Summary
Background
Birt-Hogg-Dubé (BHD) syndrome, caused by germline alteration of folliculin (FLCN) gene, develops hybrid oncocytic/chromophobe tumour (HOCT) and chromophobe renal cell carcinoma (ChRCC), whereas sporadic ChRCC does not harbor FLCN alteration. To date, molecular characteristics of these similar histological types of tumours have been incompletely elucidated.
Methods
To elucidate renal tumourigenesis of BHD-associated renal tumours and sporadic renal tumours, we conducted whole genome sequencing (WGS) and RNA-sequencing (RNA-seq) of sixteen BHD-associated renal tumours from nine unrelated BHD patients, twenty-one sporadic ChRCCs and seven sporadic oncocytomas. We then compared somatic mutation profiles with FLCN variants and RNA expression profiles between BHD-associated renal tumours and sporadic renal tumours.
Findings
RNA-seq analysis revealed that BHD-associated renal tumours and sporadic renal tumours have totally different expression profiles. Sporadic ChRCCs were clustered into two distinct clusters characterized by L1CAM and FOXI1 expressions, molecular markers for renal tubule subclasses. Increased mitochondrial DNA (mtDNA) copy number with fewer variants was observed in BHD-associated renal tumours compared to sporadic ChRCCs. Cell-of-origin analysis using WGS data demonstrated that BHD-associated renal tumours and sporadic ChRCCs may arise from different cells of origin and second hit FLCN alterations may occur in early third decade of life in BHD patients.
Interpretation
These data further our understanding of renal tumourigenesis of these two different types of renal tumours with similar histology.
Funding
This study was supported by JSPS KAKENHI Grants, RIKEN internal grant, and the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research.
Keywords: Birt-Hogg-Dubé (BHD) syndrome, Chromophobe renal cell carcinoma (ChRCC), Folliculin (FLCN), Renal tumour predisposition syndrome
Research in context.
Evidence before this study
In our previous whole exome sequencing (WES) study, very few variants were commonly observed in BHD-associated renal tumour. Transcriptomic intra-tumour heterogeneity was observed in our single-cell RNA sequencing (scRNA-seq) study of BHD-associated renal tumour. To date, there is no study comparing BHD-associated renal tumourigenesis with renal tumourigenesis of sporadic ChRCC.
Added value of this study
Some molecular characteristics in common were observed between BHD-associated renal tumour and sporadic ChRCCs in bulk tissue or at single-cell level including heterogeneous molecular characteristics conferred by mutually exclusive expressions of L1CAM and FOXI1, distinguishing markers for principal cell and intercalated cell, respectively. BHD-associated renal tumours harbor mitochondrial DNA (mtDNA) with higher copy numbers and fewer variants compared to sporadic ChRCCs. BHD-associated renal tumours and sporadic ChRCCs may arise from different cells of origin and second hit FLCN alterations may occur in early third decade of life in BHD patients.
Implications of all the available evidence
Inter-tumour or intra-tumour heterogeneity in BHD-associated renal tumours and sporadic ChRCCs may be associated with the normal nephron developmental machinery. This study denotes that these two histologically similar renal tumours are different types of tumours arising from different cells of origin and distinct therapeutical approaches are needed for these tumours.
Introduction
Birt-Hogg-Dubé (BHD) syndrome is a renal tumour predisposition syndrome, in which affected individuals develop a variety of histological types of renal tumours, pulmonary cysts and cutaneous fibrofolliculomas.1, 2, 3, 4 The folliculin (FLCN) gene, which is responsible for BHD syndrome, is a metabolic gene which regulates oxidative phosphorylation, amino acid sensing and kidney cell growth rate in collaboration with its interacting partners, folliculin-interacting proteins 1 (FNIP1) and 2 (FNIP2).5, 6, 7, 8, 9
The most frequent histology of BHD-associated renal tumour is either hybrid oncocytic/chromophobe tumour (HOCT) or chromophobe renal cell carcinoma (ChRCC). Although some of the pathological observations including intratumour peripheral small papillary tufts or strong GPNMB staining in cytoplasm have been reported to distinguish BHD-associated renal tumour from sporadic ChRCC, renal tumourigenesis of these histologically similar types of tumours remains elusive.10,11 BHD-associated renal tumourigenesis is triggered by biallelic loss of the FLCN gene, whereas sporadic ChRCC frequently harbors tumour protein p53 (TP53) and/or phosphatase and tensin homolog (PTEN) alterations without FLCN alteration and in classical types generally has copy loss of chromosomes 1, 2, 6, 10, 13 and 17.1,12, 13, 14, 15, 16 However, these tumours share some molecular features including increased oxidative phosphorylation with upregulation of PPARG coactivator 1 alpha (PPARGC1a), an important co-activator for mitochondrial metabolism, as well as increased forkhead box I1 (FOXI1) expression, a transcription factor critical for collecting duct differentiation, suggesting that these shared molecular features may lead to similar histologies in these genetically different tumours.5,17,18
Renal tumours do not develop as a result of a single gene alteration; however, our previous whole exome sequencing (WES) study of twenty-nine BHD-associated renal tumours demonstrated that very few genes were commonly mutated in BHD-associated renal tumour except for FLCN alteration.16 The lack of commonly mutated genes in BHD-associated renal tumours in addition to FLCN implies that an underlying molecular mechanism may trigger BHD-associated renal tumourigenesis.
