Abstract
The Arc two-component signal transduction system of Escherichia coli regulates the expression of numerous operons in response to respiratory growth conditions. Cellular redox state or proton motive force (Δμ̄H+) has been proposed to be the signal for the membrane-associated ArcB sensor kinase. This study provided evidence for a short ArcB periplasmic bridge that contains a His47. The dispensability of this amino acid, the only amino acid with a pK in the physiological range, renders the Δμ̄H+ model unlikely. Furthermore, results from substituting membrane segments of ArcB with counterparts of MalF indicate that the region does not play a stereospecific role in signal reception.
The Arc two-component signal transduction system of Escherichia coli regulates the expression of more than 30 operons, depending on the redox conditions of growth (20, 22, 30, 31). The system consists of ArcB, the membrane-bound sensor kinase, and ArcA, the cognate response regulator. ArcB (17, 23, 27, 48) (see Fig. 1A) belongs to the tripartite sensor kinase subfamily (1, 16, 25, 35, 37, 44, 49), is attached to the cytoplasmic membrane by two transmembrane segments (TM1 and TM2) near the N-terminal end (24), and catalyzes a phosphorelay via His292, Asp576, and His717 of ArcB to Asp54 of ArcA (14). The autophosphorylation step is stimulated by effectors, such as d-lactate, pyruvate, and acetate. These metabolites accumulate when exogenous electron acceptors limit respiration during growth (13, 18). Dephosphorylation of ArcA-P occurs by a reverse phosphorelay from the Asp54 of ArcA to the His717 and Asp576 of ArcB. The phosphoryl group is then released as Pi (12).
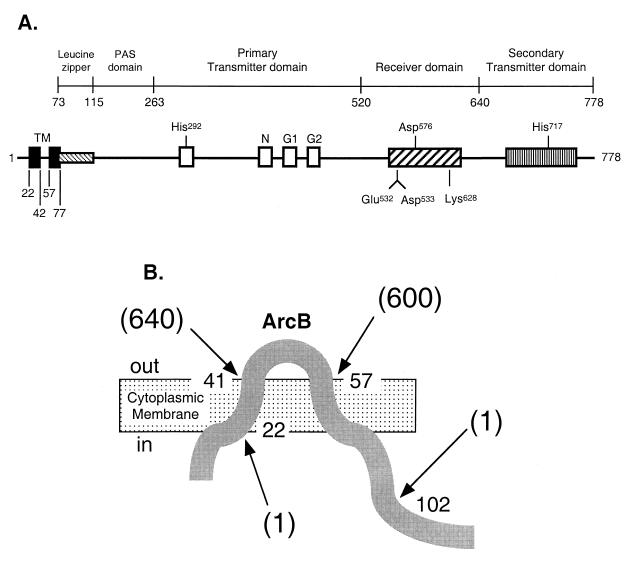
FIG. 1.
The ArcB sensor kinase and its transmembrane topology. (A) Schematic representation of ArcB. The putative leucine zipper (12) and the PAS domain (46) are based on amino acid sequence homology. The primary transmitter domain contains the conserved His292 and the catalytic determinants N, G1, and G2. The G1 and G2 sequences typify nucleotide-binding motifs. The receiver domain contains the conserved Asp576, and the secondary transmitter domain contains the conserved His717. (B) Schematic representation of ArcB transmembrane topology. Arrows point to ArcB-PhoA fusions constructed according to a two-step PCR procedure (41). The first PCR amplifications were performed, using plasmid pBB25 (23) as a template and BPH-N and either BPH-22, BPH-41, BPH-57, or BPH-102 as primers (Table 2). Each purified product was used as a primer with BPH-C for the second PCR, using the plasmid pDHB5747 that bears phoA (D. Boyd, unpublished data) as a template. The second PCR products were digested with BamHI and HindIII and cloned between the corresponding sites of vector pUC18, resulting in pBP22 (Φ[arcB1-22-′phoA]), pBP41 (Φ[arcB1-41-′phoA]), pBP57 (Φ[arcB1-57-′phoA]), and pBP102 (Φ[arcB1-102-′phoA]). Each Φ(arcB′-′phoA) plasmid was transformed into strain DHB4 (ΔphoA) and assayed for alkaline phosphatase activity as described previously (6). The alkaline phosphatase activity units are represented by numbers in parentheses and are averages of four experiments, with standard deviations of less than 10%.
