Abstract
Background
Though detection of transmission clusters of methicillin-resistant Staphylococcus aureus (MRSA) infections is a priority for infection control personnel in hospitals, the transmission dynamics of MRSA among hospitalized patients with bloodstream infections (BSIs) has not been thoroughly studied. Whole genome sequencing (WGS) of MRSA isolates for surveillance is valuable for detecting outbreaks in hospitals, but the bioinformatic approaches used are diverse and difficult to compare.
Methods
We combined short-read WGS with genotypic, phenotypic, and epidemiological characteristics of 106 MRSA BSI isolates collected for routine microbiological diagnosis from inpatients in 2 hospitals over 12 months. Clinical data and hospitalization history were abstracted from electronic medical records. We compared 3 genome sequence alignment strategies to assess similarity in cluster ascertainment. We conducted logistic regression to measure the probability of predicting prior hospital overlap between clustered patient isolates by the genetic distance of their isolates.
Results
While the 3 alignment approaches detected similar results, they showed some variation. A gene family–based alignment pipeline was most consistent across MRSA clonal complexes. We identified 9 unique clusters of closely related BSI isolates. Most BSIs were healthcare associated and community onset. Our logistic model showed that with 13 single-nucleotide polymorphisms, the likelihood that any 2 patients in a cluster had overlapped in a hospital was 50%.
Conclusions
Multiple clusters of closely related MRSA isolates can be identified using WGS among strains cultured from BSI in 2 hospitals. Genomic clustering of these infections suggests that transmission resulted from a mix of community spread and healthcare exposures long before BSI diagnosis.
Keywords: Staphylococcus aureus, bloodstream infections, hospital epidemiology, outbreak detection, infection prevention
Multiple clusters of closely related methicillin-resistant Staphylococcus aureus bloodstream infections were identified using whole genome sequencing in 2 hospitals by 3 bioinformatic workflows. Genomic epidemiology suggests that transmission resulted from a mix of community spread and healthcare exposures long before symptom onset.
Staphylococcus aureus caused nearly 119 000 bloodstream infections (BSIs) and 20 000 associated deaths in 2019 [1]. These infections are exacerbated by the emergence of methicillin-resistant S. aureus (MRSA) strains that are resistant to treatment with conventional β-lactam antibiotics. Concerted national infection control efforts have decreased MRSA healthcare-associated infections (HAIs) in the United States (US), particularly BSIs caused by MRSA strains historically associated with HAIs. However, the decrease in MRSA BSIs in the US has slowed since 2013, and community-onset infections have recently made up the largest proportion of cases [1].
Onset of a clinically significant infection is influenced by bacterial virulence, human host factors, and triggers such as skin trauma or underlying illnesses that predispose patients to opportunistic infections [2]. Asymptomatic S. aureus carriage is a risk factor for infection and can be harbored in sites across the body [2, 3], complicating elimination since detecting carriage or transmission can occur long after exposure. Consequently, hospital [4] and community [5, 6] outbreaks of S. aureus result from direct or indirect contact with colonized individuals, contamination of an intermediate person such as a healthcare worker [7], or through environmental reservoirs. Though detecting transmission clusters of MRSA is an infection control priority in hospital settings, the transmission dynamics of MRSA among hospitalized patients with BSIs has not been thoroughly studied.
Whole genome sequencing (WGS) of bacterial genomes provides high resolution of genetic relationships between MRSA isolates and possible recent transmission. Improved access and ease of use of open-source bioinformatic resources, lower costs, and expansion of publicly available DNA sequences increases the feasibility of routine genomic analysis for cluster detection [8, 9]. Of great importance for detection is maximizing gene homology through genome alignments. Alignment creation includes reference-free or reference-dependent methods, which have unique trade-offs for sensitivity, specificity, and completeness of genetic data [10].
Epidemiological investigations use genetic thresholds between S. aureus isolates to identify or rule out clusters of related infections [4, 11, 12]. Commonly, single-nucleotide polymorphisms (SNPs) are quantified to compare isolate sequences, create multisequence alignments for phylogenetic reconstruction, and estimate the likelihood of a recent common ancestor and possible transmission given a SNP threshold [13, 14]. Reference choice, sample genetic diversity, and bioinformatic tools all impact which and how many SNPs are detected in a sample set, and necessitate exploration of the consistency of genomic alignments used to infer transmission clusters.
To elucidate transmission of MRSA BSI, we conducted a retrospective analysis of MRSA BSI at 2 hospitals in one university system over 12 months. We compared core-genome sequences from the isolates to detect putative transmission events between BSI patients and examined epidemiological and molecular traits of isolates shared between cluster patients. We also tested the consistency of detectable SNP differences between isolates using different sequence alignment pipelines.
