Abstract
Acid-producing bacteria are one kind of crucial species for Baijiu fermentation. The strain BJN0003 with the ability of producing butyric acid was isolated from the cellar mud of Baijiu, and the 16S rRNA gene sequence similarity was 94.2% to its most closely related type species Caproicibacterium lactiferaments JNU-WLY1368T, less than the threshold value of 94.5% for distinguishing genera. Furthermore, the genome of BJN0003 showed a length of 2,458,513 bp and a DNA G + C content of 43.3% through high throughput sequence. BJN0003 exhibited whole-genome average nucleotide identity value of 68.9% to the most closely related species, while the whole-genome digital DNA-DNA hybridization value was only 23.1%, which were both below the delineation thresholds of species. These results indicated BJN0003 could represent a potential novel species of a new genus of the family Oscillospiraceae, and was proposed the name as Butyriproducens baijiuensis. In addition, gene annotation and metabolic analysis showed that BJN0003 harbored the metabolic pathway of converting glucose to butyric acid. The discovery of the new species provided bacterial resource for Baijiu production and the revealing of genetic characteristics would promote the investigation of acid synthesis during Baijiu manufacturing process.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03624-w.
Keywords: Baijiu, Butyric acid, Fermentation, Microorganism, New genus
Introduction
Baijiu is one of Chinese national beverages with unique flavor, and produced through microbial co-fermentation followed by distillation using grains as the raw materials (Xu et al. 2022b). Microbial metabolism is an important source of flavor chemicals in Baijiu products, and microbes also degrade some harmful substances in Baijiu raw materials, thus have a significant impact on the quality of Baijiu (Xu et al. 2020a). Most types of Baijius are produced by solid-state fermentation, and microorganisms from tools, operators, water, air and fermentation starter will participate in the fermentation process (Liu and Sun 2018). There are hundreds of microbial species reported in Baijiu, and these microbial metabolites contribute numerous flavor chemicals to Baijiu products (Wang et al. 2021; Zhang et al. 2022). Flavor analysis shows that there are more than 2000 flavor chemicals in Baijiu, including alcohols, aldehydes, acids, esters, nitrogen compounds and sulfur compounds (Hong et al. 2021). Among them, acids play an important contribution to the flavor of Baijiu.
More than 120 kinds of acids are reported in Baijiu products, including straight chain and branched chain fatty acids with short, medium and long chains, unsaturated fatty acids and acids with benzene rings (Niu et al. 2020). Among them, the concentrations of straight chain and branched chain fatty acids with short and medium chain length are relatively high, such as acetic acid, propionic acid, butyric acid, pentanoic acid, hexanoic acid, 2-methylpropionic acid, and 2-methylbutanoic acid (Wu et al. 2022). Acids, especially short chain fatty acids, have important contributions to the taste and smell of Baijiu. For example, they can give Baijiu a refreshing feeling in taste, and sour, fruity and creamy in smell (Wei et al. 2020). As important flavor chemicals in Baijiu, the synthesis of acids has drew greatly attention during the Baijiu manufacturing process.
Microbial metabolism contributes significantly to the synthesis of flavor chemicals. Abundant microorganisms are existed in cellar mud of Baijiu fermentation pit, especially acid-producing microorganisms, which are the main source of fatty acids in Baijiu (Xu et al. 2023). However, due to the special environment of the cellar mud, the stress of high acidity and high ethanol has resulted the special metabolic characteristics of the enriched microorganisms in the cellar mud. Studies have isolated a series of novel species from cellar mud and fresh strains are frequently reported. Therefore, identifying novel microorganisms from cellar mud can enrich the microbial resources, and revealing the metabolic characteristics of novel microorganisms derived from Baijiu will help to clarify the synthesis of fatty acids in Baijiu.
Recent studies identified the new species Caproicibacterium amylolyticum and Caproicibacterium lactatifermentans of the Oscillospiraceae family, and showed the ability for synthesizing fatty acids such as butyric acid and caproic acid, and were recognized as important bacteria for acid production in Baijiu fermentation process (Gu et al. 2021; Wang et al. 2022). In our previous studies, a potential new species was identified from Baijiu with the ability of converting glucose to acetic acid and butyric acid (Xu et al. 2022a). For the metabolic pathways of acid chemicals and the metabolic characteristics of acid-producing microorganisms, studies indicate that the linear chain fatty acids in Baijiu are closely related to the metabolism of carbohydrates, while the branched chain fatty acids are intrinsically related to the metabolism of amino acids (Xu et al. 2022b). Branched chain amino acid transaminase from microorganisms can catalyze the conversion of amino acids such as isoleucine to produce 3-methylbutanol-CoA, and then generate 2-methylbutyric acid through CoA transferase or thioesterase (Elsden and Hilton 1978). Compared with branched chain fatty acids, linear chain fatty acids show relatively higher concentrations in Baijiu (Wu et al. 2022). The key intermediate of glycolysis is pyruvate, and catalyzed by lactate dehydrogenase to produce lactic acid, followed by pyruvate decarboxylase to produce acetaldehyde, and finally generated acetic acid or ethanol (Xu et al. 2020b). Acid-producing microorganisms in Baijiu, such as Clostridium spp., use lactic acid, or acetic acid and ethanol as the substrates to produce short chain fatty acids like butyric acid and caproic acid. Previous studies found that Clostridium kluyveri showed the ability to synthesize caproic acid using acetic acid and ethanol as substrates, and promoted the synthesis of ethyl caproate (the characteristic chemical of strong flavor Baijiu), which was considered as an important acid-producing microorganism in Baijiu (Yan and Dong 2018). Other studies found that Clostridium sp. utilized lactic acid as the substrate to produce butyric acid and caproic acid (Zhu et al. 2015). These works indicate that there are many novel microorganisms with the ability to produce acids in Baijiu.
