Abstract
Background
Patients with bacteremia due to carbapenem-resistant Enterobacterales (CRE) experience delays until appropriate therapy and high mortality rates. Rapid molecular diagnostics for carbapenemases and new β-lactam/β-lactamase inhibitors may improve outcomes.
Methods
We conducted an observational study of patients with CRE bacteremia from 2016 to 2018 at 8 New York and New Jersey medical centers and assessed center-specific clinical microbiology practices. We compared time to receipt of active antimicrobial therapy and mortality between patients whose positive blood cultures underwent rapid molecular testing for the Klebsiella pneumoniae carbapenemase (KPC) gene (blaKPC) and patients whose cultures did not undergo this test. CRE isolates underwent antimicrobial susceptibility testing by broth microdilution and carbapenemase profiling by whole-genome sequencing. We also assessed outcomes when ceftazidime-avibactam and polymyxins were used as targeted therapies.
Results
Of 137 patients with CRE bacteremia, 89 (65%) had a KPC-producing organism. Patients whose blood cultures underwent blaKPC PCR testing (n = 51) had shorter time until receipt of active therapy (median: 24 vs 50 hours; P = .009) compared with other patients (n = 86) and decreased 14-day (16% vs 37%; P = .007) and 30-day (24% vs 47%; P = .007) mortality. blaKPC PCR testing was associated with decreased 30-day mortality (adjusted odds ratio: .37; 95% CI: .16–.84) in an adjusted model. The 30-day mortality rate was 10% with ceftazidime-avibactam monotherapy and 31% with polymyxin monotherapy (P = .08).
Conclusions
In a KPC-endemic area, blaKPC PCR testing of positive blood cultures was associated with decreased time until appropriate therapy and decreased mortality for CRE bacteremia, and ceftazidime-avibactam is a reasonable first-line therapy for these infections.
Keywords: carbapenem-resistant Enterobacterales, Klebsiella pneumoniae carbapenemase, rapid diagnostics, ceftazidime-avibactam
In this multicenter observational study of bacteremias caused by carbapenem-resistant Enterobacterales, use of a molecular assay that detects the Klebsiella pneumoniae carbapenemase gene directly from positive blood culture broths was associated with shorter delays in appropriate therapy and decreased mortality.
Carbapenem-resistant Enterobacterales (CRE) are a global public health threat because they cause infections that are associated with high mortality rates and few antimicrobial therapeutic options [1, 2]. CRE became endemic in New York and New Jersey (NY/NJ) 2 decades ago due to the emergence of Klebsiella pneumoniae carbapenemase (KPC), an enzyme that confers resistance to carbapenems and most β-lactam agents [3]. KPC-producing CRE then spread globally and KPC is now the most common carbapenemase among Enterobacterales in the United States, Europe, and Latin America [4–7].
We previously conducted a multicenter study of CRE bacteremias in 2013 in NY/NJ and found that there was a median of 47 hours from bacteremia onset until initiation of active antimicrobial therapy and a 49% 30-day mortality rate [8]. Most patients in that study received polymyxin- and tigecycline-based therapies. Since then, important advances in CRE diagnostics and therapeutics have emerged that might improve outcomes of CRE-infected patients. First, molecular panels are available that detect the KPC gene (blaKPC) within 1–2 hours of blood culture positivity [9, 10]. These tests rapidly detect carbapenem resistance in KPC-producing organisms, compared with a 2–3-day delay with conventional culture and antimicrobial susceptibility testing. However, the impact of these assays on shortening the time to administration of effective therapies and improving outcomes of infected patients is unknown. Second, new β-lactam/β-lactamase inhibitors are approved for the treatment of CRE infections [11–13]. Although prior studies demonstrated improved outcomes with these newer agents [11, 14], these studies primarily compared these agents with colistin-based regimens, not to regimens with polymyxin B, which has more favorable pharmacological properties than colistin [15].
Here, we present a follow-up study of CRE bacteremias in NY/NJ to evaluate the potential benefits of rapid molecular diagnostics and novel β-lactam/β-lactamase inhibitors for CRE infections. Our primary objectives were to determine if the use of a molecular assay that detects blaKPC directly from positive blood culture broths and/or treatment with ceftazidime-avibactam was associated with improved outcomes in patients with CRE bacteremia.
METHODS
Study Cohort
We conducted an observational study of patients with CRE bacteremia from January 2016 to June 2018 at 8 academic medical centers in NY/NJ. Institutional review board approval was obtained at each site. Patients were initially enrolled based on detection of carbapenem resistance at local clinical microbiology laboratories and only the first episode of CRE bacteremia per patient was included. The final cohort consisted of patients whose CRE bloodstream isolates underwent antimicrobial susceptibility testing and whole-genome sequencing (WGS) by a central laboratory, and whose isolates were carbapenem-resistant based on central laboratory testing [16].
Clinical Data Collection
Clinical data were abstracted from electronic medical records at each study site and recorded in a central database [17], including patient demographics, comorbidities [18], clinical status, and Acute Physiologic Assessment Chronic Health Evaluation II (APACHE II) and Pitt Bacteremia scores at the time of bacteremia onset [19, 20]. We also reviewed diagnostic methods and the time from blood culture collection until CRE detection in the local clinical microbiology laboratories. Finally, we recorded antimicrobial therapies, time until receipt of active therapy (an agent to which the bloodstream CRE pathogen[s] tested susceptible in the central laboratory), bacteremia source and source control (determined by an infectious diseases physician at each site), 14- and 30-day mortality, and acute kidney injury (AKI) [21]. Bacteremia onset was defined as the time of collection of the first blood culture from which CRE was recovered. Initial targeted therapy was defined as antimicrobial agents that were administered within 1 day after the availability of antimicrobial susceptibility testing results and that were continued for 2 or more days.
Microbiologic Analyses
Bloodstream isolates were shipped to a central laboratory. There, isolates were identified to the species level by matrix-associated laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonics, Billerica, MA) and antimicrobial susceptibility testing was performed for 18 antimicrobial agents by reference broth microdilution (ThermoFisher Scientific, Waltham, MA) [22]. Interpretive criteria of the Clinical and Laboratory Standards Institute were applied [23], except isolates for which colistin minimum inhibitory concentrations (MICs) were 2 µg/mL or less were considered susceptible to colistin and polymyxin B and those for which tigecycline MICs were 2 µg/mL or less (the US Food and Drug Administration [FDA] susceptible breakpoint) were considered tigecycline susceptible.
