Abstract
Pancreatic cancer (pancreatic ductal adenocarcinoma, PDAC) is aggressive and lethal. Although there is an urgent need for effective therapeutics in treating pancreatic cancer, none of the targeted therapies tested in clinical trials to date significantly improve its outcome. PORCN inhibitors show efficacy in pre-clinical models of Wnt-addicted cancers including RNF43-mutant pancreatic cancers and have advanced to clinical trials. In this study, we aimed to develop drug combination strategies to further enhance the therapeutic efficacy of the PORCN inhibitor ETC-159. To identify additional druggable vulnerabilities in Wnt-driven pancreatic cancers, we performed an in vivo CRISPR loss-of-function screen. CTNNB1, KRAS, and MYC were re-identified as key oncogenic drivers. Notably, glucose metabolism pathway genes were important in vivo but less so in vitro. Knockout of multiple genes regulating PI3K/mTOR signaling impacted the growth of Wnt-driven pancreatic cancer cells in vivo. Importantly, multiple PI3K/mTOR pathway inhibitors in combination with ETC-159 synergistically suppressed the growth of multiple Wnt-addicted cancer cell lines in soft agar. Furthermore, the combination of the PORCN inhibitor ETC-159 and the pan-PI3K inhibitor GDC-0941 potently suppressed the in vivo growth of RNF43-mutant pancreatic cancer xenografts. This was largely due to enhanced suppressive effects on both cell proliferation and glucose metabolism. These findings demonstrate that dual PORCN and PI3K/mTOR inhibition is a potential strategy for treating Wnt-driven pancreatic cancers.
Keywords: Wnt-addicted cancer, pancreatic cancer (PDAC), RNF43, in vivo CRISPR screen, PORCN inhibitor ETC-159, PI3K/mTOR inhibitor, drug synergy, glucose metabolism
Introduction
Pancreatic cancer is one of the most lethal cancers worldwide. Its five-year survival rate is only ~5%, mainly due to late diagnosis and lack of effective therapies 1,2. Currently, the therapeutic modalities of surgery and chemotherapy only provide slight survival benefit for these patients 3–5. It is projected that pancreatic cancer will become the second leading cause of cancer related deaths in the United States by 2030 6. There is therefore a pressing need to identify novel targets and develop new therapeutic strategies for treating pancreatic cancer.
Recent studies have identified a subset of pancreatic cancers that harbor loss of function RNF43 mutations, accounting for 5–10% of all pancreatic cancers 7–11. RNF43 encodes a transmembrane E3 ubiquitin ligase that down-regulates cell surface Wnt receptor levels 12,13. Inactivating mutations of RNF43 lead to increased abundance of Wnt receptors on the cell membrane, making the cells hypersensitive to Wnt ligands in the environment. This mediates Wnt-driven tumorigenesis, and generates a druggable addiction to Wnt ligands in these pancreatic cancers 8.
Porcupine (PORCN) is an O-acyltransferase that catalyzes the palmitoleation of Wnt proteins, a post-translational modification that is necessary for their secretion and activity 14–18. Blocking the biogenesis of Wnt ligands by PORCN inhibitors suppresses the growth of RNF43-mutant pancreatic cancers in pre-clinical models 8,19. This vulnerability therefore offers an opportunity to treat this subset of genetically defined pancreatic cancer patients with PORCN inhibitors. In an effort to exploit this, four small molecule PORCN inhibitors (LGK974, ETC-159, RXC004, and CGX1321) have advanced to clinical trials for treating cancer patients (ClinicalTrials.gov NCT01351103, NCT02278133, NCT02521844, NCT03447470, NCT02675946, and NCT03507998).
Drug resistance is an inevitable challenge in cancer therapy. Intrinsic resistance to PORCN inhibitor has been observed in some pancreatic cancer cell lines harboring inactivating mutations of RNF43 although the resistance mechanism remains unclear 8. It was also reported that long term exposure to a PORCN inhibitor in vitro selected for a AXIN1-mutant clone in a RSPO3-translocated colorectal cancer cell line and the AXIN1 mutation drove acquired resistance to PORCN inhibitor 20. Moreover, on-target adverse effects of Wnt pathway blockade by PORCN inhibitors, including bone loss and dysgeusia, were observed in pre-clinical study and clinical trials 21–23. One strategy to overcome drug resistance and reduce adverse effects is synergistic drug combinations through which lower drug doses can be used to achieve improved anticancer efficacy. It is therefore important to identify novel druggable vulnerabilities that can be used to develop combinational treatment coordinating with PORCN inhibitors.
CRISPR-Cas9 gene knockout technology enables fast and efficient genome editing and large-scale genetic screens in human cells 24,25. This provides an approach to address the above questions in a high-throughput way. While CRISPR knockout and RNAi knockdown screens have been performed in cancer cell lines to identify cancer fitness genes, most of these screens were performed in 2D cell culture system 26,27. However, it was reported that cancer cells show differential dependency on certain genes in vivo versus in vitro 28,29, and the importance of the tumor microenvironment is well established 30. For example, our recent study found that Wnt-target genes are more robustly regulated in in vivo models of Wnt dependent tumors 31. Therefore, in this study, we performed a more physiologically relevant in vivo CRISPR loss-of-function screen in a RNF43-mutant pancreatic cancer xenograft model. We found a series of glucose metabolism genes that were neglected in previous in vitro approaches to be essential for in vivo tumor growth. The in vivo CRISPR screen highlighted PI3K/mTOR signaling as a druggable vulnerability in Wnt-driven pancreatic cancer. PI3K/mTOR inhibitors and PORCN inhibitor synergistically suppressed Wnt-driven pancreatic cancer growth in vitro and in vivo, largely due to enhanced suppressive effects on cell proliferation and metabolism.
Results
Validation of a targeted in vivo CRISPR screen
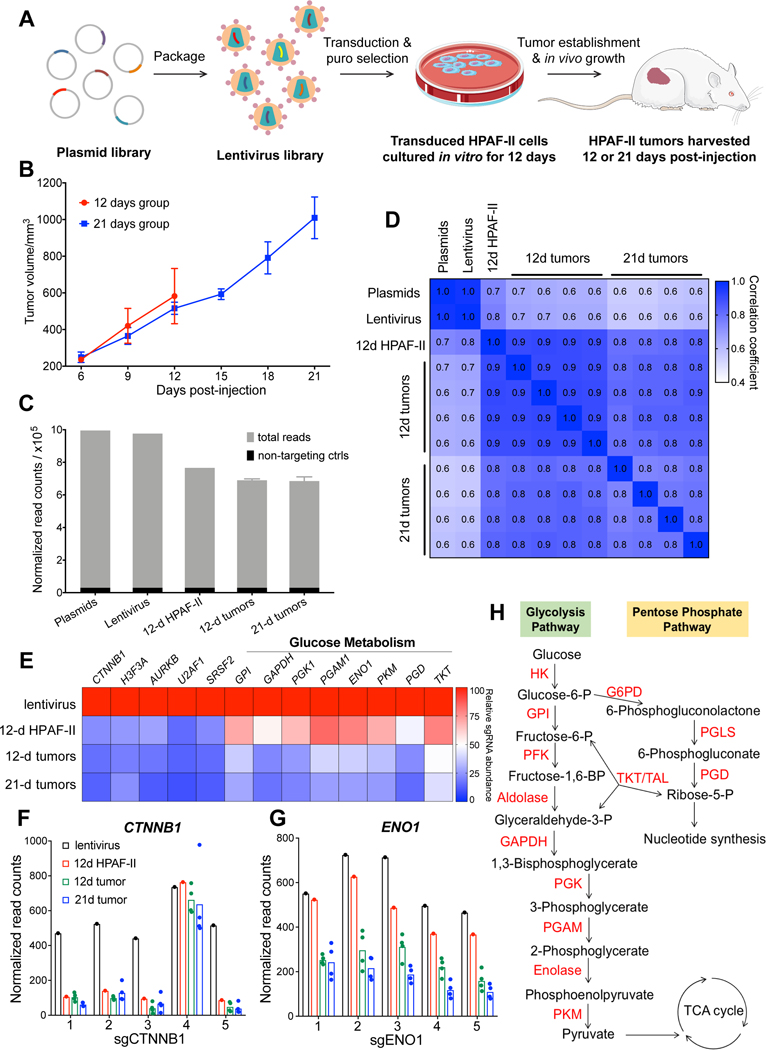
To identify fitness genes and pathways for genetically defined Wnt-driven pancreatic cancer, we performed an in vivo CRISPR loss-of-function screening in HPAF-II xenografts (Fig. 1A). HPAF-II is a RNF43-mutant Wnt-addicted pancreatic cancer cell line sensitive in vivo to PORCN inhibition 8,19. For optimal identification of negative selection acting on targeted fitness genes, an initial robust representation of all sgRNAs of the library in the cell population is a prerequisite. However, it is difficult to get large libraries well represented in one tumor because of the limited number of cells injected for tumor initiation, the low frequency of tumor initiating cells, and tumor heterogeneity. We therefore used a curated CRISPR library targeting 378 cancer related genes with five sgRNAs per gene alongside 50 non-targeting control sgRNAs, as described previously 32. The sgRNA and Cas9 are encoded in the same lentiviral plasmid that also confers puromycin resistance. HPAF-II cells were transduced with this lentiviral library at a low multiplicity of infection (MOI = 0.3) to ensure that majority of the transduced cells expressed a single sgRNA. The transduced cells were selected with puromycin for seven days and cultured in vitro for another five days, and then implanted subcutaneously in immunocompromised mice. 107 cells were injected per tumor which covered the ~2000-sgRNA library 5000 times. The tumors were harvested 12 days or 21 days after injection (Fig. 1B).
