Abstract
The FimB protein is a site-specific recombinase that inverts the fimS genetic switch in Escherichia coli. Based on amino acid sequence analysis alone, FimB has been assigned to the integrase family of tyrosine recombinases. We show that amino acid substitutions at positions R47, H141, R144, and Y176, corresponding to highly conserved members of the catalytic motif of integrase proteins, render FimB incapable of inverting the fimS element in vivo. The arginine substitutions reduced the ability of FimB to bind to fimS in vivo or in vitro, while the substitution R144Q resulted in a protein unable to bind independently to the half sites located at the left end of fimS in phase-on bacteria. These data confirm that FimB is an integrase and suggest that residue R144 has a role in binding to a specific component of the fim switch.
FimB protein is one of two site-specific recombinases that invert the fim switch (fimS) in Escherichia coli (20, 24). Inversion of the 314-bp switch is the basis of phase-variable expression of type 1 fimbriae (1, 21). FimB can invert it in either direction with approximately equal facility, whereas FimE (the other recombinase) inverts the switch predominantly in the on-to-off direction (17, 26, 38). When expressed in the same strain, FimE is completely dominant to FimB under standard growth conditions (16).
The FimB and FimE proteins are closely related in amino acid sequence (48% identical) and are encoded by tandemly arranged genes on the chromosome located adjacent to the switch (Fig. 1) (25). The fimS element harbors the fimAp promoter for transcription of the fimA gene encoding the subunit protein of type 1 fimbriae. When fimAp is directed toward fimA, the bacteria are fimbriate (phase on); when it is directed away, the bacteria are afimbriate (phase off) (Fig. 1). Both FimB and FimE require the accessory proteins integration host factor (IHF) and the leucine-responsive regulatory protein (Lrp) for efficient inversion of the switch (7, 11, 14, 35). The nucleoid-associated protein H-NS plays a poorly understood inhibitory role, at least in FimB-promoted switching (10, 23, 24, 31, 37).
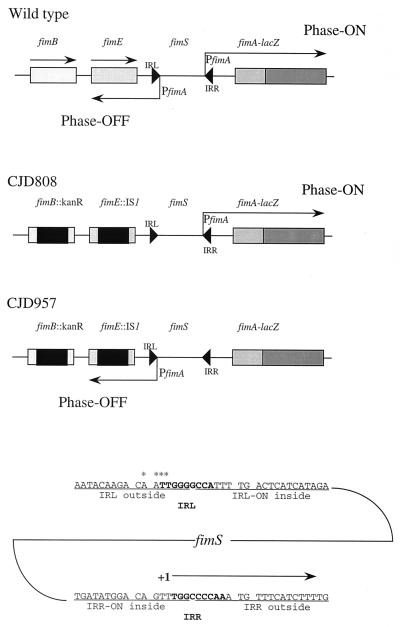
FIG. 1.
The fimS invertible element and its surroundings. The relevant section of the fim locus is shown in wild-type, CJD808 (fimS locked on), and CJD957 (fimS locked off) strains. The fimA promoter is represented by the angled arrow labeled PfimA. In phase on, this arrow is directed towards fimA; in phase off, it is directed the other way. The regulatory genes fimB and fimE are insertionally inactivated in strains CJD808 and CJD957, locking the switch in those genetic backgrounds. The converging arrowheads that bound the fimS element represent the IRL and IRR. Below, the DNA sequences of the IRL and IRR are shown, together with those of the half sites to which the Fim recombinases bind. The 9-bp inverted repeats are in boldface type, and the half sites (identified by Gally et al. [17]) are underlined. For clarity, spaces have been introduced 5′ and 3′ to the invariant CA dinucleotide motif within each half site. The asterisks indicate the 4 bp (5′-AATT-3′) deleted in the IRL of strain CJD1353. The transcription start site of fimA lies immediately to the left of the IRR and is represented by “+1” at the start of a horizontal arrow.
Inversion of fimS involves site-specific recombination between two 9-bp inverted repeats that border the switch (Fig. 1). Each repeat is flanked by two Fim recombinase binding sites (known as half sites), each of which contains the essential core dinucleotide 5′-CA (17). Previous work has shown that there are four binding sites for FimB (and FimE) at fimS (17). These half sites flank the 9-bp inverted repeats and can be designated IRL (left-hand inverted repeat)-outside, IRL-inside, IRR (right-hand inverted repeat)-outside, and IRR-inside. The IRL-outside and IRR-outside sites are constant in phase-on and phase-off cells, while the inside sites vary as fimS inverts (Fig. 1). This variation is important for the directionality of the FimB- and FimE-promoted DNA inversion event (26).