METHODS
Patient Cohort
We identified all patients diagnosed with a MRSA BSI between July 2018 and June 2019, admitted to either of 2 hospitals of the University of Pennsylvania hospital system. The Hospital of the University of Pennsylvania (HUP) is a 625-bed academic tertiary and quaternary care medical center in West Philadelphia with approximately 32 000 patient admissions, 633 000 outpatient visits, and 40 000 emergency department (ED) visits annually. The Penn Presbyterian Medical Center (PMC) is a 324-bed urban community hospital in West Philadelphia with 12 000 admissions, 130 000 outpatient visits, and 26 000 ED visits annually. A single case of MRSA BSI was defined as a MRSA isolate collected from blood of any patient at HUP or PMC during the study period. Each subject was only included once. The study was approved by the University of Pennsylvania Institutional Review Board and given a waiver of consent, as the study was retrospective and no data or samples were collected specifically for research purposes.
Isolate Selection and DNA Sequencing
Isolates were obtained from a biobank of clinical MRSA isolates cultured for routine diagnosis in the HUP Clinical Microbiology Laboratory during the study period. Isolates were screened for phenotypic antibiotic resistance using the Vitek 2 automated system, and assigned susceptibility/resistance in accordance with Clinical and Laboratory Standards Institute protocols [15]. A 1-μL loopful of frozen isolate was streaked onto blood agar and incubated overnight at 37°C, and a single representative colony was grown under the same conditions on a new plate. A 10-μL loopful of each isolate was then frozen in a bead beating tube and underwent WGS using an Illumina MiSeq at the Penn/Children’s Hospital of Philadelphia Microbiome Center. Sequencing libraries were prepared using the Illumina Nextera library preparation kit. Sequences were made publicly available through the Sequence Read Archive (Bioproject PRJNA751847).
Bioinformatic Pipelines
Paired-end 150 bp FASTQ files were passed through Bactopia workflow to assess data quality, assemble contigs, and call multilocus sequence type, SCCmec type, antibiotic resistance, and virulence genes [16]. To compare SNP-based core-genome multiple-sequence alignments, the total number of assembled contigs or subsets grouped by clonal complex (CC) was passed through 3 pipelines: (1) randomly fragmenting assembled genomes to create “pseudoreads” and mapping these to a reference genome using Snippy (version 4.6.0) [17] (pseudoread pipeline); (2) mapping assembled genomes to a reference using Parsnp (version 1.5.6) [18] (assembly pipeline); and (3) evaluating reads with Markov cluster analysis, identifying overlapping gene clusters, and aligning core genes using the Bactopia Tools pangenome workflow (gene-family pipeline). The Gene-family pipeline included PIRATE [19], ClonalFrameML [20], and maskrc-svg (version 0.5) (https://github.com/kwongj/maskrc-svg) to identify and mask possible recombinant regions within the core-genome alignment. For the 2 reference-based pipelines, we used strain N315 (GCF_000009645.1) as reference for non-CC-specific alignments (all 104 available sequences regardless of CC) and CC5-specific alignments (n = 40). For the CC8-specific alignments (n = 55), we used NCTC 8325 as reference (GCF_000013425.1). Pairwise SNP distances of the core-genomes were calculated using snp-dists [21]. Maximum likelihood trees were created with IQ-Tree (version 2.1.2) [22] using a general time reversible model allowing for invariant sites and unequal base frequencies and midpoint-rooted and visualized using ggTree [23]. Bootstrap values were calculated for 1000 repetitions. Phylogenetic similarity across pipelines was measured by calculating cophenetic correlation [24] between SNP distance matrices and estimated phylogeny tip distance, and assessing Robinson-Foulds distances [25] between different alignment trees and randomly generated trees using ape (version 5.5) [26].
Epidemiological Investigation of Clustered Isolates
A transmission cluster was defined as 2 or more subjects whose isolates’ core genomes differed from one another by 35 or fewer SNPs, based on the approximate cutoff for within-patient vs between-patient BSI lineages in a hospital setting [5, 27]. We also examined a threshold of 15 SNPs, a proposed threshold for recent interpatient MRSA transmission [11]. Demographic data, comorbidities, Pitt Bacteremia Score, source of BSI, and inpatient mortality were abstracted from the electronic medical record (EMR), summarized, and assessed for association with CC using Fisher exact test or Student t test. BSIs were considered healthcare-associated if the index blood culture was drawn >48 hours after hospital admission; healthcare-associated, community-onset if the index culture was obtained <48 hours after admission or in the community setting, and if the subject had 1 or more previous healthcare risk factors (hospitalization, surgery, hemodialysis, or nursing home/residential medical facility stay in the previous year; or presence of an indwelling intravascular catheter at time of culture); and community-associated if the index culture was obtained <48 hours after admission or in the community setting and the subject lacked these healthcare risk factors. The EMR was examined for evidence of overlap or sequential hospital/unit stays among cluster-subjects, and visualized using vistime (https://github.com/shosaco/vistime). Admission and discharge dates were recorded for each cluster-subject for all hospital stays at any of 4 networked hospitals within 1 year before the first collected BSI isolate in a cluster and 1 year after the last collected BSI isolate in the cluster. These included HUP, PMC, Pennsylvania Hospital, and a single University of Pennsylvania long-term acute care hospital in Philadelphia. Pennsylvania Hospital is a 481-bed urban community hospital located in the Society Hill district of Philadelphia with >27 000 hospital admissions, >24 000 ED visits, and 201 000 outpatient visits annually.