Figure 1. In vivo CRISPR screen identifies vulnerabilities of Wnt-driven pancreatic cancer and reveals the in vivo dependency on glucose metabolism.
A, Schematic representation of the CRISPR screen in a RNF43-mutant pancreatic cancer model.
B, Tumor growth curve of CRISPR library-transduced HPAF-II cells injected subcutaneously in NSG mice. The mice were randomized into two groups and tumors were harvested for analysis 12 days or 21 days after injection. Error bars indicate SEM. C, The total sgRNA read counts were reduced in HPAF-II cells in culture and further reduced following injection into mice. The graph represents the sum of read counts of all the sgRNAs normalized to the read counts of the non-targeting control sgRNAs from the respective samples.
D, Pearson correlation analysis of the normalized read counts of sgRNAs in all the samples. The Pearson correlation coefficients are indicated in the heatmap.
E, Depletion of sgRNAs targeting essential genes during the proliferation of HPAF-II cells in vitro and in vivo. The values indicated are the relative median abundance of all the five sgRNAs targeting a specific gene calculated using MAGeCK algorithm. The values are set to 100 for lentivirus, and 0 refers to 100% depletion of the sgRNAs.
F and G, sgRNAs targeting CTNNB1 and ENO1 are depleted in HPAF-II cells in culture and in tumors. Normalized read counts of the five individual sgRNAs are depicted.
H, Diagrammatic representation of the glucose metabolism pathway with the enzymes catalyzing the respective steps marked in red.
All samples (the plasmid pool, lentivirus pool, transduced HPAF-II cells after 12-day culture in vitro, and tumors harvested after 12-day or 21-day in vivo growth) were subject to deep sequencing to detect the frequency of all sgRNAs in each sample. Due to the small scale of the CRISPR library, unbalanced sgRNA depletion and enrichment could affect the relative frequency of other sgRNAs and lead to false increase or decrease of these sgRNAs including the neutral ones. Therefore, we normalized the sgRNA read counts to the sum of read counts of all non-targeting sgRNAs. The normalized total read counts (the sum of read counts of all sgRNAs) of the lentivirus pool were comparable to the plasmid pool, while a modest decrease in the normalized total read counts was observed in the cells cultured in vitro and following in vivo growth (Fig. 1C). This suggests that knockout of some genes in the library resulted in significant antiproliferative effects causing the loss of those sgRNAs. Consistent with the normalized total read counts, Pearson correlation analysis shows that the sgRNA distribution in the lentivirus pool was almost the same as the plasmid pool, suggesting that the lentivirus packaging process did not affect the complexity of the library (Fig. 1D). Compared to the lentiviral pool, the Pearson correlation coefficient gradually decreased in the cancer cells grown in vitro and in vivo, suggesting that an increasing number of sgRNAs were depleted or enriched. Notably, the replicates of tumor samples were highly correlated with each other, which allowed a high-resolution statistical analysis of sgRNA depletion and enrichment.
The MAGeCK algorithm 33 was used to identify significantly depleted or enriched sgRNAs and corresponding genes (Supplementary Table S2 and S3). Consistent with the Wnt/β-catenin dependency, four out of the five sgRNAs targeting CTNNB1 were depleted substantially at the cell culture stage even before the cells were injected into mice (Fig. 1E and 1F). A similar sgRNA depletion pattern was observed for other genes involved in essential biological processes, such as H3F3A (histone), AURKB (chromosome segregation), U2AF1 and SRSF2 (RNA splicing) (Fig. 1E). These results validated the screen.
Glucose metabolism is a specific in vivo vulnerability
We noted that many genes involved in glucose metabolism, specifically glycolysis and the pentose phosphatase pathway, were required in HPAF-II xenografts but this vulnerability was not evident in the in vitro cultured cells. The sgRNAs targeting these genes were depleted robustly only after the cells started to grow in vivo (Fig. 1E and 1H). For example, the five sgRNAs targeting ENO1 were depleted only by 20% on average in vitro but were depleted more than 50% in vivo compared to sgRNAs targeting CTNNB1 that were depleted substantially in vitro (Fig. 1F and 1G). Our finding that glucose metabolism genes are not identified as a sensitivity in vitro is in agreement with the results from a previously reported RNAi screen27. That RNAi screen was performed in a large panel of cancer cell lines in vitro, using cell viability as a readout following shRNA-mediated gene expression knockdown for two weeks. From that dataset we extracted the fitness score of specific genes of interest in pancreatic and colorectal cancer cell lines (Supplementary Fig. S1). As expected, in the in vitro RNAi screen knockdown of KRAS and CTNNB1 had lethal effects only in specific genetically sensitive cell lines, while knockdown of components of DNA polymerase, RNA polymerase, the proteasome, ribosomes, and the spliceosome showed general toxicity across all pancreatic and colorectal cancer cell lines. Consistent with our data, knockdown of genes involved in the glycolysis and pentose phosphate pathway had minimal effect on the in vitro cell viability in almost all the cell lines tested. These findings indicate that the importance of glycolysis and pentose phosphate pathways in survival of cancer cells may be underestimated in in vitro studies.
PI3K/mTOR signaling is a vulnerability in Wnt-driven pancreatic cancer
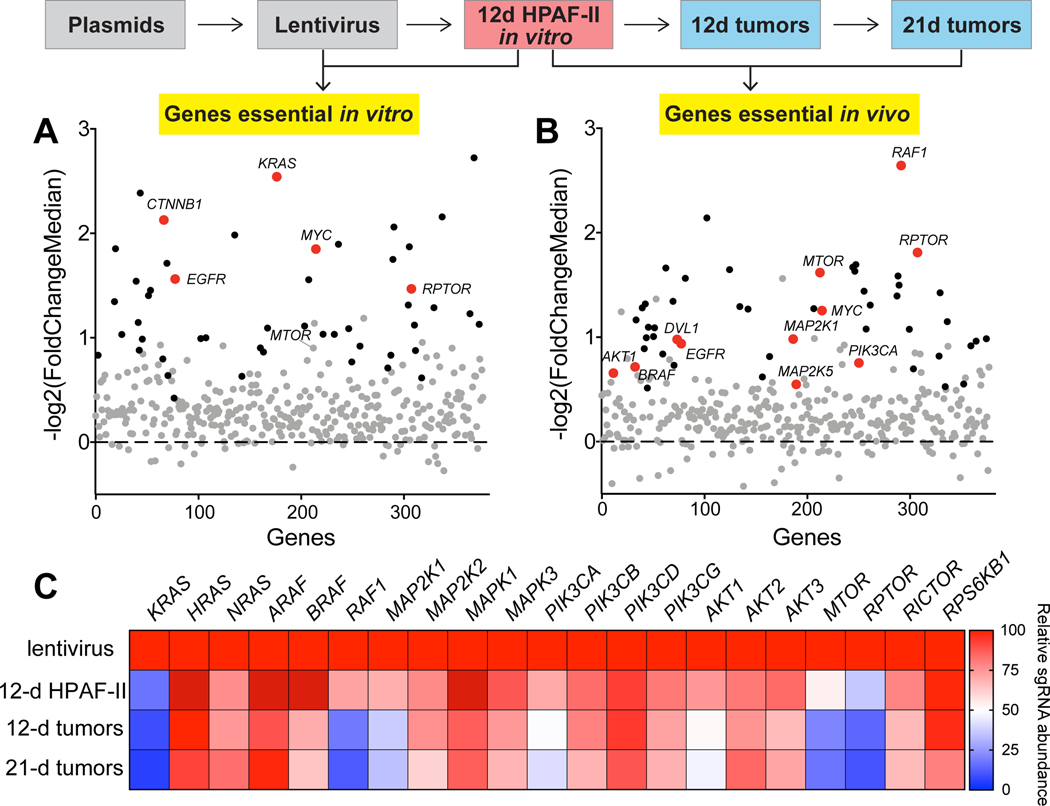
Our CRISPR screen showed that several known oncogenes and housekeeping genes are essential in vitro or in vivo. Genes involved in growth factor and Wnt signaling pathways identified to be essential based on the analysis of sgRNA depletion in our screen are highlighted in Fig. 2A and 2B (Supplementary Table S3). We note that the change in sgRNA abundance for several genes of interest failed to reach statistical significance in vitro, but their essentiality was clearly manifested in the in vivo screen. For example, HPAF-II cells showed strong dependency on RAF1 and moderate dependency on BRAF only in vivo. These results further highlight the importance of examining gene functions both in vitro and in vivo.