Most sensor kinases receive their signal from the periplasmic domain, resulting in conformation changes that trigger autophosphorylation. As a sensor kinase, ArcB is unusual in having a short putative periplasmic bridge (24, 29). Relatively little is known about the nature of the signal for ArcB and what role the membrane-associated region plays in signal reception, except that autophosphorylation seems to be activated by excessive reducing equivalents (7, 11, 21, 22, 24, 38). Two studies involving growth of cells at high pH and treatment of cells by protonophores during growth, however, led to the suggestion that ArcB kinase is activated by a decrease in proton motive force (PMF) across the cytoplasmic membrane (2, 5). Here, we confirm the transmembrane topology of ArcB by genetic analysis and probe the function of the membrane region by replacing the chromosomal arcB+ by a single copy of a mutant allele (Fig. 2; Tables 1 and 2).
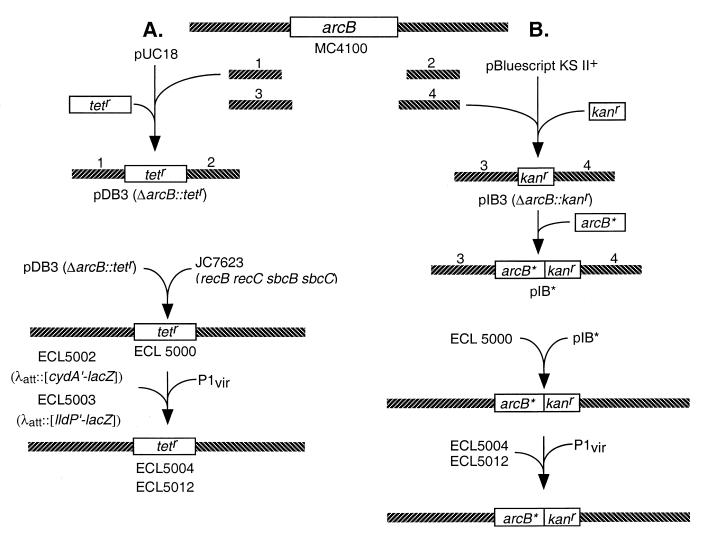
FIG. 2.
Construction of strains. (A) To construct the ΔarcB::Tetr strain, the 5′- and 3′-flanking DNA fragments of arcB (fragments 1 and 2) were prepared by PCR, using chromosomal DNA from strain MC4100 as template and, respectively, the primer pairs DAB-5N/DAB-5C and DAB-3N/DAB-3C (Table 2). The products were cloned into pUC18. A Tetr cassette isolated from pNK81 (50) was then inserted between the two arcB-flanking fragments, generating pDB3. This plasmid was transformed into strain JC7623 (36) to create ΔarcB::Tetr strain (ECL5000) by homologous recombination. The ΔarcB::Tetr allele was then P1 transduced into strain ECL5002 or ECL5003, respectively, resulting in ECL5004 and ECL5012. (B) To introduce the modified arcB sequence into the ΔarcB::Tetr strain, the 5′- and 3′-flanking DNA fragments of arcB (fragments 3 and 4) were prepared by PCR, using chromosomal DNA from strain MC4100 as template and, respectively, the primer pairs IAB-5N/IAB-5C and IAB-3N/IAB-3C (Table 2). The products were cloned into pBluescript KS II(+). A Kanr cassette, isolated from pUC4-KIXX (3), was then inserted between the two arcB-flanking fragments, generating pIB3. The 5′-flanking fragment includes the arcB promoter, ribosome-binding site, and introduced NdeI site which included the initiation codon of arcB followed by a HindIII site. Modified arcB sequences (arcB∗) were cloned into the pIB3 between the NdeI site and HindIII site, generating pIB∗. This plasmid was transformed into strain ECL5000 to replace ΔarcB::Tetr allele with arcB∗::Kanr by homologous recombination. Recombinants were selected by the Tets Kanr Amps phenotype and confirmed by the PCR. The arcB∗::Kanr was then P1 transduced into strains ECL5004 or ECL5012. Not illustrated is the construction of the reporter fusions. To construct the Φ(cydA′-lacZ) operon fusion, a 1.0-kb BamHI-EcoRI fragment of plasmid pBTKScyd1 (31) was ligated into BamHI-EcoRI-digested lacZ operon fusion vector pRS528 (42), resulting in pCAZ1. To generate the Φ(lldP′-lacZ) operon fusion, a 3.6-kb PstI-BamHI fragment of plasmid pLCT2 (9) was subcloned into pBluescript SK(−) (Stratagene), resulting in pLLD2. A 0.8-kb EcoRI-BglII fragment of pLLD2 was then ligated into the EcoRI-BamHI-digested lacZ operon fusion vector pRS415 (42), resulting in pLPZ1. The Φ(cydA′-lacZ) and Φ(lldP′-lacZ) were then transferred to the λ transducing phage λRS45 (42), yielding, respectively, λCAZ1 and λLPZ1. Lysates with high titers of λCAZ1 and λLPZ1 were used to lysogenize strain MC4100, and single lysogens were selected (28), yielding, respectively, strains ECL5001 and ECL5003. The Δfnr::Tn9 (Cmr) allele of strain JRG1728 (43) was P1 transduced into strain ECL5001 (yielding strain ECL5002) in order to avoid the transcriptional repression of Φ(cydA′-lacZ) by Fnr (8).
