Abstract
During the stage of engulfment in the Bacillus subtilis spore formation pathway, the larger mother cell engulfs the smaller forespore. We have tested the role of forespore-specific gene expression in engulfment using two separate approaches. First, using an assay that unambiguously detects sporangia that have completed engulfment, we found that a mutant lacking the only forespore-expressed engulfment protein identified thus far, SpoIIQ, is able to efficiently complete engulfment under certain sporulation conditions. However, we have found that the mutant is defective, under all conditions, in the expression of the late-forespore-specific transcription factor ςG; thus, SpoIIQ is essential for spore production. Second, to determine if engulfment could proceed in the absence of forespore-specific gene expression, we made use of a strain in which activation of the mother cell-specific sigma factor ςE was uncoupled from forespore-specific gene expression. Remarkably, engulfment occurred in the complete absence of ςF-directed gene expression under the same conditions permissive for engulfment in the absence of SpoIIQ. Our results demonstrate that forespore-specific gene expression is not essential for engulfment, suggesting that the machinery used to move the membranes around the forespore is within the mother cell.
Recent studies have demonstrated that bacterial cells share with eukaryotic cells the ability to actively move macromolecules within their cytoplasm. The rapid separation of the bacterial origin of chromosomal replication and of plasmids just prior to the onset of cell division provided the first evidence that this was the case (14–16, 33, 52–54). The second example was provided by the MinC and MinD proteins of Escherichia coli, which are required for division site selection and which rapidly oscillate from one cell pole to the other (40, 41). An oscillating localization pattern has also been observed for the Bacillus subtilis Soj protein (29, 39), which regulates the onset of sporulation (3, 18). The mechanisms for these dynamic events remain elusive, as bacteria lack a characterized cytoskeleton, although they have distant homologues of tubulin and actin (11, 51). However, these proteins do not appear to be required for either chromosome segregation or MinCD oscillation.
Another example of the movement of macromolecules within a bacterial cell is provided by the spore formation pathway of B. subtilis, in a process known as engulfment. Engulfment mediates a dramatic reorganization of the sporangium, from two adjacent sister cells to a sporangium in which one sister cell lies within the cytoplasm of the other (Fig. 1) (48). Engulfment superficially resembles phagocytosis, as it occurs by the migration of membrane projections from one cell (the mother cell) around the other cell (the forespore) and culminates in the engulfed cell being enclosed in three membranes, the forespore cytoplasmic membrane, a separate membrane derived from the engulfing mother cell membrane, and the outermost mother cell cytoplasmic membrane. However, thus far, there is no evidence for a cytoplasmic contribution to engulfment analogous to the role of actin in assembling membrane projections during phagocytosis and motility in eukaryotic organisms. Indeed, all of the engulfment proteins identified thus far are either integral membrane or exported proteins (48). It therefore seems likely that engulfment will provide a novel mechanism by which phagocytosis-like events can occur.
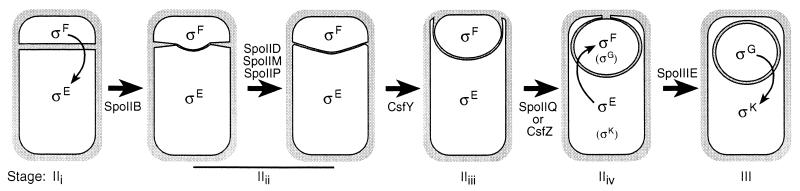
FIG. 1.
Initially, a layer of peptidoglycan (gray) lies between the two membranes separating the smaller forespore (top cell) and the larger mother cell (bottom cell; stage IIi). Engulfment starts with the thinning of this peptidoglycan, starting in the middle of the septum (stage IIii); this step is temporally and spatially regulated by SpoIIB (35). Proteins listed above the arrows are essential for engulfment under all conditions, while those below the arrows are not essential for engulfment, either because they contribute only to the efficienty of engulfment (i.e., SpoIIB) or because they are required only under certain culture conditions (i.e., SpoIIQ and two putative proteins under the control of ςF, CsfY and CsfZ, which are required only when sporulation is induced by nutrient exhaustion). The completion of septal thinning to the edge of the septum requires SpoIID, SpoIIM, and SpoIIP; this step allows the mother cell membrane to migrate up the side of the forespore (stage IIiii), a process that under certain conditions requires an unknown protein under the control of ςF, CsfY. The mother cell membrane then migrates across the cell pole, in a step that, under certain sporulation conditions, requires either SpoIIQ or a second protein under ςF control that depends on SpoIIQ for its expression, CsfZ. Ultimately the engulfing membranes meet at the cell pole (stage IIiv) and fuse to fully enclose the forespore (stage III). This membrane fusion event is the final step of engulfment. After synthesis of the asymmetrically positioned septum, the forespore-specific transcription factor ςF becomes active in the smaller forespore, thereby allowing activation of ςE in the larger mother cell. Production of inactive ςG in the forespore required both ςF activity and ςE activity. Following the completion of engulfment, ςG becomes active in the forespore, allowing activation of ςK in the mother cell. The stages of engulfment are designated by lowercase Roman numerals as previously described (17, 44).
Genetic studies have defined several stages of engulfment (Fig. 1) (17, 36, 48), starting with septal thinning, during which septal cell wall material is either removed or isomerized, resulting in the septal membranes of the mother cell and forespore being very close to one another, rather than being separated by a thick layer of peptidoglycan (Fig. 1, stage IIi to stage IIii). The spatial and temporal regulation of this process is conferred by SpoIIB, a protein made early in sporulation (28, 35). Three proteins expressed in the mother cell are necessary for septal thinning to proceed to the edges of the septum, SpoIID, SpoIIM, and SpoIIP (13, 26, 45). During the next step, the mother cell membrane migrates up the sides of the forespore and across the cell pole until the migrating membrane projections meet and fuse (Fig. 1, stage IIii to stage III). This membrane fusion event is the final step of engulfment and can readily be assessed in living sporangia, allowing genetic dissection of membrane fusion in bacteria (44).
A comparison of engulfment to phagocytosis raises the question of what role the forespore, which is being engulfed, plays in this process. If engulfment is similar to phagocytosis, then the forespore may play a more passive role in engulfment than the mother cell. Thus far, only one forespore-specific engulfment protein has been identified, SpoIIQ, which was reported to be essential for migration of the mother cell membrane across the cell pole at late stages of engulfment (24). SpoIIQ is a membrane protein with a large external domain that exhibits significant identity to endopeptidases, including proteins from Staphylococcus species that cleave peptide cross-bridges in the cell wall. This suggests that SpoIIQ could play an enzymatic role in engulfment, such as breaking the bridges between the forespore membrane and the cell wall, thereby allowing engulfment to proceed.