Integrases are usually associated with integration and excision events or the resolution of dimeric structures through recombination between directly repeated copies of specific sequences. The Fim integrases are unusual in promoting an inversion event, something that is more often associated with members of the invertase family of site-specific recombinases. The fim system is unusual in having two integrases. In other cases where two are found (such as the Xer-dif system), the two proteins cooperate to promote recombination (6). In the case of fim, FimB and FimE operate independently of one another (25). Finally, at approximately 200 amino acids, the Fim proteins are the smallest members of the Int family (over 100 recombinases), making them an attractive subject for structure-function studies.
Based on their amino acid sequences, FimB and FimE have been assigned to the integrase family of tyrosine site-specific recombinases (11, 14). Although the family members display a great deal of sequence heterogeneity, each integrase possesses a catalytic motif of amino acids that are almost absolutely conserved throughout the family. Originally, it was believed that there were just four of these: two arginines, a histidine, and a tyrosine that occurred in the order RHRY to form a catalytic tetrad for DNA cleavage, strand exchange, and religation during the recombination event (2, 3, 15, 18, 30, 39). Recently, a fifth catalytic residue (a lysine) has been identified (8). In this investigation, we sought to identify the four amino acids in FimB that correspond to the RHRY members of the catalytic domain by mutation and to study the interaction of wild-type and mutant FimB proteins with the fimS element in vivo and in vitro.
Identification of four members of the FimB catalytic motif.
Amino acid sequence alignment with the integrase family was used to identify candidates for the RHRY members of the catalytic motif of FimB (11). These residues were R47, H141, R144, and Y176. The substitutions R47A, H141A, R144Q, and Y176F were made by site-directed mutagenesis using either the QuikChange kit (Stratagene) (R47A and H141A) or the unique-site elimination method (Pharmacia Amersham Biotech) (R144Q and Y176F). For unique-site elimination mutagenesis, a unique HindIII site was eliminated using primer H3Nsi (Table 1). For mutagenesis of the fimB gene, plasmids pSLD203 (FimB) and pSGS223 (maltose binding protein [MBP]-FimB fusion) (10 ng) (Table 2) were used as substrates. The oligonucleotides (100 pmol each) used for mutagenesis were obtained from MWG Biotech and are listed in Table 1. Standard recombinant DNA techniques were employed throughout this work (36).
TABLE 1.
PCR and DNA sequencing oligonucleotidesa
| OL 4 | 5′-GACAGAACAACGATTGCCAG-3′ |
| OL 20 | 5′-CTTTAAGGAGTGATTCGATTTC-3′ |
| R47AF | 5′-CATGGTTTCgcGGCGAGTGAAATTTGTCG-3′ |
| R47AR | 5′-CGACAAATTTCACTCGCCgcGAAACCATG-3′ |
| H141AF | 5′-GAGATTCATCCGgcCATGTTACGCCATTC-3′ |
| H141AR | 5′-GAATGGCGTAACATGgcCGGATGAATCTC-3′ |
| R144 | 5′-CACATGTTAcagCATTCGTG-3′ |
| Y176 | 5′-GTCTGGTtTACCGCCAGCAATG-3′ |
| H3Nsi | 5′-CAGGCATGCATGGCACTGGCCG-3′ |
| FimBR | 5′-AACAGGATCCTGGTATCTCAAC-3′ |
| FimBL | 5′-GGGAAAACATATGAAGAATAAGGCT-3′ |
The positions of nucleotide changes corresponding to amino acid substitutions are indicated in lowercase. The positions of restriction sites (NsiI, BamHI, and NdeI) within primers are indicated in boldface type.