TABLE 1.
E. coli K-12 strains, bacteriophages, and plasmids used in this studya
| Strain, phage, or plasmid | Relevant characteristic(s) or genotype | Reference or source |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC ptsF25 rbsR | 42 |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ traD36 proAB+ lacIqlacZΔM15 | Promega |
| JRG1728 | Δfnr::Tn9(Cmr) | 43 |
| DHB4 | araD139 Δ(ara-leu)7697 ΔlacX74 ΔphoA(PvuII) phoRΔmalF3 galE galK thi rpsL/F′ lacIqpro | 6 |
| JC7623 | recB21 recC22 sbcB15 sbcC201 | 36 |
| ECL5000 | JC7623 but ΔarcB::Tetr | This study |
| ECL5001 | MC4100 but Φ(cydA′-lacZ) | This study |
| ECL5002 | MC4100 but Φ(lldP′-lacZ) | This study |
| ECL5003 | MC4100 but Δfnr::Tn9(Cmr) Φ(cydA′-lacZ) | This study |
| ECL5004 | ΔarcB::Tetr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5005 | ΔarcB::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5006 | arcB+::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5007 | arcBH47Q::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5008 | arcBH47R::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5009 | Φ(arcB1-22-malF17-35-arcB42-778)::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5010 | Φ(arcB1-57-malF40-58-arcB78-778)::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5011 | Φ(arcB1-22-malF17-39-arcB58-778)::Kanr Φ(cydA′-lacZ) Δfnr::Tn9(Cmr) | This study |
| ECL5012 | ΔarcB::Tetr Φ(lldP′-lacZ) | This study |
| ECL5013 | ΔarcB::Kanr Φ(lldP′-lacZ) | This study |
| ECL5014 | arcB+::Kanr Φ(lldP′-lacZ) | This study |
| ECL5015 | arcBH47Q::Kanr Φ(lldP′-lacZ) | This study |
| ECL5016 | arcBH47R::Kanr Φ(lldP′-lacZ) | This study |
| ECL5017 | Φ(arcB1-22-malF17-35-arcB42-778)::Kanr Φ(lldP′-lacZ) | This study |
| ECL5018 | Φ(arcB1-57-malF40-58-arcB78-778)::Kanr Φ(lldP′-lacZ) | This study |
| ECL5019 | Φ(arcB1-22-malF17-39-arcB58-778)::Kanr Φ(lldP′-lacZ) | This study |
| Phages | ||
| λRS45 | ′bla ′lacZ lacY+ | 42 |
| λLPZ1 | Φ(lldP′-lacZ+) lacY+ | This study |
| λCAZ1 | Φ(cydA′-lacZ+) lacY+ | This study |
| P1vir | Laboratory stock | |
| Plasmids | ||
| pUC18 | Cloning vector | Stratagene |
| pBluescript SK(−) | Cloning vector | Strategene |
| pBluescript KS II(+) | Cloning vector | Stratagene |
| pNK81 | Tetr | 50 |
| pUC4-KIXX | Kanr | 3 |
| pBB25 | arcB+ | 23 |
| pDHB5747 | ′phoA | D. Boyd |
| pDHB32 | malF+ | 6 |
| pBP22 | Φ(arcB1-22-′phoA) | This study |
| pBP41 | Φ(arcB1-41-′phoA) | This study |
| pBP57 | Φ(arcB1-57-′phoA) | This study |
| pBP102 | Φ(arcB1-102-′phoA) | This study |
| pLCT2 | lldPRD+ | 9 |
| pLLD2 | lldPR+ | This study |
| pRS415 | lacZ+ lacY+ bla+ | 42 |
| pLPZ1 | Φ(lldP′-lacZ) | This study |
| pBTKScyd1 | cydA′ | 31 |
| pRS528 | lacZ+ lacY+ bla+ | 42 |
| pCAZ1 | Φ(cydA′-lacZ) | This study |
| pDB3 | ΔarcB::Tetr | This study |
| pIB3 | ΔarcB::Kanr | This study |
| pQE30ArcB78-778 | His6-ArcB78-778 | 14 |
| pABW | arcB+ in pBluescript KS II(+) | This study |
| pABS | arcB78-778 in pBluescript KS II(+) | This study |
| pIBW | arcB+::Kanr in pIB3 | This study |
| pIBHQ | arcBH47Q::Kanr in pIB3 | This study |
| pIBHR | arcBH47R::Kanr in pIB3 | This study |
| pIBM1 | Φ(arcB1-22-malF17-35-arcB42-778)::Kanr in pIB3 | This study |
| pIBM2 | Φ(arcB1-57-malF40-58-arcB78-778)::Kanr in pIB3 | This study |
| pIBM3 | Φ(arcB1-22-malF17-39-arcB58-778)::Kanr in pIB3 | This study |
Luria-Bertani broth and agar (17 g/liter) were used for routine growth. When used, ampicillin, tetracycline, kanamycin, and chloramphenicol were provided at final concentrations of 50, 12, 40, and 20 μg/ml, respectively.