Identification of novel species of acid-producing microorganisms from Baijiu not only helps to reveal the characteristics of microbial acid metabolism in Baijiu, but also indicates that Baijiu manufacturing, as a specific fermentation system, contains various acid-producing microbial resources to be systematically and comprehensively investigated (Gu et al. 2021; Wang et al. 2022). Acid-producing bacteria is an important group of functional microorganisms to be studied in Baijiu, and the revealing of relevant microbial resources and the analysis of genetic information will help to better understand the acid synthesis in Baijiu. In this study, an acid-producing bacteria was isolated from the Baijiu cellar mud. The 16S rRNA gene sequence showed a similarity lower than the threshold value of 94.5% for distinguishing genera to the most closely related species Caproicibacterium lactatifermentans JNU-WLY1368T (Gu et al. 2021), indicating this species was a potential novel species of a new member of the family Oscillospiraceae. This work revealed the genetic information and acid production ability of the strains, which provided novel microbial resources for investigating acid-producing microorganisms in Baijiu.
Materials and methods
Samples and strain isolation
The samples in this work were obtained from a Baijiu company in Hebei province, China. Cellar mud samples were collected from the pit (usually called Jiaochi in Chinese), and about 2.0 g of sample was mixed with 10 ml of sterile NaCl water solution (0.9%, w/v), and diluted with NaCl water solution. The diluted solutions were spread on GAM (Glucose-acetate medium) solid plate and cultured at 37 °C for 8–10 days under anaerobic conditions. A single colony was purified several times and one of the isolated pure strains was numbered as BJN0003. The strain was stored in the China General Microbiological Culture Collection Center (CGMCC) with the no. CGMCC1.18021.
Cultivation conditions
The 19 carbon sources utilization test of strain BJN0003 was performed according to previous studies (Wang et al. 2022). To explore the substrate consumption and chemical production of BJN0003, culture media containing different types and concentrations of carbon sources were prepared. The basic culture medium contained tryptone (10 g/L), yeast extract (10 g/L), sodium acetate (3.0 g/L), (NH4)2SO4 (2.0 g/L), NaH2PO4 (1.0 g/L), K2HPO4 (0.50 g/L), MgSO4·7H2O (0.10 g/L), FeSO4·7H2O (0.015 g/L), MnSO4·H2O (0.010 g/L), CaCl2 (0.010 g/L), CoCl2 (0.0020 g/L) and ZnSO4 (0.0020 g/L) according to previous studies (Gu et al. 2021). In addition, glucose, lactic acid and ethanol were added as carbon resources. As for the abbreviation of the medium, G was glucose, A was sodium acetate, L was lactic acid and E was ethanol. The GAM and LAM were additionally added with 15 g/L glucose and lactic acid, respectively. The GLAM was additionally added with 7.5 g/L glucose and 7.5 g/L lactic acid. The GAEM was GAM supplemented with 3 g/L ethanol. All culture media were adjusted to the pH value of about 6.5, sterilized and transferred to the anaerobic incubator (AW300SG, ELECTROTEK, England).
Genome sequence and annotation
Genome of BJN0003 was extracted by bacterial genomic DNA Extraction Kit (Solarbio, Beijing, China) according to standard protocol. The primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-GGTACTTGTTACGACT-3') were used to amplify the 16S rRNA gene of BJN0003 through polymerase chain reaction (PCR) (Yin et al. 2016). The genome sequence of BJN0003 was analyzed through high throughput sequencing by the Beijing Genomics Institute (BGI) Co. (Beijing, China) according to the standard protocol. High throughput sequencing was performed using PacBio RSII sequencing platform (Rhoads and Au 2015), and de novo assembly was followed based on PacBio sequencing results. Single nucleotide correction of genomic DNA was performed using Illumina HiSeq™ 2000 (Illumina, San Diego, CA, USA) sequencing platform generated reads. Thereafter, the genes were predicted based on the genome sequence and annotated by Clusters of Orthologous Groups of proteins (COG) (Galperin et al. 2019) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2022), respectively.
Phylogenetic analysis of BJN0003 with 16S rRNA gene sequence and comparative genome analysis
Phylogenetic analysis of the 16S rRNA gene sequence of BJN0003 was performed using the online web server EzBioCloud (https://www.ezbiocloud.net) (Kim et al. 2012). The 16S rRNA gene sequences of the type species with closely related relationship were identified and downloaded from the EzTaxon database. These sequences were aligned together with the 16S rRNA gene sequence of BJN0003, followed by phylogenetic tree construction using a neighbor-joining method with bootstrap value of 1000 of MEGA 7.0 software (Kumar et al. 2016). The whole genome sequence of these reference type species were also compared with the genome of BJN0003 through DNA-DNA hybridization (DDH) and average nucleotide identity (ANI) to reveal the genetic relationships between species. The ANI values of BJN0003 with the reference strains were investigated using software OrthoANI (Yoon et al. 2017), and the DDH values were calculated with software GGDC v2.1 (Meier-Kolthoff et al. 2013).
Metabolic property investigation of strain BJN0003
Strain BJN0003 was cultured and inoculated into GAM, LAM, GLAM and GAEM, respectively, and the fermented media were taken every 24 h. High performance liquid chromatography (HPLC, Watres e2685) was used to analyze the metabolites in the fermented media, and the pH values of the fermentation broth were determined using a pH meter (Mettler Toledo). Cell density was determined by measuring the absorbance at 600 nm using an ultraviolet spectrophotometer (PERSEE TU-1901, Jiangsu, China).