To elucidate renal tumourigenesis and compare molecular characteristics of BHD-associated renal tumours and sporadic ChRCC, we conducted whole genome sequencing (WGS) of BHD-associated renal tumours and RNA-sequencing (RNA-seq) of these tumours. Using WGS data, somatic variants, tumour mutation burden (TMB), structural variants (SV), copy number alterations (CNA), mutational signature matrix and mitochondrial genome (mtDNA) alterations were investigated. In addition, we analyzed the putative origin of tumour cells and mutation acquisition timing using WGS data.
Methods
Patients and tumour histology
Nine unrelated patients known to be affected with BHD syndrome were evaluated in the Department of Urology and Department of Molecular Pathology, Yokohama City University, Yokohama, Japan.16 Each BHD patient received genetic counseling, was confirmed to have a germline FLCN variant, and was evaluated for clinical manifestations of BHD syndrome with a dermatologic examination, computed tomography scan (CT scan) and magnetic resonance imaging (MRI) (Table 1). Pathological diagnosis was done prior to the molecular analysis by three independent pathologists. The BHD-associated renal tumours and adjacent normal kidney specimens were frozen with liquid nitrogen and stored in −80 °C freezer until analysis. Some of the samples have already been analyzed in our previous whole exome sequencing (WES) study.16 Sporadic ChRCCs and sporadic oncocytomas used in the RNA-sequencing (RNA-seq) study were also surgically removed in the Department of Urology, Yokohama City University. Clinical and histological information for these samples is available in Table 2 and Supplementary Dataset 1.
Table 1.
BHD patients and BHD-associated renal tumours analyzed in this study.
| Patient No. | Sex | Germline FLCN mutation | Mutation type | Sample No. | Age at surgery | Histology | Size (mm) | Stage | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F42 | female | c.1285dupC | Frameshift | F42-T1 | 49 | ChRCC | 30 | T1a |
| F42-T2 | 49 | ChRCC | 13 | T1a | |||||
| F42-T3 | 53 | ChRCC | 20 | T1a | |||||
| 2 | F43 | male | c.1285dupC | Frameshift | F43-T1 | 54 | ChRCC | 24 | T1a |
| 3 | F59 | male | c.1528_1530delGAG | Amino acid deletion | F59-T1 | 50 | ChRCC | 43 | T1b |
| F59-T2 | 50 | ChRCC | 14 | T1a | |||||
| F59-T8 | 50 | HOCT | 32 | T1a | |||||
| F59-T11 | 50 | ChRCC | 10 | T1a | |||||
| F59-T12 | 50 | ChRCC | 8 | T1a | |||||
| 4 | F107 | male | c.1347_1353dupCCACCCT | Frameshift | F107-T1 | 56 | Clear cell | 42 | T3b |
| 5 | F123 | female | c.1285dupC | Frameshift | F123-TR1 | 34 | HOCT | 27 | T1a |
| 6 | F124 | male | c.1285dupC | Frameshift | F124-T1 | 64 | HOCT | 26 | T1a |
| 7 | F133 | female | c.1285dupC | Frameshift | F133-T1 | 66 | Unclassified | 30 | T1a |
| F133-T2 | 66 | ChRCC | 10 | T1a | |||||
| 8 | F135 | female | c.1347_1353dupCCACCCT | Frameshift | F135-T1 | 40 | HOCT | 7 | T1a |
| 9 | F192 | female | c.404delC | Frameshift | F192-T1 | 54 | ChRCC | 15 | T1a |
Table 2.
Summary of renal tumours analyzed in this study.
| BHD-associated renal tumours | Sporadic ChRCCs | Sporadic oncocytomas | |
|---|---|---|---|
| Number of patients | |||
| Female | 5 | 11 | 3 |
| Male | 4 | 10 | 4 |
| Age (median) years of age | 34-66 (50) | 35-73 (61) | 62-77 (69) |
| Histology | |||
| ChRCC | 10 | 21 | – |
| HOCT | 4 | – | – |
| Clear cell RCC | 1 | – | – |
| Unclassified | 1 | – | – |
| Oncocytoma | 0 | – | 7 |
| Size (median) mm | 7-43 (22) | 10-110 (25) | 11-60 (18) |
| Stage | |||
| T1a | 14 | 16 | – |
| T1b | 1 | 4 | – |
| T3a | 0 | 1 | – |
| T3b | 1 | 0 | – |
Ethics
This study was approved by the Institutional Review Board of Yokohama City University (A200100004) and RIKEN (H20-11), and each patient has provided written informed consent for publication.