Here we demonstrate that SpoIIQ is dispensable for engulfment under certain sporulation conditions, although it is required for the expression of some forespore-specific genes normally transcribed by ςF-containing RNA-polymerase. The ςF factor plays a key role in sporulation, as it is the first transcription factor whose activity is cell specific, and it sets into motion the entire cascade of compartmentalized gene expression (Fig. 1). Using a strain that bypasses the normal requirement for ςF to activate the mother cell transcription factor ςE (56), we found that forespore-specific gene expression is dispensable for engulfment under the same conditions permissive for engulfment in spoIIQ mutants. These results raise the possibility that SpoIIQ is not directly involved in engulfment but rather, is involved in forespore-specific gene expression. They also suggest that although the forespore contributes to engulfment under certain conditions, the essential engulfment machinery is expressed in the mother cell, demonstrating one mechanistic similarity between engulfment and phagocytosis: the relative passivity of the cell that is being engulfed.
MATERIALS AND METHODS
Bacterial strains and genetic manipulations.
All strains used in this study are derivatives of wild-type strain PY79 (55), and their genotypes are listed in Table 1. The mutations were introduced into PY79 by transformation (8). All lacZ fusions were transformed into their respective host strains with selection for chloramphenicol resistance.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| PY79 | 55 | |
| KP9 | amyE::spoIID-lacZ | 47 |
| KP11 | sspB-lacZ | 50 |
| KP51 | spoIIAC561 | 12 |
| KP66 | spoIIIG-lacZ spoIIIGΔ1 | 49 |
| KP86 | amyE::sspE(2G)-lacZ | 50 |
| KP234 | spoIIAC(VA233) spoIIIGΔ1 sspB-lacZ | 27 |
| KP331 | spoIIAC::kan amyE::spoIID-lacZ PspoIIE-spoIIR | 38, 56 |
| KP558 | spoIIQ::spc sspB-lacZ | This work |
| KP559 | spoIID::Tn917ΩHU298 sspB-lacZ | This work |
| KP560 | spoIIQ::spc spoIIIG-lacZ spoIIIGΔ1 | This work |
| KP561 | amyE::spoIIIG-lacZ | 50 |
| KP562 | spoIIQ::spc amyE::spoIIIG-lacZ | This work |
| KP564 | amyE::spoIIIG-lacZ spoIIIGΔ1 | 50 |
| KP564 | spoIIQ::spc amyE::spoIIIG-lacZ spoIIIGΔ1 | This work |
| KP565 | spoIID::Tn917ΩHU298 amyE::sspE(2G)-lacZ | This work |
| KP566 | spoIIQ::spc amyE::sspE(2G)-lacZ | This work |
| KP569 | amyE::spoIIR-lacZ | 56 |
| KP570 | spoIIQ::spc amyE::spoIIR-lacZ | This work |
| KP571 | spoIIQ::spc amyE::spoIID-lacZ | This work |
| KP572 | spoIIQ::spc spoIIAC(VA233) spoIIIGΔ1 sspB-lacZ | This work |
| KP574 | spoIID::Tn917ΩHU298 spoIIIG-lacZ spoIIIGΔ1 | This work |
| KP575 | spoIIQ::spc | 24 |
| KP576 | amyE::csfB-lacZ spoIIIGΔ1 | 6 |
| KP577 | csfA-lacZ spoIIIGΔ1 | 6 |
| KP578 | gpr-lacZ spoIIIGΔ1 | 27 |
| KP579 | spoIIQ::spc amyE::csfB-lacZ spoIIIGΔ1 | This work |
| KP580 | spoIIQ::spc csfA-lacZ spoIIIGΔ1 | This work |
| KP581 | spoIIQ::spc gpr-lacZ spoIIIGΔ1 | This work |
Growth conditions.
B. subtilis cells were grown at 37°C and induced to sporulate either by the resuspension method (46) or by nutrient exhaustion in Difco Sporulation Medium (DSM) (42). Samples were taken and analyzed at various times after the start of sporulation. Sporulation efficiency was monitored after 40 h of sporulation, by heating the cultures to 80°C for 10 min and determining the number of heat-resistant spores per milliliter.
Fusion assay.
The membrane fusion assay was performed essentially as described previously (44). Briefly, a 0.5-ml sample of each culture was taken, centrifuged, and resuspended in 0.15 ml of the original culture supernatant. A 2-μl volume of the concentrated sample was added to 1 μl of sporulation salts containing a 5-μg/ml concentration of the membrane-impermeable stain N- (3-triethylammoniumpropyl)-4-{6-[4-(diethylamino)phenyl]-hexatrienyl}py-ridinium dibromide (FM 4-64; Molecular Probes, Eugene, Oreg.), a 1-μg/ml concentration of the DNA stain 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), and a 40-μg/ml concentration of the membrane-permeable stain MitoTracker Green FM (Molecular Probes) on a microscope slide and immobilized with a poly-l-lysine-treated coverslip (44). All stains were obtained from Molecular Probes and resuspended as suggested by the manufacturer.
Microscopy and image analysis.
To visualize the cells, an Applied Precision (Issaquah, Wash.) optical sectioning microscope (described in reference 37) was used to collect images spaced 0.1 to 0.2 μm apart throughout the specimen. The images were deconvolved using the Delta Vision deconvolution software as previously described (37). Deconvolved images were saved in TIFF format and imported into Adobe Photoshop.
β-Galactosidase assay.
Samples (1 ml) from sporulating cultures were harvested, briefly centrifuged to pellet the bacteria, and frozen at −70°C. After thawing, the cells were resuspended in Z buffer, lysozyme treated, and permeabilized with Triton X-100, essentially as described previously (38). β-Galactosidase activity was measured as described by Miller (31) with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate.
RESULTS
SpoIIQ is required for engulfment only under certain conditions.