Analytical methods
Glucose, ethanol, acetic acid, lactic acid and butyric acid were determined by HPLC equipped with an Aminex HPX-87H column (300 × 7.8 mm) (Bio-Rad). The fermentation broth were centrifuged at 13,000×g for 2 min, and the supernatant was filtered by 0.22 μm filter membrane. HPLC was performed with the mobile phase of H2SO4 water solution (0.006 M) under a temperature of 65 °C with a flow rate of 0.9 mL/min. Sample were detected for 20 min under the isocratic elution condition and equipped with a 2414 refractive index detector.
Results and discussion
Baijiu is one of the six distilled beverages in the world with unique flavor. Flavor analysis shows that the total amount only accounted for 2%, but flavor chemicals determine the quality of Baijiu (Hong et al. 2021). There are many kinds of flavor chemicals, and among which acids make significant contributions to the smell and taste of products (Wu et al. 2022). In recent studies, a series of acid-producing microorganisms have been excavated, especially the species for synthesizing important fatty acids, providing valuable resources for scientific investigation of microbial acid synthesis in Baijiu fermentation process (Gu et al. 2021; Wang et al. 2022). Systematic identification of acid-producing microorganisms in Baijiu and analysis of their acid metabolic properties are of great significance for revealing the microbial metabolic synthesis of acids, and needed to be continuously investigated. The strain numbered BJN0003 was isolated from cellar mud of Baijiu, and colonies of the strain BJN0003 were 2–3 mm in diameter on GAM agar plate under 37 °C for 5 days and showed opaque white circular colonies with regular edges (Fig. S1). The strain was strictly anaerobic and gram positive with cell morphology of short and rod-shaped or approximately coccus-shaped (Fig. S1).
Phylogenetic analysis of the 16S rRNA gene
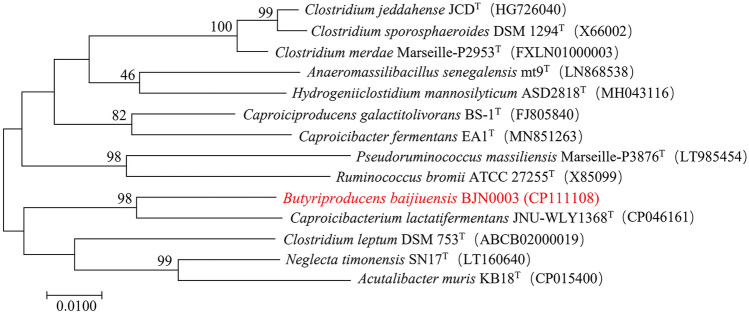
Evolutionary relationship analysis of the 16S rRNA gene showed that the sequence of BJN0003 with the most closely related type species Caproicibacterium lactatifermentans JNU-WLY1368T was only 94.2%, and sequence identity with other closely related type species Pseudoruminococcus massiliensis Marseille-P3876T (Afouda et al. 2020), Caproiciproducens galactitolivorans BS-1T (Kim et al. 2015), Anaeromassilibacillus senegalensis mt9T (Guilhot et al. 2016), Neglecta timonensis SN17T (Bessis et al. 2016) and Hydrogeniiclostidium mannosilyticum ASD2818T (Chaplin et al. 2020) were 92.9%, 92.8%, 92.6%, 92.4% and 92.3%, respectively (Fig. 1), which were all lower than the threshold value (94.5%) for distinguishing genera (Yarza et al. 2014). As it was suggested that the strain BJN0003 might be a potential novel species of a new genus of the family Oscillospiraceae, the whole genome of strain BJN0003 was sequenced and compared with those of the closely related type species at the molecular level to further identify the taxonomic status of strain BJN0003.
Fig. 1.
Phylogenetic analysis of strain BJN0003 with closely related type species based on 16S rRNA gene sequence analysis. 16S rRNA gene sequence analysis was performed using MEGA software with the neighbor-joining method and a bootstrap value of 1000. Bootstrap values > 50% (1000 replicates) are shown at branch nodes. Bar, 0.01 substitutions per nucleotide position. The accession numbers of the reference 16S rRNA gene sequences are shown in parentheses
Genome sequence and comparison
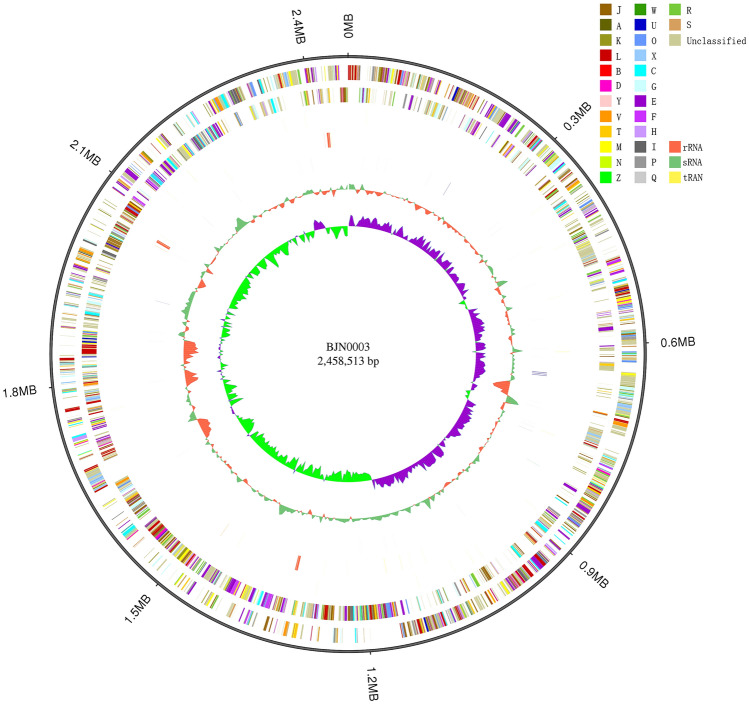
The circular genome DNA of strain BJN0003 showed a length of 2,458,513 bp with a DNA G + C content of 43.3% (Fig. 2), and annotated with 2460 genes, including 2359 protein encoding genes, 54 tRNAs and 9 rRNAs (Table 1). With the development of high-throughput sequencing technology, more and more bacterial genomes were sequenced and recognized as helpful for microbial identification (Feng et al. 2019). Genome scale DDH and ANI analysis of BJN0003 was performed with the type species which had closely related evolutionary relationships according to sequence similarities of 16S rRNA.