RNA sequencing (RNA-seq)
In this study, RNA-seq of twenty-one sporadic ChRCCs and seven oncocytomas was performed (Supplementary Dataset 1). Bulk RNA-seq of sixteen BHD-associated renal tumours and five adjacent normal kidneys from ten BHD patients has been carried out in our previous study.17 RNAs were extracted from frozen tissues using the RNA extraction reagent ISOGEN (Nippon-gene) following the manufacturer's protocol, qualified and quantified by Bioanalyzer (Agilent Technologies). Library preparation was done using TruSeq Stranded mRNA Library Prep Kit and TruSeq RNA CD Index Kit according to the manufacturer protocol (Illumina). Sequencing was performed with HiSeq 2500 system with 126-bp reads (Illumina). Using STAR (v2.7.9a), the FASTQ files were mapped to a reference sequence generated from the GRCh37 fasta and GENCODE Release 37 comprehensive gene annotation for reference chromosomes (gencode.v19.annotation.gtf). Aligned SAM files were sorted and duplicates were marked with picard MarkDuplicates. Mapped reads were counted by featureCounts. Based on the read count data, gene expression levels were calculated as fragments per kilobase of exon per million mapped reads (fpkm). For the series of analyses from FASTQ file data to gene expression level normalization, the riboduct pipeline was used (https://github.com/msfuji/riboduct), and the details of the commands and parameter settings are available in the GitHub page. Then, these read count data and normalized data were used for further expression analysis. Gene Set Enrichment Analysis (GSEA) was done using GSEA software (v.4.2.3) (https://www.gsea-msigdb.org/gsea/index.jsp); default parameters were applied.19 Plots of principal component analysis (PCA) and L1CAM and FOXI1 expressions were created using iDEP (integrated Differential Expression and Pathway analysis) (v.0.96) (http://bioinformatics.sdstate.edu/idep96/), a tool for RNA-seq analysis on a web browser.20 In the iDEP analyses, raw count data created from featureCounts was used, and default settings were used for each parameter.
Analysis of single-cell transcriptomes
Using the single-cell RNA-seq data from our previous work, we performed gene expression analysis in BHD-associated renal tumour cells.17 The R package Seurat (v3.1.2) was used for the analysis.21 Parameter settings for clustering were resolution 2.0 for the FindClusters function, dims 1:4 for the RunUMAP function, and the package default values for the other parameters. BHD-associated HOCT cells were clustered into L1CAM expressing cell cluster (cL1CAM) or FOXI1 expressing cell cluster (cFOXI1), and gene expressions in each cell cluster were investigated.
Whole genome sequencing (WGS)
WGS was performed on sixteen BHD-associated renal tumours and nine adjacent normal kidneys from nine BHD patients (Table 1 and Supplementary Dataset 1). Genomic DNA was extracted from frozen tissues using the QIAGEN DNA Mini Kit (QIAGEN) following the manufacturer's protocol, qualified and quantified by Bioanalyzer (Agilent Technologies). The library construction was done using the TruSeq Nano DNA Library Prep Kit (Illumina) following the manufacturer's protocol. Paired-end sequencing of 150-bp reads was performed using NovaSeq6000. Reads mapping and further analyses were done using Genomon2 pipeline (v.2.6.3) (https://github.com/Genomon/genomon). Briefly, sequence reads were mapped to the reference human genome GRCh37 using BWA-mem. GATK (The Genome Analysis Toolkit) was used to realign bam files. For the Genomon2 pipeline analysis, the default settings were used for each parameter, and a control panel of normal samples was used to eliminate germline mutations and errors. We obtained the BAM file data of 49 samples of sporadic ChRCC from the data resource of The Pan-Cancer Analysis of Whole Genomes (PCAWG) study (KICH).12 In tumour mutation burden (TMB) analysis and mutational signature analysis, for the comparison of BHD-associated renal tumours and KICH, the output data (VCF files) using the same method described above were used. In TMB analysis, SP123975 was excluded because of the exceptionally higher number of mutations (54,708 mutations) due to DNA mismatch repair deficiency.