Figure 2. PI3K/mTOR signaling is a vulnerability in Wnt-driven pancreatic cancer.
A and B, Scatter plots of the sgRNA depletion for all genes targeted in the library during the in vitro and in vivo selection. Each dot represents one gene, and genes are arranged in alphabetical order on the x-axis. The average depletion of the five sgRNAs targeting a gene is represented as -log2 transformed median fold change of the read counts of the five sgRNAs. Genes whose sgRNAs were significantly depleted are represented as black dots, and those without statistically significant changes are grey dots. Genes that changed significantly and are involved in KRAS signaling or Wnt signaling pathways are marked as red dots.
C, sgRNAs targeting genes involved in KRAS signaling are depleted during the proliferation of HPAF-II cells in vitro and in vivo. Heatmap representing the relative median abundance of all the five sgRNAs targeting the indicated gene calculated by the MAGeCK algorithm. Values are set to 100 for lentivirus, and 0 refers to 100% depletion of the sgRNAs.
Activating KRAS mutation occurs in ~95% of pancreatic cancer including RNF43-mutant pancreatic cancers 9,11. Our CRISPR screen showed that KRAS is indispensable even in the RNF43-mutant pancreatic cancers that are driven by hyperactive Wnt signaling. Currently, it is difficult to target mutant KRAS directly. One alternative strategy is targeting the effector signaling downstream of mutant KRAS 34. We examined the abundance of sgRNAs targeting these components of KRAS signaling with a focus on the RAF/MEK/ERK cascade and PI3K/AKT/mTOR cascade in our CRISPR screen (Fig. 2C). The single knockout of MAP2K2 (MEK2) and MAPK1/3 (ERK1/2) did not affect cell viability significantly either in vitro or in vivo, which could be due to the isoform redundancy or feedback regulation within the signaling cascade. Single knockout of PIK3CA and AKT1 showed moderate but significant antiproliferative effect, and knockout of the other isoforms had minimal effect, which could be due to redundancy of the isoforms. Notably, single knockout of MTOR and RPTOR suppressed HPAF-II cell growth potently both in vitro and in vivo. Both mTOR and Raptor are key components of mTOR complex 1 that is regulated by upstream PI3K/AKT signaling, implicating PI3K/mTOR signaling as a key vulnerability in Wnt-driven pancreatic cancer.
A drug combination study reveals the synergy between PORCN inhibitor and PI3K/mTOR inhibitors
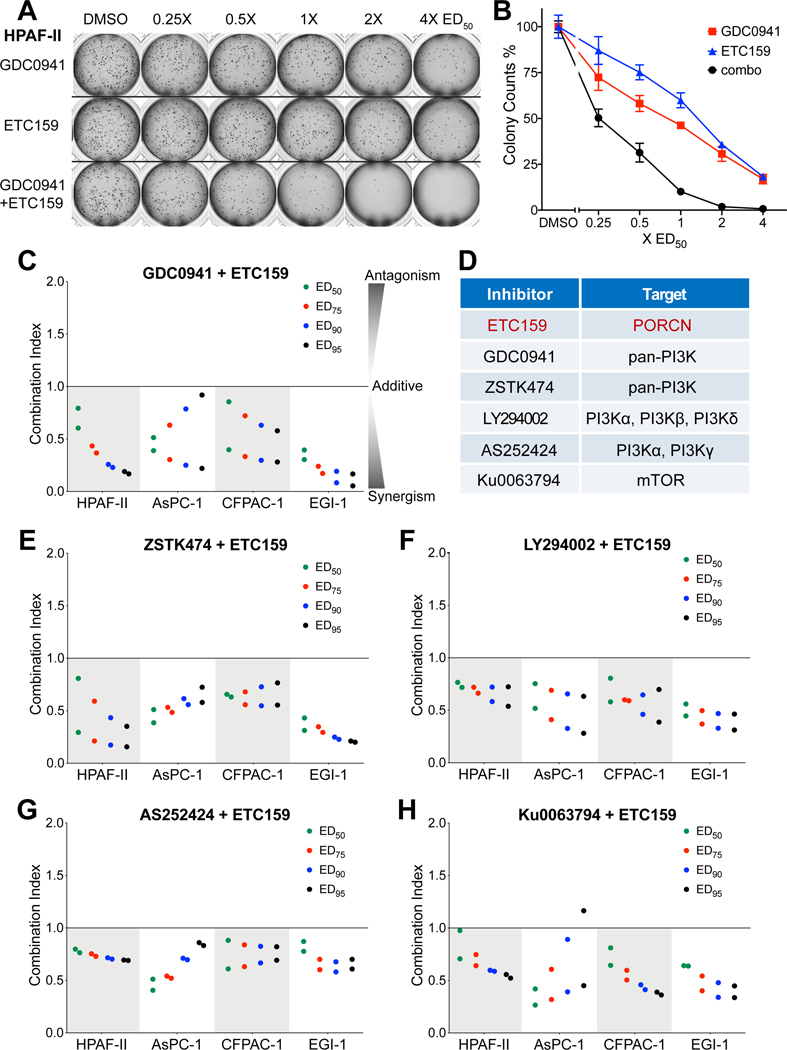
Our genetic screen showed that PI3K/mTOR signaling is essential in a Wnt-driven pancreatic cancer model known to be sensitive to PORCN inhibitors. To test the feasibility of combining PI3K/mTOR pathway inhibitors with a PORCN inhibitor for treating Wnt-driven pancreatic cancer, we performed drug combination studies using the PORCN inhibitor ETC-159 with five commercially available PI3K/mTOR pathway inhibitors in multiple cell lines. The assays were performed with a soft agar colony formation assay in Wnt-addicted pancreatic cancer cell lines. HPAF-II and AsPC-1 harbor inactivating RNF43 mutations. CFPAC-1 has no known mutation in Wnt pathway but is sensitive to β-catenin knockdown (Supplementary Fig. S1) as well as PORCN inhibitor 35. EGI-1, a cholangiocarcinoma cell line with a RSPO3 translocation that confers Wnt dependency, was also included in the drug combination study (Supplementary Fig. S2). Of note, most of the tested inhibitors alone could not suppress colony formation by 100% in the four cell lines even on treatment with high concentrations (10–100x ED50) for around two weeks, reflecting the resistance to single drug treatment (Supplementary Table S4).
As shown in Fig 3A, in HPAF-II cells the combination of ETC-159 and the pan-PI3K inhibitor GDC-0941 was more effective than using a two-fold increased dose of either of the single inhibitors and together could suppress colony formation by 100% (Fig. 3B). The drug combination index for ETC-159 and GDC-0941 determined using the Chou-Talalay CompuSyn algorithm 36 showed a synergistic effect between the two inhibitors over a large dose range (Fig. 3C). Importantly, the PORCN inhibitor ETC-159 and all five of the tested PI3K/mTOR inhibitors showed synergism in inhibiting colony formation in all four of the Wnt-driven cancer cell lines used in this study (Fig. 3C–3H). Notably, the mTOR inhibitor similarly demonstrated synergy with the PORCN inhibitor, consistent with an important role for the PI3K/mTOR axis in the Wnt-addicted cancers. We also tested the drug combination in two Wnt-independent pancreatic cancer lines, MIA PaCa-2 and PANC-1, which are resistant to ETC-159 treatment. Indeed, adding ETC-159 had no effect on their sensitivity to PI3K/mTOR inhibitors, confirming that the drug synergy is based on Wnt inhibition by the PORCN inhibitor and is not an off-target effect or synergistic toxicity (Supplementary Fig. S3).
Figure 3. PORCN inhibitor ETC-159 synergizes with PI3K/mTOR inhibitors in suppressing colony formation of Wnt-addicted cancer cells.
A, Pan-PI3K inhibitor GDC-0941 and PORCN inhibitor ETC-159 synergistically suppress the growth of HPAF-II colonies in soft agar. HPAF-II cells were seeded in soft agar, and treated with varying concentrations of GDC-0941 or ETC-159 alone or combination of the two inhibitors at an equivalent dose (i.e. cells in the three wells of the second column in the image were treated with 0.25xED50 of GDC-0941, 0.25xED50 of ETC-159, and 0.25xED50 of GDC-0941 + 0.25xED50 of ETC-159, respectively) for around two weeks. Representative images of the drug combination effect are shown.
B, Quantification of the colonies shown in A. Each data point represents the mean ± SD of technical duplicates in one experiment.