Because SpoIIQ is the only known engulfment protein required for a late stage of engulfment and also the only engulfment protein expressed by ςF and thereby forespore specific, we investigated further its involvement in engulfment using a membrane fusion assay that unambiguously detects sporangia that have completed engulfment (44). Prior to the completion of engulfment, the forespore membrane is still in contact with the external medium and the engulfing membrane remains contiguous with the mother cell cytoplasmic membrane (e.g., Fig. 1, Stage IIiv). Therefore, both membranes are accessible to membrane-impermeable fluorescent membrane stains such as FM 4-64 (Fig. 2A, arrow 1). However, following the membrane fusion event that is the final step of engulfment, both the forespore membrane and the engulfing membrane are isolated from the external environment (e.g., Fig. 1, Stage III) and therefore remain unstained with FM 4-64, which is unable to cross the mother cell cytoplasmic membrane (Fig. 2A, arrow 2). In contrast, the membrane-permeable stain MitoTracker Green FM is able to stain these membranes both before and after the completion of engulfment (Fig. 2B, arrow 2). Only sporangia that have successfully completed engulfment will exclude FM 4-64 from the forespore membranes while retaining MitoTracker Green FM staining of these membranes.
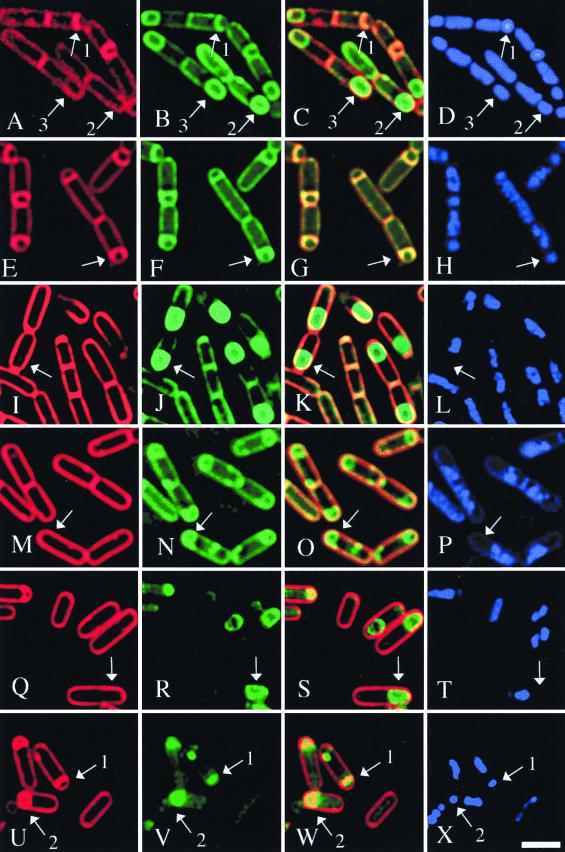
FIG. 2.
Fusion assay for the completion of engulfment in the wild-type strain and the spoIIQ-null mutant. Sporulation was induced as indicated, and samples were stained with FM 4-64 (red) (A, E, I, M, Q, and U). MitoTracker Green FM (green) (B, F, J, N, R, and V) and DAPI (blue) (D, H, L, P, T, and X), as described in the text (Materials and Methods). Overlays of FM 4-64 and MitoTracker Green FM are also shown (C, G, K, O, S, and W). (A to D) The wild-type strain (PY79) at t2.0 in a resuspension sporulation. Arrows 1 indicate the forespore of a sporangium at an early stage of engulfment; the mother cell membrane has begun to curve around the smaller forespore. Arrows 2 show a sporangium that has completed engulfment and membrane fusion; FM4-64 fails to stain the two membranes surrounding the forespore (A), while MitoTracker Green continues to stain membranes (B). Arrows 3 indicate a sporangium in which the forespore stains very faintly with FM4-64, but we consider the sporangium to have completed membrane fusion, as the forespore staining is less bright than that of the cytoplasmic membrane, rather than two- to threefold more bright as predicted based on the number of membranes surrounding the forespore (see Fig. 1). This faint staining of the forespore membranes is only rarely observed and could represent a limited exchange of FM 4-64 between the mother cell cytoplasmic membrane and the outer forespore membrane. (E to H) The spoIIQ-null mutant at t2.0 in a resuspension sporulation. Very few sporangia have completed engulfment and membrane fusion, and in most sporangia the membranes have reached, but not traversed, the cell pole (arrows). (I to L) The wild-type strain at t3.0 in a resuspension sporulation; most sporangia have complete membrane fusion (arrows), and the forespore chromosome fails to stain with DAPI, for reasons that are as yet unclear. (M to P) The spoIIQ mutant at t3.0 in a resuspension sporulation. Engulfment and membrane fusion are complete in many sporangia (arrows). (Q to T) The wild-type strain at t3.0 in a DSM nutrient exhaustion. Most sporangia have completed engulfment and membrane fusion (arrows). (U to X) The spoIIQ-null mutant at t3.0 in a DSM nutrient exhaustion. Very few sporangia have completed engulfment. Some sporangia are blocked prior to the migration of the engulging membranes across the cell pole (arrows 2), and sporangia with flat polar septa are also visible (arrows 1). Bar = 2 μm.
Both the wild-type strain (PY79) and the spoIIQ-null mutant strain (KP575) were induced to sporulate by the resuspension method, during which the bacteria are grown in a defined medium, centrifuged during exponential growth, and resuspended in a solution of mineral salts to induce sporulation. In the wild-type strain, the completion of engulfment and membrane fusion was evident 2 h after the onset of sporulation (t2.0) (44), as many forespores were inaccessible to FM 4-64 but stained with MitoTracker Green FM (Fig. 2A to D, arrow 2). In contrast, by t2.0 most spoIIQ mutant sporangia had not yet completed engulfment, and the engulfing membranes appeared to have reached but not migrated across the forespore pole in most sporangia (Fig. 2E to H, arrows). However, 1 h later (at t3.0) many spoIIQ mutant sporangia had completed membrane fusion, demonstrating the completion of engulfment (Fig. 2M to P, arrows). Indeed, 41% of spoIIQ mutant sporangia had completed engulfment by t3.0 (scored in Fig. 3D, gray bar), compared to 66% for the wild-type strain (scored in Fig. 3C, gray bar). This relatively efficient completion of engulfment was in contrast to the results of a previous study, which indicated that the spoIIQ mutant failed to complete engulfment by t4.5, suggesting that SpoIIQ is required for migration of the engulfing membranes across the cell pole (24). Thus, under these conditions, SpoIIQ is not essential for the completion of engulfment, although it contributes to speed or efficiency of membrane migration across the cell pole.
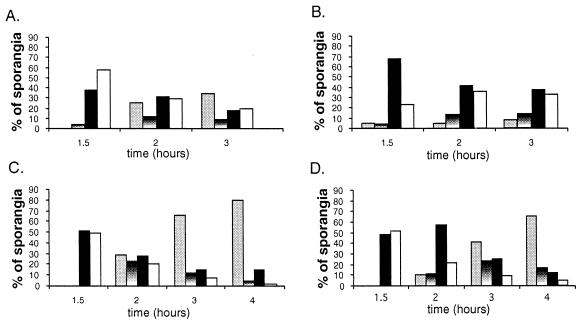
FIG. 3.