Isolates also underwent WGS using previously described methods [5]. In brief, genomes were sequenced using an Illumina Hiseq platform, followed by raw reads quality control check, filter, and assembly. Bacterial species were examined by Mash 2.3 using the NCBI RefSeq genome database [24]. Antimicrobial resistance genes were determined using Kleborate v2.0.4, AMRFinderPlus v3.10.5, and ARIBA v2.14.6 [25–27]. Multilocus sequence typing (MLST) was performed using mlst v2.19.0 (https://github.com/tseemann/mlst) utilizing pubmlst (https://pubmlst.org/databases/) databases. The raw reads of the sequenced genomes were deposited in GenBank bioproject accession no. PRJNA549322.
Statistical Analyses
We first compared baseline characteristics and treatments of patients who died within 30 days of bacteremia onset with those of patients who survived. Chi-square and Fisher’s exact tests were used for categorical variables, and the Wilcoxon rank-sum test was used for continuous variables. P < .05 indicated statistical significance. We then compared time to detection of CRE bacteremia, time to receipt of active antimicrobial therapy, and 14- and 30-day mortality between patients whose positive blood culture broths underwent blaKPC polymerase chain reaction (PCR) and those whose blood cultures did not undergo this test. We also made these comparisons among patients with KPC-producing CRE bacteremia (wherein blaKPC testing might yield benefit) and non–KPC-producing CRE bacteremia (when we did not expect benefit).
We then estimated adjusted associations between blaKPC PCR testing and 30-day mortality using Targeted Maximum Likelihood Estimation (TMLE), a method that leverages flexible regression for both the outcome and propensity score, in order to achieve robustness to misspecification of either model [28] (Supplementary Methods and Supplementary Figure 1). This technique helps ensure associations between mortality and blaKPC PCR testing are not driven by differences in baseline characteristics between patients whose blood cultures did and did not undergo blaKPC PCR testing. The TMLE analysis was conducted in R, version 4.0.3, and using the open-source package ltmp [29]. We also describe 14- and 30-day mortality based on initial targeted therapies.
RESULTS
Study Cohort
Of 178 patients initially enrolled, 17 were excluded because their CRE bloodstream isolate was not available for central laboratory analysis and 24 because their isolate was not carbapenem-resistant at the central laboratory, leaving 137 patients in the study cohort. The median age was 64 years, 61% were male, and cancer (42%) and diabetes (34%) were the most common comorbidities (Table 1). The most common sources of infection were intra-abdominal (33%), vascular catheters (13%), and the respiratory (13%) and urinary (12%) tracts.
Table 1.
Baseline Characteristics and Treatments of CRE Bacteremias and Associations With 30-Day Mortality
| Total (n = 137) | Survivors (n = 85) | Died Within 30 Days (n = 52) | P | |
|---|---|---|---|---|
| Patient characteristics | ||||
| Demographics | ||||
| Age, median (IQR), years | 64 (51–76) | 63 (49–76) | 67 (56–74) | .84 |
| Male gender | 83 (61) | 54 (64) | 29 (56) | .37 |
| Comorbidities | ||||
| Myocardial infarction | 11 (8) | 7 (8) | 4 (8) | 1.00 |
| Congestive heart failure | 18 (13) | 10 (12) | 8 (15) | .54 |
| Peripheral vascular disease | 16 (12) | 10 (12) | 6 (12) | .97 |
| Cerebrovascular disease | 17 (12) | 14 (16) | 3 (6) | .065 |
| Dementia | 16 (12) | 11 (13) | 5 (10) | .56 |
| COPD | 22 (16) | 16 (19) | 6 (12) | .26 |
| Peptic ulcer disease | 10 (7) | 5 (6) | 5 (10) | .50 |
| Liver disease | 9 (7) | 4 (5) | 5 (10) | .30 |
| Diabetes | 46 (34) | 32 (38) | 14 (27) | .20 |
| Moderate or severe kidney disease | 29 (21) | 19 (22) | 10 (19) | .66 |
| Cancer | 58 (42) | 29 (34) | 29 (56) | .013 |
| Solid tumor | 35 (26) | 20 (24) | 15 (29) | .49 |
| Hematologic malignancy | 25 (18) | 9 (11) | 16 (31) | .003 |
| HIV infection | 5 (4) | 3 (4) | 2 (4) | 1.00 |
| Charlson Comorbidity Index score [18] | 5 (3–7) | 5 (3–7) | 5 (4–8) | .53 |
| Transplant recipient | 27 (20) | 18 (21) | 9 (17) | .58 |
| Solid-organ transplant | 14 (10) | 13 (15) | 1 (2) | .012 |
| Hematopoietic cell transplant | 13 (9) | 5 (6) | 8 (15) | .078 |
| Place patient admitted from | ||||
| Home | 80 (58) | 50 (59) | 30 (58) | .90 |
| Rehabilitation or long-term care facility | 36 (26) | 21 (25) | 15 (29) | .59 |
| Transfer from a different hospital | 21 (15) | 14 (16) | 7 (14) | .64 |
| Variables at time of bacteremia onset | ||||
| Outpatient | 41 (30) | 30 (35) | 11 (21) | .079 |
| Medical ward, non-ICU | 52 (38) | 32 (38) | 20 (39) | .92 |
| Surgical ward, non-ICU | 13 (9) | 11 (13) | 2 (4) | .13 |
| ICU | 31 (23) | 12 (14) | 19 (37) | .002 |
| Days from hospital admission until BSI onset | 14 (1–31) | 13 (0–35) | 14 (2–30) | .85 |
| Receiving RRT | 26 (19) | 11 (13) | 15 (29) | .021 |
| Baseline creatinine clearance (mL/minute) among patients not on RRT [30] | 59 (31–87) | 59 (29–87) | 62 (43–100) | .53 |
| Neutropenia | 21 (15) | 12 (14) | 9 (17) | .62 |