C, The combination indices (CI) of GDC-0941 and ETC-159 were calculated using the Chou-Talalay CompuSyn software based on the colony counts in four Wnt-addicted cancer lines tested in the soft agar colony formation assay. The CI values corresponding to the drug combination doses that lead to 50% (ED50), 75% (ED75), 90% (ED90) and 95% (ED95) colony formation inhibition in the combination treatment group are shown. The CI values from two independent experiments are shown. CI < 1, = 1, and > 1 indicate synergism, additive effect, and antagonism, respectively. The smaller the CI value, the stronger the synergism.
D, The small molecule inhibitors used in the drug combination study and their corresponding targets.
E-H, PORCN inhibitor ETC-159 in combination with multiple PI3K/mTOR inhibitors synergistically suppresses the colony formation of four Wnt-addicted cancer lines in soft agar. The CI values of ETC-159 and indicated PI3K/mTOR inhibitors in the four cell lines tested in soft agar colony formation assay are shown. The CI values from two independent experiments are shown.
Dual Wnt and PI3K/mTOR pathway inhibition potently blocks in vivo tumor growth
Several small molecule inhibitors that can potently suppress the PI3K/mTOR signaling show efficacy in pre-clinical models of pancreatic cancer. However, although these inhibitors can be well tolerated by patients, none of them improved the survival of pancreatic cancer in clinical trials to date 38–40. Among the five PI3K/mTOR pathway inhibitors tested in drug combination, GDC-0941 has advanced the furthest and has completed phase II clinical trials 37. Based on our finding of synergy between ETC-159 and PI3K/mTOR inhibitors in Wnt-addicted pancreatic cancers in the 3D in vitro system, we selected the pan-PI3K inhibitor GDC-0941 for further in vivo studies.
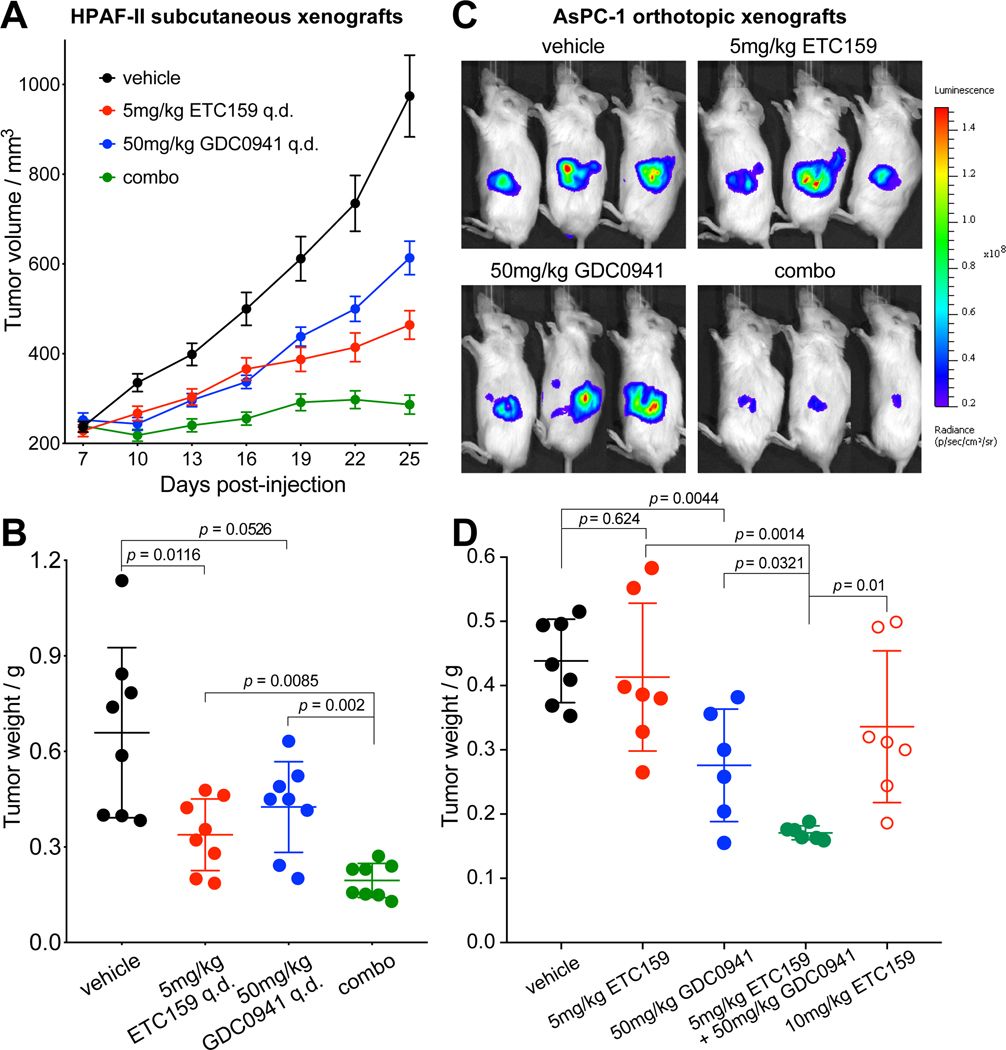
We investigated the tumor growth inhibition effect of ETC-159 and GDC-0941 combinational treatment in the HPAF-II subcutaneous xenograft mouse model (Fig. 4A and 4B). As one goal of the study was to minimize the on-target side effects of PORCN inhibitor, we used a relatively low dose (5 mg/kg/day) of ETC-159 based on our prior studies 19,21. 5 mg/kg ETC-159 and 50 mg/kg GDC-0941 as single-agent treatment suppressed tumor growth by 68% and 51% respectively, while the combination treatment suppressed tumor growth by 94%. Importantly, while tumors in the monotherapy groups were still progressing, the combination treatment kept the tumors in a “stable disease” status.
Figure 4. Dual Wnt and PI3K/mTOR pathway inhibition potently prevents growth of Wnt-driven pancreatic xenografts.
A, ETC-159 and GDC-0941 potently prevent growth of HPAF-II xenografts. NSG mice bearing HPAF-II subcutaneous xenograft were randomized into four groups and gavaged daily with vehicle, 5 mg/kg ETC-159, 50 mg/kg GDC-0941, or 5 mg/kg ETC-159 + 50 mg/kg GDC-0941 (combo). The treatment was started at 7th day post-injection and lasted for 19 days except the 16th day post-injection. The tumors were harvested six hours after the last dose for analysis. Error bars indicate SEM.
B, Weights of HPAF-II tumors shown in A on the last day of the study. Error bars indicate SD.
C and D, ETC-159 and GDC-0941 potently prevent growth of AsPC-1 orthotopic xenografts. NSG mice bearing luciferase-expressing AsPC-1 orthotopic xenograft were randomized into five groups and gavaged daily with vehicle, 5 mg/kg ETC-159, 50 mg/kg GDC-0941, 5 mg/kg ETC-159 + 50 mg/kg GDC-0941 (combo), or 10 mg/kg ETC-159. The treatment was started two weeks post-injection and lasted for one month, using a “6 days on, 1 day off” dosing schedule. Tumor growth was monitored by measuring luciferase activity. Representative luminescence images on the last day of the study are shown in C. Weights of AsPC-1 tumors harvested on the last day of the study are shown in D. Error bars indicate SD. One-way ANOVA was used to analyze statistical significance for data shown in this figure.
To test if the Wnt/PI3K/mTOR synergy was conserved in an independent Wnt-addicted cancer, we tested the drug combination in the AsPC-1 orthotopic xenograft mouse model (Fig. 4C and 4D). AsPC-1 tumors showed moderate sensitivity to GDC-0941 but were less sensitive to ETC-159. However, combining ETC-159 with GDC-0941 potently inhibited AsPC-1 tumor growth more effectively than the monotherapies. Notably, the combinational treatment also showed significantly better efficacy than single-agent treatment using a two-fold increased dose of ETC-159, consistent with the synergistic antitumor effect observed in the 3D in vitro system. Mice tolerated the combination treatment well with only a minor decrease of body weight and no significant change in their gut architecture (Supplementary Fig. S4). These results suggest that combining ETC-159 with a PI3K/mTOR inhibitor is an effective and safe therapeutic strategy for treating Wnt-driven pancreatic cancers.
