Abstract
The homolog of the chromosomally encoded stationary-phase sigma factor RpoS in Borrelia burgdorferi was inactivated using gyrBr as a selectable marker. Two-dimensional nonequilibrium pH gradient electrophoresis of stationary-phase cell lysates identified at least 11 differences between the protein profiles of the rpoS mutant and wild-type organisms. Wild-type B. burgdorferi had a growth phase-dependent resistance to 1 N NaCl, similar to the stationary-phase response reported for other bacteria. The B. burgdorferi rpoS mutant strain was less resistant to osmotic stress in stationary phase than the isogenic rpoS wild-type organism. The results indicate that the B. burgdorferi rpoS homolog influences protein composition and participates in stationary-phase-dependent osmotic resistance. This rpoS mutant will be useful for studying regulation of gene expression in response to changing environmental conditions.
Lyme disease is the most common arthropod-borne disorder in the United States, with a variable spectrum of clinical manifestations ranging from localized infection to systemic disease (1, 11, 54, 55). Borrelia burgdorferi, the spirochetal agent of Lyme disease, is normally maintained in an enzootic cycle between wild mammals and Ixodes ticks (30). In ticks and in mammals, the bacteria are present only in low numbers throughout most of the infectious cycle. However, periods of rapid growth occur in ticks after blood engorgement (13, 43). Although little is known about the growth kinetics of B. burgdorferi within the mammalian host, spirochetes appear to be present in considerably lower densities in chronically infected mice than in acutely infected animals (4). B. burgdorferi, similar to other bacterial pathogens, undoubtedly has a repertoire of adaptive molecular responses to environmental signals to assist survival within and successful transmission between its hosts (16, 35, 36). For example, in vivo, differential synthesis of membrane proteins occurs during tick transmission (3, 52) and mammalian infection (19, 41). Temperature, pH, and growth phase also have been shown to modify the protein profile of cultivated B. burgdorferi (7, 8, 12, 23, 37, 44, 52, 53, 56; J. G. Frye, B. K. Kremer, T. R. Hoover, and F. C. Gherardini, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. D/B-259, p. 259).
Despite this evidence for the presence of adaptive responses in B. burgdorferi, the molecular mechanisms responsible for regulation of gene expression in response to environmental changes are unknown. This lack is due to the difficulty in studying the physiology of a fastidious organism and the limited availability of genetic tools to manipulate B. burgdorferi.
An important mechanism involved in regulation of bacterial gene expression in response to environmental signals is the use of alternative sigma factors to alter RNA polymerase specificity (59). Three genes encoding sigma factor homologs are present in the genome of B. burgdorferi: rpoS (ςS), ntrA (ς54), and rpoD (ς70). In Escherichia coli, RpoS controls a regulon of more than 30 genes positively or negatively regulated in response to starvation and transition to stationary phase (15, 31). In the natural environment, B. burgdorferi and other bacteria often encounter nutrient limitations, resulting in periods of negligible or absent growth. E. coli and other bacteria respond to nutrient starvation by entering a metabolic state referred to as stationary phase (25, 26, 42, 58), allowing them to survive environmental stresses such as oxidative stress, heat, high salt, and near-UV radiation. The stationary-phase response depends in part on the expression of the sigma factor rpoS (22, 31). Several bacterial pathogens, including Salmonella (14, 29), Yersinia enterocolitica (24), Vibrio cholerae (60), and Legionella pneumophila (20), have RpoS homologs with various roles. For example, in Salmonella enterica serovar Typhimurium RpoS regulates chromosomal and plasmid-encoded virulence genes, and the 50% lethal dose in mice is 1,000-fold higher for a Salmonella serovar Typhimurium rpoS mutant than for the wild-type strain (14, 29).
Here, we begin a study to elucidate the role of RpoS in B. burgdorferi biology. As a first step, we inactivated the rpoS locus by allelic exchange with gyrBr, a mutated form of the B subunit of DNA gyrase, as a selectable marker. Although this technique has been used previously to inactivate several B. burgdorferi genes located on a 26-kb circular plasmid (cp26) (5, 57), rpoS is the first chromosomal gene to be inactivated in B. burgdorferi. Stationary-phase cells of the isogenic B. burgdorferi rpoS mutant have an altered protein composition compared with the rpoS wild-type organism and are more sensitive to osmotic stress. These results indicate that RpoS participates in the stationary-phase-related adaptive response.
MATERIALS AND METHODS
Bacterial strains.
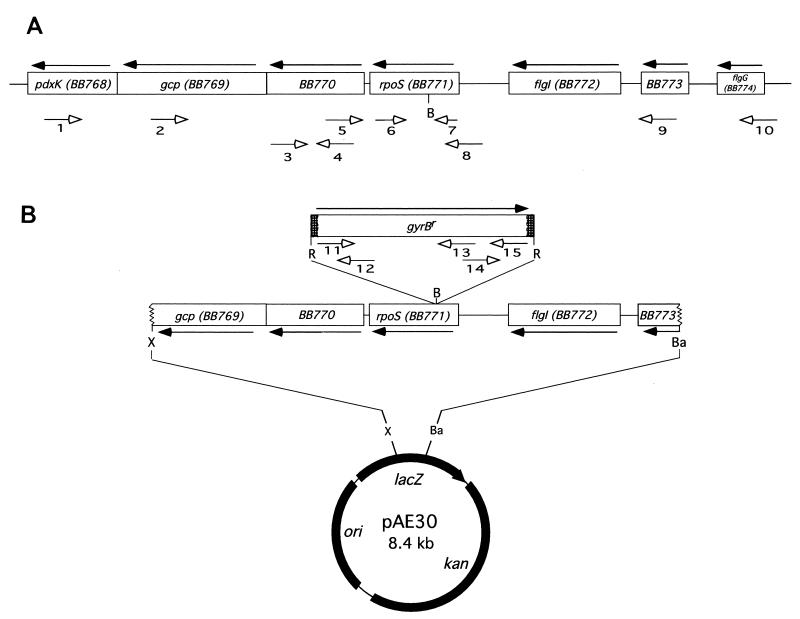
The B. burgdorferi strains used in this study are listed in Table 1. Bacteria were grown at 35°C in liquid BSK-H medium (Sigma, St. Louis, Mo.) supplemented with 6% rabbit serum (Sigma) (2) or in solid BSK medium (45, 46). B31 (ATCC 35210) was originally isolated from a tick collected on Shelter Island, N.Y. (6). B31 clone A (B31-A) is a clone derived from culture-attenuated, noninfectious B31. B31 clone 4A (B31-4A) is a low-passage (P3) infectious clone derived from B31. Coumermycin A1-resistant clones B31-A59 (A59), B31-A74 (A74), and B31-A29 (A29) were derived from B31-A by transformation with the recombinant plasmid pAE30 (described below). This plasmid contains a copy of gyrBr, the mutated B subunit of DNA gyrase from B31-NGR (45), conferring resistance to coumermycin A1, inserted in a 4.2-kb B. burgdorferi chromosomal DNA fragment.