Logistic regression assessed the predictive power of SNP distances and likelihood of patient hospitalization overlaps. Goodness of fit was assessed using a receiver operating characteristic (ROC) curve and measuring the area under the curve. All analyses were conducted in R studio (version 1.4.1106) [28] run with R version 4.0.4, and final figures were labeled in InkScape (version 0.92.5) [29]. Analysis code is available at: https://github.com/Read-Lab-Confederation/MRSA_bloodstream_clusters.
RESULTS
Patient Demographics and Isolate Characteristics
We screened all patients diagnosed with a MRSA BSI at 2 academic hospitals between July 2018 and June 2019, identifying 106 qualifying subjects. Of the BSI source sites that could be identified from EMR, skin site infections made up 19% and central venous catheter infections made up 14% (Table 1). Among included subjects, 17% died while hospitalized. From each individual, single MRSA isolates were sequenced, of which 105 had sufficient coverage for further analysis and 104 isolates were S. aureus. One isolate was identified by WGS as Staphylococcus argenteus and was excluded. Among the 104 genomes, 55 were assigned to CC8, 49 of which were USA300 strains; 40 were assigned to CC5; and the remaining 9 were assigned CC30, CC72, and CC78. No significant association emerged between the 2 most common CCs (CC5 and CC8) and sex, age group, race, ethnicity, BSI source site, hospital death, Pitt bacteremia score, or hospital of diagnosis (Supplementary Tables 1 and 2).
Table 1.
Demographics and Clinical Outcomes of Subjects With Methicillin-Resistant Staphylococcus aureus Bloodstream Infection (N = 106)
| Characteristic | No. (%) of Patients (N = 106) |
|---|---|
| Demographic characteristics | |
| Age group, y | |
| 20–29 | 12 (11%) |
| 30–39 | 13 (12%) |
| 40–49 | 16 (15%) |
| 50–59 | 18 (17%) |
| 60–69 | 32 (30%) |
| ≥70 | 15 (14%) |
| Sex | |
| Female | 51 (48%) |
| Male | 55 (52%) |
| Race | |
| Asian | 1 (1%) |
| White | 50 (48%) |
| Black | 49 (46%) |
| Other/unknown | 6 (6%) |
| Ethnicity | |
| Hispanic/Latino | 2 (2%) |
| Non-Hispanic/Latino | 99 (93%) |
| Unknown | 5 (5%) |
| Clinical characteristics | |
| Total | 106 |
| Source of BSI | |
| Arteriovenous graft | 4 (4%) |
| Central venous catheter infection | 15 (14%) |
| Device infection | 4 (4%) |
| Respiratory source | 2 (2%) |
| Skin site | 20 (19%) |
| Surgical site | 4 (4%) |
| Other | 3 (3%) |
| Unknown | 52 (49%) |
| Hospital of BSI diagnosis | |
| Hospital A | 65 (61%) |
| Hospital B | 41 (39%) |
| Infection setting | |
| HA | 22 (21%) |
| CA | 11 (10%) |
| HACO | 71 (68%) |
| In-hospital deatha | |
| No | 88 (83%) |
| Yes | 18 (17%) |
| Pitt Bacteremia Score | |
| Mean (SD) | 2.1 (2.6) |
| Median (range) | 1.00 (0–10.0) |
Abbreviations: BSI, bloodstream infection; CA, community-associated; HA, healthcare-associated; HACO, healthcare-associated, community-onset; SD, standard deviation.
Indicates death prior to discharge during the index methicillin-resistant Staphylococcus aureus BSI hospitalization.
Assessment of Sequence Alignment Pipelines
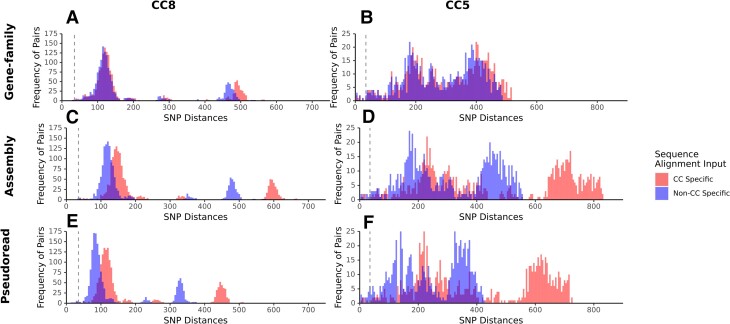
We generated multiple alignments of all isolate sequences using 3 approaches to determine their effect on pairwise SNP distances. Alignments generated with all 104 isolates had lower distances compared to CC-specific alignments. SNP distances produced by the gene-family pipeline were consistent between CC groups and whole species alignments (Figure 1A and 1B), whereas the SNP distances produced by the pseudoread and assembly pipelines were greater when isolates of the same CC were the input (Figure 1C–F). Pipeline choice on phylogenetic structure was assessed by comparing tree topology and SNP matrices across pipelines and sequence input groupings (Table 2). The cophenetic correlation showed the highest correlation for alignments produced from CC-specific inputs, though all alignment pipelines and inputs produced a value >0.90. Tree topology across pipelines suggested that trees are highly similar to one another compared to a random tree.