TABLE 2.
Oligonucleotides used in this studya
| Primer | Sequenceb |
|---|---|
| BPH-N | 5′-CCCGGATCCGGATGCGGTGCTGGATCTGC-3′ |
| BamHI | |
| BPH-22 | 5′-CGCTACTTGTGTATAAGAGTCCGGGCGCACCAGACCTAACTTCATC-3′ |
| BPH-41 | 5′-CGCTACTTGTGTATAAGAGTCCGGCGCCATTTGTACCACAATGG-3′ |
| BPH-57 | 5′-CGCTACTTGTGTATAAGAGTCCGGACGAATAACATCAATGCTTTCGACC-3′ |
| BPH-102 | 5′-CGCTACTTGTGTATAAGAGTCCGGCAAATCGCGCTCGCGCATCTCC-3′ |
| BPH-C | 5′-TCAGCAAGCTTGCGCCCGTGATCTGCC-3′ |
| HindIII | |
| DAB-3N | 5′-TCGGTCGACAGATCTCTGCGCCAACACCAGGG-3′ |
| SalI BglII | |
| DAB-3C | 5′-CCACTGCAGGTCGCCAAATTCGG-3′ |
| PstI | |
| DAB-5N | 5′-TGCGAGCTCCCTGCCTTGAACTG-3′ |
| SacI | |
| DAB-5C | 5′-GTCAGATCTCCCCTCAACGACCTACTCCG-3′ |
| BglII | |
| IAB-5N | 5′-AGCGATATCGAACTGACGACAAAACCAGC-3′ |
| EcoRV | |
| IAB-5C | 5′-AGCAAGCTTCATATGGGAATTCCTTCACGACAACC-3′ |
| HindIII NdeI | |
| IAB-3N | 5′-ACTGTCGACCCGGGGTGCGCGAATACTGC-3′ |
| SalI SmaI | |
| IAB-3C | 5′-TCACTCGAGGATCCCCAGCTACGCCCATCCC-3′ |
| XhoI BamHI | |
| B5NDE | 5′-CCCGGATCCCATATGAAGCAAATTCGTCTGCTGGCGC-3′ |
| BamHI NdeI | |
| B3NRU | 5′-GTAATGTCGCGACCAAAGCCCATCAAACCG-3′ |
| BH47Q | 5′-GCGGTAACCATGGTGCTGCAGGGTCAGGTCGAAAGCATTGATG-3′ |
| BH47R | 5′-ATGGCGGTAACCATGGTGCTCCGCGGTCAGGTCGAAAGCATTG-3′ |
| BMF-22 | 5′-CCGAGCAGACCTAGCACTGACCAGCGCACCAGACCTAACTTCATC-3′ |
| BMF-35 | 5′-GCAGCACCATGGTTACGTACATTAAAACAACAAGGTAACC-3′ |
| BMF-39 | 5′-CAGCAAACCAAAGAAGATAGATTCCCCTTGTGCGTACATTAAAAC-3′ |
| BMF-40 | 5′-GCGTGGTAATGGCGAACAGGTAACGAATAACATCAATGC-3′ |
| BMF-58 | 5′-GTCGTGACTCTCCAGTTGCTCGGCGAAAATATACAGCCCCGC-3′ |
Oligonucleotides were synthesized by either Oligos Etc. or Integrated DNA Technologies Inc. PCRs were carried out by using the TaqPlus Precision PCR system (Stratagene). Sequence verification of PCR-amplified DNA was performed at Micro Core Facility of the Department of Microbiology and Molecular Genetics, Harvard Medical School.