Germline and somatic variant calling
Single-nucleotide variants (SNV), multi-nucleotide variants (MNV) and small insertion-deletions (Indels) were called using SAGE v2.8 (Somatic Alterations in Genome) from the Hartwig Medical Foundation (HMF: https://github.com/hartwigmedical/hmftools/releases/tag/sage-v2.8). Germline calling parameters were based on the "germline mode" in the SAGE documentation. The panel-of-normals (PON) VCFs used in the pipeline for annotation and filtering of somatic variants called by SAGE were created from 149 Japanese germline samples.22 Genome-wide somatic variant calls were performed with a quality cutoff of 70 and then filtered using bcftools with a quality cutoff of 70 for hotspots and 100 for other genomic locations. The maximum acceptable variant allele frequency (VAF) for the reference sample was set at 4% and the minimum VAF for the tumour was set at 2.5%. The output VCFs were filtered for PON variant sites, sites that were variant or likely fixed (MAF >0.01 and AF < 0.99; AF ≥ 0.99) in dbSNP or ALFA population samples.22 The resulting VCFs were annotated using ANNOVAR and SnpEff.
Analyses of gene and structural variants (SV), and copy number alterations (CNA)
Structural Variation (SV) was called using GRIDSS 2.12.0 (the Genomic Rearrangement IDentification Software Suite)23 with the BAM file for each sample as the input. Scripts were used from GRIDSS- PURPLE-LINX (GPL) pipeline (v.1.3.2) (https://github.com/hartwigmedical/gridss-purple-linx). Analysis of copy number alteration (CNA) was done using CNApp (https://tools.idibaps.org/CNApp/), a tool for the quantification of copy number alterations.24 TUMOUR.purple.cnv.somatic.tsv, output files from the PURPLE, were used as input files. In order to minimize the copy number noise, the optimal solution for ploidy and purity in the PURPLE was selected and the noise was removed. After this process, F43-T1, F59-T11 and F59-T12 were excluded because of the high copy number noise. In the CNApp analysis, the default settings were used for each parameter.
Mutational signature analysis
Analysis of somatic mutation signatures was performed using SigProfiler (v.3.3) (https://cancer.sanger.ac.uk/signatures/tools/) with the VCFs created from the Genomon pipeline. Somatic mutation signature profiles were decomposed to COSMIC 3.2 signatures.25 The default settings were used for each parameter in all of analyses.
Putative cell-of-origin (COO) estimation
COOBoostR is a xgboost-based machine learning algorithm which aims to predict putative tissue-of-origin (TOO) or cell-of-origin (COO) (https://github.com/SWJ9385/COOBoostR).26 In our study, COOBoostR was applied to predict putative COO of BHD-associated renal tumours and ChRCCs. Briefly, 1-megabase window-based somatic point mutation frequencies from WGS data were calculated for individual tumour samples along with processing single-cell ATAC-seq data from normal kidney tissue27 by calculating 1-megabase window sequencing count density per each cell type (prespecified inside the metadata). For the parameters, default values were used except for the eta and depth (set to 0.3 and 10).
Putative variant acquisition timing analysis
R package MutationTimeR (v.1.0.2) (https://github.com/gerstung-lab/MutationTimeR) was used to examine the timing of a second hit occurrence of the remaining FLCN allele in BHD-associated renal tumours.28 MutationTimeR is an R package used by the PCAWG consortium to calculate mutation times for 2778 whole genome sequence samples.28 VCFs of BHD-associated renal tumour were used and each parameter was set as default setting. A small “clusters” data.frame, which includes the estimated number of subclonal clusters, SNVs in each cluster and clonal proportions of each cluster, created from Clip pipeline (v.1.2.1) (CliP: Clonal structure identification through penalizing pairwise differences) (https://github.com/wwylab/CliP) (BioRxiv: https://doi.org/10.1101/2021.03.31.437383) was used as an input for MutationTimeR.
Mitochondrial DNA analysis
Mitochondrial genome (mtDNA) copy number per cell in tumour and normal tissues was calculated as follows. Germline SAGE variants were extracted, the mean depth of coverage for variant sites on autosomes (mean.DPauto) or on chrM (mean.DPchrM) was calculated, and the mtDNA copy number in each tissue was estimated using the formula.29
This formula scales as:
for purity (f: 0–1 for tumours, 1 for normal tissues) and ploidy (2 in normal tissues).