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant details | Reference or source |
|---|---|---|
| Strains | ||
| CJD808 | VL386; fimB::Kanr; phase locked on | 12 |
| CJD931 | VL386; fimB::Kanrihf::cml lrp::Tn10 | 12 |
| CJD957 | VL386; fimB::Kanr; phase locked off | 38 |
| CJD1353 | CJD808; recD::Tn10; 4-bp deletion in IRL | This work |
| DPB271 | recD::Tn10Δ16Δ17 | 5 |
| VL386 | Φ(fimA-lacZ)λpL(209) fimE::IS1 | 1 |
| VL386recD | VL386; recD::Tn10Δ16Δ17 | 38 |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZ M15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pACYC184 | p15A replicon; Cmr Tetr | New England Biolabs |
| pLSB110 | pSLD203(R47A) | This work |
| pLSB111 | pSLD203(H141A) | This work |
| pLSB121 | pSGS501; phase on | This work |
| pLSB122 | pLSB121; 4-bp deletion in IRL | This work |
| pLSB201 | pSGS223(R144Q) | This work |
| pLSB202 | pSGS223(R47A) | This work |
| pLSB203 | pSGS223(H141A) | This work |
| pMalc-2 | Vector for creation of gene fusions to malE | New England Biolabs |
| pSGS223 | Plasmid encoding MBP-FimB protein fusion | This work |
| pSGS224 | pSGS223(Y176F) | This work |
| pSGS226 | pSLD203(Y176F) | This work |
| pSGS227 | pSLD203(R144Q) | This work |
| pSGS411 | fimE cloned in pUC18 | 38 |
| pSGS501 | fimB::KanrfimE::IS1 Φ(fimA-lacZ) in pACYC184 fimS; phase off | 38 |
| pSLD203 | fimB gene cloned in pUC18; Apr | 12 |
| pUC18 | ColE1 replicon; Apr | 40 |
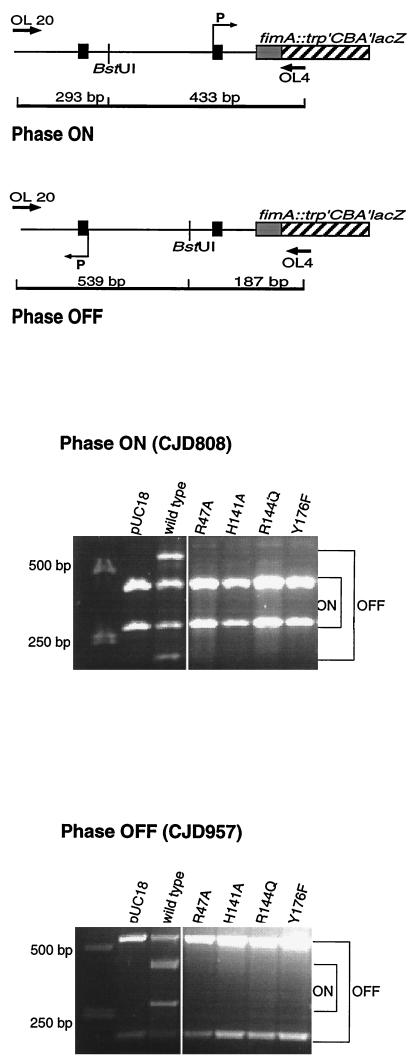
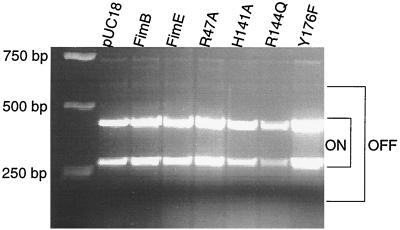
Each mutant FimB protein was compared with the wild type in an in vivo recombination assay in which inversion of the fimS element was measured on the chromosome by PCR as previously described (38). In this assay, cells that were phase on gave rise to two DNA bands of 293 and 433 bp; phase-off cells gave bands of 539 and 187 bp. Mixed populations of phase-on and phase-off cells displayed bands of all four sizes. The in vivo assay for FimB function involved complementation of a fimB gene knockout mutation. Plasmids harboring wild-type or mutant fimB genes were introduced to E. coli tester strains (4) in which fimS was locked in either the on or the off orientation (CJD808 and CJD957, respectively) (Fig. 1). The bacterial strains were grown as described previously (12, 38). CJD808 (on) and CJD957 (off) each had knockout insertion mutations in both the fimB and fimE genes on their chromosomes. In the locked-on strain, the fimA promoter was directed towards the fimA-lacZ reporter gene fusion, and this strain produced red colonies on MacConkey lactose indicator plates. In the locked-off strain, the fimA promoter was directed away from the fimA-lacZ reporter gene fusion, and this strain produced white colonies on the indicator plates. When incoming plasmids expressed functional FimB protein, they complemented the fimB knockout mutation and restored phase-variable fimA-lacZ expression, giving rise to sectored colonies on the MacConkey lactose plates. The data obtained showed that while the plasmid with the wild-type fimB gene could reactivate inversion in both the phase-on and the phase-off fimB fimE tester strains, the plasmids with the mutant fimB gene copies could not (Fig. 2). These data confirmed that the four residues that had been substituted were indeed critical for the DNA inversion activity of the FimB recombinase. Since it was possible that the amino acid substitutions had disabled the DNA binding activity of the mutants, it was necessary to examine them in more detail in order to distinguish between defects in DNA binding and catalytic defects which, while permitting binding, prevented site-specific recombination from proceeding.
FIG. 2.