Restriction enzymes whose sites were introduced for subsequent cloning are indicated.
Transmembrane topology of ArcB.
To test the suggested topology based on hydrophobicity analysis, we constructed four phoA protein fusions (32) of arcB (Fig. 1B). The PhoA fusions at residues 22 and 102 of ArcB exhibited very low levels of alkaline phosphatase activity. In contrast, the PhoA fusions at residues 41 and 57 of ArcB showed very high levels of the enzyme activity (Fig. 1B). On the basis of this genetic analysis and a more recent algorithm for determining membrane-spanning regions (40), we suggest that a periplasmic bridge of ArcB is flanked by TM1 delimited by residues 23 to 41 and TM2 delimited by residues 58 to 77.
Testing the periplasmic His47 as a possible PMF sensor.
In order for ArcB to sense Δμ̄H+, at least one amino acid residue on each side of the plasma membrane with pK values within biological range would be required. The only periplasmic candidate would be His47. We therefore tested the phenotypes of His47Gln and His47Arg on the expression of positively controlled λ::Φ(cydA′-lacZ) or negatively controlled λ::Φ(lldP′-lacZ) (8, 19). Neither substitution resulted in any significant change in the expression of Φ(cydA′-lacZ) or Φ(lldP′-lacZ) (data not shown).
Testing the amino acid sequence of membrane regions.
To examine the function of various segments of the ArcB membrane region, we replaced them by a corresponding section of MalF (a subunit of maltose permease). MalF was chosen because its periplasmic bridge between the first and second transmembrane segments is also short (45) and because of the lack of any sequence homology with ArcB. In each hybrid construct, a portion of the ArcB N terminus was retained (Fig. 3 and 4). The reason for this measure is that when the cytosolic N-terminal segment of ArcB was replaced by the corresponding segment of MalF, the level of the hybrid protein diminished in the cell extract, as assayed by Western analysis (data not shown).
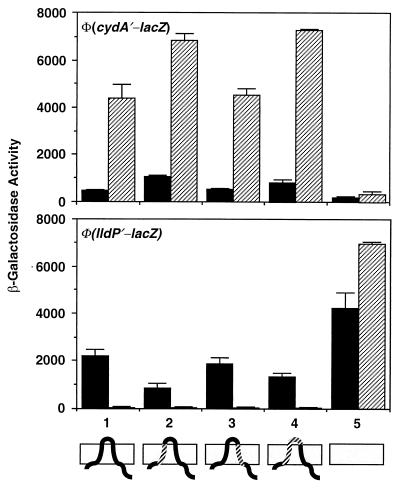
FIG. 3.
Effects of substituting segments of ArcB with MalF on the expressions of Φ(cydA′-lacZ) and Φ(lldP′-lacZ). To construct Φ(arcB1-22-malF17-35-arcB42-778), PCR was performed with pIBW as a template and BPH-N and BMF-22 as primers. The purified PCR product and BMF-35 were used as primers for the second PCR with pDHB32 (6) as a template. The PCR product and B3NRU were used as primers for a third PCR with the pABW as a template. The product was digested with NdeI and NruI and cloned between the corresponding sites of the pIBW, resulting in pIBM1. To construct Φ(arcB1-57-malF40-58- arcB78-778), PCR was performed with pIBW as a template and BPH-N and BMF-40 as primers. The purified PCR product and BMF-58 were used as primers for the second PCR with pDHB32 as a template. The PCR product and B3NRU were used as primers for a third PCR with the pABW as a template. The product was digested with NdeI and NruI and cloned between the corresponding sites of the pIBW, resulting in pIBM2. To construct a Φ(arcB1-22-malF17-39-arcB58-778), PCR was performed with pIBW as a template and BPH-N and BMF-22 as primers. The purified PCR product and BMF-39 were used as primers for the second PCR with pDHB32 as a template. The PCR product and B3NRU were used as primers for a third PCR with pABW as a template. The product was digested with NdeI and NruI and cloned between the corresponding sites of the pIBW, resulting in pIBM3. Plasmids pIBM1, pIBM2, and pIBM3 were used to integrate the modified arcB sequences into the chromosome of the reporter-bearing strains by the gene replacement techniques, as described in the legend of Fig. 2. For the β-galactosidase activity assay, the Φ(cydA′-lacZ)-bearing strains were cultured in buffered Luria-Bertani broth containing 0.1 M MOPS (morpholinepropanesulfonic acid) (pH 7.4) and 20 mM d-xylose. For the growth of Φ(lldP′-lacZ)-bearing strains, the above medium was supplemented with 20 mM l-lactate as an inducer (9). The data are averages of four experiments and the standard deviations are indicated. The different alleles of arcB are shown as follows: 1, arcB+; 2, Φ(arcB1-22-malF17-35-arcB42-778); 3, Φ(arcB1-57-malF40-58-arcB78-778); 4, Φ(arcB1-22-malF17-39-arcB58-778); 5, ΔarcB. The topology of the chimeric proteins is illustrated at the bottom: solid segments represent ArcB sequences, and hatched segments represent MalF sequences. Solid bars, aerobically grown cells; hatched bars, anaerobically grown cells.