Dual Wnt and PI3K pathway inhibition leads to enhanced cell cycle arrest and cellular senescence
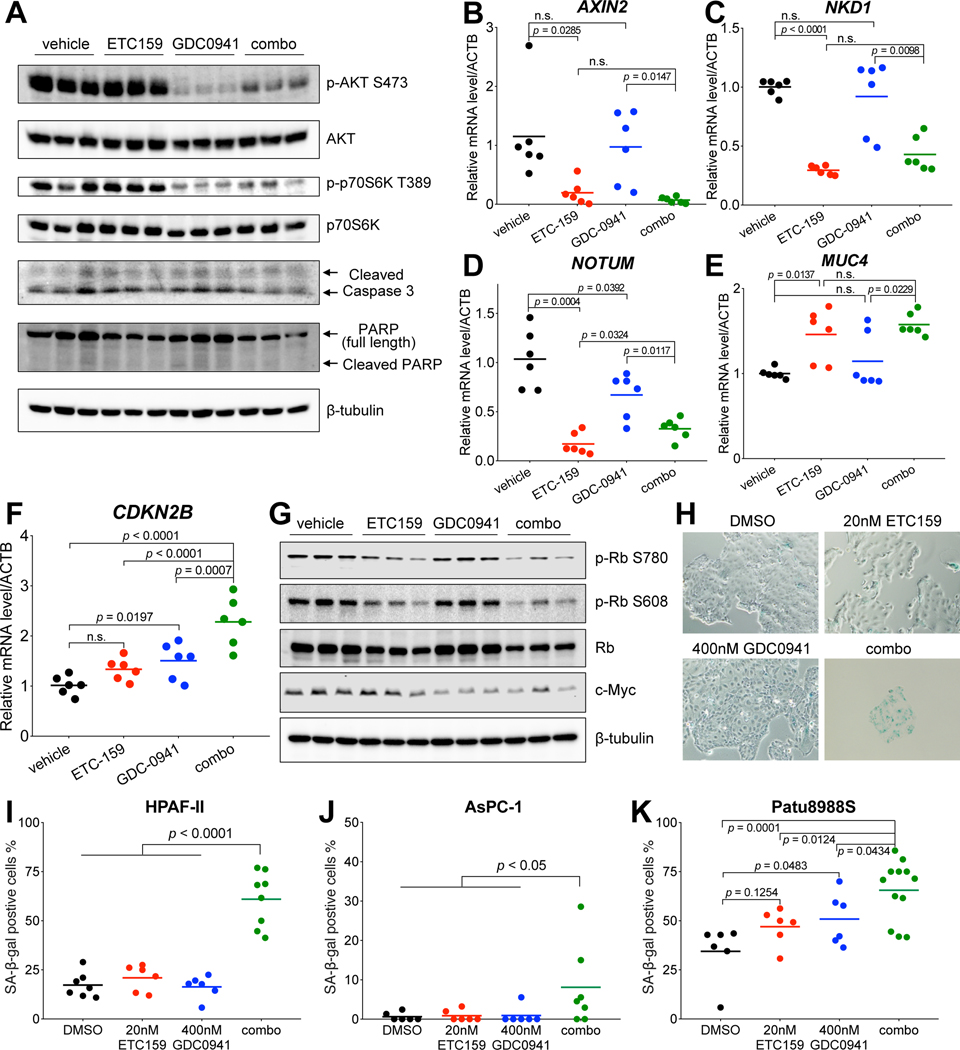
To study the mechanism underlying the synergy between PORCN inhibitor and PI3K inhibitor in Wnt-driven pancreatic cancer, we analyzed the HPAF-II tumors after treatment. As shown by western blot, PI3K downstream signaling assessed by phosphorylation of both AKT at Ser473 and p70S6K at Thr389 was potently inhibited by GDC-0941 (Fig. 5A). qPCR showed that the expression of well-established Wnt target genes including AXIN2, NKD1, and NOTUM was significantly suppressed by ETC-159 (Fig. 5B–5D). As expected, the Wnt-suppressed differentiation marker mucin genes were up-regulated by ETC-159, a finding supported by the Alcian blue staining for mucins on the tissue sections (Fig. 5E and Supplementary Fig. S5). For these targets, the inhibitors worked as expected but without evidence of direct crosstalk or synergy on inhibition of the two signaling pathways by the two inhibitors. The combined agents did not induce significant apoptosis as assessed by immunoblotting for cleaved Caspase 3 and cleaved PARP (Fig. 5A), suggesting that the drug synergy is not due to enhanced cell death.
Figure 5. Dual Wnt and PI3K/mTOR pathway inhibition leads to enhanced cell cycle arrest and cellular senescence.
A, Protein abundance of PI3K/mTOR pathway components and apoptosis related proteins in the HPAF-II tumors after drug treatment. Each lane represents an independent tumor.
B-F, Transcript levels of indicated genes determined by RT-qPCR in the HPAF-II tumors after drug treatment. Tumors from three mice of each group were used for analysis.
G, Abundance of phospho-Rb, total Rb, and c-Myc proteins in the HPAF-II tumors after drug treatment. Each lane represents an independent tumor.
H, ETC-159 and GDC-0941 combinational treatment induces senescence in HPAF-II cells. HPAF-II cells in culture were treated with DMSO, 20 nM ETC-159, 400 nM GDC-0941, or a combination of 20 nM ETC-159 + 400 nM GDC-0941 for six days, fixed, and stained for senescence-associated β-galactosidase (SA-β-gal) activity. Representative images are shown.
I, Quantification of the SA-β-gal positive cells shown in H. The data are representative of two independent experiments.
J and K, Quantification of the SA-β-gal positive cells in AsPC-1 and Patu8988S cells after drug treatment. The experiments were performed as in H. One-way ANOVA was used to analyze statistical significance for data shown in this figure. n.s., not significant.
We assessed the expression of well-established cell cycle suppressor genes CDKN2A and CDKN2B in the treated HPAF-II tumors. As CDKN2A is mutated in HPAF-II cells (COSMIC database), its transcript was not detectable in HPAF-II tumors. The expression of CDKN2B was synergistically up-regulated in the tumors treated with the combination, indicating enhanced cell cycle arrest (Fig. 5F). ETC-159 led to potent dephosphorylation of Rb when used either alone or in combination (Fig. 5G). The total Rb protein abundance was also decreased by the long-term ETC-159 treatment, perhaps due to the decreased stability of dephosphorylated Rb 41,42. Interestingly, GDC-0941 treatment significantly decreased MYC protein abundance both when used alone and in combination, while low dose ETC-159 treatment alone showed no obvious effect (Fig. 5G).
Considering that cellular senescence is a consequence of potent cell cycle arrest, we tested if the drug combination induced senescence. Treated HPAF-II cells were stained for senescence-associated β-galactosidase (SA-β-gal). As expected, cells treated in vitro with either single inhibitor had no significant increase of SA-β-gal compared to the control group (Fig. 5H and 5I). However, the cells treated with the drug combination showed strong staining for SA-β-gal activity. Similar results were observed in two other RNF43-mutant pancreatic cancer cell lines AsPC-1 and Patu8988S (Fig. 5J and 5K). Collectively, these findings indicate that dual Wnt and PI3K pathway inhibition leads to enhanced cell cycle arrest and cellular senescence, one mechanism underlying the potent synergistic inhibition of cell proliferation in Wnt-driven pancreatic cancer.
Dual Wnt and PI3K pathway inhibition synergistically suppresses glucose metabolic flux
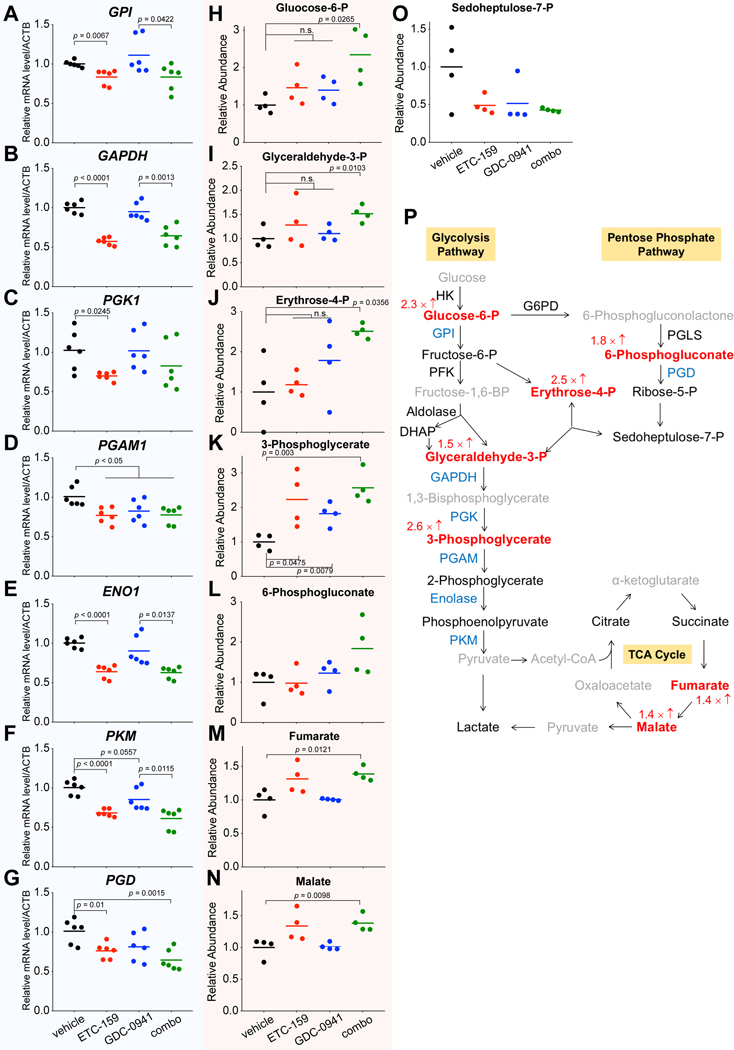
Our in vivo CRISPR screen also highlighted the importance of glucose metabolism in HPAF-II xenografts. As the PI3K/mTOR pathway is well-known for sensing environmental conditions and regulating cellular metabolism 43, and Wnt signaling can regulate glucose metabolism in colon cancer 44, we investigated the effect of dual PI3K and Wnt pathway inhibition on glucose metabolism. We assessed the expression levels of genes involved in multiple steps of glycolysis and the pentose phosphate pathway in HPAF-II tumors after 19 days of treatment. ETC-159 treatment significantly suppressed the expression of all tested glucose metabolism genes, while GDC-0941 treatment alone only suppressed the expression of PGAM1. Down-regulation of these genes was also observed in the combination treatment group (Fig. 6A–6G). Western blot for GPI, ENO1, and PGD confirmed these changes at the protein level as well (Supplementary Fig. S6). Confirming the functional significance of these changes, the abundance of multiple glucose metabolic intermediates in the treated HPAF-II tumors were altered, especially in the combination treatment group (Fig. 6H–6O). As shown in Figure 6H, 6I, and 6J, single-agent treatments with ETC-159 or GDC-0941 alone slightly increased the abundance of glucose 6-phosphate, glyceraldehyde 3-phosphate, and erythrose 4-phosphate, albeit not reaching statistical significance. However, the combination treatment showed an enhanced effect and led to a statistically significant accumulation of these metabolites.