| Bacteremia source | ||||
| Intra-abdominal | 45 (33) | 23 (27) | 22 (42) | .065 |
| Vascular catheter | 18 (13) | 14 (16) | 4 (8) | .14 |
| Urinary tract | 17 (12) | 16 (19) | 1 (2) | .004 |
| Respiratory tract | 18 (13) | 8 (9) | 10 (19) | .099 |
| Gastrointestinal translocation during neutropenia | 13 (9) | 8 (9) | 5 (10) | 1.00 |
| Skin/soft tissue | 7 (5) | 4 (5) | 3 (6) | 1.00 |
| Other | 5 (4) | 5 (6) | 0 | .16 |
| Unknown | 14 (10) | 7 (8) | 7 (13) | .33 |
| Fever (temperature ≥38.0°C) | 86 (63) | 60 (71) | 26 (50) | .016 |
| Receiving mechanical ventilation | 48 (35) | 24 (28) | 24 (46) | .033 |
| APACHE II score [19] | 20 (15–25) | 18 (14–22) | 25 (18–31) | <.0001 |
| Pitt Bacteremia score [20] | 3 (1–5) | 2 (1–4) | 4 (2–7) | .0003 |
| Treatment | ||||
| Active therapy at any time | 107 (78) | 75 (88) | 32 (62) | <.001 |
| Hours until active therapy (n = 107) | 29 (9–78) | 32 (13–86) | 25 (7–66) | .16 |
| Active therapy within 24 hours | 43 (31) | 27 (32) | 16 (31) | .90 |
| Initial targeted therapya (n = 112) | n = 84 | n = 28 | ||
| Polymyxinb | 45 (40) | 31 (37) | 14 (50) | .22 |
| Carbapenemc | 39 (35) | 28 (33) | 11 (39) | .57 |
| Ceftazidime-avibactam | 32 (29) | 26 (31) | 6 (21) | .33 |
| Fluoroquinoloned | 16 (14) | 13 (15) | 3 (10) | .76 |
| Tigecycline | 13 (12) | 10 (12) | 3 (11) | 1.00 |
| Piperacillin-tazobactam | 7 (6) | 7 (8) | 0 | .19 |
| Aminoglycosidee | 8 (7) | 5 (6) | 3 (11) | 1.00 |
| TMP-SMX | 7 (6) | 5 (6) | 2 (7) | 1.00 |
| Minocycline | 5 (4) | 4 (5) | 1 (4) | 1.00 |
| No. of active agents | ||||
| None | 21 (19) | 15 (18) | 6 (21) | .68 |
| Single active agent | 68 (61) | 56 (67) | 12 (43) | .025 |
| ≥2 active agents | 23 (21) | 13 (15) | 10 (36) | .022 |
| Infectious diseases consult | 124 (91) | 79 (93) | 45 (87) | .22 |
| Source control | 46 (34) | 36 (42) | 10 (19) | .005 |
Variables are expressed as n (% of total) or median (IQR). Bolded P values indicate statistical significance. Abbreviations: APACHE, Acute Physiologic Assessment and Chronic Health Evaluation; COPD, chronic obstructive pulmonary disease; CRE, carbapenem-resistant Enterobacterales; HIV, human immunodeficiency virus; ICU, intensive care unit; IQR, interquartile range; RRT, renal replacement therapy; TMP-SMX, trimethoprim-sulfamethoxazole.
Antimicrobial agents that are active against gram-negative bacteria and were administered within 1 day of the availability of susceptibility results for ≥2 calendar days. Only antimicrobial therapies that were administered to ≥5 patients are displayed in the table. One hundred twelve patients received initial targeted therapy; 24 patients died prior to or within 1 day after the availability of susceptibility results and 1 patient was discharged alive prior to the availability of susceptibility results.
Polymyxin targeted therapy was with polymyxin B (n = 39) and colistin (n = 6).
Carbapenem targeted therapy was with meropenem (n = 36), ertapenem (n = 2), and imipenem (n = 1). The carbapenem was administered in combination with a second agent in 26 patients and was active in vitro against the CRE bloodstream pathogen in 4 patients.
Fluoroquinolone targeted therapy was with levofloxacin (n = 11) and ciprofloxacin (n = 5).
Aminoglycoside targeted therapy was with gentamicin (n = 4), amikacin (n = 3), and tobramycin (n = 1).
Active therapy was administered to 107 (78%) patients (Table 1). Of the 30 patients who never received active therapy, 14 died or pursued comfort care prior to the availability of antimicrobial susceptibility testing results. Among patients who received active therapy, there was a median of 29 hours (interquartile range [IQR]: 9–78 hours) from blood culture collection until receipt of active therapy. Polymyxins were the most common agents used for initial targeted therapy (40% of patients), followed by carbapenems (35%) and ceftazidime-avibactam (29%). Sixty-one percent of patients who received targeted therapy received a single active agent and 21% received 2 or more active agents. Infectious diseases consultation was obtained in 91% of patients.
Forty (29%) patients died within 14 days of bacteremia onset and 52 (38%) died within 30 days. Among baseline characteristics, the presence of cancer (particularly a hematologic malignancy), onset of infection in the intensive care unit, receipt of renal replacement therapy or mechanical ventilation, and increasing APACHE II and Pitt Bacteremia scores were associated with increased 30-day mortality. Fever, receipt of a solid-organ transplant, and urinary source were associated with decreased mortality (Table 1).
Characterization of CRE Bloodstream Isolates
CRE bacteremia was most commonly caused by K. pneumoniae (64%), Escherichia coli (15%), and Enterobacter cloacae (11%; Table 2). The most common K. pneumoniae sequence type was ST258 (58% of K. pneumoniae). One hundred and six patients (77%) were infected with carbapenemase-producing CRE (CP-CRE), including 89 (65%) with blaKPC, 8 (6%) with blaOXA-48, and 7 (5%) with blaNDM. The 30-day mortality rate was 38% in patients infected with CP-CRE and 39% in patients with non-CP CRE.
Table 2.