FIG. 4.
Membrane association of the ArcB-MalF hybrid proteins. Cultures grown aerobically in Luria-Bertani broth were harvested during mid-exponential growth. The cells were washed with buffer S (50 mM Na-phosphate [pH 7.8], 300 mM NaCl, and 1 mM EDTA) by centrifugation. The cell pellet was resuspended in 3 ml of the same buffer and disrupted by sonication. Cell debris was removed by centrifugation for 10 min at 4,000 × g. The supernatant fluid was again centrifuged for 45 min at 35,000 × g to separate the cytosolic (C) and the membrane (M) fractions. The resultant supernatant fluid containing the soluble proteins was collected. The remaining pellet was resuspended in 0.5 ml of buffer M (20 mM HEPES [pH 7.5], 50 mM KCl, 1 mM EDTA, and 50% glycerol). Samples of cytosolic and membrane (containing 10 μg of protein) fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide gel) and the proteins were transferred to a Hybond-ECL filter (Amersham). The filter was equilibrated in TTBS buffer (25 mM Tris, 150 mM NaCl, and 0.05% Tween-20) for 10 min and incubated in blocking buffer (0.5% bovine serum albumin in TTBS) for 1 h at 37°C. ArcB polyclonal antibodies raised against His6-ArcB78-520 were added at a dilution of 1:10,000 to the filter and incubated for 1 h at room temperature. The bound antibody was detected by using anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase and the ECL detection system (Amersham). The topology of the chimeric proteins is depicted: solid segments represent ArcB sequences, and hatched segments represent MalF sequences.
When TM1 alone or TM1 plus the periplasmic bridge of ArcB was replaced by the counterparts of MalF, the expression of Φ(cydA′-lacZ) was increased under both aerobic and anaerobic growth conditions (Fig. 3). As to be expected, the expression of Φ(lldP′-lacZ) was partially repressed aerobically. However, because anaerobic repression of Φ(lldP′-lacZ) was already severe in the wild-type background, further repression by the chimeric ArcB proteins was not readily discernible. When TM2 was replaced, no significant changes in the expression of either reporter fusion were observed. When TM1, the periplasmic bridge, and TM2 were all replaced by the corresponding MalF region, the protein became inactive as an ArcA kinase (data not shown). However, the lack of kinase activity is difficult to interpret for the following reasons. First, there may be a failure in signal reception. Second, there may be a serious conformational distortion. Third, the protein may fail to dimerize, which is believed to be necessary for signal transmission. It might also be mentioned that when ArcB is liberated from membrane association by removal of the transmembrane domain, the truncated protein becomes constitutively active as an ArcA kinase (data not shown). This is to be expected, since purified ArcB78-778 has been shown to be highly active in vitro as an ArcA kinase and phosphatase (12, 14).
To ascertain that each ArcB-MalF hybrid protein remains membrane associated, we performed Western blot analysis on cytosolic and membrane fractions of the cells. In all cases, the hybrid proteins were found to be associated with the cytoplasmic membrane (Fig. 4).
Discussion and conclusion.
Most sensor kinases have a periplasmic domain of substantial size flanked by two TM segments for sensing signals (10, 15, 26, 33, 34, 37, 47). For many sensor kinases, however, the true signal and its input site on the protein remain unknown. According to the PMF sensing model by ArcB (2, 5), anaerobic growth diminishes the energy yield, thereby diminishing the Δμ̄H+, and activates the kinase. Results of His47 replacement experiments deprive this model of an obvious mechanism. It might be recalled that PMF was suggested as the signal primarily on the basis of protonophore effects on target gene expression. The validity of the conclusion, however, is compromised by the severe growth inhibition. Also, from a theoretical point of view, Δμ̄H+ seems not to be ideal as a signal, since its level is likely to be homeostatically controlled by the FoF1-ATPase. Moreover, even during aerobic growth, the energy source may become limiting. The resultant drop in PMF would repress the tricarboxylic acid cycle and the electron transport system in a situation when derepression would help to enhance the substrate-scavenging power of the starving cell.