Inversion of the fimS element by FimB and its mutant derivatives. Inversion of fimS was detected by a PCR assay. The structures of the phase-on and phase-off substrates are summarized at the top, together with details of the oligonucleotide primers used to amplify the inversion products. Gels with the PCR products obtained with phase-on and phase-off substrates are shown below. The positions to which PCR products diagnostic of phase-on or phase-off switches migrate are indicated to the right of each gel. Inversion of fimS is seen only in the case of the wild-type FimB protein (third lane from the left). DNA size markers are shown in the first lane; pUC18 is the vector-only control.
Binding of FimB proteins to the IRR in vivo.
In previous work it was found that when FimB was expressed from a multicopy plasmid it could repress transcription from the fimAp promoter by a promoter occlusion mechanism, and this binding could be demonstrated by in vivo footprinting (13). In other words, in phase-on bacteria, the binding of FimB to the IRR on the chromosome resulted in repression of the fimA promoter. This offered a method for assessing the binding of FimB to the IRR in vivo, since binding of the protein to this sequence simultaneously placed the protein directly on the transcription start site for the fimA gene in a phase-on strain (Fig. 1) and the effect on beta-galactosidase expression from the fimA-lacZ reporter gene fusion could be assayed readily. Furthermore, recent work with the FimE recombinase showed that a similar effect could be achieved with that protein in vivo, a result that has been validated by in vitro binding studies (38). Beta-galactosidase activity was assayed by the method of Miller (29).
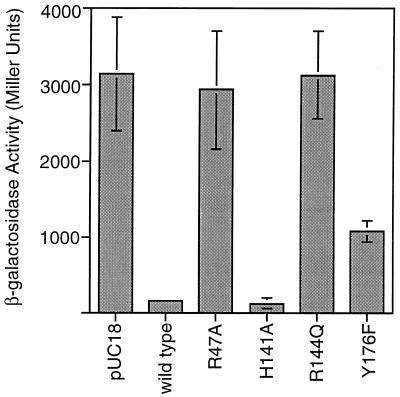
The wild-type FimB protein repressed expression of the fimA-lacZ fusion by 90% in the phase-on tester strain CJD808 (Fig. 3). Transcription repression was also seen with the mutant proteins with the H141A or Y176F substitution; little repression was seen with the proteins with the R47A or the R144Q substitution (Fig. 3). Assays were repeated at least three times. These data suggested that two of the catalytic-motif substitutions (R47A and R144Q) had significantly altered the ability of FimB to bind to the IRR in vivo. Therefore, it was decided to test the interactions of the wild-type and mutant proteins with both the left (IRL) and the right (IRR) repeats in vitro.
FIG. 3.
Indirect measurement by a promoter occlusion test of FimB binding to the IRR. The data shown are beta-galactosidase activities for cultures harboring plasmids expressing no recombinase (pUC18), wild-type FimB, or mutant derivatives of FimB. These experiments were repeated on three separate occasions; typical data are shown. The error bars indicate standard deviations.
Interaction of wild-type and mutant FimB proteins with IRL and IRR in both orientations in vitro.
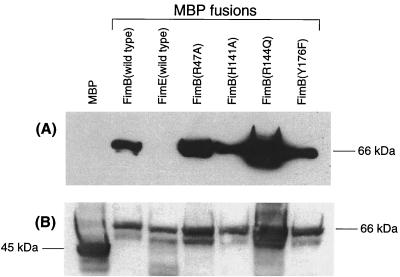
Sufficient quantities of FimB to perform electrophoretic mobility shift assays were not obtainable from lysates of strains expressing multicopy FimB. Instead, FimB was found to be expressed better as an MBP fusion. Therefore, the in vitro binding assay was carried out with MBP fusions to the wild-type and mutant FimB proteins. First, the fimB open reading frame was amplified with primers FimBR and FimBL (Table 1) and then inserted between the XmnI and BamHI sites of pMalC2, resulting in plasmid pSGS223. In this construct, the fimB open reading frame was inserted downstream and in frame with the malE gene to create an MBP-FimB fusion protein. This protein could be overexpressed and was soluble (see below). In vivo, the MBP-FimB hybrid had full wild-type FimB activity (data not shown). The expression of wild-type and mutant FimB proteins was confirmed by Western blotting using an anti-FimB antibody from New Zealand White rabbits immunized with a multiple-antigen peptide (Research Genetics) (Fig. 4). This peptide (PLLNKEVQALKNWLS) corresponded to amino acids 80 to 94 of FimB, and the procedure used for raising antibodies and performing Western immunoblot analyses was essentially identical to that described previously for FimE (38).
FIG. 4.