Fig. 2.
Circular map of strain BJN0003 based on genome sequence and annotation. From outside to inside: forward strand gene, reverse strand gene, forward strand RNAs, reverse strand RNAs, repeat, GC content, GC skew. The number in the center of the circle is the genome size of BJN0003
Table 1.
Genome sequence characteristics of strain BJN0003
| Attributes | Value |
|---|---|
| Genome assembly length (Mb) | 2.46 |
| GC content (%) | 43.30 |
| Number of genes | 2460 |
| Number of protein-coding genes | 2359 |
| Average gene length (bp) | 849.33 |
| tRNA | 54 |
| rRNA | 9 |
DDH is usually recognized as the taxonomic gold standard for species delineation of prokaryotes at the genomic level (Meier-Kolthoff et al. 2013). Generally, the DDH threshold is 70% for identifying species (Meier-Kolthoff et al. 2013). The DDH analysis results of Butyriproducens baijiuensis BJN0003 and the closely related species Caproicibacterium lactatifermentans JNU-WLY1368T, P. massiliensis Marseille-P3876T, Caproiciproducens galactitolivorans BS-1T, A. senegalensis mt9T, N. timonensis SN17T and H. mannosilyticum ASD2818T were 23.1%, 29.6%, 20.9%, 22.6%, 31.4% and 22.8%, respectively (Table 2). BJN0003 showed the highest similarity with N. timonensis SN17T (31.4%), which was far below the threshold value for species identification (70%), indicating that BJN0003 might be a potential novel species of a new genus of the family Oscillospiraceae.
Table 2.
Genome scale DNA–DNA hybridization (DDH) and average nucleotide identity (ANI) analysis of strain Butyriproducens baijiuensis BJN0003 compared with reference type species
| Analysis methods | Strains | Butyriproducens baijiuensis BJN0003 | Caproicibacterium lactatifermentans JNU-WLY1368T | Pseudoruminococcus massiliensis Marseille-P3876T | Caproiciproducens galactitolivorans BS-1T | Anaeromassilibacillus senegalensis mt9T | Neglecta timonensis SN17T | Hydrogeniiclostidium mannosilyticum ASD2818T |
|---|---|---|---|---|---|---|---|---|
| DDH (%) | BJN0003 | – | ||||||
| JNU-WLY1368T | 23.1 | – | ||||||
| Marseille-P3876T | 29.6 | 21.1 | – | |||||
| BS-1 T | 20.9 | 30.4 | 22.3 | – | ||||
| mt9T | 22.6 | 19.4 | 19.8 | 20.2 | – | |||
| SN17T | 31.4 | 27.5 | 28.4 | 24.3 | 23.4 | – | ||
| ASD2818T | 22.8 | 20.9 | 21.0 | 21.2 | 22.5 | 33.0 | – | |
| ANI (%) | BJN0003 | – | ||||||
| JNU-WLY1368T | 68.9 | – | ||||||
| Marseille-P3876T | 67.1 | 66.7 | – | |||||
| BS-1 T | 68.2 | 68.5 | 67.9 | – | ||||
| mt9T | 68.4 | 69.0 | 68.6 | 70.9 | – | |||
| SN17T | 67.5 | 67.5 | 68.0 | 68.2 | 69.3 | – | ||
| ASD2818T | 67.6 | 68.1 | 67.5 | 67.5 | 70.6 | 69.0 | – |
ANI is defined as the average base similarity between homologous segments of two microbial genomes, which showed good performance with a high degree of differentiation between closely related species (Richter and Rossello-Mora 2009). It is generally believed that the threshold value for species identification is 95% (Richter and Rossello-Mora 2009). ANI values of BJN0003 with Caproicibacterium lactatifermentans JNU-WLY1368T, P. massiliensis Marseille-P3876T, Caproiciproducens galactitolivorans BS-1T, A. senegalensis mt9T, N. timonensis SN17T and H. mannosilyticum ASD2818T were 68.9%, 67.1%, 68.2%, 68.4%, 67.5% and 67.6%, respectively (Table 2). BJN0003 and Caproicibacterium lactatifermentans JNU-WLY1368T showed the highest ANI value of 68.9%, and was much lower than the threshold value of 95%.