CRE Bloodstream Pathogens and 30-Day Mortality Rates by Pathogen Type
| Bloodstream Pathogen | No. (% of Total Cohort) | No. (%) of Patients Who Died Within 30 Days, per Pathogen |
|---|---|---|
| Klebsiella pneumoniae | 88 (64) | 38 (43) |
| ST258 | 51 (58)a | 22 (43) |
| Escherichia coli | 20 (15) | 8 (40) |
| ST131 | 7 (35)a | 3 (43) |
| Enterobacter cloacae complex | 15 (11) | 4 (27) |
| ST171 | 5 (33)a | 3 (60) |
| Klebsiella oxytoca | 5 (4) | 1 (20) |
| Serratia marcescens | 3 (2) | 0 |
| Multiple CRE | 4 (3) | 1 (25) |
| Other | 2 (2) | 2 (100) |
| Meropenem-resistant | 103 (75) | 41 (40) |
| Difficult-to-treat [31] | 89 (65) | 37 (42) |
| Carbapenemase-producerb | 106 (77) | 40 (38) |
| KPC-producerc | 89 (65) | 34 (38) |
| OXA-48–like-producerd | 8 (6) | 4 (50) |
| NDM-producere | 7 (5) | 2 (29) |
| Non–carbapenemase-producer | 31 (23) | 12 (39) |
| Polymicrobial bacteremiaf | 44 (32) | 18 (41) |
Abbreviations: CRE, carbapenem-resistant Enterobacterales; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-B-lactamase; ST, multilocus sequence type.
The denominator for this percentage is the number of patients infected with the corresponding species.
One patient was infected with a non-metallocarbapenemase class A (NMC-A)–producing E. cloacae, 1 patient with a Serratia marcescens enzyme (SME)–producing S. marcescens, and 1 patient with a Verona integron-encoded metallo-ß-lactamase (VIM)–producing E. coli. Two patients were infected with 2 different carbapenemase-producing bacteria (KPC-2–producing K. pneumoniae and KPC-3–producing E. cloacae [n = 1] and KPC-2–producing K. pneumoniae and KPC-3–producing S. marcescens [n = 1]), and 1 patient was infected with a K. pneumoniae that harbored NDM-5 and OXA-232.
Patients were infected with KPC-3–producing organisms (n = 52), KPC-2-producers (n = 37), and KPC-4 producers (n = 2). Two patients were infected with both KPC-2– and KPC-3–producing organisms.
Patients were infected with OXA-181–producing organisms (n = 4), OXA-48-producers (n = 2), and OXA-232-producers (n = 2).
Patients were infected with NDM-5–producing organisms (n = 4) and NDM-1-producers (n = 3). One patient was infected with an NDM-5–producing and OXA-232–producing K. pneumoniae.
Polymicrobial infection was defined as a non-CRE bloodstream infection (BSI) that occurred within 2 days of CRE BSI.
The most frequently active antimicrobial agents in vitro were ceftazidime-avibactam (89% susceptible), tigecycline (89%), colistin (87%), amikacin (80%), and gentamicin (64%; Table 3). Ninety-five percent of KPC-producing isolates were susceptible to ceftazidime-avibactam and 90% of non–CP-CRE and OXA-48–producing isolates were ceftazidime-avibactam susceptible.
Table 3.
Antimicrobial Susceptibilities Among CRE Bloodstream Isolates Stratified by Carbapenemase Type
| Percent Susceptible | |||||
|---|---|---|---|---|---|
| Antimicrobial Agents | All CRE (n = 143)a | KPC-Producers (n = 93) | MBL-Producersb (n = 8) | OXA-48-Producersc (n = 10) | Non-CP CRE (n = 31) |
| Amikacin | 80% | 83% | 38% | 70% | 81% |
| Aztreonam | 5% | 0% | 13% | 10% | 16% |
| Cefepime | 8% | 7% | 0% | 10% | 10% |
| Ceftazidime | 9% | 4% | 0% | 10% | 19% |
| Ceftazidime-avibactam | 89% | 95% | 13% | 90% | 90% |
| Ceftriaxone | 4% | 0% | 0% | 10% | 13% |
| Ciprofloxacin | 13% | 11% | 0% | 0% | 19% |
| Colistind | 87% | 85% | 100% | 100% | 90% |
| Ertapenem | 3% | 3% | 0% | 0% | 3% |
| Gentamicin | 64% | 67% | 13% | 40% | 71% |
| Imipenem | 9% | 1% | 0% | 10% | 35% |
| Levofloxacin | 16% | 14% | 0% | 10% | 23% |
| Meropenem | 16% | 15% | 0% | 30% | 19% |
| Minocycline | 49% | 48% | 63% | 40% | 48% |
| Piperacillin-tazobactam | 1% | 0% | 0% | 0% | 3% |
| Tigecyclinee | 89% | 90% | 88% | 90% | 81% |
| Tobramycin | 21% | 18% | 0% | 40% | 26% |
| TMP-SMX | 20% | 16% | 13% | 20% | 29% |
Abbreviations: CP, carbapenemase; CRE, carbapenem-resistant Enterobacterales; KPC, Klebsiella pneumoniae carbapenemase; MBL, metallo-β-lactamase; TMP-SMX, trimethoprim-sulfamethoxazole.
There were 143 CRE bloodstream isolates collected from the 137 patients.
Metallo-β-lactamase–producing CRE included 4 organisms with blaNDM-5, 3 with blaNDM-1, and 1 with blaVIM-1.
OXA-48–producing CRE included 5 organisms with blaOXA-181, 3 with blaOXA-48, and 2 with blaOXA-232.
An organism was considered susceptible if the colistin minimum inhibitory concentration (MIC) was ≤2 µg/mL.
An organism was considered susceptible if the tigecycline MIC was ≤2 µg/mL.
Impact of blaKPC PCR Testing on Positive Blood Culture Broths
Three of the eight study hospitals used the BioFire FilmArray Blood Culture Identification Panel (BCID; BioFire Diagnostics, Salt Lake City, UT) on positive blood culture broths during the study period (Supplementary Table 1). This assay detects blaKPC and 24 pathogens but does not detect other carbapenemase genes [9]. The bacterial species and blaKPC results from this panel were reported to clinicians at the 3 hospitals. The BCID assay detected blaKPC in 32 of 33 patients where the gene was detected by WGS of bloodstream isolates. No other molecular assays that detect carbapenemase genes from blood cultures were used at study hospitals.