The lack of evidence for the PMF model redirected our focus on the redox model and the possible functional importance of the membrane-associated portion of ArcB for signal reception. Three kinds of mechanisms may be envisaged. First, a redox-signaling element generated within the lipid bilayer may stereospecifically interact with a transmembrane or periplasmic segment of ArcB. In such a case, a drastic change in amino acid sequence should disrupt signal recognition. Second, the transmembrane region may simply serve as a mechanical anchor to ensure proximity of the rest of ArcB to the cytoplasmic membrane for signal reception. Third, one or both of the TM segments may play an entirely novel and unsuspected role in signal sensing. Results from the TM replacement experiments would favor the second or third model. An example of the second model is the Aer protein, which acts as a sensor for aerotaxis. In that case, anchorage of the protein to cytoplasmic membrane by the two TM segments is thought to allow the bound flavin adenine dinucleotide (FAD) to detect the redox state of the electron transport chains (4, 39). There is no evidence, however, that a cytosolic domain of ArcB binds to FAD. First, although everted vesicles containing ArcB catalyzed the phosphorylation of ArcA, the addition of FAD did not stimulate the reaction (18). Second, unlike the case of Aer (4), extracts of cells containing abundant ArcB (specified by a multicopy plasmid) did not exhibit a detectable absorption spectrum that is characteristic of flavins (O. Kwon, unpublished data). Eventual identification of the true signal and the characterization of its mode of reception will likely require a combined biochemical, physiological, and genetic approach and the development of rigorous in vitro assays. In the meantime, the results of our structural probing revealed an unexpected degree of robustness of apparent ArcB function to wholesale substitutions in the transmembrane region.
Acknowledgments
We thank Jon Beckwith, Peter De Wulf, and Jorge Membrillo-Hernández for helpful discussions.
This work was supported by U.S. Public Health Service Grant GM40993 from NIGMS of the National Institutes of Health.
REFERENCES
- 1.Arico B, Miller J F, Roy C, Stibitz S, Monack D, Falkow S, Gross R, Rappuoli R. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci USA. 1989;86:6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avetisyan A V, Bogachev A V, Murtasina R A, Skulachev V P. Involvement of a d-type oxidase in the Na+-motive respiratory chain of Escherichia coli growing under low Δμ̄H+ conditions. FEBS Lett. 1992;306:199–202. doi: 10.1016/0014-5793(92)80999-w. [DOI] [PubMed] [Google Scholar]
- 3.Barany F. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene. 1985;37:111–123. doi: 10.1016/0378-1119(85)90263-x. [DOI] [PubMed] [Google Scholar]
- 4.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogachev A V, Murtazina R A, Skulachev V P. Cytochrome d induction in Escherichia coli growing under unfavorable conditions. FEBS Lett. 1993;336:75–78. doi: 10.1016/0014-5793(93)81612-4. [DOI] [PubMed] [Google Scholar]
- 6.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter P A, Chepuri V, Gennis R B, Gunsalus R P. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J Bacteriol. 1990;172:6333–6338. doi: 10.1128/jb.172.11.6333-6338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter P A, Melville S B, Albrecht J A, Gunsalus R P. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 9.Dong J-M, Taylor J S, Latour D J, Iuchi S, Lin E C C. Three overlapping lct genes involved in L-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doty S L, Yu M C, Lundin J I, Heath J, Nester E W. Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens. J Bacteriol. 1996;178:961–970. doi: 10.1128/jb.178.4.961-970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu H-A, Iuchi S, Lin E C C. The requirement of ArcA and Fnr for peak expression of the cyd operon in Escherichia coli under microaerobic conditions. Mol Gen Genet. 1991;226:209–213. doi: 10.1007/BF00273605. [DOI] [PubMed] [Google Scholar]
- 12.Georgellis D, Kwon O, De Wulf P, Lin E C C. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 13.Georgellis D, Kwon O, Lin E C C. Amplification of signaling activity of the Arc two-component system of Escherichia coli by anaerobic metabolites: an in vitro study with different protein modules. J Biol Chem. 1999;274:35950–35954. doi: 10.1074/jbc.274.50.35950. [DOI] [PubMed] [Google Scholar]
- 14.Georgellis D, Lynch A S, Lin E C C. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 16.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishige K, Nagasawa S, Tokishita S-I, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iuchi S. Phosphorylation/dephosphorylation of the receiver module at the conserved aspartate residue controls transphosphorylation activity of histidine kinase in sensor protein ArcB of Escherichia coli. J Biol Chem. 1993;263:23972–23980. [PubMed] [Google Scholar]