Proteins of BJN0003 were annotated with COG and KEGG databases. A total of 1756 genes were annotated by COG database (Fig. 3a), accounting for 71.4% of the genes, and 191 belonged to the metabolic pathways were involved in amino acid transport and metabolism, 189 involved in translation, ribosomal structure and biogenesis, 146 related to transcription, 113 involved in energy production and conversion, 110 belonged to replication, recombination and repair, 93 related to carbohydrate transport and metabolism, and other genes were involved in the left 20 COG categories. Proteins of BJN0003 were also sequence aligned with the KEGG database. A total of 1433 proteins were annotated through KEGG database (Fig. 3b), accounting for 58.3% of the predicted proteins. Among them, 386 annotated genes belonged to the metabolic pathway were involved in global and overview maps, 134 related to amino acid metabolism, 130 belonged to carbohydrate metabolism, 78 belonged to energy metabolism, 78 involved in translation, 69 related to metabolism of cofactor and vitamins, and other genes were divided into the left 33 KEGG categories. The results of genome annotation were helpful to clarify all genetic information of strain BJN0003. In addition, combined with the analysis of fermentation data of the strain, the metabolic characteristics of the strain could be comprehensively revealed.
Fig. 3.
COG and KEGG annotation of genome sequence of strain BJN0003. a COG (Cluster of Orthologous Groups of proteins) annotation, b KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation
Metabolic characteristic analysis of BJN0003
The BJN0003 utilization capabilities of 19 carbohydrates were investigated (Table 3). BJN0003 grew well with cellobiose, d-glucose, d-fructose, maltose, starch, d-xylose, d-sorbitol and tartarate, but grew weakly with d-mannose, sucrose, galactitol, lactose as carbon sources. However, it could not grow when using d-galactose, glycerol, d,l-lactate, l-fucose, l-rhamnose, arabinose, pyruvate as the carbon sources. The carbon source utilization of BJN0003 differed from that of the mostly related species JNU-WLY1368T. BJN0003 could utilize cellobiose, d-xylose, d-sorbitol, tartarate whereas JNU-WLY1368T could not. In conclusion, the above results further proved that the strain BJN0003 was a type species of a potential new genus of microorganisms. The strain BJN0003 was named as Butyriproducens baijiuensis. For the genus and species names, Butyriproducens gen. nov. (Bu.tyr.i.pro.du’cens. N.L. n. acidum butyricum butyric acid; L. part. adj. producens producing; N.L. masc. n. Butyriproducens the bacteria producing butyric acid), Butyriproducens baijiuensis sp. nov. (bai.jiu.en’sis. N.L. masc. adj. baijiuensis of or belonging to baijiu, the place where the type strain was isolated).
Table 3.
Comparison of carbon source utilization characteristics between Butyriproducens baijiuensis BJN0003 and the mostly related species Caproicibacterium lactatifermentans JNU-WLY1368T
| Carbon source utilization | Butyriproducens baijiuensis BJN0003 | Caproicibacterium lactatifermentans JNU-WLY1368T |
|---|---|---|
| Cellobiose | + | – |
| d-Galactose | – | w |
| d-Glucose | + | + |
| Glycerol | – | – |
| d-Fructose | + | + |
| Maltose | + | + |
| d-Mannose | w | w |
| Sucrose | w | w |
| Starch | + | + |
| d-Xylose | + | – |
| Galactitol | W | – |
| d-Sorbitol | + | – |
| d,l-Lactate | – | + |
| Lactose | w | – |
| l-Fucose | – | + |
| l-Rhamnose | – | w |
| Arabinose | – | – |
| Pyruvate | – | w |
| Tartarate | + | – |
+ positive, w weak positive, − negative
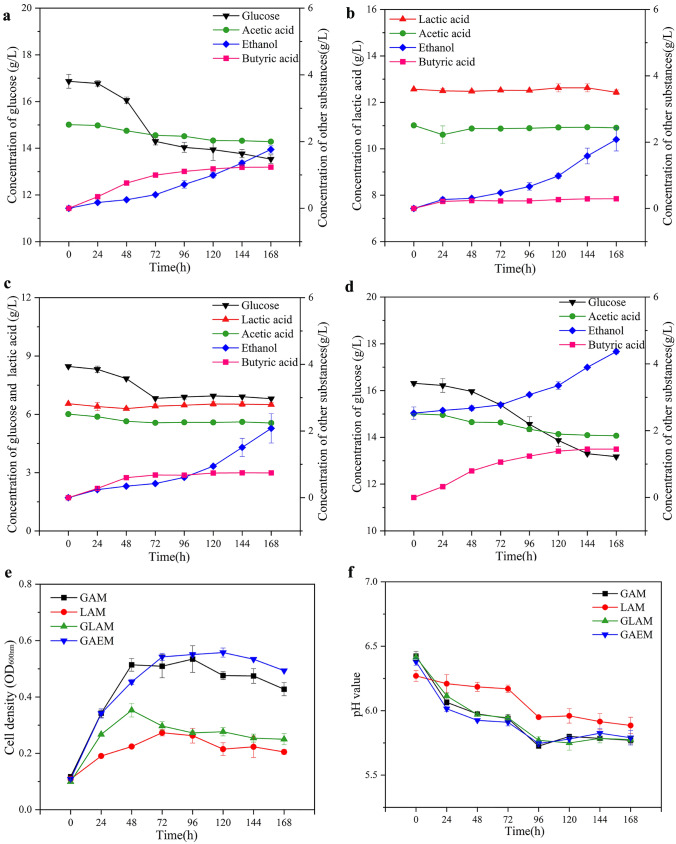
The fermentation characteristics of the strain BJN0003 were investigated with different media under anaerobic conditions. This work focused on investigating the metabolic characteristics of microorganisms under specific condition with the selected nutrients. In the Baijiu fermentation system, glucose was the main carbon source used by microorganisms. In addition, acetic acid and lactic acid were the main components in yellow water, which participated in the growth and metabolism of microorganisms in the cellar mud during the fermentation process. Therefore, glucose, acetic acid, lactic acid and ethanol were used in this study as the main carbon sources. The results showed that under GAM culture conditions, the strain could produce butyric acid and ethanol using glucose and trace amount of acetic acid as the substrates (Fig. 4a), while with LAM culture conditions, the strain consumed slight amount of lactic acid and acetic acid, and produced trace amount of butyric acid and ethanol (Fig. 4b). At the end of fermentation, butyric acid produced by BJN0003 in GAM and LAM was 1.23 ± 0.01 g/L and 0.29 ± 0.02 g/L (p < 0.05), and ethanol was 1.76 ± 0.11 g/L and 2.08 ± 0.35 g/L, indicating that the metabolic synthesis of butyric acid by the strain were different under varied