The percentage of wild-type and spoIIQ-null mutant sporangia that fall into various phenotypic classes. Samples from at least two independent experiments at t1.5, t2.0, t3.0, and t4.0 were scored for the percentage of sporangia that are in various stages of engulfment. At least 70 (and usually more than 100) sporangia were scored from each time point. Grey bar, fully engulfed and fused; shaded bar, engulfed but not fused; black bar, curved polar septa; white bar, straight polar septa. (A) Wild-type strain in DSM nutrient exhaustion; (B) spoIIQ mutant in DSM nutrient exhaustion; (C) wild-type in resuspension sporulation; (D) spoIIQ mutant in resuspension sporulation.
A second frequently used method to induce sporulation (and that used in the previous morphological study of the spoIIQ mutant) is nutrient exhaustion, whereby the bacteria are grown in a complex medium DSM until the culture enters stationary phase (defined as t0). When sporulation was induced by this method, the spoIIQ mutant was less able to complete engulfment (Fig. 2U to X, arrows 1 and 2), and only 8% of the spoIIQ mutant sporangia had completed engulfment by t3.0 (Fig. 3B, gray bar), compared to 35% for wild-type (Fig. 2Q to T, arrows, and Fig. 3A, gray bar). Indeed, by t4.0, the number of fused spoIIQ mutant sporangia decreased to 4% (data not shown), suggesting that some of the spoIIQ mutant sporangia that had finished engulfment may have lysed, perhaps accounting for the absence of completely engulfed sporangia at t4.5 in previous morphological studies (24).
Thus, our results demonstrate that SpoIIQ is only conditionally required for the completion of engulfment and that culture medium dramatically influences the engulfment phenotype of the spoIIQ mutant. However, we noted that SpoIIQ is essential for spore production, as under both engulfment-permissive and engulfment-restrictive conditions, the spoIIQ-null mutant makes very few spores (9.7 × 104 and 8.9 × 104 spores/ml, respectively, versus 3 × 108 spores/ml for the wild-type strain under both conditions). It is unlikely that the slight engulfment delay of the spoIIQ mutant under engulfment-permissive conditions is responsible for this dramatic decrease in spore production, as a mutant that is more severely delayed in the completion of engulfment, spoIIB, makes nearly wild-type levels of spores (28, 35). This suggests that SpoIIQ, in contrast to SpoIIB, has a second function that is essential for spore formation (discussed further below).
ςF is required for engulfment only under certain conditions.
SpoIIQ is the only engulfment protein identified so far that is under the control of ςF (24, 48); however, our results indicate that SpoIIQ is not essential for engulfment. If, indeed, no other ςF-dependent engulfment protein exists, then mutants defective in ςF-directed gene expression should have the same conditional engulfment defect as the spoIIQ mutant. Previous studies with a mutated form of ςF (spoIIAC561) which alters promoter specificity but allows ςE activation (17, 24) resulted in a late engulfment defect similar to that observed with the spoIIQ mutant; indeed, our results confirmed these studies. In spoIIAC561 mutant sporangia, 3 h after resuspension, the mother cell membrane has migrated up the sides of the forespore, but not across the cell pole (Fig. 4A to C, arrows 1). We also observed a higher percentage of dispores than previously reported (Fig. 4A to C, arrows 2). However, the spoIIAC561 mutation is not ideal for studying the role of ςF in engulfment, because it has pleiotropic effects on the expression of ςF-dependent genes, such that some genes are overexpressed whereas others are underexpressed (20, 25, 34). Indeed, the mutation allows ςF to recognize some ςB promoters (27), so it is possible that certain ςB-dependent genes are abnormally expressed in the forespore, perhaps interfering with engulfment.
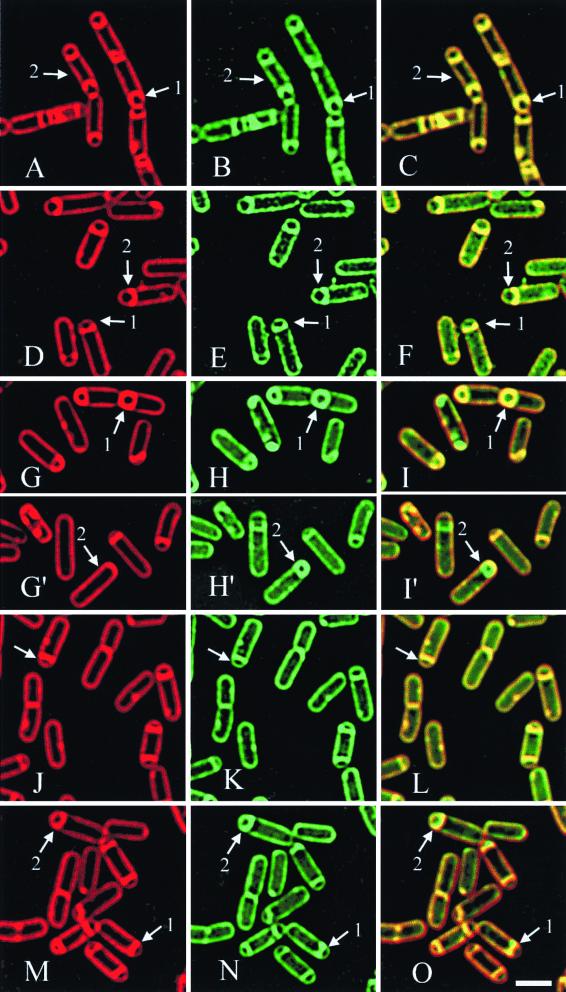
FIG. 4.