Figure 1.
Frequencies and distribution of single-nucleotide polymorphism (SNP) distances between isolates vary by alignment tool. The frequency of pairwise distances between isolates from clonal complexes (CC) 8 and 5 were quantified from distance matrices derived from alignments generated from 2 groupings of isolate input: the total number of isolates in the investigation (blue) or CC-specific isolates only (red). Isolate inputs were aligned using each of the 3 alignment pipelines, the gene-family pipeline (A and B), assembly pipeline (C and D), and pseudoread pipeline (E and F).
Table 2.
Comparability Phylogenetic Fit of Alignment Pipelines Using Cophenetic Correlation (R2), Alignment Size, and Robinson-Foulds Comparisona by Alignment Pipeline
| Pipeline | Total Isolates (N = 104) | CC5-Specific Isolates (n = 40) | CC8-Specific Isolates (n = 55) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R 2 | Alignment Size, bp | RF Values | R 2 | Alignment Size, bp | RF Value | R 2 | Alignment Size, bp | RF Values | |
| Gene family | 0.984 | 2 141 357 | 56 52 200 | 0.984 | 2 182 742 | 10, 8, 72 | 0.987 | 2 176 046 | 40, 36, 104 |
| Pseudoread | 0.983 | 2 839 469 | 46 56 202 | 0.993 | 2 839 469 | 10, 10, 72 | 0.999 | 2 821 361 | 34, 40, 104 |
| Assembly | 0.929 | 2 163 693 | 52 46 202 | 0.995 | 2 497 454 | 8, 10, 72 | 0.999 | 2 482 874 | 34, 36, 104 |
Abbreviations: CC, clonal complex; RF, Robinson-Foulds.
Row alignment pipeline compared to each other alignment pipeline and a random tree of the same number of phylogenetic tips.
Identification of Suspected Transmission Clusters
Using alignments from each pipeline containing 104 isolates, we identified 9 clusters (C1–C9) among 29 isolates that differed by 35 SNPs or fewer from at least 1 other subject isolate (Table 3). The pseudoread pipeline clustered 29 isolates, the assembly pipeline clustered 21, and the gene-family pipeline clustered 19. Five clusters contained CC5 isolates, 3 clusters were CC8, and 1 cluster was CC30. The median cluster size was 3 isolates (range, 2–6). The longest collection date difference between clustering isolates was 265 days (C1), and the shortest 12 days (C6). Median SNP differences were variable across clusters, and smaller differences did not correlate with shorter collection date differences.
Table 3.
Summary of Suspected Methicillin-Resistant Staphylococcus aureus (MRSA) Transmission Clusters Identified Through Pseudoread, Assembly, and Gene-Family Alignment Pipelines Among 104 Sequential MRSA Bloodstream Infection Patients at 2 Hospitals
| Transmission Cluster | MRSA Isolate | Clonal Cluster | No. of Isolates | Median Pairwise SNP Difference (Range) | Median Collection Date Difference, Days (Range) | ||
|---|---|---|---|---|---|---|---|
| Pseudoread Pipeline | Assembly Pipeline | Gene-Family Pipeline | |||||
| C1 | SAMN20960259, SAMN20960281, SAMN20960331 | CC5 | 3 | 11 (3–12) | 16 (4–16) | 14 (5–16) | 177 (110–265) |
| C2 | SAMN20960260, SAMN20960274 | CC5 | 2 | 6 | 7 | 7 | 61 |
| C3a | SAMN20960263, SAMN20960326, SAMN20960280, SAMN20960314, SAMN20960328 | CC5 | 5 | 35 (20–46) | 44 (26–62) | 50 (36–62)a | 128 (7–241) |
| C3b | SAMN20960280, SAMN20960314, SAMN20960328 | CC5 | 3 | 25 (20–25) | 32 (26–36) | 39 (36–39)a | 113 (37–150) |
| C4 | SAMN20960270, SAMN20960325 | CC5 | 2 | 20 | 26 | 24 | 189 |
| C5a | SAMN20960271, SAMN20960343 | CC5 | 4 | 29 (6–35) | 44 (10–53)a | 41 (33–46)a | 119 (56–237) |
| C5b | SAMN20960271, SAMN20960343 | CC5 | 2 | 29 | 42a | 33 | 237 |
| C5c | SAMN20960298, SAMN20960324 | CC5 | 2 | 6 | 10 | 7 | 78 |
| C6a | SAMN20960276, SAMN20960282, SAMN20960287, SAMN20960293, SAMN20960301, SAMN20960306 | CC8 | 6 | 29 (15–42) | 39 (21–62) | 38 (26–53)a | 54 (12–121) |
| C6b | SAMN20960276, SAMN20960282, SAMN20960293, SAMN20960301, SAMN20960306 | CC8 | 5 | 26 (15–31) | 35 (21–40) | 37 (26–43) | 66 (12–121) |
| C6c | SAMN20960276, SAMN20960282, SAMN20960293, SAMN20960306 | CC8 | 4 | 23 (15–30) | 32 (21–36) | 34 (26–38) | 63 (32–121) |
| C7 | SAMN20960299, SAMN20960305, SAMN20960334 | CC8 | 3 | 34 (30–34) | 39 (36–41) | 45 (41–50) | 80 (24–104) |
| C8 | SAMN20960313, SAMN20960323 | CC8 | 2 | 1 | 1 | 1 | 28 |
| C9 | SAMN20960316, SAMN20960337 | CC30 | 2 | 23 | 25 | 22 | 67 |
Abbreviations: CC, clonal complex; MRSA, methicillin-resistant Staphylococcus aureus; SNP, single-nucleotide polymorphism.