Also, variants in the mtDNA of BHD-associated renal tumours were analyzed using VarScan and Mutect2, from which variant unions were extracted and further filtered as described below. Filtering criteria were as follows. Position was not in (302–315, 513–525, 568–573, 956–965, 3105–3109, 5895–5899, 8270–8289, 16180–16195 (indel)). Variant reads were≥10, variant base quality score was≥20. Forward read/Total variant read ratio: >0.1 and <0.9. Mean read position: >0.15 and <0.85 (from 5′ and 3’ both). Reference read length - Variant read length was <25. Reference read mapping quality - Variant read mapping quality was <10. Reads with normal context were >80% of variant reads (except variant next to germline mutation). Variant NM - Ref NM was≤3. Remove Germline Polymorphism (germline list of SAMPLES and PCAWG (VAF >95%), dbSNP (Frequency >1%)). Calculate Noise Level in normal sample in SAMPLE and PCAWG - > remove VAF lower than noise level. Variants that passed the filter were labeled as somatic or tumour-specific dysplastic variants based on VAF differences observed in normal tissue. We analyzed mtDNA variants of sporadic ChRCCs in TCGA cohort in the same method as we stated above .12
Immunohistochemistry
Immunohistochemistry was done on 4-mm-thick formalin-fixed paraffin-embedded sections of five each of L1CAM expressing sporadic ChRCCs (gL1CAM) and FOXI expressing sporadic ChRCCs (gFOXI1). In immunohistochemistry of FOXI1 protein, the tissue slides were autoclaved in Tris–EDTA buffer (pH 6.0) for antigen retrieval, then incubated with goat polyclonal FOXI1 antibody at 1:400 dilution (#ab20454; Abcam, Cambridge, United Kingdom) for 1 h at room temperature and visualized with N-Histofine Simple Stain MAX PO(G) (NICHIREI BIOSCIENCE INC., Tokyo, Japan). The FOXI1 protein was considered to be positive when nucleic and/or cytoplasmic FOXI1 staining was observed. In immunohistochemistry of L1CAM protein, the tissue slides were autoclaved in Tris–EDTA buffer (pH 9.0) for antigen retrieval, then incubated with mouse monoclonal L1CAM antibody at 1:200 dilution (clone 2C2, #ab24345; Abcam) for 1 h at room temperature and visualized with the Real EnVision Detection System, Peroxidase/DAB+, Mouse (Agilent Technologies). The L1CAM protein was considered to be positive when membranous L1CAM staining was observed.
Statistics
Welch's two sample t-test was applied to determine whether the means of two populations were different, and differences were considered to be statistically significant at a value of P < 0.05. All statistical tests were 2-sided. For the analysis of the correlation between the two gene expressions, Pearson's correlation coefficient and p-value were calculated.
Role of the funding source
The funders played no role in study design, data collection, data analyses, interpretation and writing of the report.
Results
BHD-associated renal tumours display totally different expression profiles from sporadic ChRCCs
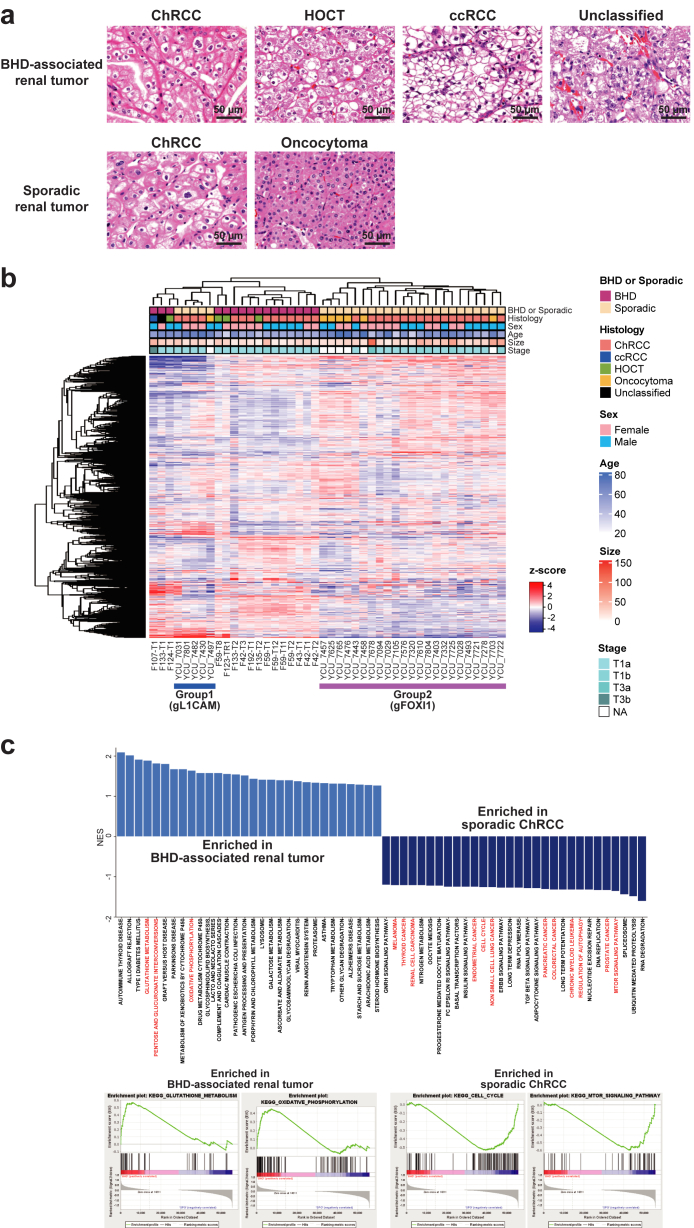
We conducted RNA-seq of sixteen BHD-associated renal tumours (ten ChRCCs, four HOCTs, one clear cell renal cell carcinoma and one unclassified renal cell carcinoma) along with twenty-one sporadic ChRCCs and seven sporadic oncocytomas (Fig. 1a). A heatmap exhibited distinctive molecular characteristics in BHD-associated renal tumours compared to sporadic renal tumours (Fig. 1b). Gene set enrichment analysis (GSEA) revealed that BHD-associated renal tumours exhibited increased oxidative phosphorylation and glutathione metabolism relative to sporadic ChRCCs, whereas sporadic ChRCCs showed upregulated pathways related to various types of cancers, cell cycle and DNA replication when compared to BHD tumours, consistent with the notion that BHD-associated renal tumours might be indolent with increased mitochondrial metabolism compared to sporadic ChRCCs (Fig. 1c) (Supplementary Dataset 2).5,30,31
Fig. 1.