TABLE 1.
B. burgdorferi strains used in this study
| Strain | rpoS locus | gyrB locus | Coumermycin A1 phenotypea | Reference |
|---|---|---|---|---|
| B31-A | wtb | wt | Sensitive | 45 |
| B31-4A | wt | wt | Sensitive | 10 |
| B31-NGR | wt | gyrBr | Resistant | 45 |
| B31-A59 | wt | gyrBr | Resistant | This study |
| B31-A74 | rpoS::gyrBr | wt | Resistant | This study |
| B31-A29 | rpoS::gyrBr | wt | Resistant | This study |
Coumermycin resistance was defined as the ability to grow in the presence of ≥0.5 μg of coumermycin A1 per ml in liquid or solid BSK.
wt, wild type.
Spirochetes were counted by dark-field microscopy with a Petroff-Hausser counting chamber. The number of spirochetes was determined once or twice daily. Stationary phase was defined as the period of reduced bacterial growth after exponential growth. The onset of stationary phase usually corresponds to a cell density of approximately 108 spirochetes/ml.
Construction of the rpoS mutant.
Recombinant plasmid pAE30 was constructed to inactivate the B. burgdorferi rpoS gene. A 4.2-kb fragment encompassing the rpoS locus of B31-4A was amplified from chromosomal DNA by PCR (38, 49) with primers 2 and 9 (Table 2 and Fig. 1A). The PCR product was cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.), which contains an ampicillin resistance gene. Ampicillin is a beta-lactam antibiotic and is related to antimicrobial agents used to treat Lyme disease. Hence, this drug resistance phenotype cannot be introduced into B. burgdorferi. Therefore, the rpoS-containing fragment was recloned into pOK12, a 2.1-kb low-copy-number plasmid that contains a kanamycin resistance gene. The gyrBr gene and its putative promoter were amplified by PCR from B31-NGR chromosomal DNA (Table 1) with primers 11 and 15 (Fig. 1B and Table 2) and cloned into pCR2.1. The gyrBr gene was excised from pCR2.1 by EcoRI digestion and inserted into the single BbsI site of rpoS cloned in pOK12, which has ends compatible with EcoRI. The resulting plasmid (pAE30) was confirmed to contain the rpoS::gyrBr construct by DNA sequencing with a 373A instrument (Applied Biosystems, Foster City, Calif.) (Fig. 1B). Plasmid DNAs were purified from E. coli with Qiagen purification kits (Qiagen, Chatsworth, Calif.). Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs (Beverly, Mass.). The sequence of the B. burgdorferi rpoS locus was obtained from the web page of the Institute for Genomic Research at http://www.tigr.org, where rpoS has accession number BB771.
TABLE 2.
Oligonucleotide primers used in this study
| Primer no.a | Designation | Sequence (5′→3′) | Gene |
|---|---|---|---|
| 1 | rpoSfla-F1 | TCCATCGTCAGCAAACACAGG | pdxK (BB768) |
| 2 | rpoSfla-F2 | TGTTCGATGTTTGGACCTCC | gcp (BB769) |
| 3 | BB770-F1 | TGTTTCCTCGTAAAGATTTAATAAGTTGTC | BB770 |
| 4 | BB770-B1 | GGATAAAAAAGCCAAAAGCAAACC | BB770 |
| 5 | rpoS-F1 | CCTATTTTTAAGTGGGGCATTTGC | BB770 |
| 6 | rpoS-F3 | GGTGCTTTTTTTGGGACTATTG | rpoS (BB771) |
| 7 | rpoS-B3 | GCAGGACAAATACAAAGAGGC | rpoS (BB771) |
| 8 | rpoS-B1 | AATTGGCACAGTTTTTGCATGG | 5′-rpoS |
| 9 | rpoSfla-B2 | TTAAGGTAAAAAGCAACAATTCATAC | BB773 |
| 10 | rpoSfla-B1 | ATCCAAAACTCAATTACAATATCTGAAGAG | 5′-flgG |
| 11 | U178+BglII | ACCAGATCTTGTTGGTTTTAGCACTATA | 5′-gyrBr |
| 12 | 246R | CTCTTCATGAATATCGGTAGG | gyrBr |
| 13 | 1580R | TTATAAAGAGGAGGCATGGC | gyrBr |
| 14 | 1696F | CTTCAGAGATATAAAGGGCTTGGG | gyrBr |
| 15 | 1905R+BclI | ACCTTGATCATTACACATCAAGATTAATTAC | gyrBr |
| 16 | Fl6 | TTCAGGGTCTCAAGCGTCTTGGACT | flaB |
| 17 | Fl7 | GCATTTTCAATTTTAGCAAGTGATG | flaB |
The numbers refer to the relative positions and orientations of primers in the rpoS flanking region and gyrBr given in Fig. 1.
FIG. 1.
Schematic diagram of the B. burgdorferi chromosomal rpoS gene and plasmid pAE30. (A) Location of rpoS in a 6-kb chromosomal region. (B) Construct pAE30 used for transformation of B. burgdorferi. The gyrBr gene is inserted in the single BbsI site of the rpoS locus. Solid arrows indicate direction of gene transcription; open arrows represent primers used in this study, and their numbers refer to the primers listed in Table 2. R, EcoRI; B, BbsI; Ba, BamHI; X, XbaI. Thick lines represent the pOK12 vector, and thin lines represent the B. burgdorferi insert.
Transformation of B. burgdorferi.
Transformation of B31-A with pAE30 by electroporation was performed as described previously (45, 51). Each transformation used 40 μg of DNA in 5 μl of distilled water. After electroporation, the bacteria were resuspended in 10 ml of liquid BSK-H and incubated overnight at 35°C. The transformed bacteria were plated in solid BSK with 0.5 μg of coumermycin A1 per ml. Plating efficiency was determined to be between 50 and 70%, as assessed by growth on solid BSK without antibiotics. The transformation frequency (2 × 10−5) was calculated as the ratio of coumermycin-resistant CFU relative to the total number of CFU in the transformed cultures.
Screening of B. burgdorferi transformants by PCR.