carbon sources. The strain BJN0003 preferred to use glucose rather than lactic acid (Fig. 4a and b). When cultured under GLAM conditions, the strain also used glucose and trace amount of acetic acid to produce butyric acid and ethanol (Fig. 4c). After 72 h of fermentation, the glucose consumption by BJN0003 reached the highest level, and remained almost unchanged during the followed fermentation. Meanwhile, the glucose consumption under GLAM culture conditions (1.34 ± 0.02 g/L) was significantly lower than that in the GAM culture conditions (2.98 ± 0.03 g/L), and the butyric acid production was relatively low (0.74 ± 0.02 g/L VS 1.23 ± 0.01 g/L, p < 0.05) (Fig. 4c). This indicated the inhibition of lactic acid on the cell growth of BJN0003 (Fig. 4e), so that reduced glucose consumption and generated less butyric acid. The pH values in the fermentation process were analyzed and the tendency of pH values of the culture media were similar, which might be caused by the different butyric acid production in the fermentation process, except for the pH values of LAM (Fig. 4f). GAEM was modified of GAM by adding ethanol in the medium, and about 1.84 ± 0.10 g/L ethanol was produced at the end of fermentation (Fig. 4d), similar to the ethanol production under other culture media, and indicated that the ethanol production of this strain showed no mutual coupling with strain growth or consumption of substrate glucose. However, cell density of BJN0003 was low under the analyzed conditions, the reason might be due to the specific characteristics of the strain, and the phenomenon was also observed in other anaerobic strains with relatively low cell density (Marounek et al. 1989; Weimer and Stevenson 2012; Liu et al. 2020).
Fig. 4.
Substrate consumption and chemical production during fermentation of strain BJN0003 under glucose-acetate medium (GAM), lactate-acetate medium (LAM), glucose-lactate-acetate medium (GLAM) and glucose-acetate-ethanol medium (GAEM). a GAM, b LAM, c GLAM, d GAEM, e cell density at 600 nm, f pH value of media
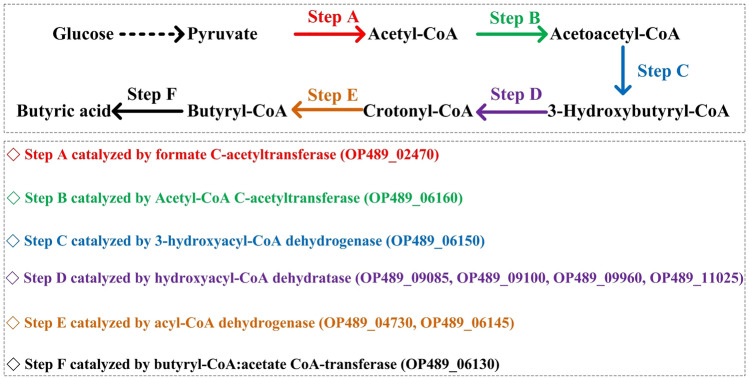
Based on the gene annotation and metabolic analysis of strain BJN0003, the metabolic pathway of butyric acid synthesis was proposed with glucose as the substrate (Fig. 5). The metabolic intermediate pyruvate, produced by glycolysis, was converted to acetyl-CoA through the catalysis of formate C-acetyltransferase (EC 2.3.1.54), then acetoacetyl-CoA was produced under the catalysis of acetyl-CoA acetyltransferase (EC 2.3.1.9), and thereafter 3-hydroxybutyryl-CoA was produced by 3-hydroxybutyryl CoA dehydrogenase (EC 1.1.1.157), followed by acetyl-CoA hydratase (EC 4.2.1.17) to catalyze the synthesis of crotonyl-CoA, and finally butyryl-CoA dehydrogenase (EC 1.3.2.1) and butyryl CoA: acetate CoA transferase (EC 2.8.3.8) catalyzed crotonyl-CoA step by step to produce butyric acid. The results of fermentation and genome comparison of strain BJN0003 showed significantly difference with that of the most closely related type species Caproicibacterium lactiferaments JNU-WLY1368T, which was a novel species belonged to a new genus of the family Oscillospiraceae (Gu et al. 2021). Subsequently, the biochemical and chemotaxonomic analysis of the strain can be performed to further identify the taxonomic status of the strain BJN0003.
Fig. 5.
The proposed metabolic pathway for butyric acid production by strain BJN0003. Numbers in brackets are GenBank accession numbers of the annotated functional enzymes of strain BJN0003 according to the whole genome sequence
Conclusions
In this study, a potential new member of the family Oscillospiraceae was isolated from Baijiu cellar mud sample. 16S rRNA sequence alignments of BJN0003 with the closely related type species indicated that it was a potential novel species of a new genus of the family Oscillospiraceae. The ANI and DDH values of BJN0003 with the closely related type species were much lower than the threshold values for species discrimination according to genome sequence. Furthermore, the strain could use glucose as the carbon source when cultured under different fermentation media to produce butyric acid, and the metabolic pathway was proposed together with the information of genome annotation. In all, the identification of strain Butyriproducens baijiuensis BJN0003 provided a new resource for investigating acid-producing microorganisms and the metabolic characteristics enriched the understanding of pit mud microorganisms from Baijiu.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
MW: methodology, investigation, and data curation. YX: methodology, investigation, data curation, writing—original draft, funding acquisition, and supervision. MD: data curation and methodology. WL: methodology and investigation. CZ: data curation and methodology. XL: review and editing, funding acquisition, and supervision. BS: review and editing, and supervision.