Figure 6. Dual Wnt and PI3K/mTOR pathway inhibition synergistically suppresses the glucose metabolic flux.
A-G, Transcript levels of genes involved in glucose metabolism as determined by RT-qPCR are reduced following ETC-159 treatment in HPAF-II tumors. Tumors from three mice of each group were used for analysis.
H-O, Abundance of glucose metabolites in HPAF-II tumors altered following drug treatment. Tumors from four mice of each group were used for gas chromatography/mass spectrometry (GC/MS) analysis.
P, Schematic diagram showing the reduced glucose metabolic flux after the combinational drug treatment. The glucose metabolic enzymes down-regulated on transcriptional level (combo vs vehicle) are marked in blue. The accumulated metabolites are marked in red with the fold change (combo vs vehicle) labelled accordingly. Metabolites with no significant change by treatment are marked as black, and metabolites not detected by GC/MS are marked as grey. One-way ANOVA was used to analyze statistical significance. n.s., not significant.
Notably, most of these accumulated metabolites are the upstream substrates of these down-regulated enzymes, suggesting a decreased glucose metabolic flux following dual PI3K and Wnt pathway inhibition (Fig. 6P). As glucose metabolism is essential for the growth of pancreatic cancers in vivo, the suppressive effect on glucose metabolism by dual PI3K and Wnt pathway inhibition may contribute to the synergistic tumor growth inhibition.
Discussion
Despite increasing research efforts over the past decades there has been little improvement of the therapeutic strategies and outcome for pancreatic cancer patients. PORCN inhibitors show efficacy in pre-clinical models of RNF43-mutant pancreatic cancer and have advanced to clinical trials. Using an in vivo genetic screen using CRISPR/Cas9 genome editing technology with a small curated CRISPR library, we identified novel druggable vulnerabilities in the PI3K/mTOR pathway. Drugs targeting this pathway synergized with the PORCN inhibitor ETC-159 both in vitro and in mouse xenograft studies. These finding may allow the development of effective combination therapies for patients with Wnt-addicted cancers.
Interestingly, our results also reveal that multiple genes regulating glucose metabolism are not required in tissue culture but are important for in vivo tumor growth. This highlights the importance of glucose metabolic pathways for survival of cancer cells in vivo. This finding is consistent with a previous study reporting an important role of PGD, a gene involved in the pentose phosphate pathway, for cancer cells only in vivo but not in vitro 45. The differences in dependence on glucose metabolism in vitro versus in vivo could result from the differences in the nutrient availability in tissue culture where cells are cultured in media rich in glucose, glutamine and pyruvate compared to the harsher tumor microenvironment. Moreover, we also found that sgRNAs targeting several other essential genes were more robustly depleted in vivo than in vitro. Taken together, these findings highlight that the in vivo CRISPR screen has unique abilities to identify cancer vulnerabilities. While curated CRISPR sgRNA libraries have the limitation that they do not survey the entire transcriptome, they are an important complement to in vitro surveys that lack a realistic tumor microenvironment.
Pancreatic cancer is driven by dysregulated KRAS signaling and our CRISPR screen confirmed that KRAS is indispensable in the subset of pancreatic cancers that are Wnt-driven. Almost all efforts targeting KRAS downstream signaling including the MAPK cascade and PI3K/mTOR cascade in pancreatic cancer have failed in clinical trials 39,40,46–48. In our study, although the genetic screen identified PI3K/mTOR signaling to be essential in Wnt-driven pancreatic cancer, treatment with the pan-PI3K inhibitor GDC-0941 alone at a dose that substantially blocked the PI3K pathway only moderately delayed the tumor progression in the HPAF-II xenograft model. This suggests that targeting multiple pathways simultaneously using combination therapies may provide added benefit to cancer patients.
We showed that ETC-159, a PORCN inhibitor in current clinical trials, synergized with multiple PI3K/mTOR inhibitors in suppressing Wnt-driven pancreatic cancer proliferation in vitro. The combination of ETC-159 and GDC-0941 led to reduced cell proliferation in vivo due to modulation of cell cycle regulators including MYC. Our previous study showed that Wnt pathway blockade via ETC-159 led to dramatic change of the transcriptome in Wnt-driven cancer through both Wnt/β-catenin transcriptional regulation and modulation of protein stability of important transcription factors including MYC. Wnt inhibition regulated both cell cycle progression and ribosome biogenesis in part through MYC regulation 31. Here using a suboptimal dose of ETC-159 (5mg/kg/day) we still observed potent inhibition of a subset of canonical Wnt/β-catenin target genes including AXIN2 and NKD1 as well as suppression on the cell cycle but with a lesser effect on MYC expression. The addition of the pan-PI3K inhibitor GDC-0941 further decreased MYC protein abundance, potentially via both the translational control of MYC by mTOR signaling 49 and a PI3K/AKT/GSK3 control of MYC stability 50,51.
The sensitivity of the Wnt-addicted cells to knockout of glycolytic pathway genes in vivo led us to examine this pathway in some depth. The combinational treatment led to potential suppression of glucose metabolic flux in vivo. Mechanistically, inhibition of Wnt signaling by ETC-159 globally suppressed the expression of multiple genes involved in glucose metabolism, including those identified in the CRISPR screen. But as the gene expression analysis was performed after long term drug treatment, the decreased expression of glucose metabolism genes could be a consequence of decreased metabolic demand due to the cell cycle arrest and decreased ribosome biogenesis. The PI3K inhibitor alone did not have a major effect on the mRNA and protein abundance of most of the tested glucose metabolism genes with the exception of PGAM1. It is possible that inhibition of the PI3K/mTOR signaling down-regulated the translation or activity of additional metabolic enzymes that we did not directly assess 52. Collectively, dual Wnt and PI3K/mTOR pathway inhibition showed significant suppressive effects on glucose metabolism. This is consistent with the CRISPR screen that found that glucose metabolic genes were essential for cancer proliferation in vivo. Glucose metabolism is an interesting vulnerability, demonstrated by the requirement for genes such as enolase (ENO1) and phosphogluconate dehydrogenase (PGD) in vivo. Pharmacological inhibition of glucose metabolism enzymes for cancer therapy is challenging, considering the potential general toxicity 53. Pharmacological inhibition of Wnt and PI3K/mTOR pathway could be an alternative strategy to target the glucose metabolism in Wnt-driven pancreatic cancer.
In summary, this study demonstrates that dual PORCN inhibitor and PI3K/mTOR inhibitor treatment is a potential strategy for treating Wnt-driven pancreatic cancer. Notably, as we also observed the drug synergy in a RSPO3-translocated cholangiocarcinoma cell line, this drug combination strategy may apply to other cancer types with alterations in upstream Wnt pathway and hyperactive PI3K/mTOR signaling.
Materials and Methods
Cell lines.
HPAF-II, CFPAC-1, MIA PaCa-2, and PANC-1 were from Duke Cell Culture Facility. AsPC-1 was from ATCC. Patu8988S and EGI-1 were from DSMZ. HPAF-II and EGI-1 were grown in Eagle’s Minimum Essential Medium (EMEM). AsPC-1 was grown in Roswell Park Memorial Institute (RPMI) 1640 Medium. CFPAC-1 was grown in Iscove’s Modified Dulbecco’s Medium (IMDM). Patu8988S, MIA PaCa-2, and PANC-1 were grown in Dulbecco’s Modified Eagle’s Medium (DMEM). All culture media contained 10% FBS, 1 mM sodium pyruvate, 2 mM L-glutamine, and 1% penicillin/streptomycin. Cells were cultured in a humidified incubator with 5% CO2. All cell lines were regularly tested for mycoplasma contamination and confirmed to be mycoplasma free.