Coumermycin-resistant colonies were screened for allelic exchange at the rpoS locus by PCR. Individual colonies picked with sterile toothpicks were added to tubes containing a 20-μl PCR mix, and oligonucleotides 5 and 8 (Table 2 and Fig. 1A) were used to amplify a fragment that spanned the rpoS gene. Reaction conditions were 94°C for 1 min and then 30 cycles with 94°C for 30 s, 55°C for 45 s, and 68°C for 3 min in a GeneAmp 9600 DNA Thermal Cycler (Perkin-Elmer, Norwalk, Conn.) with 96-well PCR plates. E. coli colonies that contained plasmid pAE30 or a pOK12 derivative with a 6-kb rpoS-spanning fragment were used as positive controls. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. To obtain clonal mutants, individual colonies were aspirated with a sterile Pasteur pipette and grown in 5 ml of liquid BSK containing coumermycin A1 (0.5 μg/ml). Appropriate dilutions of cultures were replated in solid BSK-H with coumermycin A1 (0.5 μg/ml), and individual colonies were picked, grown up, and subjected to PCR and Southern blot analysis to confirm insertion of gyrBr within rpoS (see below).
SspI digestion of the PCR-amplified gyrB locus from potential transformants.
To determine the proportion of Cour colonies that were “true” transformants, i.e., that had a copy of the B31-NGR gyrBr in their chromosome, the gyrB locus in 42 randomly chosen Cour colonies was amplified by PCR with primers 11 and 13 (Table 2 and Fig. 1B) and digested with SspI. The gyrBr from B31-NGR contains one SspI site, which is due to a dinucleotide change in codon 133 (45; D. S. Samuels, unpublished data) and is not present in gyrBr of spontaneous mutants. The PCR products from 38 of the 42 colonies were cut by SspI, indicating that approximately 90% of the Cour colonies were transformants and 10% were spontaneous resistance mutants.
Southern blot analysis.
B. burgdorferi total genomic DNA was isolated, digested with restriction endonucleases, and separated by pulsed-field gel electrophoresis in a 0.8% (wt/vol) agarose gel under field inversion conditions (47) with a PPI-200 programmable power inverter (MJ Research, Watertown, Mass.). Bidirectional transfer to Biotrans nylon membranes (ICN, Irvine, Calif.), DNA hybridization with radiolabeled probes, and visualization by autoradiography were performed as described previously (47).
Northern blot analysis.
Total borrelial RNA from stationary-phase cultures grown in liquid BSK-H was extracted with the ULTRASPECII RNA Isolation System (Biotecx, Houston, Tex.) according to the manufacturer's instructions. After denaturation with glyoxal and dimethyl sulfoxide, 10 μg of total RNA was electrophoresed in a 1% (wt/vol) agarose gel in 10 mM sodium phosphate buffer, pH 7.0 (50). RNA transfer to nylon membranes (MSI, Westboro, Mass.), hybridization with radiolabeled probes, and visualization by autoradiography were performed as previously described (5).
Protein analysis.
B. burgdorferi was grown in liquid BSK-H to stationary phase and harvested by centrifugation (6,000 × g, 10 min, 4°C). For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (27), cells were washed twice in 100 mM sodium chloride–20 mM N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid, pH 7.3 (HEPES buffer), and lysed by a previously described protocol (9). Protein concentrations were determined by a modified Lowry protein assay (33). Equivalent amounts of cell lysates were solubilized by boiling in Laemmli sample buffer for 5 min (50) and separated through 12.5% polyacrylamide gels in a Hoefer SE600 gel apparatus (Hoefer Scientific, San Francisco, Calif.). For two-dimensional nonequilibrium pH gradient electrophoresis (2D-NEPHGE), stationary-phase cultures were harvested and washed as described above. Pellets were solubilized directly in NEPHGE sample buffer (9 M urea, 4% NP-40, 2% β-mercaptoethanol, 2% ampholytes in distilled H2O) for 2 h at room temperature, followed by ultracentrifugation at 100,000 × g for 1 h at room temperature. A total of 108 cells were loaded onto 10-cm tube gels, and 2D-NEPHGE was performed as previously described (9, 40). Preblended ampholytes (pH 3.5 to 9.5) were purchased from Pharmacia Biotech (Piscataway, N.J.). Proteins were visualized by staining with Coomassie R250 or by silver stain with the Silver Stain Plus kit (Bio-Rad, Hercules, Calif.). Integrated density values were measured with an Alphaimager 2000 digital imaging system (Alpha Innotech Corporation, San Leandro, Calif.).
Osmotic shock assay.
Borrelia cultures were inoculated at an initial concentration of 105 organisms/ml and grown in BSK-H medium. Starting at a density of 107 bacteria per ml, which corresponds to mid-log phase, daily aliquots were divided into two equal portions that contained identical numbers of bacteria. An appropriate amount of 5 N NaCl was added to one aliquot to increase the osmolarity by 1 M, whereas the corresponding control aliquot received an identical volume of fresh BSK. At 10, 40, and 100 min after addition of NaCl, 200 μl of each mixture was centrifuged at 5,000 × g for 5 min at 4°C, and the pellets were resuspended in 0.6 to 1 ml of HEPES buffer. Spirochetes were then assayed for viability with the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Wilsonville, Oreg.) according to the manufacturer's instructions. Viable bacteria with intact membranes stain fluorescent green, whereas dead bacteria with damaged membranes stain fluorescent red. The stained cells were counted with a Petroff-Hausser chamber under fluorescence microscopy (Zeiss, Jena, Germany). The limits of detection with this technique were between 5 × 104 and 1 × 105 cells/ml. In independent experiments, the total numbers of bacteria prior to NaCl exposure varied between 5 × 107 and 9 × 107/ml in log-phase cultures and 1 × 108 and 3 × 108/ml in stationary-phase cultures. The percentage of survivors was calculated as the number of live (green-fluorescent) bacteria in the aliquot exposed to 1 N NaCl divided by the number of live bacteria in the control aliquot, multiplied by 100.
In preliminary experiments, we tested the effect of centrifugation and resuspension in buffer on the viability of untreated log- and stationary-phase cultures and the effectiveness of the washing step in removing free nucleic acids that might result in variation in staining. Centrifugation and washing in various buffers did not alter cell viability (data not shown). Likewise, the addition of up to 25 μg of purified plasmid DNA did not have an effect on the staining character (data not shown).
To confirm that the BacLight stain accurately reflects spirochetal viability, growth endpoint determinations were performed with a microdilution assay. A log-phase B31-A culture was counted, divided into two aliquots, and treated as described above for 40 min. After resuspension in HEPES buffer, the bacterial suspensions were diluted 1:10 with fresh BSK-H into a microtiter plate well to a final volume of 260 μl, corresponding to 3.5 × 105 bacteria prior to treatment. Duplicate twofold serial dilutions were then performed. The microtiter plates were sealed with a gas-permeable sealing membrane (Breathe Easy; Diversified Biotech, Boston, Mass.) and incubated at 35°C in a humidified environment with 1% CO2. The number of spirochetes surviving treatment with 1 N NaCl was determined from the dilution at which no bacterial growth was observed (growth endpoint) as determined by lack of color change of the medium (48) and detectable spirochetes. After incubation for 25 days, no further color change of the medium was observed, and each well was examined by dark-field microscopy for the presence of spirochetes.