Expression analyses highlighted unique molecular characteristics of BHD-associated renal tumours. (a) Hematoxylin and eosin staining of renal tumours analyzed in this study. Images are shown at 400× magnification. Scale bars represent 50 μm. (b) Heatmap shows expression profiling of sixteen BHD-associated renal tumours, twenty-one sporadic ChRCC and seven sporadic oncocytomas. Sporadic ChRCC and sporadic oncocytomas was further clustered into two distinct subclusters; Group1(gFOXI1) and Group2 (gL1CAM); further analyses are done in Fig. 2 gL1CAM: group of sporadic ChRCCs with predominant expression of L1CAM. gFOXI1: group of sporadic ChRCCs with predominant expression of FOXI1. (c) Bar plot shows gene set enrichment analysis (GSEA) results comparing sixteen BHD-associated renal tumours and twenty-one sporadic ChRCCs (upper panel). Some of the GSEA enrichment plots are shown (lower panels).
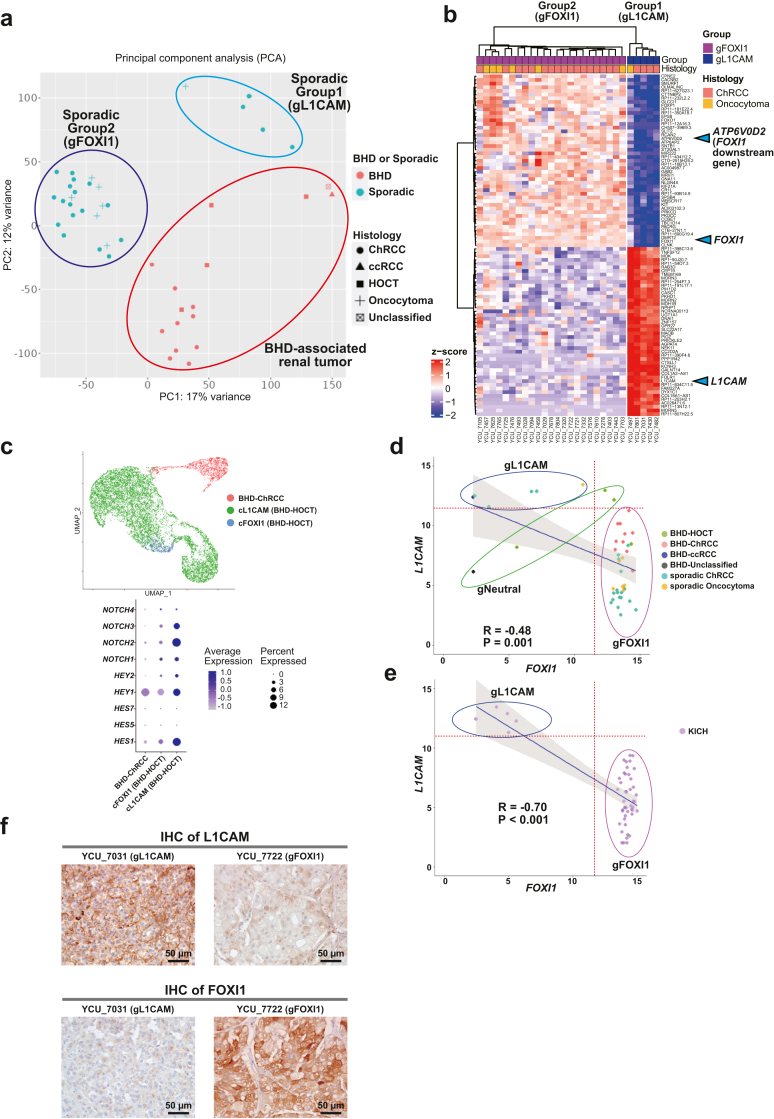
Importantly, sporadic ChRCCs and sporadic oncocytomas were further clustered into two distinct clusters, and differentially expressed genes (DEGs) analysis identified L1CAM, FOXI1 and its downstream gene, ATP6V0D2 as molecular markers for these distinct clusters (Group1 and 2) (Fig. 1, Fig. 2b, Fig. 1, Fig. 2a and Fig. 1, Fig. 2b) (Supplementary Dataset 3). In renal development, Notch signalling regulates a differentiation of ureteric bud either into L1CAM-positive principal cells or into FOXI1-positive intercalated cells.18,16 Of note, our single-cell transcriptome analysis has uncovered transcriptomic intra-tumour heterogeneity in BHD-associated HOCT, comprised of two distinct cell clusters of L1CAM expressing cells and FOXI1 expressing cells in which expression patterns of Notch signalling-associated genes were different, suggesting that expression patterns of L1CAM and FOXI1 determined by Notch signalling may associate with diverse molecular characteristics of renal tumours (Fig. 2c) (Supplementary Fig. S1a).17 In addition, we observed opposing