Funding
This work was supported by National Natural Science Foundation of China (No. 32072165, 31830069), Beijing Municipal Natural Science Foundation & Beijing Municipal Education Commission (No. KM202110011003, KZ202010011018).
Data availability
The genome sequence was submitted to National Center for Biotechnology Information with GenBank accession No. CP111108.1. The BioProject accession number was PRJNA899669, and BioSample accession number was SAMN31666078.
Declarations
Conflict of interest
All authors have read the submitted version of the manuscript. The authors have no conflict of interest in relation to this work.
Research involving human/animal participants
There are no human or animal subjects in this work.
Consent for publication
This work has not been published in whole or in part elsewhere. The manuscript is not currently being considered for publication in another journal. All authors have been personally and actively involved in substantive work leading to the manuscript, and will hold themselves jointly and individually responsible for its content.
Footnotes
Mengqin Wu and Youqiang Xu have contributed equally to this work.
Contributor Information
Youqiang Xu, Email: xuyouqiang@btbu.edu.cn.
Xiuting Li, Email: Lixt@btbu.edu.cn.
References
- Afouda P, Dubourg G, Nguyen TT, Raoult D, Fournier P-E. Pseudoruminococcus massiliensis gen. nov., sp. nov., a new bacterium isolated from the human gut microbiota. New Microbes New Infect. 2020;34:100645. doi: 10.1016/j.nmni.2019.100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alain K, Querellou J. Cultivating the uncultured: limits, advances and future challenges. Extremophiles. 2009;13(4):583–594. doi: 10.1007/s00792-009-0261-3. [DOI] [PubMed] [Google Scholar]
- Bessis S, Ndongo S, Lagier J-C, Raoult D, Fournier P-E. ‘Neglecta timonensis’ gen. nov., sp. nov., a new human-associated species. New Microbes New Infect. 2016;13:13–14. doi: 10.1016/j.nmni.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin AV, Sokolova SR, Shcherbakova VA, Suzina NE, Kochetkova TO, Goltsov AY, Trofimov DY, Efimov BA. Hydrogeniiclostidium mannosilyticum gen. nov., sp. nov. isolated from human faeces. Int J Syst Evol Microbiol. 2020;70(2):1210–1216. doi: 10.1099/ijsem.0.003900. [DOI] [PubMed] [Google Scholar]
- Elsden SR, Hilton MG. Volatile acid production from threonine, valine, leucine and isoleucine by clostridia. Arch Microbiol. 1978;117(2):165–172. doi: 10.1007/bf00402304. [DOI] [PubMed] [Google Scholar]
- Feng GD, Chen MB, Zhang XJ, Wang DD, Zhu HH. Whole genome sequences reveal the presence of 11 heterotypic synonyms in the genus Sphingobium and emended descriptions of Sphingobium indicum, Sphingobium fuliginis, Sphingobium xenophagum and Sphingobium cupriresistens. Int J Syst Evol Microbiol. 2019;69(7):2161–2165. doi: 10.1099/ijsem.0.003432. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Kristensen DM, Makarova KS, Wolf YI, Koonin EV. Microbial genome analysis: the COG approach. Brief Bioinform. 2019;20(4):1063–1070. doi: 10.1093/bib/bbx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Zhu XJ, Lin F, Shen CH, Li Y, Ao L, Fan WL, Ren C, Xu Y. Caproicibacterium amylolyticum gen. nov., sp. nov., a novel member of the family Oscillospiraceae isolated from pit clay used for making Chinese strong aroma-type liquor. Int J Syst Evol Microbiol. 2021 doi: 10.1099/ijsem.0.004789. [DOI] [PubMed] [Google Scholar]
- Guilhot E, Alou MT, Diallo A, Raoult D, Khelaifia S. Anaeromassilibacillus senegalensis gen. nov., sp. nov., isolated from the gut of a child with kwashiorkor. New Microbes New Infect. 2016;12:59–60. doi: 10.1016/j.nmni.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JX, Zhao DR, Sun BG. Research progress on the profile of trace components in Baijiu. Food Rev Int. 2021;137:109695. doi: 10.1080/87559129.2021.1936001. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kim BC, Seung Jeon B, Kim S, Kim H, Um Y, Sang BI. Caproiciproducens galactitolivorans gen. nov., sp. nov., a bacterium capable of producing caproic acid from galactitol, isolated from a wastewater treatment plant. Int J Syst Evol Microbiol. 2015;65(12):4902–4908. doi: 10.1099/ijsem.0.000665. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Sun BG. Effect of fermentation processing on the flavor of Baijiu. J Agric Food Chem. 2018;66(22):5425–5432. doi: 10.1021/acs.jafc.8b00692. [DOI] [PubMed] [Google Scholar]
- Liu B, Popp D, Müller N, Sträuber H, Harms H, Kleinsteuber S. Three novel Clostridia isolates produce n-caproate and iso-butyrate from lactate: comparative genomics of chain-elongating bacteria. Microorganisms. 2020;8(12):1970. doi: 10.3390/microorganisms8121970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marounek M, Fliegrova K, Bartos S. Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl Environ Microbiol. 1989;55(6):1570–1573. doi: 10.1128/AEM.55.6.1570-1573.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YW, Zhang J, Xiao ZB, Zhu JC. Evaluation of the perceptual interactions between higher alcohols and off-odor acids in Laimao Baijiu by sigma-tau plot and partition coefficient. J Agric Food Chem. 2020;68(50):14938–14949. doi: 10.1021/acs.jafc.0c05676. [DOI] [PubMed] [Google Scholar]