RESULTS
Construction of a B. burgdorferi rpoS mutant.
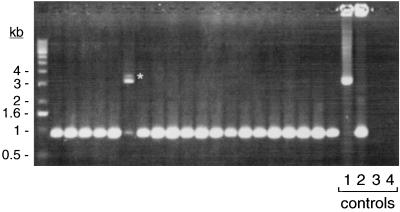
The plasmid pAE30 used for disruption of the B. burgdorferi rpoS gene contains a 4-kb fragment of borrelial chromosomal DNA with gyrBr inserted into the rpoS locus in the reverse orientation (Fig. 1B). This disrupts rpoS in the first quarter of the gene and leaves approximately 2 kb of flanking sequence on either side of gyrBr to mediate allelic exchange. Clone B31-A was transformed with pAE30 DNA by electroporation. Seven mutants with an rpoS::gyrBr genotype were identified by PCR screening of 576 Cour colonies with primers that flank the gyrBr insertion within rpoS (5 and 8, Table 2 and Fig. 2). An estimated 90% of the Cour colonies were transformants, as determined by SspI digestion of the gyrB gene (see Materials and Methods), whereas 10% resulted from spontaneous resistance mutations. Hence, an estimated 1.4% of the transformants were rpoS mutants.
FIG. 2.
Agarose gel of Cour transformants screened by PCR for insertion of gyrBr in the chromosomal rpoS gene. The rpoS gene from individual B. burgdorferi colonies was amplified by PCR with primers 5 and 8 (Table 2 and Fig. 1A). The asterisk indicates the position of the mutant PCR fragment containing gyrBr. The additional wild-type PCR product was not present in clonal mutants (Fig. 3). Controls: 1 and 2, E. coli colonies containing plasmid pAE30 and a pOK12 derivative with an rpoS-flanking fragment, respectively; 3, blank spot on transformation plate; 4, reagent blank. The positions of DNA size standards are indicated on the left.
The mutant colonies were picked from solid medium for growth in liquid BSK-H with coumermycin A1 (0.5 μg/ml) and replated on solid BSK to ensure clonality. The homozygous rpoS::gyrBr genotype of individual clones was confirmed by PCR with primers 5 and 8. The additional wild-type PCR product present in the initial screen (Fig. 2) was not present in clonal mutants. Two rpoS mutants (A74 and A29) and A59, a Cour transformant with a wild-type rpoS locus in which allelic exchange had occurred at the gyrB locus, were chosen for subsequent analyses.
Confirmation of the structure of the mutant rpoS gene.
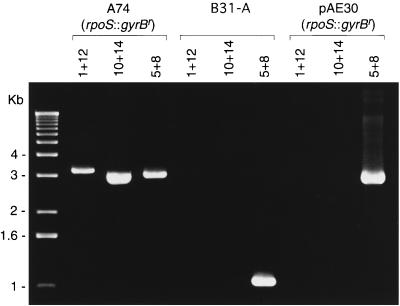
To confirm that the rpoS mutants were the result of allelic exchange at the chromosomal rpoS locus, we amplified chromosomal sequences flanking the cloned 4-kb fragment from pAE30 with primers 1 and 10 (Table 2 and Fig. 1A), in combination with internal gyrB primers 12 and 14 (Table 2 and Fig. 1B). The PCR results were only compatible with allelic exchange occurring at the rpoS locus (Fig. 3).
FIG. 3.
Confirmation of gyrBr insertion in the rpoS chromosomal locus by PCR. Genomic DNA from B. burgdorferi clones B31-A (rpoS wild type) and transformant A74 (rpoS::gyrBr) and pAE30 plasmid DNA were amplified by PCR with the primer sets indicated above the lanes, analyzed by agarose gel electrophoresis, and visualized with ethidium bromide. Primer numbers refer to Table 2 and Fig. 1. The positions of DNA size standards are indicated on the left.
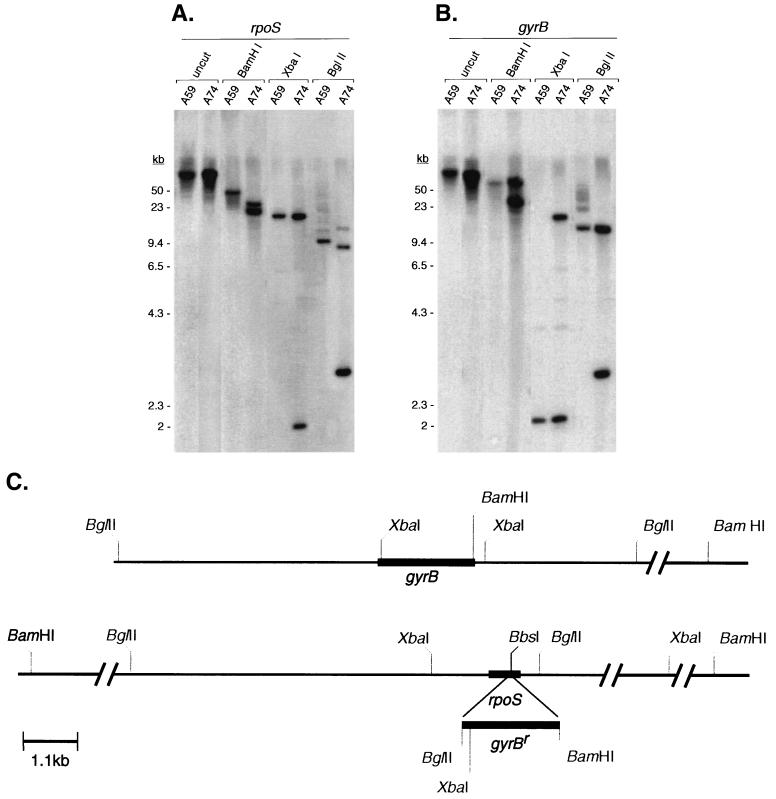
Southern blot analysis provided further evidence for the presence of gyrBr within the rpoS locus (Fig. 4). Hybridization with a probe specific for rpoS resulted in restriction patterns consistent with the presence of a single rpoS gene and the integration of gyrBr into the rpoS locus (Fig. 4A and C). Similarly, a probe specific for gyrBr identified additional bands in A74 (rpoS::gyrBr) compared to A59 (rpoS+ wild type) due to the presence of two gyrBr copies in the chromosome (Fig. 4B and C).
FIG. 4.