correlation between L1CAM expressions and FOXI1 expressions both in our cohort and in the TCGA ChRCC cohort (KICH), further suggesting that mutually exclusive expressions of L1CAM and FOXI1 may confer intra-tumour and inter-tumour heterogeneity on BHD-associated HOCT cells and sporadic renal tumours, respectively (Fig. 2d, e and f) (Supplementary Fig. S1b).
Fig. 2.
Sporadic ChRCCs and sporadic oncocytomas formed two distinct subclusters characterized by L1CAM and FOXI1 expressions. (a) Scatter plot of principal component analysis (PCA) for Fig. 1b is shown. gL1CAM: group of sporadic ChRCCs and sporadic oncocytoma with predominant expression of L1CAM. gFOXI1: group of sporadic ChRCCs and sporadic oncocytomas with predominant expression of FOXI1. (b) Heatmap shows top fifty genes differentially expressed in Group1 (gL1CAM) and Group2 (gFOXI1). Blue triangles indicate L1CAM, FOXI1 and its downstream gene, ATP6V0D2. (c) Uniform Manifold Approximation and Projection (UMAP) shows cell clusters of BHD-associated ChRCC and BHD-associated HOCT. BHD-associated HOCT is comprised of two distinct cell clusters characterized by L1CAM expression (cL1CAM) or by FOXI1 expression (cFOXI1) (upper panel). Dot plot shows differentially expressing patterns of Notch genes and downstream genes in BHD-associated renal tumours (lower panel). (d) Scatter plots show an inverse correlation of FOXI1 and L1CAM expression levels (upper panel). gL1CAM: group of sporadic ChRCCs with predominant expression of L1CAM. gFOXI1: group of sporadic ChRCCs with predominant expression of FOXI1. gNeutral; group of sporadic ChRCCs which were clustered into neither gL1CAM nor gFOXI1. Pearson's correlation coefficient and p-value are shown. (e) Scatter plot shows an inverse correlation of FOXI1 and L1CAM expressions (lower panel). Pearson's correlation coefficient and p-value are shown. (f) Immunohistochemistry (IHC) of L1CAM protein (upper panel) or FOXI1 protein (lower panel) in sporadic ChRCCs. Scale bars represent 50 μm.