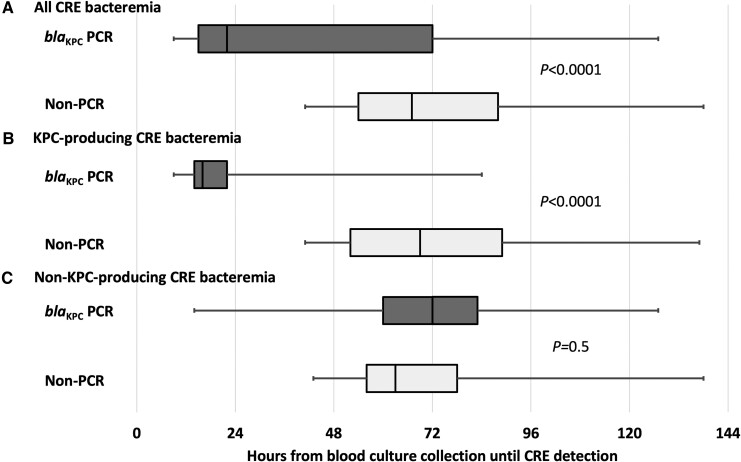
Characteristics of the 51 patients whose blood cultures underwent blaKPC PCR testing (PCR patients) were similar to those of the 86 patients whose blood cultures did not undergo this test (non-PCR patients), except PCR patients were less likely to have bacteremia onset as an outpatient, had a longer duration of hospitalization prior to bacteremia onset, and were more likely to receive initial targeted therapy with ceftazidime-avibactam (35% vs 16%; P = .011; Supplementary Table 2). The PCR patients had a median of 22 hours (IQR: 15–72 hours) from blood culture collection until detection of CRE bacteremia, compared with 67 hours (IQR: 54–88 hours) in non-PCR patients (P < .0001; Figure 1A). Decreased time to CRE bacteremia detection with blaKPC PCR testing occurred exclusively in KPC-producing CRE bacteremia, where the median time until CRE bacteremia detection was 16 hours (IQR: 14–22 hours; Figure 1B and 1C).
Figure 1.
Hours from blood culture collection until CRE detection in patients whose positive blood culture broths underwent blaKPC PCR testing (blaKPC PCR) and those whose blood culture broths did not undergo blaKPC PCR testing (non-PCR). Results are displayed for (A) all CRE bacteremia, (B) KPC-producing CRE bacteremia, and (C) non–KPC-producing CRE bacteremia. Boxes represent 25th and 75th percentiles, and the vertical line in the box represents the median. The range is represented by the whiskers. P values compare blaKPC PCR patients with non-PCR patients. Abbreviations: CRE, carbapenem-resistant Enterobacterales; KPC, Klebsiella pneumoniae carbapenemase; PCR, polymerase chain reaction.
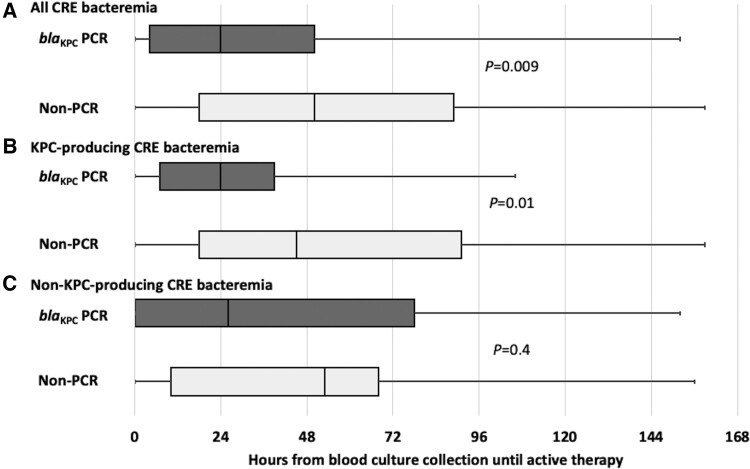
PCR patients were more likely to receive active antimicrobial therapy within 24 hours (22/51 [43%] vs 21/86 [24%]; P = .02) and within 48 hours (32/51 [63%] vs 32/86 [37%]; P = .004) after bacteremia onset than non-PCR patients. Among patients who received active therapy, the median time from blood culture collection until active therapy was 24 hours (IQR: 4–50 hours) in PCR patients and 50 hours (IQR: 18–89 hours) in non-PCR patients (P = .009; Figure 2A). This decreased time to receipt of active therapy in PCR patients compared with non-PCR patients was only observed in patients with KPC-producing CRE bacteremia (Figure 2B and 2C).
Figure 2.
Hours from blood culture collection until receipt of active therapy in patients whose positive blood culture broths underwent blaKPC PCR testing (blaKPC PCR) and those whose blood culture broths did not undergo blaKPC PCR testing (non-PCR). Results are displayed for (A) all CRE bacteremia, (B) KPC-producing CRE bacteremia, and (C) non–KPC-producing CRE bacteremia. Boxes represent 25th and 75th percentiles, and the vertical line in the box represents the median. The range is represented by the whiskers. P values compare blaKPC PCR patients with non-PCR patients. Only data from the 107 patients who received active therapy are included. Abbreviations: CRE, carbapenem-resistant Enterobacterales; KPC, Klebsiella pneumoniae carbapenemase; PCR, polymerase chain reaction.
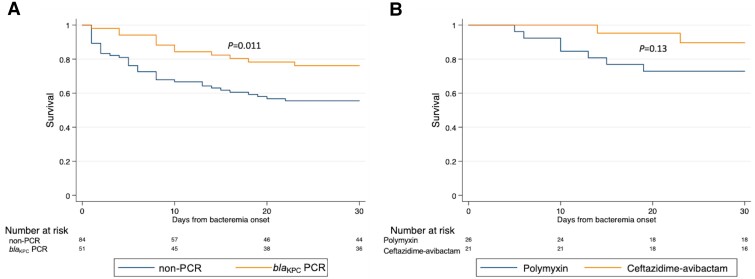
Fourteen- and 30-day mortality rates were lower among PCR patients compared with non-PCR patients (Figure 3A): 14-day: 8 of 51 (16%) versus 32 of 86 (37%) (P = .007); 30-day: 12 of 51 (24%) versus 40 of 86 (47%) (P = .007). This decrease in mortality in PCR patients was only observed in patients with KPC-producing CRE bacteremia (Supplementary Figure 2A and 2B). blaKPC PCR testing was associated with decreased unadjusted odds of 30-day mortality using logistic regression (odds ratio: .35; 95% confidence interval [CI]: .16–.75; P = .01) and decreased adjusted odds of 30-day mortality using TMLE (adjusted odds ratio: .37; 95% CI: .16–.84; P = .02).