Southern blot analysis of Cour transformants. Total borrelial DNA of the Cour rpoS wild-type A59 and the rpoS mutant A74 was digested with restriction endonucleases BglII, XbaI, and BamHI. (A) Hybridization with an rpoS probe (generated with primers 6 and 7 [Table 2 and Fig. 1A]). (B) Hybridization with a probe specific for gyrB (generated with primers 11 and 13 [Table 2 and Fig. 1B]). A59, Cour rpoS wild-type strain; A74, rpoS::gyrBr. DNA source, restriction enzyme, and probe are indicated above the lanes. DNA size standards are indicated on the left. (C) Relevant BglII, XbaI, and BamHI restriction sites at the chromosomal loci for gyrB (upper line) and rpoS (lower line). The rpoS gene of rpoS mutant A74 was disrupted by the insertion of gyrBr into the BbsI site. A size bar (1.1 kb) is indicated. Hatch marks indicate discontinuity to distal restriction sites.
Transcriptional analysis.
To study rpoS transcription, we performed Northern blot analysis of total borrelial RNA from stationary-phase cultures of B31-4A and B31-A. The 798-bp rpoS gene is located between nucleotides 812442 and 813239 on the minus strand of the B. burgdorferi chromosome (18). The gene is flanked upstream by flgI, encoding a flagellar P-ring protein homolog, and downstream by an 879-bp open reading frame that lacks homology with genes of known function (BB770, Fig. 1A) (18). The 454-bp intergenic region between flgI and rpoS does not contain a sequence with more than 60% identity to a consensus ς70 promoter (determined with the program MacTargsearch). A 54-bp sequence which lacks apparent rho-independent terminators lies between the 3′ end of rpoS and the start codon of BB770.
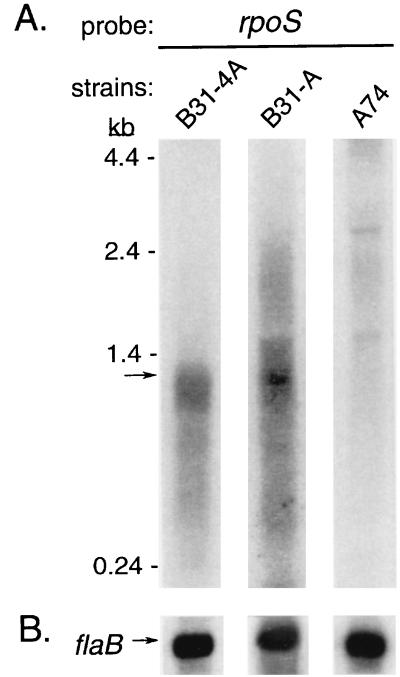
A specific rpoS transcript of approximately 1 kb was identified in B31-A and B31-4A, a size that matches the predicted length of a transcript encompassing only the rpoS gene (Fig. 5A). In contrast, no detectable rpoS transcript was observed in the rpoS mutant A74 (Fig. 5A). No distinct transcript for BB770 was identified in B31-A, B31-4A, and A74 with a probe specific for this gene (data not shown). The rpoS blots were reprobed with an internal flaB probe, and the results confirmed that equivalent amounts of RNA were analyzed for all three strains (Fig. 5B).
FIG. 5.
Northern blot analysis of the B. burgdorferi rpoS transcript. Total borrelial RNA from stationary-phase cultures was extracted and transferred to nylon membranes. (A) Hybridization with a probe specific for rpoS (PCR fragment amplified with primers 6 and 7 [Table 2 and Fig. 1A]). The arrow indicates the rpoS transcript. (B) Rehybridization with a flaB probe (PCR fragment amplified with primers 16 and 17 [Table 2]) after decay of the rpoS probe confirmed the presence of equivalent amounts of RNA in all three lanes. RNA sources are indicated above the lanes. RNA size standards are indicated on the left.
Growth phase-dependent resistance of wild-type B. burgdorferi to osmotic stress.
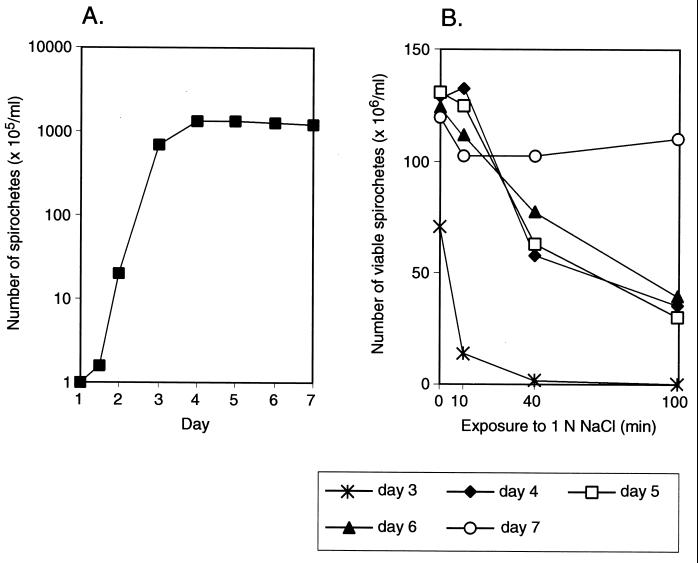
RpoS participates in stationary-phase-mediated resistance to environmental stresses in many bacteria (17, 22, 34). Next, we tested whether growth phase affects the ability of wild-type B. burgdorferi to survive treatment with a high salt concentration. A representative experiment in which resistance to high salt was determined for clone B31-A with a wild-type rpoS allele is shown in Fig. 6. With an initial inoculum of 105 bacteria/ml, stationary phase was reached after 4 days and the number of viable bacteria remained constant through day 7. Log-phase and stationary-phase organisms were exposed to 1 N NaCl for 10, 40, and 100 min, and the number of viable spirochetes was determined. Greater than 90% of the log-phase bacteria were dead after the 10-min exposure, and no viable bacteria were detected by microscopic examination after 100 min (Fig. 6B). Between days 4 and 7, greater than 90% of the bacteria survived 1 N NaCl for 10 min, but only 30% were viable after 100 min (Fig. 6B). However, on day 7 the majority of spirochetes remained viable after 100 min of exposure to 1 N NaCl (Fig. 6B). The distinct patterns of osmotic resistance in log phase and early and late stationary phase were confirmed by repetition of the experiment five times. In all experiments less than 0.2% (0.005 to 0.14%) of log-phase spirochetes survived 70 to 100 min of exposure to 1 N NaCl. Survival of bacteria in late stationary phase varied between 74 and 105% after 10 to 100 min in 1 N NaCl.
FIG. 6.
Growth phase-dependent resistance of rpoS wild-type B31-A to 1 N NaCl. (A) Growth curve of B31-A. (B) Survival of B31-A grown for 3 to 7 days in 1 N NaCl. The number of viable spirochetes was determined at the indicated times before and after addition of NaCl.