Detection of hybrid MBP-FimB protein by Western blotting. Polypeptides expressed from multicopy recombinant plasmids were detected by antibody against FimB (A) or MBP (B). The MBP-FimE fusion protein is included as a negative control in panel A (third lane from the left). The positions of native MBP (44 kDa) and MBP fusions (66 kDa) are indicated.
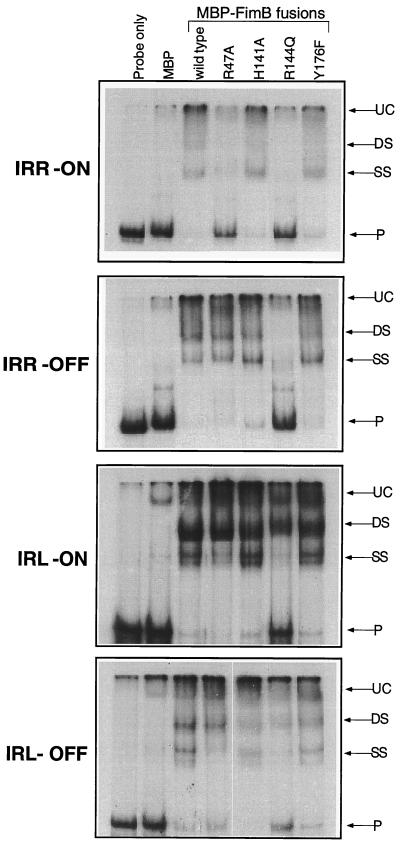
The strain CJD931, which is deficient in the fimS-binding proteins FimB, FimE, IHF, and Lrp (Table 2), was transformed with plasmids expressing wild-type or mutant MBP-FimB protein fusions. Lysates were prepared and used in electrophoretic mobility shift assays (38) with radiolabeled DNA fragments consisting of the IRL or IRR sequence amplified from fimS in the on or off orientation. The wild-type FimB protein shifted the mobilities of all four DNA fragments, producing two shifted species called SS and DS (Fig. 5). The MBP protein alone did not affect the mobilities of the fimS DNA fragments. The upper band (DS) was consistent with occupation of both half sites at each inverted repeat (Fig. 1), while the lower band (SS) represented occupation of just one half site. These results were consistent with previous work showing that two shifted complexes arise when the Fim recombinases interact with fimS in vitro (17, 38). In the IRL-on experiment, the SS species ran as a doublet, perhaps reflecting the previously described ability of integrases to bend their DNA substrates (20). In agreement with the in vivo data from the fimAp promoter occlusion experiments (see above), the amino acid substitutions R47A and R144Q each altered the ability of FimB to bind to IRR-on in vitro (compare Fig. 3 and 5). However, while R47A could still bind to IRR-off, R144Q could not. Therefore, R144Q could not bind to IRR in either the on or the off orientation.
FIG. 5.
Electrophoretic mobility shift analysis of FimB-containing lysate interactions with IRR or IRL sequences from fimS. Data are shown for the IRR and IRL amplified from phase-on or phase-off fimS elements. Lanes 1, no added protein; lanes 2, MBP alone; lanes 3, MBP-FimB; lanes 4, MBP-FimB R47A; lanes 5, MBP-FimB H141A; lanes 6, MBP-FimB R144Q; lanes 7, MBP-FimB Y176F. Species corresponding to complexes with two bound protein protomers (DS) and a single bound protomer (SS) are shown. Also indicated are an unknown complex (UC) that migrated near the top of all lanes and the unbound radiolabeled probe (P). The IRL DNA fragment was 111 bp, while the IRR DNA fragment was 205 bp.
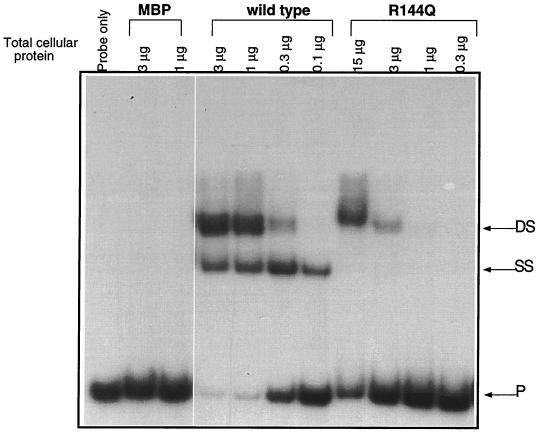
At IRL, only R144Q was affected in its ability to bind. At IRL-off, R144Q produced little electrophoretic mobility shift (Fig. 5), while at IRL-on it created the shifted species that was consistent with occupation of both half sites (DS) but not the species indicative of single-half-site occupancy (SS) (Fig. 5). This interaction with IRL-on was investigated in greater detail by comparing the abilities of wild-type FimB and R144Q to bind as their concentrations were increased (Fig. 6). While the wild-type protein produced two electrophoretically shifted species indicative of single-half-site and dual-half-site occupancy at IRL-on, the R144Q mutant protein could only produce the species indicative of binding to the two IRL-on half sites, and much more of the mutant protein was needed to form this complex. This was examined in more detail by mutagenizing one of the half sites at IRL-on.