Partial or no detection of isolates as part of the cluster.
Phylogenetic Analysis of Isolates
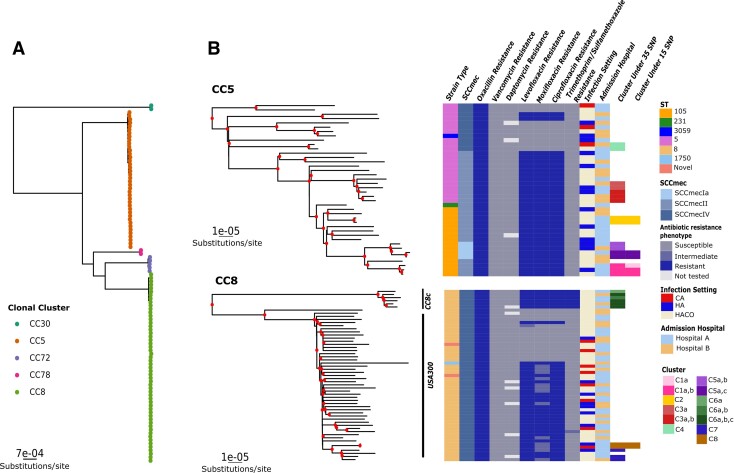
To assess phylogenetic relationships within clusters, we created a representative tree using the gene-family pipeline of the 104 isolates. This tree was selected because it had the strongest cophenetic correlation, tree structure similarity, and conservation of SNP distances between pipelines for the 104 isolates together (Table 2; Figure 1A and 1B). BSI isolates occupied significantly divergent clades of CCs (Shimodaira-Hasegawa approximate likelihood ratio test and ultrafast bootstrap values >70) (Figure 2A). Candidate transmission clusters arose from distinct sublineages (Figure 2B). The largest cluster, C5, diverged significantly from other CC8 isolates, and isolates were identified as part of the CC8c lineages [30]. Cluster and noncluster isolates had varied distributions for infection setting, with most BSIs categorized as healthcare-associated, community-onset (68%). At a 15-SNP threshold, only isolates in clusters C1, C2, C5(a, c), and C8 remained clustered. All isolates were susceptible to vancomycin and daptomycin, but isolates in both the CC5 and CC8 clades showed resistance to multiple β-lactams and quinolones. Thus, multiple lineages of MRSA associated with BSI could transmit multiclass-resistant strains between patients.
Figure 2.
Suspected transmission clusters fall into distinct clonal groups. Maximum likelihood trees were generated from the PIRATE alignment of 104 isolates and visualized using ggtree. A, Tree indicating clades containing individual clonal complexes (CCs). B, Subtrees from the complete maximum likelihood trees for the 2 most abundant CCs. Nodes with bootstrap values ≥70 are marked in red. Heat maps show strain type (ST), SCCmec element type, and resistance phenotype for indicated antibiotics per sequence, infection setting (healthcare-associated [HA], community-associated [CA], and healthcare-associated community-onset [HACO]), admission hospital, and transmission cluster at a threshold of 35 single-nucleotide polymorphisms (SNPs) or 15 SNPs.
Genomic Similarity Predicts Overlapping Hospital Stay in Transmission Clusters
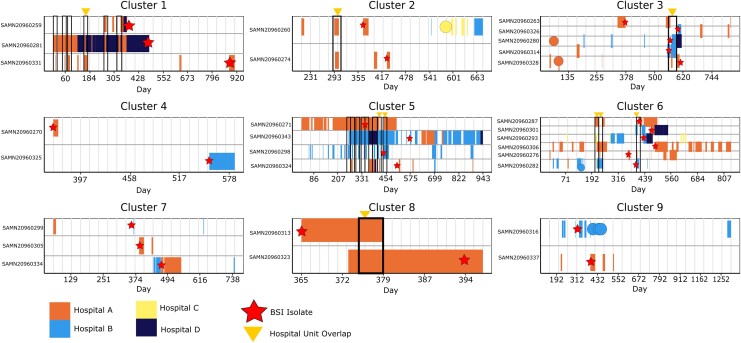
For every cluster-subject we examined hospitalization history at 4 networked hospitals in the University of Pennsylvania system 1 year before the first index BSI isolate and 1 year after the last patient index isolate per cluster. Six clusters included subjects with overlapping hospital stays, of which 3 had median SNP distances between 1 and 16 with corresponding hospital unit overlaps (Table 3; Figure 3). Cluster C5c had a median SNP difference of 7 (range, 6–10 SNPs across pipelines) with no common hospital overlap. In comparison, cluster C4 had no subjects with overlapping hospital admissions prior to their index BSI, but a median SNP distance range of 20–26 SNPs across pipelines.