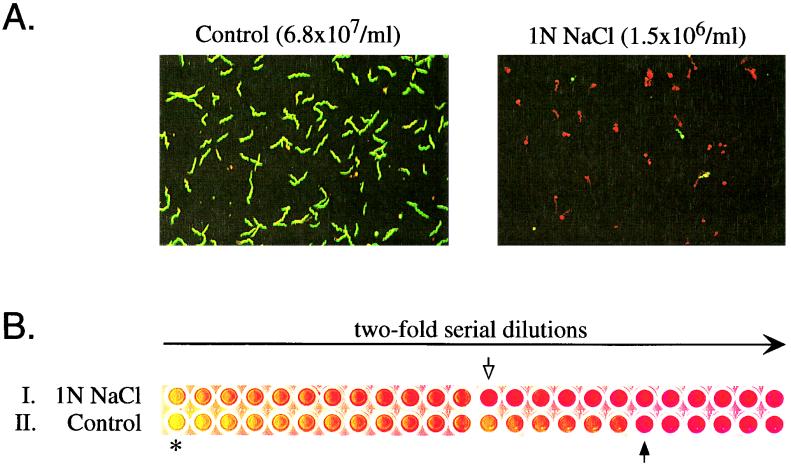
To determine the ability of the LIVE/DEAD BacLight bacterial viability kit to accurately reflect cell viability, we compared data from this assay with the results of a growth endpoint determination obtained with a microdilution assay (Fig. 7). In the microdilution assay, the growth endpoint of B31-A log-phase bacteria was determined at a dilution corresponding to 1.3 spirochetes (Fig. 7B). After 40 min of exposure to 1 N NaCl, the growth endpoint occurred at a dilution corresponding to 87 spirochetes in the untreated control culture (Fig. 7B). Thus, exposure of a log-phase culture to 1 N NaCl for 40 min resulted in greater than 90% reduction in the number of viable organisms, as assessed by limiting dilution. This value is comparable to the results obtained with the LIVE/DEAD BacLight bacterial viability kit (Fig. 6 and 8). These results also demonstrate a good correlation between the LIVE/DEAD BacLight bacterial viability kit and the growth endpoint determination by limiting dilution with respect to absolute numbers of viable bacteria.
FIG. 7.
Susceptibility of rpoS wild-type B31-A to 1 N NaCl. (A) Live/dead stain with the LIVE/DEAD BacLight bacterial viability kit of a B31-A log-phase culture without and 40 min after addition of 1 N NaCl. (B) Susceptibility of B31-A to 1 N NaCl determined with a microdilution assay. Successive wells represent twofold dilutions. Row I, after 40 min of exposure to 1 N NaCl. Row II, control culture not exposed to NaCl. The first well of the control received an initial inoculum of 3.5 × 105 bacteria (row II, asterisk). An equivalent inoculum was treated with 1 N NaCl for 40 min prior to addition to the first well (row I). Solid arrow, growth endpoint of the untreated control culture, corresponding to a calculated number of 1.3 spirochetes in the initial inoculum; open arrow, growth endpoint of the NaCl-exposed culture. A >90% reduction in viability was calculated as the inverse of the ratio of the number of viable spirochetes as assessed by growth endpoint following salt treatment relative to the untreated control culture.
FIG. 8.
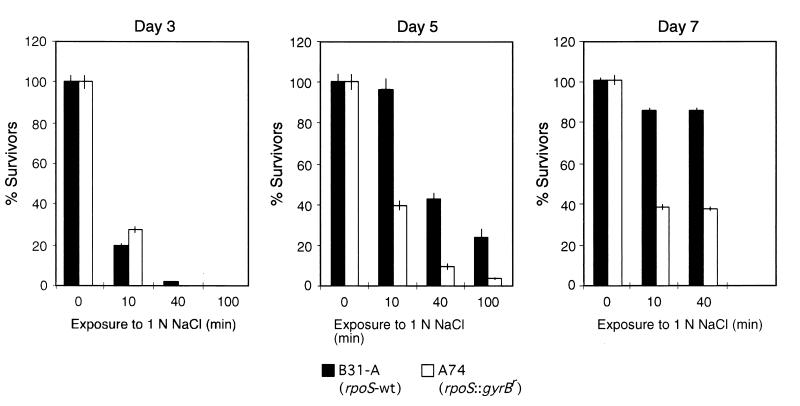
Comparison of rpoS wild-type (wt) B31-A and rpoS mutant A74 in their resistance to 1 N NaCl. Spirochetes were grown for 3, 5, and 7 days in liquid culture. At days 3, 5, and 7, the number of viable spirochetes was determined without (0 min) and 10, 40, and 100 min after addition of NaCl. The percent survivors represents the number of viable spirochetes in salt-treated cultures relative to that in comparable sham controls. Error bars indicate the standard error obtained from three independent countings.
Survival of the isogenic rpoS mutant under high-salt conditions.
Our results indicate that osmotic resistance in wild-type B31-A is growth phase dependent and increases during stationary phase. To assess the role of RpoS in resistance to osmotic stress, we compared the survival of B31-A and the rpoS mutant A74 after addition of 1 N NaCl to the medium. Wild-type and mutant bacteria in log phase died shortly after NaCl was added (Fig. 8, day 3). However, in early stationary phase, the rpoS mutant was more susceptible to 1 N NaCl than wild-type B31-A (Fig. 8, day 5). Although both the wild-type and mutant organisms were more resistant to prolonged osmotic shock in late stationary phase, survival was decreased by more than 50% in the rpoS mutant relative to B31-A (Fig. 8, day 7). These results indicate that RpoS participates in growth phase-dependent osmotic resistance in B. burgdorferi.
Due to the limited set of genetic methods for B. burgdorferi, we were not able to use genetic complementation to confirm that the observed phenotype of A74 was due to inactivation of rpoS. However, we tested another rpoS mutant (A29, Table 1) for osmotic resistance and obtained comparable results (data not shown). The Cour rpoS wild-type A59 (Table 1) showed a growth phase-dependent survival in high-salt medium similar to that of B31-A (data not shown). Therefore, it is unlikely that the presence of the altered B subunit of the DNA gyrase in rpoS mutants A74 and A29 accounted for their increased susceptibility to osmotic stress.
Protein analysis of the rpoS wild-type and mutant strains.