- 19.Iuchi S, Aristarkhov A, Dong J-M, Taylor J S, Lin E C C. Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked L-lactate dehydrogenase. J Bacteriol. 1994;176:1695–1701. doi: 10.1128/jb.176.6.1695-1701.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iuchi S, Cameron D C, Lin E C C. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989;171:868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iuchi S, Chepuri V, Fu H-A, Gennis R B, Lin E C C. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol. 1990;172:6020–6025. doi: 10.1128/jb.172.10.6020-6025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iuchi S, Lin E C C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iuchi S, Lin E C C. Mutational analysis of signal transduction by ArcB: a membrane sensor protein for anaerobic expression of operons involved in the central aerobic pathways in Escherichia coli. J Bacteriol. 1992;174:3972–3980. doi: 10.1128/jb.174.12.3972-3980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iuchi S, Matsuda Z, Fujiwara T, Lin E C C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 25.Jourlin C, Ansaldi M, Méjean V. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS sensor in Escherichia coli. J Mol Biol. 1997;267:770–777. doi: 10.1006/jmbi.1997.0919. [DOI] [PubMed] [Google Scholar]
- 26.Kaspar S, Perozzo R, Reinelt S, Meyer M, Pfister K, Scapozza L, Bott M. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol Microbiol. 1999;33:858–872. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- 27.Kato M, Mizuno T, Shimizu T, Hakoshima T. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell. 1997;88:717–723. doi: 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- 28.Kwon O, Hudspeth M E S, Meganathan R. Anaerobic biosynthesis of enterobactin in Escherichia coli: regulation of entC gene expression and evidence against its involvement in menaquinone (vitamin K2) biosynthesis. J Bacteriol. 1996;178:3252–3259. doi: 10.1128/jb.178.11.3252-3259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;154:367–382. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Lynch A S, Lin E C C. Responses to molecular oxygen. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1526–1538. [Google Scholar]
- 31.Lynch A S, Lin E C C. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 33.Miller J F, Johnson S A, Black W J, Beattie D T, Mekalanos J J, Falkow S. Constitutive sensory transduction mutations in the Bordetella pertussis bvgS gene. J Bacteriol. 1992;174:970–979. doi: 10.1128/jb.174.3.970-979.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mowbray S L, Koshland J D E. Mutations in the aspartate receptor of Escherichia coli which affect aspartate binding. J Biol Chem. 1990;265:15638–15643. [PubMed] [Google Scholar]
- 35.Nagasawa S, Tokishita S, Aiba H, Mizuno T. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol. 1992;6:799–807. doi: 10.1111/j.1365-2958.1992.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 36.Oden K L, Deveaux L C, Vibat C R, Cronan J E, Jr, Gennis R B. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene. 1990;30:29–36. doi: 10.1016/0378-1119(90)90337-q. [DOI] [PubMed] [Google Scholar]
- 37.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 38.Reams S G, Lee N, Mat-Jan F, Clark D. Effect of chelating agents and respiratory inhibitors on regulation of the cadA gene in Escherichia coli. Arch Microbiol. 1997;167:209–216. doi: 10.1007/s002030050437. [DOI] [PubMed] [Google Scholar]
- 39.Rebbapragdala A, Johnson M S, Harding G P, Zuccarilli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 42.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 43.Spiro S, Guest J R. Activation of the lac operon of Escherichia coli by a mutant FNR protein. Mol Microbiol. 1987;1:53–58. doi: 10.1111/j.1365-2958.1987.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 44.Stevens A M, Sanders J M, Shoemaker N B, Salyers A A. Genes involved in production of plasmid-like forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J Bacteriol. 1992;174:2935–2942. doi: 10.1128/jb.174.9.2935-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapia M I, Mourez M, Hofnung M, Dassa E. Structure-function study of MalF protein by random mutagenesis. J Bacteriol. 1999;181:2267–2272. doi: 10.1128/jb.181.7.2267-2272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokishita S, Kojima A, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the periplasmic domain of the membrane-located protein kinase, EnvZ. J Biol Chem. 1991;266:6780–6785. [PubMed] [Google Scholar]
- 48.Tsuzuki M, Ishege K, Mizuno T. Phosphotransfer circuitry of the putative multi-signal transducer, ArcB, of Escherichia coli: in vitro studies with mutants. Mol Microbiol. 1995;18:953–962. doi: 10.1111/j.1365-2958.1995.18050953.x. [DOI] [PubMed] [Google Scholar]
- 49.Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene. 1994;140:73–77. doi: 10.1016/0378-1119(94)90733-1. [DOI] [PubMed] [Google Scholar]
- 50.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]