National Key R&D Program of China (projects number 2021YFC2301200).
Declaration of Competing Interest
All authors declare no conflict of interest in this study.
Acknowledgments
The author thanks all the researchers, doctors, nurses, medical technicians, front-line workers, and public health officials for their hard work during this pandemic. We also thank the Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College; Suzhou Institute of Systems Medicine for helping with the pseudovirus neutralization assay.
Contributor Information
Yanjun Zhang, Email: yjzhang@cdc.zj.cn.
Jinsong Huang, Email: huangjinsongyz@126.com.
Keda Chen, Email: chenkd@zjsru.edu.cn.
Data availability
Data will be made available on request.
References
- NIH . National Institutes of Health (US)[CE: Author name is missing in reference 1. NIH inserted dummy, Pleaes check and update in reference as well as in the text citation]; Bethesda (MD): 2021. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. [PubMed] [Google Scholar]
- Bošnjak B., Stein S.C., Willenzon S., Cordes A.K., Puppe W., Bernhardt G.…Förster R. Low serum neutralizing anti-SARS-CoV-2 S antibody levels in mildly affected COVID-19 convalescent patients revealed by two different detection methods. Cell Mol. Immunol. 2021;18(4):936–944. doi: 10.1038/s41423-020-00573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K.…Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602(7898):664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A.…Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H.…Krammer F. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H.…Sigal A. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602(7898):654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Gonzales-Wartz K.K., Huang D., Yuan M., Peterson M., Liang J.…Tan J. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021;13(616):eabj5413. doi: 10.1126/scitranslmed.abj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais F., Gantner P., Bruel T., Velay A., Planas D., Wendling M.J.…Fafi-Kremer S. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. 2021;71 doi: 10.1016/j.ebiom.2021.103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M.…Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L.…Balazs A.B. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185(3):457–466. doi: 10.1016/j.cell.2021.12.033. e454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M.…Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A.…Pöhlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185(3):447–456. doi: 10.1016/j.cell.2021.12.032. e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wei X.Y., Xiang J., Peng P., Xu F.L., Wu K.…Huang A.L. Reduced neutralization of SARS-CoV-2 B1.617 variant by convalescent and vaccinated sera. Genes Dis. 2021 doi: 10.1016/j.gendis.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 2022;94(4):1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 Variants. J. Mol. Biol. 2021;433(15) doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Guo Y., Fang Z., Zhang H., Zhang Y., Chen K. Analysis of the protective efficacy of approved COVID-19 vaccines against various mutants. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.804945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A.…Screaton G.R. Reduced neutralization of SARS-CoV-2 B1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220–4236. doi: 10.1016/j.cell.2021.06.020. e4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Chen X., Mei F., Xiong Q., Liu Q., Dong L.…Lan K. Drastic decline in sera neutralization against SARS-CoV-2 Omicron variant in Wuhan COVID-19 convalescents. Emerg Microbes Infect. 2022;11(1):567–572. doi: 10.1080/22221751.2022.2031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S.…Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375(6582):760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Zhou S.…Subramaniam S. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep. 2021;37(12) doi: 10.1016/j.celrep.2021.110156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C.…Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332–2347. doi: 10.1016/j.cell.2021.03.028. e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J.…Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J.…Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P.…Ng W.L. Coronavirus RNA Proofreading: molecular Basis and Therapeutic Targeting. Mol. Cell. 2020;79(5):710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 Omicron variant neutralization in serum from vaccinated and convalescent persons. N. Engl. J. Med. 2022;386(7):698–700. doi: 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S.…Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M.…Screaton G.R. Reduced neutralization of SARS-CoV-2 B1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184(8):2201–2211. doi: 10.1016/j.cell.2021.02.033. e2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L.…Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Yu L., Jin Z., Wang Y., Gao M., Ding H.…Lv H. Predictive analysis of the neutralization activity in convalescent plasmas from COVID-19 recovered patients in Zhejiang Province. Front. Cell Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.650487. China, January-March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J.…Schreiber G. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6(9):1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F.…Beer M. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zhu X., Mannar D., Srivastava S.S., Berezuk A.M., Demers J.P., Saville J.W.…Subramaniam S. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19(4) doi: 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.