The engulfment defects of ςF mutants, as assessed by the membrane fusion assay. FM 4-64 (red) (A, D, G, G′, J, and M) and MitoTracker Green FM (green) (B, E, H. H′, K, and N) staining are shown, as is an overlay of both membrane stains (C, F, I, I′, L, and O). (A to C) The spoIIAC561 mutant strain KP51 was induced to sporulate by resuspension, and samples were harvested at t3.0. Arrows 1 point to a sporangium in which engulfment has proceeded to, but not across, the cell pole. Arrows 2 point to a sporangium in which division has occurred at both polar division sites, producing a disporic sporangium. The absence of ςF-directed gene expression affects engulfment, as shown in panels D to O. Strain KP331 (P spoIIE-spoIIR spoIIAC::kan) was used to allow activation of ςE in the absence of ςF. (D to F) Samples harvested at t2.0 in a resuspension sporulation. Arrows 1 show a sporangium with a flat septum, while arrows 2 indicate a sporangium with a curved septum. (G to I and G′ to I′) Two separate fields showing samples harvested at t3.0 in a resuspension sporulation. Arrows 1 indicate a sporangium in which the engulfing membrane has migrated around the forespore but membrane fusion is not complete. Arrows 2 indicate a sporangium in which membrane fusion is complete, as indicated by the exclusion of FM 4-64 from the forespore membranes, which are stained with the membrane-permeable MitoTracker Green FM. (J to L) Samples harvested at t2,0 in a DSM nutrient exhaustion. The arrows indicate a sporangium with a flat septum. (M to O) Samples harvested at t3.0 in a DSM nutrient exhaustion. Arrows 1 indicate a sporangium with a flat septum, while arrows 2 indicate a sporangium with a curved septum. Several disporic sporangia (those containing two forespores) are also evident; such sporangia are formed in mutants that fail to activate ςE. The quantitative analysis of these engulfment phenotypes is shown in Fig. 5. Bar = 2 μM.
To further investigate the role of ςF in engulfment, we utilized a ςF-null strain (ΔspoIIAC) in which spoIIR, the ςF-transcribed gene required for ςE activation, is expressed from the spoIIE promoter prior to the onset of polar septation (56). Remarkably, under these conditions many sporangia have normal compartmentalization of ςE activity (25, 56). When sporulation was induced by resuspension, at t2.0, the majority of the sporangia have either straight septa (Fig. 4D to F, arrows 1) or slightly curved septa (Fig. 4D to F, arrows 2), with straight septa being the predominant phenotype (81%) (Fig. 5). By t4.0, however, sporangia that have completed membrane migration across the cell pole (18%) (Fig. 4G to I, arrows 1, and Fig. 5) and that have complete membrane fusion are observed (Fig. 4 G′ to I′, arrows 2), although the frequency of sporangia that have clearly completed membrane fusion is low (5%) (Fig. 5). However, it is possible that membrane fusion occurred at a higher frequency than is shown in Fig. 4 and 5, as some cells with FM 4-64 staining of the cytoplasmic membrane contained diffuse, but bright, MitoTracker Green FM staining in the cytoplasm near the expected location of a fully engulfed forespore (not shown), suggesting that the forespore had lysed after membrane fusion. Consistent with this suggestion, the percentage of sporulating cells in the culture declined by t4.0, suggesting that either the sporangia had lysed or that they were no longer recognized as sporangia due to the lack of a forespore. Thus, ςF is not essential for migration of the mother cell membrane around the forespore during a resuspension sporulation but is required for the efficient production of fully engulfed (i.e., fused) forespores.
FIG. 5.
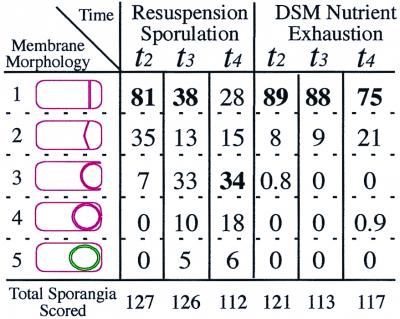
Quantitative analysis of the engulfment phenotype of strain KP311 (ΔspoIIAC PspoIIE-spoIIR) under different sporulation conditions. The numbers shown are the percentages of nondisporic sporangia at given time points within the identical morphology class; bold type indicates the most prevalent phenotypic class for a given time point. Class 1 sporangia have flat septa (Fig. 4J, arrow); class 2 sporangia have slightly curved septa with little or no migration of the membrane towards the pole (Fig. 4D, arrow 2); in class 3 sporangia, the engulfing membrane has moved up the side of the forespore but not across the pole (Fig. 4A, arrow 1); in class 4 sporangia, the migrating membranes have moved around the forespore, but membrane fusion is not complete (Fig. 4G, arrow 1), whereas membrane fusion is complete in class 5 sporangia (Fig. 4G′, arrow 2). The frequency of sporangia was 30% at t2.0, 42% at t3.0, and 31% at t4.0 in a resuspension sporulation and 14% at t2.0, 16% at t2.0, and 20% at t4.0 in a DSM nutrient exhaustion. Disporic sporangia comprised between 2 and 6% of the cells in the culture, likely reflecting the inefficient activation of ςE when SpoIIR production is uncoupled from forespore-specific gene expression. Such sporangia were excluded from the analysis of engulfment phenotypes because dispores lack a mother cell chromosome, which eliminates all mother cell-specific gene expression, thereby preventing engulfment.
In contrast, engulfment required ςF-directed gene expression when sporulation was induced by nutrient exhaustion and migration of the mother cell membrane around the forespore was almost completely blocked, similar to previous results (24). At both t2.0 and t4.0, the majority of the sporangia had straight septa (89% and 75%, respectively) (Fig. 5), and less than 10% of the sporangia had slightly curved septa (Fig. 4M to O, arrows 2, and Fig. 5). No completely engulfed (fused) sporangia were observed. Thus, there appears to be at least one unidentified protein under the control of ςF that is required for migration of the mother cell membrane up the sides of the sporangium when sporulation is induced by nutrient exhaustion. However, the requirement for forespore-specific gene expression can be bypassed when sporulation is induced by resuspension, suggesting that the essential engulfment machinery resides within the mother cell.
spoIIQ-null mutant lacks ςG activity.
To determine why SpoIIQ is required for sporulation even when it is not required for engulfment, we investigated the activities of late sporulation-specific sigma factors (ςG and ςK) in the spoIIQ-null mutant. Activation of ςG appears to depend on the completion of engulfment (34), so using the sspB-lacZ fusion that is under the control of ςG, we first investigated whether spoIIQ mutants have normal ςG activity when they can complete engulfment. Surprisingly, the spoIIQ mutant, under conditions in which engulfment has been completed, shows a reduction in ςG activity significantly more severe than that seen with a spoIID mutant, which never completes engulfment (Fig. 6A). Similar results were obtained when the protein products of the ςG-dependent sspB and sspA genes, the α/β-type small, acid-soluble proteins (SASP) were assayed by immunofluorescence microscopy (data not shown). Thus, the spoIIQ null mutant appears to almost completely lack ςG activity.
FIG. 6.