Figure 3.
Comparisons of 30-day survival after bacteremia onset between (A) patients whose positive blood culture broths underwent blaKPC PCR testing (blaKPC PCR) and those whose blood culture broths did not undergo blaKPC PCR testing (non-PCR) and (B) patients who received ceftazidime-avibactam monotherapy as initial targeted therapy and patients who received polymyxin monotherapy. P values compare survival curves by log-rank testing. Abbreviation: PCR, polymerase chain reaction.
Initial Targeted Therapy With Ceftazidime-Avibactam and Mortality
There were 112 patients who survived to receive 2 or more days of targeted therapy. Six (19%) of 32 patients who received ceftazidime-avibactam as part of their initial targeted therapy died within 30 days, compared with 22 (28%) of 80 patients who did not receive ceftazidime-avibactam (P = .33; Table 1). None of the 21 patients who received ceftazidime-avibactam monotherapy as initial targeted therapy died within 14 days and 2 (10%) died within 30 days (Table 4; Figure 3B). In contrast, 5 (19%) of 26 patients who received polymyxin monotherapy as initial targeted therapy died within 14 days and 8 (31%) died within 30 days. Patients whose initial targeted therapy consisted of 2 or more active agents (combination therapy) had higher mortality rates than patients who received 1 active agent (monotherapy). Risk of AKI was similar among treatment regimens.
Table 4.
Initial Targeted Therapies and Clinical Outcomes
| Antimicrobial Therapy | No. | 14-Day Mortality | 30-Day Mortality | AKIa |
|---|---|---|---|---|
| Monotherapy (1 active agent) | 68 | 9% | 18% | 22% |
| Polymyxinb | 26 | 19% | 31% | 26% |
| Ceftazidime-avibactamc | 21 | 0% | 10% | 22% |
| Fluoroquinolone | 12 | 8% | 17% | 22% |
| Otherd | 9 | 0% | 0% | 13% |
| Combination therapy (≥2 active agents) | 23 | 17% | 43% | 29% |
Abbreviation: AKI, acute kidney injury.
AKI was defined by RIFLE criteria and the denominator included only patients who did not require renal replacement therapy at the time of bacteremia onset [21].
Polymyxin monotherapy consisted of polymyxin B (n = 24) and colistin (n = 2). Additional inactive agents were administered to 17 (65%) of these patients, including 12 patients who also received a carbapenem and 3 patients who received a non-carbapenem β-lactam agent.
Additional inactive agents were administered to 4 (19%) of these patients.
Patients were treated with the following monotherapy regimens: minocycline (n = 2), tigecycline (n = 2), ertapenem (n = 1), gentamicin (n = 1), trimethoprim-sulfamethoxazole (n = 1), cefepime (n = 1), and piperacillin-tazobactam (n = 1).
DISCUSSION
Our study of CRE bacteremia in 2013 in NY/NJ identified long delays until active therapy and a 49% 30-day mortality rate [8]. We conducted this follow-up study to evaluate the impact of 2 new interventions: the availability of blaKPC PCR testing on positive blood culture broths and of ceftazidime-avibactam for treatment. We found the 30-day mortality rate among patients with CRE bacteremia had decreased to 38%. However, this decreased mortality was observed primarily in patients whose blood cultures underwent blaKPC PCR testing, in whom the 30-day mortality rate was 24%, but not in patients whose blood cultures did not undergo this test, for whom the 30-day mortality rate was 47%. We believe that the mortality reduction with blaKPC PCR testing was related to the earlier initiation of active therapy observed in these patients compared with non-PCR patients. Earlier initiation of active antimicrobial therapy is consistently associated with decreased mortality in patients with sepsis [32, 33].
To our knowledge, this is the first study to identify an association between blaKPC PCR testing on positive blood culture broths and decreased mortality. Given that this was not a randomized trial, and patients at hospitals that used blaKPC PCR testing may differ from those at other study hospitals, this association warrants careful assessment for confounding. We compared characteristics of PCR and non-PCR patients and did not find baseline characteristics of PCR patients that would predispose to lower mortality, including similar APACHE II, Pitt Bacteremia, and Charlson Comorbidity Index scores (Supplementary Table 2). PCR patients were more likely to receive ceftazidime-avibactam, but this choice in therapy may have been related to detection of blaKPC, providing clinicians with confidence to use this agent given its reliable in vitro activity against KPC-producing Enterobacterales [34]. The association between blaKPC PCR testing and decreased 30-day mortality also persisted in a propensity score–adjusted analysis. Furthermore, if other characteristics of the 3 study hospitals that used blaKPC PCR testing were responsible for the improved outcomes, then one would expect to identify improved outcomes with both KPC-producing and non–KPC-producing CRE bacteremia at these centers. However, the decreases in time to active therapy and mortality were only observed in patients with KPC-producing CRE bacteremia. Based on these considerations, we believe that our finding of improved outcomes with blaKPC PCR testing was not due to confounding. Ultimately, we believe that a multicenter clinical trial with randomization to PCR testing at the individual or hospital level would be ideal to confirm our findings.
Although not statistically significant, the numerically lower 30-day mortality with ceftazidime-avibactam compared with polymyxin monotherapy (10% vs 31%) is consistent with prior studies that documented decreased mortality with this new β-lactam/β-lactamase inhibitor compared with polymyxins [11, 14]. In our study, polymyxin B was the predominant polymyxin used, whereas prior studies compared ceftazidime-avibactam with colistin. Polymyxin B has favorable pharmacological properties compared with colistin because it does not require conversion into its active form [15], yet this study suggests that it is also unlikely to be as effective as ceftazidime-avibactam for non–metallo-β-lactamase-producing CRE bacteremia. Even though ceftazidime-avibactam was available throughout the study period, polymyxins were still used more than ceftazidime-avibactam for initial targeted therapy. This finding is consistent with national data that showed that colistin was used more than ceftazidime-avibactam during the 2 years after FDA approval of ceftazidime-avibactam [35]. In addition to leading to more rapid administration of appropriate therapy, another potential advantage of using a molecular assay that detects blaKPC is that knowledge of the carbapenem resistance mechanism could lead to increased use of ceftazidime-avibactam and other newer agents for KPC-producing organisms and decreased use of polymyxins.