- Rhoads A, Au KF. PacBio Sequencing and its applications. Genom Proteom Bioinform. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WH, Xu YQ, Huang HQ, Pang ZM, Fu ZL, Niu JL, Zhang CN, Li WW, Li XT, Sun BG. Correlation between microbial communities and flavor compounds during the fifth and sixth rounds of sauce-flavor baijiu fermentation. Food Res Int. 2021;150:110741. doi: 10.1016/j.foodres.2021.110741. [DOI] [PubMed] [Google Scholar]
- Wang HL, Gu Y, Zhao D, Qiao ZW, Zheng J, Gao JJ, Ren C, Xu Y. Caproicibacterium lactatifermentans sp. nov., isolated from pit clay used for the production of Chinese strong aroma-type liquor. Int J Syst Evol Microbiol. 2022 doi: 10.1099/ijsem.0.005206. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zou W, Shen CH, Yang JG. Basic flavor types and component characteristics of Chinese traditional liquors: a review. J Food Sci. 2020;85(12):4096–4107. doi: 10.1111/1750-3841.15536. [DOI] [PubMed] [Google Scholar]
- Weimer PJ, Stevenson DM. Isolation, characterization, and quantification of Clostridium kluyveri from the bovine rumen. Appl Microbiol Biotechnol. 2012;94:461–466. doi: 10.1007/s00253-011-3751-z. [DOI] [PubMed] [Google Scholar]
- Wu YS, Hou YX, Chen H, Wang JS, Zhang CS, Zhao ZG, Ao R, Huang H, Hong JX, Zhao DR, Sun BG. "Key factor" for Baijiu quality: research progress on acid substances in Baijiu. Foods. 2022;11(19):2959. doi: 10.3390/foods11192959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YQ, Minhazul K, Wang XC, Liu X, Li XT, Meng Q, Li HH, Zhang CN, Sun XT, Sun BG. Biodegradation of phthalate esters by Paracoccus kondratievae BJQ0001 isolated from Jiuqu (Baijiu fermentation starter) and identification of the ester bond hydrolysis enzyme. Environ Pollut. 2020;263(Pt B):114506. doi: 10.1016/j.envpol.2020.114506. [DOI] [PubMed] [Google Scholar]
- Xu YQ, Zhu Y, Li XT, Sun BG. Dynamic balancing of intestinal short-chain fatty acids: the crucial role of bacterial metabolism. Trends Food Sci Technol. 2020;100:118–130. doi: 10.1016/j.tifs.2020.02.026. [DOI] [Google Scholar]
- Xu YQ, Wu MQ, Niu JL, Huang HQ, Nie Z, Fu ZL, Zhang CS, Zhao ZG, Lu HY, Li XT, Sun BG. Clostridium btbubcensis BJN0001, a potentially new species isolated from the cellar mud of Chinese strong-flavor baijiu, produces ethanol, acetic acid and butyric acid from glucose. 3 Biotech. 2022;12(9):203. doi: 10.1007/s13205-022-03271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YQ, Zhao JR, Liu X, Zhang CS, Zhao ZG, Li XT, Sun BG. Flavor mystery of Chinese traditional fermented baijiu: the great contribution of ester compounds. Food Chem. 2022;369:130920. doi: 10.1016/j.foodchem.2021.130920. [DOI] [PubMed] [Google Scholar]
- Xu YQ, Wu MQ, Zhao D, Zheng J, Mq D, Xt Li, Ww Li, Cn Z, Bg S. Simulated fermentation of strong-flavor baijiu through functional microbial combination to realize the stable synthesis of important flavor chemicals. Foods. 2023;12(3):644. doi: 10.3390/foods12030644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SB, Dong D. Improvement of caproic acid production in a Clostridium kluyveri H068 and Methanogen 166 co-culture fermentation system. AMB Express. 2018;8:175. doi: 10.1186/s13568-018-0705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12(9):635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- Yin Q, Tao Y, Zhu XY, Zhou Y, He XH, Cheng L, Huang Y, Li DP. Clostridium liquoris sp nov., isolated from a fermentation pit used for the production of Chinese strong-flavoured liquor. Int J Syst Evol Microbiol. 2016;66:749–754. doi: 10.1099/ijsem.0.000787. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek Int J Gen Mol Microbiol. 2017;110(10):1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu SP, Sun HL, Jiang ZF, Xu YZ, Mao JQ, Qian B, Wang L, Mao J. Metagenomics-based insights into the microbial community profiling and flavor development potentiality of Baijiu Daqu and Huangjiu wheat Qu. Food Res Int. 2022;152:110707. doi: 10.1016/j.foodres.2021.110707. [DOI] [PubMed] [Google Scholar]
- Zhu XY, Tao Y, Liang C, Li XZ, Wei N, Zhang WJ, Zhou Y, Yang YF, Bo T. The synthesis of n-caproate from lactate: a new efficient process for medium-chain carboxylates production. Sci Rep. 2015;5:14360. doi: 10.1038/srep14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence was submitted to National Center for Biotechnology Information with GenBank accession No. CP111108.1. The BioProject accession number was PRJNA899669, and BioSample accession number was SAMN31666078.