FIG. 6.
Comparison of wild-type and mutant R144Q binding to IRL-on in vitro. Shown is an electrophoretic mobility shift assay in which increasing concentrations of lysates containing MBP, wild-type MBP-FimB, and the MBP-FimB mutant R144Q were incubated with radiolabeled DNA amplified from the IRL component of fimS. Species corresponding to complexes with two bound recombinase protomers (DS) and a single bound protomer (SS) are shown. P is the unbound DNA probe (111 bp).
A 4-bp deletion was engineered in the outside half site at IRL-on. This was done with plasmid pSGS501 (38), which contains an 8.5-kb fragment that includes inactive copies of the fimB and fimE genes (interrupted by a kanamycin resistance cassette and IS1 insertion element, respectively) and fimS in the off orientation, inserted into the EcoRV site of pACYC184 (Table 2). This plasmid was cotransformed into strain CH1483 with multicopy fimB (pSLD203) to invert fimS in plasmid pSGS501 to the on orientation. Transformants were screened using the PCR switch assay (38), and plasmid DNA was isolated from one transformant that contained fimS in the on orientation. This DNA was digested with SacI and KpnI, which cut within plasmid pSLD203 but not within plasmid pSGS501. The digested DNA was transformed into the IHF-deficient strain CH1483. DNA from one transformant was isolated (pLSB121), and analysis by PCR switch assay showed that it contained fimS in the on orientation. (All plasmid constructs were maintained in the strain CH1483, as the absence of IHF was found to prevent random DNA rearrangements.) Plasmid pLSB121 was digested with MfeI, which cuts only once in plasmid pLSB121, within the IRL. The digested DNA was treated with mung bean nuclease to remove the 4-bp overhang resulting from MfeI digestion, ligated, and transformed into CH1483. Plasmid DNA (pLSB122) was isolated and sequenced (MWG Biotech) to confirm that it contained a 4-bp deletion within the IRL and that fimS was locked in the on orientation (Fig. 1).
The mutant IRL was crossed onto the chromosome of a strain lacking a functional fimE gene (VL386recD). Plasmid pLSB122 was digested with EcoRV, and an 8.5-kb fragment, encompassing DNA upstream of the fimB gene (interrupted by a kanamycin resistance gene cassette) through to the lacZ gene, was isolated from a gel. Approximately 2 μg of this fragment was electroporated into strain VL386recD (38), and transformants were selected on MacConkey lactose agar containing kanamycin (25 μg ml−1). Candidate allele replacement mutants were then screened to confirm that they did not contain plasmid DNA. PCR was used to confirm that they were phase on, and it was confirmed that the mutant IRL sequence could not be cleaved with MfeI. Finally, the 4-bp deletion within the IRL and the integrity of the rest of fimS were confirmed by sequencing. Site-specific inversion of fimS mediated by either FimE or FimB could not be detected in this background (Fig. 7). Plasmids expressing the four mutant FimB derivatives were introduced, and the strains were tested for inversion of fimS by the PCR assay. The results obtained showed that in no case could any of the mutant FimB proteins invert fimS (Fig. 7).
FIG. 7.
The mutant fimS element is not invertible. Shown is a DNA inversion assay in which the fimS derivative with a 4-bp deletion in the outer half site at IRL-on is amplified by PCR from cells expressing wild-type and mutant FimB proteins. pUC18 is the vector-only control; FimE is wild-type FimE recombinase, a protein that inverts the multicopy wild-type switch rapidly from the on to the off phase. The switch remained in the on orientation in all samples.
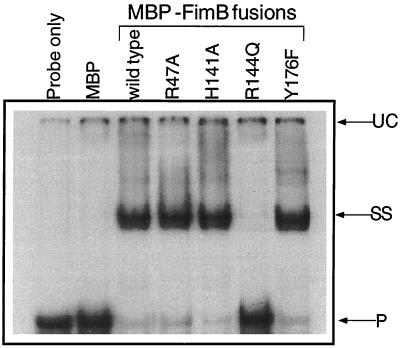
Each of the half sites at the IRL (and IRR) probably binds one Fim recombinase protomer. It is possible that the R144Q mutation has altered the way that FimB binds to the IRL half sites, making binding at one site contingent on occupation of the other. This could account for the lack of single-half-site occupation demonstrated for the R144Q mutant at the IRL. Thus, the R144Q mutant would be unable to produce the SS mobility-shifted species. The requirement for two half sites at IRL-on for R144Q to bind was confirmed when mobility shift assays were performed with the mutant IRL. The wild type and mutants R47A, H141A, and Y176F could all form the SS shifted species with the mutated IRL sequence (Fig. 8). This showed that these proteins did not require both halves of the IRL site in order to bind. In contrast, mutant FimB R144Q could not bind to this mutant IRL at all, showing that it required both halves to be intact in order for it to bind to the IRL.