This study identified changes in the organisms causing CRE bacteremia in the NY/NJ area. In 2013, 90% of CRE bacteremias were caused by K. pneumoniae and 92% were KPC-producers [8]. In this study, only 64% of CRE bacteremias were caused by K. pneumoniae, only 65% were KPC-producers, and OXA-48- and NDM-producing organisms were more common than in the earlier study. We found similar 30-day mortality rates between CP-CRE and non–CP-CRE bacteremias. This contrasts with findings from a single-center study that identified an increase in mortality with CP-CRE [36] but is consistent with more recent studies that have not demonstrated increased mortality with CP-CRE [5, 37]. In this study, the presence of blaKPC may have permitted rapid diagnosis and earlier appropriate therapy of CRE bacteremia in PCR patients, and this may have decreased the overall mortality with CP-CRE.
This study has strengths and limitations. Among its strengths include its multicenter design and use of reference antimicrobial susceptibility testing and genotyping on all isolates to provide comprehensive assessments of carbapenemases and of the activity of antibacterial therapies. A limitation is that all centers are from NY/NJ, and the epidemiology and clinical impact of blaKPC PCR testing on positive blood cultures may be different in other geographic areas where KPC-producing CRE are less prevalent [38]. Molecular panels are now available that detect not only blaKPC but also genes that encode other carbapenemases [39–41]. We encourage future investigations of clinical outcomes associated with use of these panels in other geographic areas where other carbapenemases are more common. Given the diversity in antimicrobial therapies in this study, we had limited power to detect differences in outcomes by treatment regimen. Furthermore, the study was not designed to compare outcomes with combination therapy versus monotherapy, and we suspect that the unexpected finding of worse outcomes with combination therapy may have been due to confounding by indication. This study also predated the use of other new β-lactam/β-lactamase inhibitors, such as meropenem-vaborbactam and imipenem-relebactam, and thus we were unable to assess treatment outcomes with these agents.
In conclusion, we found that PCR testing for blaKPC in positive blood culture broths was associated with more prompt administration of effective therapy and decreased mortality among patients with CRE bacteremia in a geographic area where KPC production was the most common carbapenem resistance mechanism. This study suggests that rapid molecular assays have a role in improving outcomes in regions where CP-CRE are prevalent pathogens. We also found that ceftazidime-avibactam use led to favorable outcomes in patients with CRE bacteremia, and thus it should be considered as a first-line agent for non–metallo-β-lactamase-producing CRE bacteremia.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support . This work was supported by an investigator-initiated grant from Allergan (CAZ-IT-21), and grants from the National Institute of Allergy and Infectious Diseases (5R01AI090155; to B. N. K.), the National Center for Advancing Translational Sciences (UL1TR002384), and the National Cancer Institute (P30CA008748; to S. K. S. and Y.-W. T.) of the National Institutes of Health.
Supplementary Material
Contributor Information
Michael J Satlin, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine, New York, New York, USA; Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, New York, USA.
Liang Chen, Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, New Jersey, USA; Department of Medical Sciences, Hackensack Meridian School of Medicine, Nutley, New Jersey, USA.
Angela Gomez-Simmonds, Division of Infectious Diseases, Department of Medicine, Columbia University Irving Medical Center, New York, New York, USA.
Jamie Marino, Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, New York, USA.
Gregory Weston, Division of Infectious Diseases, Department of Medicine, Montefiore Medical Center, Albert Einstein College of Medicine, Bronx, New York, USA.
Tanaya Bhowmick, Division of Allergy, Immunology, and Infectious Diseases, Department of Medicine, Rutgers Robert Wood Johnson Medical School, New Brunswick, New Jersey, USA.
Susan K Seo, Infectious Diseases Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
Steven J Sperber, Division of Infectious Diseases, Hackensack Meridian School of Medicine, Nutley, New Jersey, USA; Division of Infectious Diseases, Department of Medicine, Hackensack University Medical Center, Hackensack, New Jersey, USA.
Angela C Kim, Division of Infectious Diseases, Donald and Barbara Zucker School of Medicine at Hofstra/Northwell, Manhasset, New York, USA.
Brandon Eilertson, Division of Infectious Diseases, Department of Medicine, State University of New York Downstate, Brooklyn, New York, USA.
Sierra Derti, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
Stephen G Jenkins, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine, New York, New York, USA; Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, New York, USA.
Michael H Levi, Department of Pathology, Albert Einstein College of Medicine, Bronx, New York, USA.
Melvin P Weinstein, Division of Allergy, Immunology, and Infectious Diseases, Department of Medicine, Rutgers Robert Wood Johnson Medical School, New Brunswick, New Jersey, USA; Department of Pathology and Laboratory Medicine, Rutgers Robert Wood Johnson Medical School, New Brunswick, New Jersey, USA.
Yi-Wei Tang, Department of Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
Tao Hong, Department of Pathology, Hackensack University Medical Center, Hackensack, New Jersey, USA.
Stefan Juretschko, Department of Pathology, Northwell Health, Manhasset, New York, USA.
Katherine L Hoffman, Division of Biostatistics, Department of Population Health Sciences, Weill Cornell Medicine, New York, New York, USA.
Thomas J Walsh, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
Lars F Westblade, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine, New York, New York, USA; Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, New York, USA.
Anne-Catrin Uhlemann, Division of Infectious Diseases, Department of Medicine, Columbia University Irving Medical Center, New York, New York, USA.
Barry N Kreiswirth, Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, New Jersey, USA; Department of Medical Sciences, Hackensack Meridian School of Medicine, Nutley, New Jersey, USA.
References
- 1. Centers for Disease Control and Prevention . Antibiotic resistance threats in the United States. Atlanta, GA: Centers for Disease Control and Prevention, 2019. Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 19 October 2021. [Google Scholar]
- 2. World Health Organization . Global Priority list of antibiotic-resistant bacteria to guide research discovery, and development of new antibiotics. Available at: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. Accessed 19 October 2021.