Figure 3.
Hospitalization history among patients in genomic bloodstream infection (BSI) clusters. Hospitalization history at 4 study hospitals (A, B, C, and D) up to 365 days before the date of the earliest methicillin-resistant Staphylococcus aureus (MRSA) bloodstream isolate culture in each cluster (relative day 0) and up to 365 days after the latest MRSA bloodstream isolate in the cluster. Note that BSIs were only included at hospitals A and B. Rows represent the hospitalization history of each patient associated with a sequenced cluster isolate. Colored rectangles and circular marks represent individual hospitalization durations (rectangles) or 1-day admissions (circles); the color indicates hospital A, B, C, or D. Black outlined boxes represent areas where 2 or more patients overlapped in the same hospital at the same time. Stars indicate the date of collection of the sequenced BSI isolate for each patient. Triangles indicate a hospitalization where 2 or more patients overlapped in the same hospital unit.
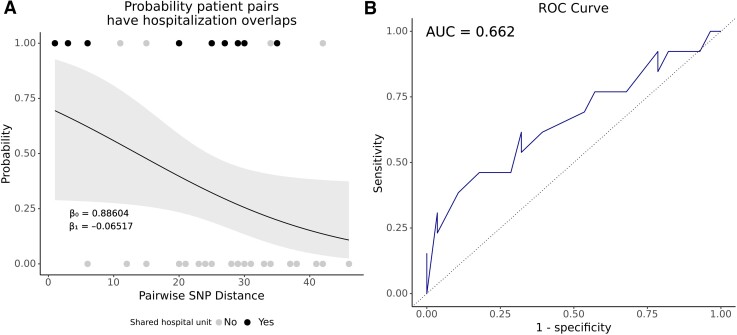
We performed a logistic regression to measure the association between likely hospital exposure and SNP difference assessing a SNP threshold range (Figure 4). The log odds of clustered patient pairs overlapping in the same hospital decreases by 0.065 with every increase of 1 SNP (P = .05), and showed that with 13 SNPs the likelihood that any 2 patients in a cluster overlapped in a hospital was 50%, with a trend toward no overlap at higher SNP differences (Figure 4A). The ROC area under the curve classified known prior overlapping hospitalizations 66% of the time from the SNP difference (Figure 4B).
Figure 4.
Higher single-nucleotide polymorphism (SNP) distances trend toward ruling out hospital overlaps between clustering patients. A, Logistic regression model indicating the relationship between patient pairs overlapping in the same hospital at the same time (prior to the diagnosis of an index methicillin-resistant Staphylococcus aureus bloodstream infection) and the pairwise SNP distance. Points indicate the true result for each pair as overlapping (1.0) or not overlapping (0). The color of the points indicates whether hospital overlap patient pairs also overlapped (black) or did not overlap (gray) in the same hospital unit. Gray ribbon indicates the 95% confidence interval. B, Receiver operating characteristic (ROC) curve of the logistic model in A. Area under the curve (AUC) = 0.662.
DISCUSSION
We combined clinical and genome data to describe a cohort of 104 US patients with MRSA BSI. The predominant genetic background of MRSA isolates in this study is consistent with known prevalence of CC8 and CC5 MRSA strains causing healthcare- and community-associated infections in the US [31]. The resolution of WGS was critical for identifying clusters of BSIs that would have otherwise gone unnoticed in the hospital setting. It is well established that WGS is useful for S. aureus outbreaks in hospitals [4, 8, 12, 27, 32, 33], though many reports focus on its use in emergent, point-source outbreaks, such as those occurring in neonatal intensive care units with an identifiable index case [27, 32, 33]. In other instances, WGS confirmed related cases of MRSA infection only after initial outbreak detection by other means, including an unusual antibiograms [32] or uncommon strain types [8]. Collectively, these investigations identified an epidemiologically significant core-genome SNP difference as small as 13 SNPs [11] to as large as 40 SNPs [34] among outbreak isolates.
A SNP threshold <35 was effective for cluster detection with evidence of prior hospital overlaps among adult patients in a population where transmission pathways are difficult to identify. Four clusters showed pairwise differences between 1 and 25 SNPs and patients with diagnosis date within 3 months. Considering estimates of S. aureus neutral mutation of approximately 5–6 SNPs per genome per year [35], a likely scenario is a recent common exposure in a healthcare setting several weeks to months prior to BSI onset for clustered subjects. However, clusters lacking evidence of a hospital overlap also had small SNP difference ranges, suggesting alternative routes of MRSA transmission among BSI patients, such as hospital environmental reservoirs like equipment [7, 36] or a community reservoir of patients carrying MRSA [37], possibly reintroducing bacteria to the hospital. We demonstrated that it is reasonable to investigate healthcare histories for patients at or below 13 SNPs to find sources of transmission associated with hospital settings.