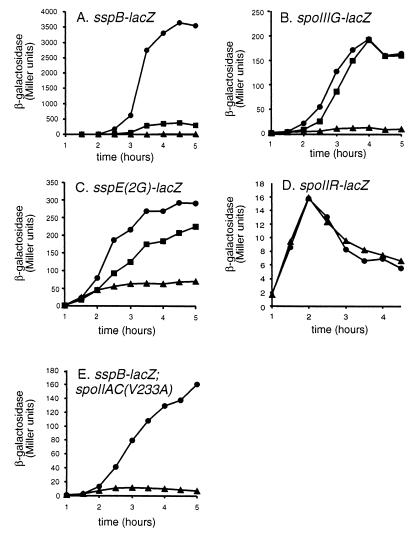
The effect of spoIID and spoIIQ mutations on the expression of several ςF-dependent lacZ fusions. (A to D) Strains containing sspB-lacZ (A), spoIIIG-lacZ (B), sspE(2G)-lacZ (C), and spoIIR-lacZ (D) chromosomal lacZ fusions were induced to sporulate by resuspension sporulation. Samples were taken every 30 min, and β-galactosidase activity was measured. ●, wild-type strain; ■, spoIID mutant; ▴, spoIIQ mutant. Strains used for panels B through D also contain the spoIIIG mutation to eliminate β-galactosidase activity contributed by ςG. (E) Strain KP234 (●) [spoIIGΔ1 spoIIIAC(V233A) sspB-lacZ] in which the promoter specificity of ςF has been changed to that of ςG. The spoIIIG mutation inactivates ςG, allowing use of the sspB-lacZ fusion (which is normally dependent on ςG) to monitor ςF activity. Strain KP572 (▴) is KP234 plus spoIIQ::spc.
spoIIQ null mutant lacks ςF-directed expression of the gene encoding ςG.
Because ςG is under both transcriptional and posttranslational control (19, 30), we investigated whether the lack of ςG activity in a spoIIQ mutant is due to a lack of expression of the gene encoding it, spoIIIG, or to a lack of activation of ςG following completion of engulfment. The spoIIIG gene is initially transcribed by ςF (49, 50); however, ςG is autoregulatory and thus, once activated, can recognize the spoIIIG promoter and maintain its own synthesis (19, 49). We therefore utilized a ςG-null background (spoIIIGΔ1 or spoIIIG::neo) to avoid β-galactosidase units contributed by ςG. Using a translational spoIIIG-lacZ fusion that is well recognized by ςF-containing RNA polymerase (49), we found that ςF-directed expression of spoIIIG is abolished in the spoIIQ mutant (Fig. 6B). The same effect is observed with this spoIIIG-lacZ fusion integrated at the amyE locus (data not shown). In contrast, the spoIID mutant showed normal expression of spoIIIG (Fig. 6B), suggesting that the decreased ςG activity of this mutant is due not to a failure to express the protein but rather to a failure to fully activate ςG, possibly as a consequence of a failure to complete engulfment. The spoIIIG gene is unique among ςF-transcribed genes, as it also requires a factor under the control of ςE for its expression (34). We therefore investigated ςE activity in the spoIIQ mutants, using the spoIID-lacZ reporter gene, and found normal ςE activity (not shown) as previously reported (24). Thus, the defect in ςG activity in the spoIIQ mutant appears to be due to the lack of ςF-directed expression of the gene encoding ςG, spoIIIG.
spoIIQ-null mutant affects the expression of some, but not all, ςF-dependent genes.
We next investigated if the spoIIQ mutant affected expression of other ςF-dependent genes or if the phenomenon was specific to the spoIIIG promoter. We made use of another ςF-dependent promoter, sspE(2G)-lacZ (50). The sspE promoter is normally recognized by ςG; however, with the substitution of two guanines at the −15 and −16 positions, it can be transcribed by ςF-containing RNA polymerase and is well transcribed in a mutant lacking ςG protein (50). The spoIIQ mutant shows 24% the level of expression of sspE(2G)-lacZ of the wild-type strain, a less severe effect than that seen with expression of spoIIIG (Fig. 6C). A spoIID mutant has slightly decreased expression of sspE(2G)-lacZ relative to the wild-type strain (77% at t5.0), although the reduction is less than that caused by the spoIIQ mutation (24% at t5.0) (Fig. 6C). A third promoter affected by the spoIIQ mutation is gpr, a promoter recognized by both ςF and ςG, whose ςF-dependent transcription was therefore monitored in a spoIIIG mutant background. The spoIIQ mutant showed about 45% the level of β-galactosidase activity as the wild-type strain at t4.0 (data not shown). However, several other ςF-dependent promoters were unaffected by the spoIIQ mutation, including spoIIR-lacZ (Fig. 6D), csfA-lacZ, and csfB-lacZ (reference 6 and data not shown). Therefore, the spoIIQ mutation has no effect on expression of some ςF-dependent genes (spoIIR, csfA, and csfB), while it has moderate [gpr and sspE(2G)] or strong (spoIIIG) effects on others.
SpoIIQ is required for the ςF-dependent expression of genes normally under the control of ςG.
The decrease in ςF-dependent gene expression in the spoIIQ mutant might be due to a requirement for SpoIIQ to modify the activity of an activator or repressor of certain ςF-dependent promoters, or it might be due to a requirement for SpoIIQ to allow high levels of ςF-directed gene expression. To distinguish between the first and second possibilities, we made use of a strain with a mutation in the gene encoding ςF (spoIIAC V233A), which allows ςF to recognize promoters normally recognized by ςG (27, 43). We inactivated ςG (with a spoIIIGΔ1 mutation) and used a ςG-dependent promoter (sspB-lacZ) to monitor activity of the altered ςF. If SpoIIQ acts via a repressor or activator of transcription of ςF-dependent genes, then it seems unlikely that it would be required for expression of an artificial ςF gene such as sspB-lacZ. However, the spoIIQ-null mutant showed a dramatic decrease in the ςF-directed expression of this gene (Fig. 6E). A similar though somewhat weaker effect is observed with another unnatural promoter (sspE(2G), as described earlier (Fig. 6C). Because SpoIIQ is required for expression from two unnatural ςF promoters, it seems unlikely that SpoIIQ decreases ςF-directed transcription via an activator or repressor of transcription, unless this transcription factor acted on both ςF- and ςG-dependent promoters. It is therefore possible that SpoIIQ is required for the full activation of the very sigma factor required for its expression. However, it is also possible that SpoIIQ does not play a direct role in forespore-specific gene expression but rather is required for some aspect of forespore physiology essential for the expression of certain forespore-specific genes.