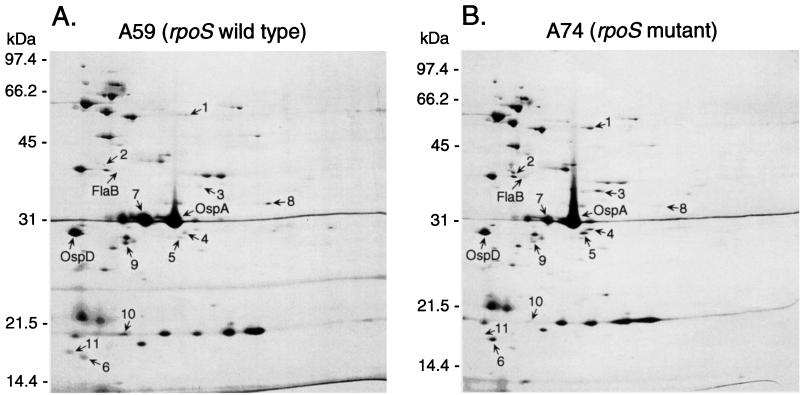
2D-NEPHGE (9, 39) was used to assess differences in protein composition between rpoS mutant A74 and isogenic rpoS wild-type A59. Representative silver-stained 2D gels containing protein lysates from 108 spirochetes grown to stationary phase are shown in Fig. 9. Two independent cultures of each clone were analyzed, and 11 protein spots with three- to eightfold differences in abundance, as determined by densitometry, were repeatedly detected (Fig. 9). The lysates from rpoS wild-type strain A59 had five protein spots that were either absent or markedly decreased in the lysates from rpoS mutant strain A74 (Fig. 9, spot numbers 7, 8, 9, 10, and 11). In contrast, six protein spots were present in the lysates from rpoS mutant A74 that were either absent from or markedly decreased in lysates from rpoS wild-type A59 (Fig. 9, spot numbers 1, 2, 3, 4, 5, and 6). The levels of OspA, OspD (39), and flagellin (FlaB) were not significantly different in the isogenic strains (Fig. 9).
FIG. 9.
Silver-stained gels of total borrelial lysates from stationary-phase cultures separated by 2D-NEPHGE. (A) A59 (rpoS wild type). (B) A74 (rpoS mutant). Acidic end is on the left. Protein spots that differ more than threefold in abundance as determined by densitometry are indicated by arrows. The numbers indicate corresponding protein spots. Protein size standards are indicated on the left of each panel. The locations of OspA, OspD, and flagellin (FlaB) are indicated by an arrow. Flagellin does not stain with silver but is easily detectable with Coomassie blue staining (data not shown).
DISCUSSION
Borrelia burgdorferi survives within and is transmitted between two very different host environments, the tick vector and the mammalian host. In many bacteria, alternative sigma factors regulate gene expression in response to environmental conditions. The genome of B. burgdorferi contains two genes encoding homologs of alternative sigma factors, rpoS and ntrA, but nothing is known about their function. Here we describe the site-directed inactivation of rpoS in B. burgdorferi and the characterization of the rpoS mutant.
By using 2D gel electrophoresis of stationary-phase spirochetes, we identified 11 proteins that differed at least threefold in abundance in the rpoS mutant compared with the rpoS wild-type strain. Interestingly, 6 of the 11 proteins were made in higher amounts by the rpoS mutant than by the isogenic wild-type organism. In E. coli, RpoS mainly acts as a positive regulator of a group of genes expressed in stationary-phase cells (21, 22), and only a small number of genes have been reported to be negatively regulated (15). In contrast, our results suggest that the borrelial RpoS participates in both up- and downregulation of gene expression. The natural environment and life cycle of B. burgdorferi differ greatly from those of members of the Enterobacteria, for which the role of RpoS has been studied most extensively (21, 22, 28, 31). Therefore, we anticipate that many genes regulated by RpoS in B. burgdorferi will have functions and expression patterns distinct from those of E. coli and related enteric organisms. Synthesis of most proteins, including OspA, OspD, and flagellin (FlaB), was not significantly altered in the rpoS mutant relative to the rpoS wild type, suggesting that either ς70 or ς54 is responsible for transcription of the respective genes.
The designation of BB771 as an RpoS homolog is based on sequence similarity with RpoS of other bacteria. The most closely related RpoS homolog is found in Pseudomonas aeruginosa (34% identity and 58% similarity using the BLASTP program, National Center for Biotechnology Information) (18). Preliminary results indicate that the B. burgdorferi rpoS gene partially complements a Shigella flexneri rpoS mutant in an acid resistance assay (data not shown). Similar to E. coli, B. burgdorferi wild-type organisms had an increased osmotic resistance in the stationary phase relative to the exponential growth phase, and our results indicate that the borrelial RpoS homolog participates in this stationary-phase response. This is consistent with the previous demonstration of RpoS induction in stationary-phase spirochetes (Frye et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999). However, RpoS is not solely responsible for these changes, since the osmotic resistance of the rpoS mutant also increased during stationary phase, although to a lesser extent. Growth phase-related factors, such as pH change of the BSK medium, could influence osmotic resistance independently of RpoS. RpoS in B. burgdorferi is not required for resistance to oxidative stress, because survival of rpoS mutant organisms exposed to 15 mM hydrogen peroxide in stationary phase was unaltered compared with the isogenic rpoS wild-type strain (data not shown). We note that in Legionella pneumophila, RpoS does not participate in stationary-phase-dependent resistance to environmental factors such as oxidative stress but is required for growth within the protozoan host Acanthamoeba castellanii (20).
Further characterization of the rpoS mutant will provide information about how B. burgdorferi adapts to variable environmental conditions. Identification of the 11 proteins differentially made by rpoS wild-type and mutant spirochetes will address which B. burgdorferi genes are regulated by RpoS, either directly or indirectly. In this regard, several borrelial proteins have previously been shown to be differentially regulated in log phase versus stationary phase (23, 44; Frye et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999), but their dependence upon RpoS expression has not been investigated. Osmotolerance requires de novo protein synthesis (25), and some of the proteins made in greater abundance in the rpoS wild-type than in the rpoS mutant could be responsible for the higher osmotic resistance. For example, proX, proW, and proV on the chromosome of B. burgdorferi encode an ABC transporter for osmoprotectants such as proline and betaine (ProU). In E. coli, ProU participates in osmoregulation and is regulated by RpoS (32).
RpoS is important for the survival of Salmonella serovar Typhimurium and L. pneumophila in their hosts and regulates virulence genes in Salmonella (14, 20, 29) and V. cholerae (60). Inactivation of rpoS in a low-passage, infectious B. burgdorferi strain will enable us to directly test the function of RpoS in the infectious cycle of this important pathogen.
ACKNOWLEDGMENTS
We thank J. Battisti, B. J. Hinnebusch, J. M. Musser, and T. G. Schwan for critical review of the manuscript; K. Matteson for assistance in manuscript preparation; G. Hettrick and R. Evans for artwork and photography; and J. Frye, F. Gherardini, and T. Hoover for helpful discussions.
ADDENDUM IN PROOF
While this article was in press, Knight et al. reported the disruption of the gac gene located on the chromosome of B. burgdorferi (S. W. Knight, B. J. Kimmel, C. H. Eggers, and D. S. Samuels, J. Bacteriol. 182:2048–2051, 2000).