Small molecule compounds.
ETC-159 was manufactured by Experimental Therapeutics Centre, A*STAR, Singapore. GDC-0941, ZSTK474, LY294002, AS252424, Ku-0063794, and Rapamycin were purchased from SynMedChem.
Lentiviral packaging.
The lentiviral vector was co-transfected with the packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) in the HEK293FT cells using Opti-MEM and lipofectamine 2000. The Opti-MEM was replaced with DMEM six hours after transfection. The virus-containing culture media were collected 48 hours and 72 hours after transfection, filtered through 0.45 μm filter membrane, and stored at −80 °C.
In vivo CRISPR loss-of-function screen.
The CRISPR screen library used in this study was described previously 32. The plasmid library was packaged into lentivirus. HPAF-II cells were transduced with this lentivirus pool at MOI = 0.3 in the presence of 7.5 μg/ml polybrene. 2 × 106 HPAF-II cells were used for transduction of this CRISPR library that contains ~2,000 sgRNAs to maintain the library coverage of ~300 cells/sgRNA given the MOI. The transduced HPAF-II cells were selected with 0.8 μg/ml puromycin for seven days and expanded in vitro for another five days. This expanded HPAF-II pool carrying the CRISPR library was injected subcutaneously into the two flanks of NOD scid gamma (NSG) mice. 107 cells were injected per flank to get library coverage of 5000 cells/sgRNA. 106 cells of this expanded HPAF-II population were also harvested and stored as cell pellet at −80 °C for deep sequencing. Tumor-bearing mice were sacrificed 12 days or 21 days after injection, and tumors were harvested and stored at −80 °C. The tumors were homogenized in liquid nitrogen with a mortar and pestle. Genomic DNA from the frozen cell pellets and homogenized tumor tissues were extracted using a homemade salt precipitation protocol similar to the QIAGEN Gentra Puregene Kit. The viral RNA was extracted using the QIAamp Viral RNA Kit and converted into cDNA using Thermo Scientific RevertAid First Strand cDNA Synthesis Kit and viral specific primers. The sgRNA cassette that can integrate into the HPAF-II genome was amplified from the plasmid, lentiviral cDNA, and genomic DNA of HPAF-II cells and tumors by PCR using specific primers. Illumina sequencing adaptors and index sequences were added on the PCR products from each sample by a second round PCR. The barcoded PCR products were purified using the QIAGEN Gel Purification Kit, quantified using the KAPA Library Quantification Kit, and sequenced on the Illumina MiSeq platform.
Data analysis of the CRISPR screen results.
The fastq file generated by the Illumina MiSeq was processed by R. The “processAmplicons” package from the “edgeR” library was used to decode all the samples based on their specific barcodes, align the sequences, and count the number of the reads for each sgRNA in each sample. The sgRNA read counts were normalized to the total read counts of those non-targeting control sgRNAs in each sample. The Python based MAGeCK algorithm 33 was used to compare different sample groups with each other and filter out the statistically significantly depleted or enriched sgRNAs and the corresponding genes.
Mouse xenografts.
NOD scid gamma (NSG) mice were purchased from InVivos, Singapore or Jackson Laboratories, Bar Harbor, Maine. All animal studies were approved by the Duke-NUS Institutional Animal Care and Use Committee. Mice were housed in standard cages and were allowed access ad libitum to food and water. For HPAF-II subcutaneous implantation, HPAF-II cells were harvested using TrypLE, washed with cold PBS, resuspended in ice cold 50% Matrigel in PBS, and subcutaneously injected into the two flanks of NSG mice. For the CRISPR screen, 107 HPAF-II cells carrying the library resuspended in 200 μl Matrigel/PBS were injected per flank. For the combinational drug treatment study, 5 × 106 HPAF-II cells resuspended in 200 μl Matrigel/PBS were injected per flank. The tumor dimensions were measured with a caliper routinely, and the tumor volumes were calculated as 0.5 × length × width × width. When the tumor volumes reached ~200 mm3 seven days after injection, the tumor bearing mice were randomized into four groups (n = 8/group). The small molecule inhibitors ETC-159 and GDC-0941 were formulated in 50% PEG400 (vol/vol) in water and administered by oral gavage at a dosing volume of 10 μl/g mice body weight once every day. The four groups of mice were treated with vehicle, ETC-159, GDC-0941, and combo of the two inhibitors, respectively. Drug treatment was lasted for 19 days except the 16th day post-injection. For AsPC-1 orthotopic implantation, AsPC-1 cells stably expressing luciferase were resuspended in ice cold 50% Matrigel in PBS. 1.4 × 106 AsPC-1 cells resuspended in 50 μl Matrigel/PBS were injected into the tail of the pancreas of NSG mice. Orthotopic tumors in the living animals were monitored weekly by intra-peritoneally injecting 150 mg/kg D-luciferin (PerkinElmer #122799) into the mice and imaged using a PerkinElmer In Vivo Imaging System (IVIS). Two weeks after injection, the tumor-bearing mice were randomized into five groups (n = 6–7/group) and treated with the indicated inhibitors or vehicle for one month using a “six days on one day off” dosing schedule. All mice were sacrificed six hours after the last dosing. The tumors were resected, weighed, and snap frozen in liquid nitrogen or fixed in 10% neutral buffered formalin.
Soft agar colony formation assay and drug combination study.
HPAF-II, AsPC-1, CFPAC-1, EGI-1, MIA PaCa-2, or PANC-1 cells were plated in 48-well suspension cell culture plates. 5000 cells/well mixed with 0.35% agar in 200 μl complete culture media were layered on top of 0.5% agar in 200 μl complete culture media. After solidification of the agar layers, 500 μl complete culture media containing small molecule inhibitors or DMSO control was added to each well. The cancer cells were allowed to form colonies and grow for 1–2 weeks. The colonies were then stained with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and counted by GelCOUNT (Oxford Optronix, Abingdon, UK). For the drug combination study, the ED50 (the dose that leads to 50% of the maximum colony formation suppressive effect in soft agar) of the inhibitors was determined first. Then based on the ED50, cells seeded in soft agar were treated with a concentration gradient of either ETC-159, or a PI3K/mTOR inhibitor, or the combination of the two inhibitors. Synergism between the two inhibitors in suppressing colony formation was calculated using the Chou-Talalay CompuSyn software 36.
RNA isolation and quantitative PCR (qPCR).
Total RNA was isolated from frozen tumor tissues using the QIAGEN RNeasy Kit. Total RNA was reverse transcribed into cDNA using the BioRad iScript cDNA Synthesis Kit. Real time qPCR was performed using BioRad SsoFast EvaGreen Supermix Kit on a BioRad CFX96 real time cycling machine. Primer sequences used for qPCR are described in Supplementary Table S1.
Western blot.
Tumor tissues were homogenized in 4% SDS in water using Polytron Homogenizer. Protein concentration of the lysate was quantified using BCA assay. Equal amount of proteins for different samples was resolved by 10% SDS-PAGE and transferred to PVDF membranes. Western blot was performed following standard protocol. Antibodies used for western blot include anti-pan-AKT (CST 4691), anti-phospho-AKTS473 (CST 9271), anti-p70S6K (sc-230), anti-phospho-S6KT389 (CST 9234), anti-cleaved Caspase-3 (CST 9661), anti-PARP (BD 556494), anti-Rb (CST 9309), anti-phospho-RbS780 (CST 8180), anti-phospho-RbS608 (CST 2181), anti-c-Myc (Abcam 32072), anti-GPI (sc-365066), anti-α Enolase (sc-100812), anti-PGD (sc-398977), and anti-β-tubulin (Abcam 52623).
Histological analysis.
The senescence-associated β-galactosidase (SA-β-gal) staining was performed in tissue culture using the Senescence Detection Kit (ab65351, Abcam) following manufacture’s protocol. Tumors were fixed in 10% neutral buffered formalin and embedded in paraffin. Tumor sections were deparaffinized and rehydrated. For Alcian blue mucin staining, the deparaffinized and rehydrated sections were stained using the Alcian blue (pH 2.5) Mucin Stain Kit (ab150662, Abcam). All stained sections were dehydrated and mounted with DPX. Bright field images were acquired using a Nikon Eclipse Ni-E microscope.
Quantification of glucose metabolites in tumor tissues.
The frozen tumor tissues were homogenized in a solution consisting of 50% acetonitrile containing 0.3% formic acid. Proteins in the samples were crashed out by methanol and the supernatant was retrieved for further analysis. Methoxyamine and N-Methyl-N-(trimethylsilyl)trifluoroacetamide were added and incubated above 50 °C sequentially for methoxyamination and derivatization of samples. The trimethylsilyl derivatives were separated by gas chromatography on an Agilent Technologies HP 7890A and quantified by selected ion monitoring on a 5975C mass spectrometer using stable isotope dilution.