- 3. Bratu S, Landman D, Haag R, et al. . Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med 2005; 165:1430–5. [DOI] [PubMed] [Google Scholar]
- 4. Munoz-Price LS, Poirel L, Bonomo RA, et al. . Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Duin D, Arias CA, Komarow L, et al. . Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 2020; 20:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grundmann H, Glasner C, Albiger B, et al. . Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective multinational study. Lancet Infect Dis 2017; 17:153–63. [DOI] [PubMed] [Google Scholar]
- 7. Kazmierczak KM, Karlowsky JA, de Jonge BLM, Stone GG, Sahm DF. Epidemiology of carbapenem resistance determinants identified in meropenem-nonsusceptible Enterobacterales collected as part of a global surveillance program, 2017 to 2017. Antimicrob Agents Chemother 2021; 65:e0200020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Satlin MJ, Chen L, Patel G, et al. . Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother 2017; 61:e02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salimnia H, Fairfax MR, Lephart PR, et al. . Evaluation of the FilmArray Blood Culture Identification Panel: results of a multicenter controlled trial. J Clin Microbiol 2016; 54:687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tojo M, Fujita T, Ainoda Y, et al. . Evaluation of an automated rapid diagnostic assay for detection of gram-negative bacteria and their drug-resistance genes in positive blood cultures. PLoS One 2014; 9:394064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Duin D, Lok JJ, Earley M, et al. . Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 2018; 66:163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wunderink RG, Giamarellos-Bourboulis EJ, Rahav G, et al. . Effect and safety of meropenem-vaborbactam versus best-available therapy in patients with carbapenem-resistant Enterobacteriaceae infections: the TANGO II randomized clinical trial. Infect Dis Ther 2018; 7:439–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Motsch J, Murta de Oliveria C, Stus V, et al. . RESTORE-IMI 1: a multicenter, randomized, double-blind trial comparing efficacy and safety of imipenem/relebactam vs colistin plus imipenem in patients with imipenem-nonsusceptible bacterial infections. Clin Infect Dis 2020; 70:1799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shields RK, Nguyen MH, Chen L, et al. . Ceftazidime-avibactam is superior to other treatment regimens against carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother 2017; 25:e00883-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nation RL, Velkov T, Li J. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis 2014; 59:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. Healthcare-associated infections: carbapenem-resistant Enterobacterales (CRE): CRE technical information. Available at: https://www.cdc.gov/hai/organisms/cre/technical-info.html#Definition. Accessed 19 October 2021.
- 17. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 19. Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med 1985; 13:818–29. [PubMed] [Google Scholar]
- 20. Paterson DL, Ko WC, Von Gottberg A, et al. . International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial infections. Ann Intern Med 2014; 140:26–32. [DOI] [PubMed] [Google Scholar]
- 21. Bellomo R, Kellum JA, Ronco C. Defining and classifying acute renal failure: from advocacy to consensus and validation of the RIFLE criteria. Intensive Care Med 2007; 33:409–13. [DOI] [PubMed] [Google Scholar]
- 22. Clinical and Laboratory Standards Institute (CLSI) . Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 11th ed. CLSI Standard M07. Wayne, PA: CLSI, 2018. [Google Scholar]
- 23. Clinical and Laboratory Standards Institute (CLSI) . Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI Supplement M100. Wayne, PA: CLSI, 2020. [Google Scholar]
- 24. Ondov BD, Treangen TJ, Melsted P, et al. . Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 2016; 17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotypic tool for Klebsiella pneumoniae and its related species complex. Nat Commun 2021; 12:4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feldgarden M, Brover V, Haft DH, et al. . Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 2019; 63:e00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunt M, Mather AE, Sánchez-Busó L, et al. . ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017; 3:e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schuler MS, Rose S. Targeted Maximum Likelihood Estimation for causal inference in observational studies. Am J Epidemiol 2017; 185:65–73. [DOI] [PubMed] [Google Scholar]
- 29. Williams NT, Díaz I. lmtp: Non-parametric casual effects of feasible interventions based on modified treatment policies. 2020. R package version 1.0.0.5001. Available at: https://github.com/nt-williams/lmtp. Accessed on 25 Mar 2022.
- 30. Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron 1976; 16:31–41. [DOI] [PubMed] [Google Scholar]
- 31. Kadri SS, Adjemian J, Lai YL, et al. . Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis 2018; 67:1803–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar A, Roberts D, Wood KE, et al. . Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34:1589–96. [DOI] [PubMed] [Google Scholar]
- 33. Garnacho-Montero J, Garcia-Garmendia JL, Barrero-Almodovar A, Jimenez-Jimenez FJ, Perez-Paredes C, Ortiz-Leyba C. Impact of adequate empirical antibiotic therapy on the outcome of patients admitted to the intensive care unit with sepsis. Crit Care Med 2003; 31:2742–51. [DOI] [PubMed] [Google Scholar]
- 34. Castanheira M, Mendes R, Sader HS. Low frequency of ceftazidime-avibactam resistance among Enterobacterales isolates carrying blaKPC collected in U.S. hospitals from 2012 to 2015. Antimicrob Agents Chemother 2017; 61:e02369–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stritch JR, Ricotta E, Warner S, et al. . Pharmacoepidemiology of ceftazidime-avibactam use: a retrospective cohort analysis of 210 US hospitals. Clin Infect Dis 2021; 72:611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamma PD, Goodman KE, Harris AD, et al. . Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 2017; 64:257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hovan MR, Narayanan N, Cedarbaum V, Bhowmick T, Kirn TJ. Comparing mortality in patients with carbapenemase-producing carbapenem-resistant Enterobacterales and non-carbapenemase-producing carbapenem-resistant Enterobacterales bacteremia. Diagn Microbiol Infect Dis 2021; 101:115505. [DOI] [PubMed] [Google Scholar]
- 38. Guh AY, Bulens SN, Mu Y, et al. . Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 2015; 314:1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mancini N, Infurnari L, Ghidoli N, et al. . Potential impact of a microarray-based nucleic acid assay for rapid detection of Gram-negative bacteria and resistance markers in positive blood cultures. J Clin Microbiol 2014; 52:1242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wolk DM, Young S, Whitfield NN, et al. . A multicenter clinical study to demonstrate the diagnostic accuracy of the GenMark Dx ePlex blood culture identification gram-negative panel. J Clin Microbiol 2021; 59:e0248420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cortazzo V, D’Inzeo T, Giordano L, et al. . Comparing BioFire FilmArray BCID2 and BCID panels for direct detection of bacterial pathogens and antimicrobial resistance genes from positive blood cultures. J Clin Microbiol 2021; 59:e03163-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.