REFERENCES
- 1.Asbrink E, Hovmark A. Early and late cutaneous manifestations in Ixodes-borne borreliosis (erythema migrans borreliosis, Lyme borreliosis) In: Benach J L, Bosler E M, editors. Lyme disease and related disorders. New York, N.Y: New York Academy of Sciences; 1988. pp. 4–15. [DOI] [PubMed] [Google Scholar]
- 2.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour A G, Tessier S L, Todd W J. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthold S W, de Souza M S, Janotka J L, Smith A L, Persing D H. Chronic Lyme borreliosis in the laboratory mouse. Am J Pathol. 1993;143:959–972. [PMC free article] [PubMed] [Google Scholar]
- 5.Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 6.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 7.Carreiro M M, Laux D C, Nelson D R. Characterization of the heat shock response and identification of heat shock protein antigens of Borrelia burgdorferi. Infect Immun. 1990;58:2186–2191. doi: 10.1128/iai.58.7.2186-2191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll J A, Garon C F, Schwan T G. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll J A, Gherardini F C. Membrane protein variations associated with in vitro passage of Borrelia burgdorferi. Infect Immun. 1996;64:392–398. doi: 10.1128/iai.64.2.392-398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control. Lyme disease—United States, 1996. Morbid Mortal Weekly Rep. 1997;46:531–535. [PubMed] [Google Scholar]
- 12.Cluss R G, Boothby J T. Thermoregulation of protein synthesis in Borrelia burgdorferi. Infect Immun. 1990;58:1038–1042. doi: 10.1128/iai.58.4.1038-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Silva A M, Fikrig E. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- 14.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 16.Finlay B B, Falkow S. Common themes in microbial pathogenicity. Microbiol Rev. 1989;53:210–230. doi: 10.1128/mr.53.2.210-230.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer D, Teich A, Neubauer P, Hengge-Aronis R. The general stress sigma factor ςS of Escherichia coli is induced during diauxic shift from glucose to lactose. J Bacteriol. 1998;180:6203–6206. doi: 10.1128/jb.180.23.6203-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidmann J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 19.Fung B P, McHugh G L, Leong J M, Steere A C. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect Immun. 1994;62:3213–3221. doi: 10.1128/iai.62.8.3213-3221.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hales L M, Shuman H A. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol. 1999;181:4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 22.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 23.Indest K J, Ramamoorthy R, Sole M, Gilmore R D, Johnson B J B, Philipp M T. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect Immun. 1997;65:1165–1171. doi: 10.1128/iai.65.4.1165-1171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins D E, Chaisson S A, Matin A. Starvation-induced cross protection against osmotic challenge in Escherichia coli. J Bacteriol. 1990;172:2779–2781. doi: 10.1128/jb.172.5.2779-2781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King G J, Swanson J. Studies on gonococcus infection. XV. Identification of surface proteins of Neisseria gonorrhoeae correlated with leukocyte association. Infect Immun. 1978;21:575–584. doi: 10.1128/iai.21.2.575-584.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 29.Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane R S, Piesman J, Burgdorfer W. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol. 1991;36:587–609. doi: 10.1146/annurev.en.36.010191.003103. [DOI] [PubMed] [Google Scholar]
- 31.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 32.Manna D, Gowrishankar J. Evidence for involvement of proteins HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia coli. J Bacteriol. 1994;176:5378–5384. doi: 10.1128/jb.176.17.5378-5384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markwell M A, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 34.McCann M P, Kidwell J P, Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J F, Mekalanos J J, Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 37.Montgomery R R, Malawista S E, Feen K J M, Bockenstedt L K. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J Exp Med. 1996;183:261–269. doi: 10.1084/jem.183.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp Quant Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 39.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 41.Padula S J, Sampieri A, Dias F, Szczepanski A, Ryan R W. Molecular characterization and expression of p23 (OspC) from a North American strain of Borrelia burgdorferi. Infect Immun. 1993;61:5097–5105. doi: 10.1128/iai.61.12.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peak M J. Some observations on the lethal effects of near-ultraviolet light on Escherichia coli, compared with the lethal effects of far-ultraviolet light. Photochem Photobiol. 1970;12:1–8. doi: 10.1111/j.1751-1097.1970.tb06031.x. [DOI] [PubMed] [Google Scholar]
- 43.Piesman J, Oliver J R, Sinsky R J. Growth kinetics of the Lyme disease spirochete (Borrelia burgdorferi) in vector ticks (Ixodes dammini) Am J Trop Med Hyg. 1990;42:352–357. doi: 10.4269/ajtmh.1990.42.352. [DOI] [PubMed] [Google Scholar]
- 44.Ramamoorthy R, Philipp M T. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosa P, Samuels D S, Hogan D, Stevenson B, Casjens S, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosa P A, Hogan D. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants. In: Munderloh U G, Kurtti T J, editors. First international conference on tick-borne pathogens at the host-vector interface: an agenda for research. St. Paul: University of Minnesota; 1992. pp. 95–103. [Google Scholar]
- 47.Rosa P A, Schwan T G. A specific and sensitive assay for the Lyme disease spirochete Borrelia burgdorferi using the polymerase chain reaction. J Infect Dis. 1989;160:1018–1029. doi: 10.1093/infdis/160.6.1018. [DOI] [PubMed] [Google Scholar]
- 48.Sadziene A, Thompson P A, Barbour A G. In vitro inhibition of Borrelia burgdorferi growth by antibodies. J Infect Dis. 1993;167:165–172. doi: 10.1093/infdis/167.1.165. [DOI] [PubMed] [Google Scholar]
- 49.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Samuels D S. Electrotransformation of the spirochete Borrelia burgdorferi. In: Nickoloff J A, editor. Methods in molecular biology, vol. 47: electroporation protocols for microorganisms. Totowa, N.J: Humana Press, Inc.; 1995. pp. 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamm L V, Gherardini F C, Parrish E A, Moomaw C R. Heat shock response of spirochetes. Infect Immun. 1991;59:1572–1575. doi: 10.1128/iai.59.4.1572-1575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 55.Steere A C, Grodzicki R L, Kornblatt A N, Craft J E, Barbour A G, Burgdorfer W, Schmid G P, Johnson E, Malawista S E. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 56.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 58.Tuveson R W, Jonas R B. Genetic control of near-UV (300–400 nm) sensitivity independent of the recA gene in strains of Escherichia coli K12. Photochem Photobiol. 1979;30:667–676. doi: 10.1111/j.1751-1097.1979.tb07197.x. [DOI] [PubMed] [Google Scholar]
- 59.Wösten M M. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 60.Yildiz F H, Schoolnik G K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]