Statistical analysis.
Group differences were analyzed by one-way ANOVA using GraphPad Prism 8. The statistical significance threshold was p value < 0.05.
Supplementary Material
Acknowledgements
This study is supported in part by the National Research Foundation Singapore and administered by the Singapore Ministry of Health’s National Medical Research Council under the STAR Award Program to D.M. Virshup. B. Madan acknowledges the support of the Singapore Ministry of Health’s National Medical Research Council Open Fund-Independent Research Grant. We acknowledge the assistance from Yun Ka Wong and other members of the Virshup lab and Experimental Therapeutics Centre. We thank Prof. Stuart Cook and Edmund Pua for help with the MiSeq; Muhammad Idris Bin Kachi Mydin for help with bioinformatic analysis; Duke-NUS Metabolomics Facility, Dr. Jianhong Ching, and Dr. Zing Tan for help with the glucose metabolite quantification, and Profs. Shazib Pervaiz and Patrick Tan for useful suggestions on this study. Figure 1A was adapted from Servier Medical Art, licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com).
Footnotes
Disclosure of Potential Conflicts of Interest: Babita Madan and David M. Virshup have a financial interest in ETC-159.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017; 67: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M Pancreatic cancer. N Engl J Med 2010; 362: 1605–1617. [DOI] [PubMed] [Google Scholar]
- 3.Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997; 15: 2403–2413. [DOI] [PubMed] [Google Scholar]
- 4.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol 2007; 25: 1960–1966. [DOI] [PubMed] [Google Scholar]
- 5.Hammel P, Huguet F, van Laethem J-L, Goldstein D, Glimelius B, Artru P et al. Effect of Chemoradiotherapy vs Chemotherapy on Survival in Patients With Locally Advanced Pancreatic Cancer Controlled After 4 Months of Gemcitabine With or Without Erlotinib: The LAP07 Randomized Clinical Trial. JAMA 2016; 315: 1844–1853. [DOI] [PubMed] [Google Scholar]
- 6.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. PNAS 2011; 108: 21188–21193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X, Hao H-X, Growney JD, Woolfenden S, Bottiglio C, Ng N et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. PNAS 2013; 110: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017; 32: 185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012; 488: 665–669. [DOI] [PubMed] [Google Scholar]
- 13.Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012; 485: 195–200. [DOI] [PubMed] [Google Scholar]
- 14.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell 2006; 11: 791–801. [DOI] [PubMed] [Google Scholar]
- 15.Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK et al. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J Cell Sci 2010; 123: 3357–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proffitt KD, Virshup DM. Precise regulation of porcupine activity is required for physiological Wnt signaling. J Biol Chem 2012; 287: 34167–34178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM et al. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation 2012; 84: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science 2012; 337: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 2016; 35: 2197–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picco G, Petti C, Centonze A, Torchiaro E, Crisafulli G, Novara L et al. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO3 fusions. EMBO Molecular Medicine 2017; 9: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madan B, McDonald MJ, Foxa GE, Diegel CR, Williams BO, Virshup DM. Bone loss from Wnt inhibition mitigated by concurrent alendronate therapy. Bone Res 2018; 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janku F, Connolly R, LoRusso P, de Jonge M. Abstract C45: phase I study of WNT974, a first-in-class Porcupine inhibitor, in advanced solid tumors. 2015. doi: 10.1158/1535-7163.TARG-15-C45. [DOI] [Google Scholar]
- 23.Ng M, Tan DS, Subbiah V, Weekes CD, Teneggi V, Diermayr V et al. First-in-human phase 1 study of ETC-159 an oral PORCN inhbitor in patients with advanced solid tumours. Journal of Clinical Oncology 2017; 35: 2584–2584. [Google Scholar]
- 24.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014; 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015; 163: 1515–1526. [DOI] [PubMed] [Google Scholar]
- 27.McDonald ER, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 2017; 170: 577–592.e10. [DOI] [PubMed] [Google Scholar]
- 28.Possik PA, Müller J, Gerlach C, Kenski JCN, Huang X, Shahrabi A et al. Parallel in vivo and in vitro melanoma RNAi dropout screens reveal synthetic lethality between hypoxia and DNA damage response inhibition. Cell Rep 2014; 9: 1375–1386. [DOI] [PubMed] [Google Scholar]
- 29.Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 2017; 547: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muir A, Vander Heiden MG. The nutrient environment affects therapy. Science 2018; 360: 962–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madan B, Harmston N, Nallan G, Montoya A, Faull P, Petretto E et al. Temporal dynamics of Wnt-dependent transcriptome reveal an oncogenic Wnt/MYC/ribosome axis. J Clin Invest 2018; 128: 5620–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson GR, Winter PS, Lin KH, Nussbaum DP, Cakir M, Stein EM et al. A Landscape of Therapeutic Cooperativity in KRAS Mutant Cancers Reveals Principles for Controlling Tumor Evolution. Cell Rep 2017; 20: 999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papke B, Der CJ. Drugging RAS: Know the enemy. Science 2017; 355: 1158–1163. [DOI] [PubMed] [Google Scholar]
- 35.Madan B, Ke Z, Lei ZD, Oliver FA, Oshima M, Lee MA et al. NOTUM is a potential pharmacodynamic biomarker of Wnt pathway inhibition. Oncotarget 2016; 7: 12386–12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou T-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010; 70: 440–446. [DOI] [PubMed] [Google Scholar]
- 37.Schmid P, Pinder SE, Wheatley D, Macaskill J, Zammit C, Hu J et al. Phase II Randomized Preoperative Window-of-Opportunity Study of the PI3K Inhibitor Pictilisib Plus Anastrozole Compared With Anastrozole Alone in Patients With Estrogen Receptor-Positive Breast Cancer. J Clin Oncol 2016; 34: 1987–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Javle MM, Shroff RT, Xiong H, Varadhachary GA, Fogelman D, Reddy SA et al. Inhibition of the mammalian target of rapamycin (mTOR) in advanced pancreatic cancer: results of two phase II studies. BMC Cancer 2010; 10: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolpin BM, Hezel AF, Abrams T, Blaszkowsky LS, Meyerhardt JA, Chan JA et al. Oral mTOR inhibitor everolimus in patients with gemcitabine-refractory metastatic pancreatic cancer. J Clin Oncol 2009; 27: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung V, McDonough S, Philip PA, Cardin D, Wang-Gillam A, Hui L et al. Effect of Selumetinib and MK-2206 vs Oxaliplatin and Fluorouracil in Patients With Metastatic Pancreatic Cancer After Prior Therapy: SWOG S1115 Study Randomized Clinical Trial. JAMA Oncol 2017; 3: 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittaker SR, Walton MI, Garrett MD, Workman P. The Cyclin-dependent kinase inhibitor CYC202 (R-roscovitine) inhibits retinoblastoma protein phosphorylation, causes loss of Cyclin D1, and activates the mitogen-activated protein kinase pathway. Cancer Res 2004; 64: 262–272. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Wei Y, Pushel I, Heinze K, Elenbaas J, Henry RW et al. Integrated stability and activity control of the Drosophila Rbf1 retinoblastoma protein. J Biol Chem 2014; 289: 24863–24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. The EMBO Journal 2014; 33: 1454–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin R, Elf S, Shan C, Kang H-B, Ji Q, Zhou L et al. 6-Phosphogluconate dehydrogenase links oxidative PPP, lipogenesis and tumour growth by inhibiting LKB1-AMPK signalling. Nat Cell Biol 2015; 17: 1484–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Infante JR, Somer BG, Park JO, Li C-P, Scheulen ME, Kasubhai SM et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer 2014; 50: 2072–2081. [DOI] [PubMed] [Google Scholar]
- 47.Ko AH, Bekaii-Saab T, Van Ziffle J, Mirzoeva OM, Joseph NM, Talasaz A et al. A Multicenter, Open-Label Phase II Clinical Trial of Combined MEK plus EGFR Inhibition for Chemotherapy-Refractory Advanced Pancreatic Adenocarcinoma. Clin Cancer Res 2016; 22: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Cutsem E, Hidalgo M, Bazin I, Canon J-L, Poddubskaya E, Manojlovic N et al. Phase II randomized trial of MEK inhibitor pimasertib or placebo combined with gemcitabine in the first-line treatment of metastatic pancreatic cancer. Journal of Clinical Oncology 2017; 33: 344–344. [Google Scholar]
- 49.Wall M, Poortinga G, Hannan KM, Pearson RB, Hannan RD, McArthur GA. Translational control of c-MYC by rapamycin promotes terminal myeloid differentiation. Blood 2008; 112: 2305–2317. [DOI] [PubMed] [Google Scholar]
- 50.Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem 2003; 278: 51606–51612. [DOI] [PubMed] [Google Scholar]
- 51.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000; 14: 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward PS, Thompson CB. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol 2012; 4: a006783–a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012; 491: 364–373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.