DISCUSSION
The role of forespore-specific gene expression in engulfment has previously been difficult to investigate, due both to the lack of methods by which to assess the completion of engulfment and to the absolute dependence of engulfment on the mother cell transcription factor ςE, whose activity requires the forespore-specific transcription factor ςF. We have recently developed fluorescent membrane stain-based assays for engulfment (37) and for the membrane fusion event that is the final step of engulfment (44). Using these assays, we have investigated the role of forespore-specific gene expression in engulfment, making use of a strain in which ςE is able to become active in the absence of ςF (56) and also the role of the only forespore-expressed engulfment protein described to date, SpoIIQ (24). Our results indicate that forespore-specific gene expression is not essential for engulfment, which, under certain conditions, can be completed without SpoIIQ and also in the complete absence of forespore-specific gene expression (albeit inefficiently in the latter case). These results suggest that the essential components of the engulfment machinery are within the mother cell. Thus, engulfment, which bears a superficial resemblance to phagocytosis, shares certain mechanistic similarities to phagocytosis, as in both processes the engulfed cell (in this case the forespore) plays a relatively passive role, whereas the engulfing cell (in this case the mother cell) plays a more active role.
We did, however, find that under certain conditions, forespore-specific gene expression is essential for engulfment. When sporulation is induced by nutrient exhaustion, the absence of ςF-directed gene expression results in an early engulfment defect, whereas the absence of SpoIIQ results in a late engulfment defect similar to that previously described (24). These results suggest that there is at least one unidentified engulfment protein encoded by ςF, CsfY (controlled by ςF), that is required for the movement of the mother cell membrane up the sides of the forespore only when sporulation is induced by nutrient exhaustion (Fig. 1, stage IIii to stage IIiii) but not by resuspension. Because only a small proportion of the spoIIQ mutant sporangia were able to complete mother cell membrane migration across the cell pole after nutrient exhaustion, it is possible that SpoIIQ is directly required for this late step of engulfment. However, because spoIIQ mutants have defects in ςF-dependent gene expression (as discussed below), it is also possible that unidentified engulfment protein under the control of ςF whose expression depends on SpoIIQ (CsfZ) is conditionally required for this step of engulfment (Fig. 1, stage IIiii to stage IIiv).
The medium dependence of forespore-specific gene expression in engulfment is unanticipated, and may reflect differences in the barriers to the migration of the engulfing membranes under the two conditions used here. It has previously been proposed that bridges between the forespore membrane and the cell wall, such as that formed by lipoteichoic acid, would provide a steric barrier to the engulfing membranes and that such bonds would be removed prior to engulfment (36). Forespore-expressed hydrolytic enzymes would be ideally positioned for this task. The two sporulation conditions used here differ both in medium composition (that used for nutrient exhaustion is a more complex medium) and in the means by which sporulation is induced (in a nutrient exhaustion, the bacteria are gradually starved as they enter stationary phase, whereas in resuspension sporulation, starvation occurs rapidly, as exponentially growing bacteria are resuspended in a salt solution). The two methods could thereby result in an altered cell wall structure, which is modified both in response to medium composition (2, 22, 23) and during the transition to stationary phase (4, 5, 23). The future identification of genes required for engulfment when sporulation is induced by nutrient exhaustion but not by resuspension will help to elucidate the physical basis for the unusual effect of media on the requirement of forespore-specific gene expression in engulfment.
Although SpoIIQ is not essential for engulfment, it is essential for sporulation under all conditions, apparently because it is required for the expression of spoIIIG, the gene which encodes the second forespore-specific sigma factor, ςG. spoIIIG is initially transcribed by ςF-containing RNA polymerase, although the spoIIIG promoter is poorly recognized by ςF, and is later transcribed by ςG-containing RNA polymerase (49). The spoIIQ-null mutation also reduces expression of two other genes transcribed by ςF- and ςG-containing RNA polymerase [gpr and sspE(2G)] but does not affect expression of the genes transcribed solely by ςF-containing RNA polymerase that we examined [spoIIR, csfA, and csfB 6]. It is therefore possible that SpoIIQ governs the activity of a transcription factor that affects expression of promoters transcribed by both ςF and ςG, perhaps by controlling the activity of an activator or repressor of such genes or by allowing the full activation of ςF. However, it is not immediately apparent how SpoIIQ, and extacellular protein, could modify the ςF activation pathway (1, 7, 9, 10, 32) and it remains possible that the spoIIQ mutation adversely affects forespore viability or transcription and indirectly reduces expression of certain forespore specific genes.
However, regardless of the mechanism by which SpoIIQ affects forespore-specific gene expression, our results confirm the existence of two phases of ςF-dependent gene expression (34), with the initial expression of certain ςF-dependent genes such as spoIIR. During the second phase, the remaining ςF-dependent genes, such as spoIIIG are expressed (21, 34). We suggest two possible benefits of dividing ςF-directed gene expression into two phases. First, if SpoIIQ is directly involved in forespore-specific gene expression, it could contribute to the compartmentalization of ςF activity to the forespore by providing a positive feedback loop for ςF-dependent gene expression in the forespore. Second, having two phases of ςF-directed gene expression may serve as a checkpoint to delay production of ςG until the onset of engulfment, which may ensure more effective coupling of ςG and ςK activation to the completion of engulfment.
Does SpoIIQ play an active role in engulfment, or is the conditional engulfment defect of spoIIQ mutants a secondary consequence of decreased forespore-specific gene expression? SpoIIQ is a protein with a large extracellular domain that shows homology to endopeptidases such as lysostaphin, which cleaves peptide cross-bridges in the cell wall. Certainly, this potential enzymatic activity suggests an active role in engulfment, as does the observation that SpoIIQ initially accumulates in the septum and then spreads around the forespore membrane (24). It is possible that SpoIIQ plays a dual role, contributing to membrane migration as well as to forespore-specific gene expression, possibly coupling the full expression of ςF-dependent genes to the onset of engulfment. However, until it is possible either to genetically separate the two functions of SpoIIQ or to achieve full ςF-directed gene expression in the absence of SpoIIQ, this question will remain open.
ACKNOWLEDGMENTS
We are grateful to Richard Losick and to Patrick Stragier for contributing strains and useful advice, to Peter Setlow for providing the α/β-SASP antibodies, and to Joe Pogliano for his comments on the manuscript.
This work was supported by NIH grant GM-57045 to K.P., as well as by awards from the Arnold and Mabel Beckman Foundation and the Searle Scholars Program of the Chicago Community Trust.