FIG. 8.
Binding of wild-type and mutant MBP-FimB fusion proteins to the mutated fimS element. Shown is an electrophoretic mobility shift assay of interactions of lysates containing FimB and its mutant derivatives with a radiolabeled IRL sequence with a 4-bp deletion in the outer half site. The first two lanes are negative controls. SS is the shifted species that corresponds to IRL with one half site occupied by a FimB protomer. UC and P are an unknown complex found in all lanes and the unbound radiolabeled probe, respectively. The IRL DNA fragment was 107 bp.
Although the integrase family of tyrosine site-specific recombinases contains over 100 members, only a small number have been examined in detail by site-directed mutagenesis. These proteins bind to distinct sequences and recombine short repeated motifs that are unique to each integrase (30). Most of the mutations made in these proteins affect catalytic-motif residues, making the data interesting to compare with those obtained in this study of FimB. When the first arginine of the lambda integrase catalytic motif is converted to a glutamine (R212Q), the protein retains partial DNA binding activity but loses DNA recombination activity (28), a situation similar to that of R47A in FimB. Substituting glutamate for the corresponding arginine in the Flp recombinase in Saccharomyces cerevisiae leaves DNA binding activity intact while abolishing recombination (9). When the first arginine of the catalytic motif in the integron Intl1 was converted to an isoleucine, valine, or glutamate, all DNA binding activity and recombination activity was lost (19). These data show that, in addition to its role in recombination, the first arginine of the motif also contributes to DNA binding activity in at least some integrases.
Converting the second catalytic-motif arginine of the yeast Flp protein to glycine disrupted recombination but only weakened rather than abolished DNA binding activity (33). In contrast, the corresponding substitution in the integron Intl1 integrase abolished both DNA binding and recombination (19). Thus, different members of the integrase family show different responses to an equivalent substitution within the motif. In the case of FimB, the R144Q substitution had a very precise effect on the activity of the protein: it altered its interaction with a particular component of the DNA substrate and abolished DNA recombination activity.
The tyrosine residue of the catalytic motif forms a covalent bond with the cleaved DNA at the recombination site, making it an essential amino acid for recombination. Substituting the related aromatic amino acid phenylalanine for this tyrosine abolished recombinase activity in the lambda, the integron Intl1, and the Flp integrases (19, 28, 32, 34). The Y176F substitution in FimB resulted in an equivalent phenotype. In common with other well-characterized integrases, FimB was not impaired in DNA binding by mutagenesis of this tyrosine residue.
The results obtained in vitro with the R144Q mutant of FimB suggest that this integrase interacts with itself, at least at IRL-on, and that this is a DNA-dependent interaction. This specific DNA dependence may explain why we have been unable to demonstrate FimB-FimB protein-protein interactions (or FimE-FimE or FimB-FimE interactions) using the yeast two-hybrid system (S. G. J. Smith and C. J. Dorman, unpublished data). To date, the crystal structures of four integrases have been solved. These are HP1 (22), Cre (20), XerD (39), and Int (27). At least for XerD, the implied interactions for the XerC-XerD heterodimer bound at the dif site are consistent with DNA-dependent protein-protein interactions. Perhaps significantly, XerC and XerD are the integrases most closely related to FimB (and FimE) (15). Further work is in progress to elucidate in more detail the relation of structure to function in the FimB recombinase.
Acknowledgments
We thank present and past members of the Dorman laboratory for many useful discussions.
This work was supported by grants 046233/Z/95/Z from the Wellcome Trust and SC96/301 from Enterprise Ireland (Forbairt).