Most US hospitals have not yet implemented a WGS surveillance system for infection control. Hospitals can approach bioinformatic surveillance using commercial workflows with integrated processes [33] or open-source options [38], or create robust in-house surveillance methods [39]. We demonstrated that different approaches to sequence alignment detect similar SNP differences and phylogenies. However, alignment sizes and the number of clusters at the threshold of interest did differ. Choosing the most appropriate tool ideally optimizes sensitivity and comparability across investigations. The gene-family approach consistently detects similar SNP differences among alignments of mixed clonal clusters and is suited to studies comparing diverse sample sets. However, higher sensitivity can be achieved using an assembly or pseudoread pipeline because they also compare a larger portion of the genome where SNPs can accumulate. We suggest future studies use both approaches, first for general detection of clusters with highly sensitive approaches, followed by a gene-family approach to compare clusters across a broader context of transmission cluster history in a specific environment. A sliding scale [40] or a threshold range [11] could also offer a more flexible alternative for including patients in transmission investigations.
Reference-based alignments and phylogenetic reconstruction is advantageous for identifying transmission events in healthcare settings, particularly where MRSA infections are rare [13, 40]. However, S. aureus transmission from healthcare facilities into community settings and back suggest that hospitals and the surrounding community are a single reservoir of transmission [27]. Our investigation also points to the importance of long-term MRSA carriage prior to diagnosis of a BSI. Overlapping hospitalization may provide an opportunity for MRSA transmission and subsequent asymptomatic colonization in a recipient patient, but BSI onset may occur weeks or months later. Consequently, clusters are not identified after the critical moment of transmission when infection control interventions could be implemented. As WGS surveillance becomes prospectively implemented, gene-family alignments are advantageous for assessing increasingly diverse collections of isolates in a hospital or single healthcare system.
In our analysis, the long span of time between BSI onset among cluster patients and lack of an obvious transmission pathway suggests possible intermediate patients without a BSI but still carriers of the infecting MRSA strains. We did not collect isolates from the hospital environment or from healthcare workers directly, so we cannot discern the role of these intermediaries for transmission in the clusters.
We revealed MRSA BSI clusters among adults with various prior healthcare exposures in a setting with relatively high incidence of MRSA infections. We identified genetically similar clusters while the routine epidemiological signal was weak, but our investigation suggested healthcare or other shared exposures well before BSI presentation. Including WGS as a part of current routine colonization screening for MRSA in high-risk clinical settings could identify and prevent transmission events in areas of hospitals not regularly scrutinized by infection control staff. With robust and consistent cluster detection pipelines and the prospective collection of detailed exposure histories, with a focus on identifying exposures during hospitalization to specific healthcare workers, fomites, and medical procedures, outbreak sources can be better resolved before the onset of a BSI event.
Supplementary Material
Contributor Information
Brooke M Talbot, Graduate School of Biological and Biomedical Sciences, Emory University, Atlanta, Georgia, USA.
Natasia F Jacko, Division of Infectious Diseases, Department of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Robert A Petit, III, Division of Infectious Diseases, Emory University School of Medicine, Atlanta, Georgia, USA.
David A Pegues, Division of Infectious Diseases, Department of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Margot J Shumaker, Division of Infectious Diseases, Department of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Timothy D Read, Division of Infectious Diseases, Emory University School of Medicine, Atlanta, Georgia, USA.
Michael Z David, Division of Infectious Diseases, Department of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Laurel Glaser for assistance with the biobanking of isolates used in this study, and Katrina Hofstetter for support in troubleshooting R code. They also thank the Penn/Children’s Hospital of Philadelphia Microbiome Center for performing the isolate sequencing used in this analysis.
Financial support. This work was supported by the National Institutes of Health (NIH) (grant number R01 AI139188 to B. M. T., M. Z. D., and T. D. R.).