REFERENCES
- 1.Arigoni F, Pogliano K, Webb C, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- 2.Atrih A, Bacher G, Allmaier G, Williamson M P, Foster S J. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP5 in peptidoglycan maturation. J Bacteriol. 1999;181:3956–3966. doi: 10.1128/jb.181.13.3956-3966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cervin M A, Spiegelman G B, Raether B, Ohlsen K, Perego M, Hoch J A. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol. 1998;29:85–95. doi: 10.1046/j.1365-2958.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 4.Cheung H V, Vitkovic L, Freese E. Rates of peptidoglycan turnover and cell growth of Bacillus subtilis are correlated. J Bacteriol. 1983;156:1099–1106. doi: 10.1128/jb.156.3.1099-1106.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer W R, Meyer P D, Jordens C G, Kruyssen F J, Wouters J T. Cell wall turnover in growing and nongrowing cultures of Bacillus subtilis. J Bacteriol. 1982;149:977–984. doi: 10.1128/jb.149.3.977-984.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decatur A, Losick R. Identification of additional genes under the control of the transcription factor ςF of Bacillus subtilis. J Bacteriol. 1996;178:5039–5041. doi: 10.1128/jb.178.16.5039-5041.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diederich B, Wilkinson J F, Magnin T, Najafi S M A, Errington J, Yudkin M. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor ςF of B. subtilis. Genes Dev. 1994;8:2653–2663. doi: 10.1101/gad.8.21.2653. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- 9.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- 10.Duncan L, Losick R. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein ςF from Bacillus subtilis. Proc Natl Acad Sci USA. 1993;90:2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson H P. Atomic structures of tubulin and FtsZ. Trends Cell Biol. 1998;8:133–137. doi: 10.1016/s0962-8924(98)01237-9. [DOI] [PubMed] [Google Scholar]
- 12.Errington J, Mandelstam J. Variety of sporulation phenotypes resulting from mutations in a single regulatory locus, spoIIA, in Bacillus subtilis. J Gen Microbiol. 1983;129:2091–2101. doi: 10.1099/00221287-129-7-2091. [DOI] [PubMed] [Google Scholar]
- 13.Frandsen N, Stragier P. Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 15.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 16.Gordon G S, Wright A. DNA segregation: putting chromosomes in their place. Curr Biol. 1998;8:925–927. doi: 10.1016/s0960-9822(98)00011-6. [DOI] [PubMed] [Google Scholar]
- 17.Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of ςE and ςF in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ireton K, Gunther N W, Grossman A D. spoOJ is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karmazyn-Campelli C, Bonamy C, Savelli B, Stragier P. Tandem genes encoding ς-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 1989;3:150–157. doi: 10.1101/gad.3.2.150. [DOI] [PubMed] [Google Scholar]
- 20.Karow L M, Glaser P, Piggot P J. Identification of a gene, spoIIR, which links the activation of ςE to the transcriptional activity of ςF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karow M L, Piggot P J. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 22.Kruyssen F J, de Boer W R, Wouters J T. Cell wall metabolism in Bacillus subtilis subsp. niger: effects of changes in phosphate supply to the culture. J Bacteriol. 1981;146:867–876. doi: 10.1128/jb.146.3.867-876.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruyssen F J, de Boer W R, Wouters J T. Effects of carbon source and growth rate on cell wall composition of Bacillus subtilis subsp. niger. J Bacteriol. 1980;144:238–246. doi: 10.1128/jb.144.1.238-246.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Londoño-Vallejo J-A, Fréhel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- 25.Londoño-Vallejo J-A, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Diaz I, Clarke S, Mandelstam J. spoIID operon of Bacillus subtilis: cloning and sequence. J Gen Microbiol. 1986;132:341–354. doi: 10.1099/00221287-132-2-341. [DOI] [PubMed] [Google Scholar]
- 27.Margolis P, Driks A, Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 28.Margolis P S, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marston A L, Errington J. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol Cell. 1999;4:673–683. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 30.Mason J M, Hackett R H, Setlow P. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J Bacteriol. 1988;170:239–244. doi: 10.1128/jb.170.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H, editor. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. Assay of β-galactosidase; pp. 352–355. [Google Scholar]
- 32.Min K-T, Hilditch C M, Diederich B, Errington J, Yudkin M D. ςF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 33.Mohl D A, Gober J W. Cell cycle dependent polar localization of chromosome partitioning proteins in Caulobacter crecentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 34.Partridge S R, Errington J. Importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 35.Perez A R, Abanes-DeMello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogliano J, Osborne N, Sharp M, Abanes-DeMello A, Perez A R, Sun Y-L, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pogliano K, Hofmeister A E M, Losick R. Disappearance of the ςE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to the establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quisel J D, Lin D C, Grossman A D. Control of development by altered localization of a transcription factor in Bacillus subtilis. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 40.Raskin D M, de Boer P A. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raskin D M, de Boer P A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaeffer P, Millet J, Aubert J. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt R, Margolis P, Duncan L, Coppolecchia R, Moran C P, Jr, Losick R. Control of developmental transcription factor ςF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp M D, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith K, Bayer M E, Youngman P. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol. 1993;175:3607–3617. doi: 10.1128/jb.175.11.3607-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 48.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 49.Sun D, Cabrera-Martinez R M, Setlow P. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor ςG. J Bacteriol. 1991;173:2977–2984. doi: 10.1128/jb.173.9.2977-2984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D, Fajardo-Cavazos P, Sussman M D, Tovar-Roja F, Cabrera-Martinez R-M, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EςF: identification of features of good EςF-dependent promoters. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicente M, Errington J. Structure, function and controls in microbial division. Mol Microbiol. 1996;20:1–7. doi: 10.1111/j.1365-2958.1996.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 52.Webb C D, Graumann P L, Kahana J A, Teleman A A, Silver P A, Losick R. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 53.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C-H, Grossman A D, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler R T, Shapiro L. Bacterial chromosome segregation: is there a mitotic apparatus? Cell. 1997;88:577–579. doi: 10.1016/s0092-8674(00)81898-x. [DOI] [PubMed] [Google Scholar]
- 55.Youngman P, Perkins J B, Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Higgins M L, Piggot P J, Karow M L. Analysis of the role of prespore gene expression in the compartmentalization of mother cell-specific gene expression during sporulation of Bacillus subtilis. J Bacteriol. 1996;178:2813–2817. doi: 10.1128/jb.178.10.2813-2817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]