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abremski K E, Hoess R H. Evidence for the second conserved arginine residue in the integrase family of site-specific recombination proteins. Protein Eng. 1992;5:87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljundquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V L, Pierson L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Seidman J G, Smith D D, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 5.Biek D P, Cohen S N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blakely G, May G, McCullough R, Arciszewska L K, Burke M, Lovett S T, Sherratt D J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K-12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 7.Blomfield I C, Calie P J, Eberhardt K J, McClain M S, Eisenstein B I. Lrp stimulates phase variation of type 1 fimbriae in Escherichia coli K-12. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y, Hayes F. A newly identified, essential catalytic residue in a critical secondary structure element in the integrase family of site-specific recombinases is conserved in a similar element in eucaryotic type IB topoisomerases. J Mol Biol. 1999;289:517–527. doi: 10.1006/jmbi.1999.2793. [DOI] [PubMed] [Google Scholar]
- 9.Chen J-W, Evans B R, Yang S-H, Araki H, Oshima Y, Jayaram M. Functional analysis of box I mutations in yeast site-specific recombinases Flp and R: pairwise complementation with recombinase variants lacking the active site tyrosine. Mol Cell Biol. 1992;12:3757–3765. doi: 10.1128/mcb.12.9.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donato G M, Lelivelt M J, Kawula T H. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol. 1997;179:6618–6625. doi: 10.1128/jb.179.21.6618-6625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorman C J, Higgins C F. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J Bacteriol. 1987;169:3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dove S L, Dorman C J. The site-specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Mol Microbiol. 1994;14:975–988. doi: 10.1111/j.1365-2958.1994.tb01332.x. [DOI] [PubMed] [Google Scholar]
- 13.Dove S L, Dorman C J. Multicopy fimB gene expression in Escherichia coli: binding to inverted repeats in vivo, effect on fimA gene transcription and DNA inversion. Mol Microbiol. 1996;21:1161–1173. doi: 10.1046/j.1365-2958.1996.681434.x. [DOI] [PubMed] [Google Scholar]
- 14.Eisenstein B I, Sweet D S, Vaughn V, Friedman D I. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esposito D, Scocca J J. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 18.Grainge I, Jayaram M. The integrase family of recombinases: organization and function of the active site. Mol Microbiol. 1999;33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 19.Gravel A, Messier N, Roy P H. Point mutations in the integron integrase Intl1 that affect recombination and/or substrate recognition. J Bacteriol. 1998;180:5437–5442. doi: 10.1128/jb.180.20.5437-5442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo F, Gopaul D N, van Duyne G D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 21.Henderson I R, Owen P, Nataro J P. Molecular switches—the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 22.Hickman A B, Waninger S, Scocca J J, Dyda F. Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 Å resolution. Cell. 1997;89:227–237. doi: 10.1016/s0092-8674(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 23.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 24.Kawula T H, Orndorff P E. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histonelike protein H-NS. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulasekara H D, Blomfield I C. The molecular basis for the specificity of fimE in the phase variation of type 1 fimbriae of Escherichia coli K-12. Mol Microbiol. 1999;31:1171–1181. doi: 10.1046/j.1365-2958.1999.01257.x. [DOI] [PubMed] [Google Scholar]
- 27.Kwon H J, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacWilliams M P, Gumport R I, Gardner J F. Genetic analysis of the bacteriophage λ attL nucleoprotein complex. Genetics. 1996;143:1069–1079. doi: 10.1093/genetics/143.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 30.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Gara, J. P., and C. J. Dorman. Effects of local transcription and H-NS on inversion of the fim switch of Escherichia coli. Mol. Microbiol., in press. [DOI] [PubMed]
- 32.Pargellis C A, Nunes-Düby S E, Moitoso de Vargas L, Landy A. Suicide recombination substrates yield covalent λ integrase-DNA complexes and lead to identification of the active site tyrosine. J Mol Biol. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 33.Parsons R L, Evans B R, Zheng L, Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. J Biol Chem. 1990;265:4527–4533. [PubMed] [Google Scholar]
- 34.Prasad P V, Young L-J, Jayaram M. Mutations in the 2-μm circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci USA. 1987;84:2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roesch P L, Blomfield I C. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol Microbiol. 1998;27:751–761. doi: 10.1046/j.1365-2958.1998.00720.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schembri M A, Olsen P B, Klemm P. Orientation-dependent enhancement by H-NS of the activity of the type 1 fimbrial phase switch promoter in Escherichia coli. Mol Gen Genet. 1998;259:336–344. doi: 10.1007/s004380050820. [DOI] [PubMed] [Google Scholar]
- 38.Smith S G J, Dorman C J. Functional analysis of the FimE integrase of Escherichia coli K-12: isolation of mutant derivatives with altered DNA inversion preferences. Mol Microbiol. 1999;34:965–979. doi: 10.1046/j.1365-2958.1999.01657.x. [DOI] [PubMed] [Google Scholar]
- 39.Subramanya H S, Arciszewska L K, Baker R A, Bird L E, Sherratt D J, Wigley D B. Crystal structure of the site-specific recombinase, XerD. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]