References
- 1. Kourtis AP, Hatfield K, Baggs J, et al. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections—United States. MMWR Morb Mortal Wkly Rep 2019; 68:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raineri EJM, Altulea D, van Dijl JM. Staphylococcal trafficking and infection—from “nose to gut” and back. FEMS Microbiol Rev 2022; 46:fuab041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang ES, Tan J, Eells S, Rieg G, Tagudar G, Miller LG. Body site colonization in patients with community-associated methicillin-resistant Staphylococcus aureus and other types of S. aureus skin infections. Clin Microbiol Infect 2010; 16:425–31. [DOI] [PubMed] [Google Scholar]
- 4. Azarian T, Maraqa NF, Cook RL, et al. Genomic epidemiology of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. PLoS One 2016; 11:e0164397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention . Methicillin-resistant Staphylococcus aureus infections among competitive sports participants—Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000–2003. MMWR Morb Mortal Wkly Rep 2003; 52:793–5. [PubMed] [Google Scholar]
- 6. Marks LR, Calix JJ, Wildenthal JA, et al. Staphylococcus aureus injection drug use–associated bloodstream infections are propagated by community outbreaks of diverse lineages. Commun Med 2021; 1:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Montoya A, Schildhouse R, Goyal A, et al. How often are health care personnel hands colonized with multidrug-resistant organisms? A systematic review and meta-analysis. Am J Infect Control 2019; 47:693–703. [DOI] [PubMed] [Google Scholar]
- 8. Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 2014; 52:2365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Armstrong GL, MacCannell DR, Taylor J, et al. Pathogen genomics in public health. N Engl J Med 2019; 381:2569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Armstrong J, Fiddes IT, Diekhans M, Paten B. Whole-genome alignment and comparative annotation. Annu Rev Anim Biosci 2019; 7:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coll F, Raven KE, Knight GM, et al. Definition of a genetic relatedness cutoff to exclude recent transmission of meticillin-resistant Staphylococcus aureus: a genomic epidemiology analysis. Lancet Microbe 2020; 1:e328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eyre DW, Golubchik T, Gordon NC, et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2012; 2:e001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lees JA, Kendall M, Parkhill J, Colijn C, Bentley SD, Harris SR. Evaluation of phylogenetic reconstruction methods using bacterial whole genomes: a simulation based study. Wellcome Open Res 2018; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stimson J, Gardy J, Mathema B, Crudu V, Cohen T, Colijn C. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol Biol Evol 2019; 36:587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing. Wayne, PA: CLSI, 2019. [Google Scholar]
- 16. Petit RA, Read TD, Segata N. Bactopia: a flexible pipeline for complete analysis of bacterial genomes. mSystems; 5:e00190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seemann T. snippy: fast bacterial variant calling from NGS reads. 2015. Available at: https://github.com/tseemann/snippy. Accessed 23 December 2021.
- 18. Treangen TJ, Ondov BD, Koren S, Phillippy AM. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014; 15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bayliss SC, Thorpe HA, Coyle NM, Sheppard SK, Feil EJ. PIRATE: a fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience 2019; 8:giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 2015; 11:e1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seemann T. Source code for snp-dists software. Zenodo. 2018. Available at: 10.5281/zenodo.1411986. Accessed 23 December 2021. [DOI]
- 22. Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 2020; 37:1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu G, Smith DK, Zhu H, et al. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 2017; 8:28–36. [Google Scholar]
- 24. Jombart T. Assessing the quality of a phylogeny. In: Introduction to phylogenetics using R.2016. Available at: https://adegenet.r-forge.r-project.org/files/Glasgow2015/practical-introphylo.1.0.pdf. Accessed 9 December 2021.
- 25. Briand S, Dessimoz C, El-Mabrouk N, Lafond M, Lobinska G. A generalized Robinson-Foulds distance for labeled trees. BMC Genomics 2020; 21:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019; 35:526–8. [DOI] [PubMed] [Google Scholar]
- 27. Harris SR, Cartwright EJ, Török ME, et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 2013; 13:130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. RStudio Team . RStudio: integrated development environment for R.2021. Available at: http://www.rstudio.com/. Accessed 23 December 2021.
- 29. Inkscape Project . Inkscape. 2020. Available at: https://inkscape.org. Accessed 23 December 2021.
- 30. Bowers JR, Driebe EM, Albrecht V, et al. Improved subtyping of Staphylococcus aureus clonal complex 8 strains based on whole-genome phylogenetic analysis. mSphere 2018; 3:e00464-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith JT, Eckhardt EM, Hansel NB, Eliato TR, Martin IW, Andam CP. Genomic epidemiology of methicillin-resistant and -susceptible Staphylococcus aureus from bloodstream infections. BMC Infect Dis 2021; 21:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Köser CU, Holden MTG, Ellington MJ, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 2012; 366:2267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slingerland BCGC, Vos MC, Bras W, et al. Whole-genome sequencing to explore nosocomial transmission and virulence in neonatal methicillin-susceptible Staphylococcus aureus bacteremia. Antimicrob Resist Infect Control 2020; 9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popovich KJ, Green SJ, Okamoto K, et al. MRSA transmission in intensive care units: genomic analysis of patients, their environments, and healthcare workers. Clin Infect Dis 2021; 72:1879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Senn L, Clerc O, Zanetti G, et al. The stealthy superbug: the role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. mBio; 7:e02039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanamori H, Rutala WA, Weber DJ. The role of patient care items as a fomite in healthcare-associated outbreaks and infection prevention. Clin Infect Dis 2017; 65:1412–9. [DOI] [PubMed] [Google Scholar]
- 37. Mattner F, Biertz F, Ziesing S, Gastmeier P, Chaberny IF. Long-term persistence of MRSA in re-admitted patients. Infection 2010; 38:363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berbel Caban A, Pak TR, Obla A, et al. PathoSPOT genomic epidemiology reveals under-the-radar nosocomial outbreaks. Genome Med 2020; 12:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sundermann AJ, Chen J, Kumar P, et al. Whole-genome sequencing surveillance and machine learning of the electronic health record for enhanced healthcare outbreak detection. Clin Infect Dis 2022; 75:476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gorrie CL, Da Silva AG, Ingle DJ, et al. Systematic analysis of key parameters for genomics-based real-time detection and tracking of multidrug-resistant bacteria. bioRxiv [Preprint]. 25 September 25, 2020. Available from: https:/doi,org/ 10.1101/2020.09.24.310821. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.