BHD-associated renal tumours most commonly harbour biallelic FLCN alterations but fewer mutations and copy number alterations
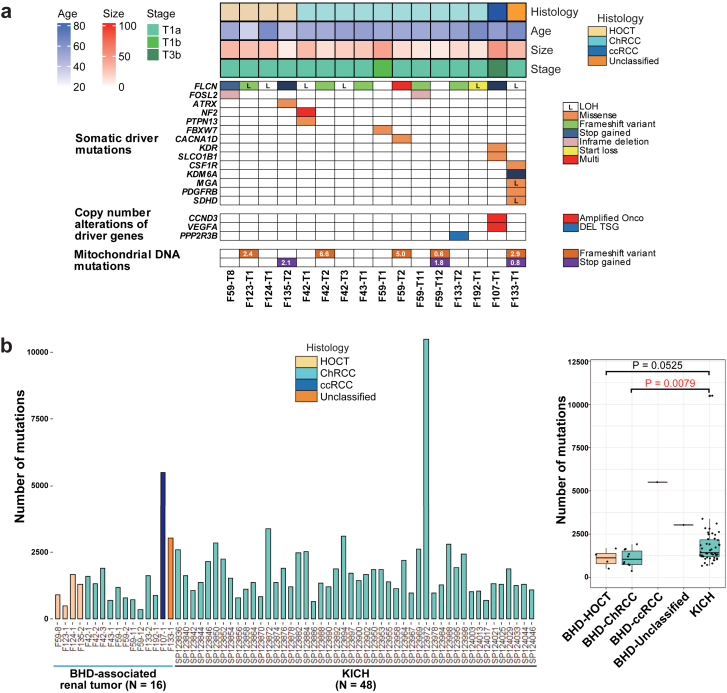
We conducted WGS analysis of sixteen BHD-associated renal tumours and nine adjacent normal kidneys from nine BHD patients (Table 1 and Supplementary Dataset 1). WGS analysis in this study, together with our previous WES analysis, confirmed somatic second hit FLCN alterations in all of the BHD-associated renal tumours (Fig. 3a) (Supplementary Dataset 4).16 In addition, WGS analysis of BHD-associated renal tumours demonstrated that very few genes were commonly mutated except for FLCN, as was observed in our previous WES study, suggesting that FLCN alteration should be the main driver for BHD-associated renal tumourigenesis.16 In F107 case, interestingly, amplification of VEGFA and CCND3 on chromosome 6 was observed, which may lead to the formation of clear cell renal cell carcinoma. F133-T1, whose histology was unclassified renal cell carcinoma, harboured a variant in succinate dehydrogenase D (SDHD), a gene responsible for SDH-associated tumour predisposition syndrome.32,33
Fig. 3.
BHD-associated renal tumours harbour less numbers of gene variants compared to sporadic ChRCCs. (a) Mutation matrices show histology, clinical characteristics (age, size and stage) and somatic variants revealed by WGS of sixteen BHD-associated renal tumours. (b) Bar plots (left panel) and box plots (right panel) show the number of mutations in sixteen BHD-associated renal tumours and forty-eight sporadic ChRCCs in TCGA cohort (KICH). Welch's two sample t-test was used.
The average number of mutations per tumour was 1107, 1090 and 1650 in BHD-associated ChRCCs, BHD-associated HOCTs and sporadic ChRCCs in the TCGA cohort (KICH), respectively, supporting that BHD-associated renal tumours have less inter-tumour heterogeneity than sporadic ChRCCs (Fig. 3b, and Supplementary Dataset 5). In fact, BHD-associated renal tumours did not show increased number of mutations in an age-dependent manner, whereas sporadic ChRCCs in the TCGA cohort did exhibit an age-dependent increase in number of mutations, further supporting the notion that BHD-associated renal tumours have less inter-tumour heterogeneity compared to ChRCCs (Supplementary Fig. S2a).
Of note, we observed no common structural variants in BHD-associated renal tumours, suggesting that structural variants may not be key drivers for BHD-associated renal tumourigenesis (Supplementary Fig. S2b). High-resolution copy number alteration (CNA) analysis using WGS data exhibited that CNAs of BHD-associated renal tumours were considerably different from those already reported in sporadic cases; fewer CNAs were observed in BHD-associated HOCT and BHD-associated ChRCC compared to those in BHD-associated clear cell renal cell carcinoma, which might reflect the indolent nature of BHD-associated HOCT and BHD-associated ChRCC. Loss of chromosomes 1, 2, 6, 10, 13 and 17, which is characteristic of sporadic ChRCC, was not observed in BHD-associated renal tumours (Supplementary Fig. S2c).1,12,16,31
BHD-associated renal tumours demonstrate unique mutational signatures
Mutational signatures are imprints of DNA damage and repair processes that have been operative during tumourigenesis and can provide insights into environmental and endogenous causes of cancer and biological implications of somatic mutations.25,34 In this study, mutational signature analysis was performed using WGS data from 16 BHD-associated renal tumours and 49 sporadic ChRCCs in TCGA cohort (Supplementary Fig. S3a). BHD-associated renal tumours and sporadic ChRCCs in the TCGA cohort generally demonstrated similar patterns of mutational signatures. However, some of the signatures were different between these tumours, including SBS1 (Spontaneous deamination of 5-methylcytosine, clock-like signature), SBS2 (APOBEC activity), SBS5 (Unknown, clock-like signature), SBS13 (APOBEC activity), insertions and deletions (ID)2 (Slippage during DNA replication of the template DNA strand) and ID8 (DBS repair by non-homologous end-joining), highlighting molecular differences of these tumours.
BHD-associated renal tumours harbour mtDNA with higher copy numbers and fewer variants
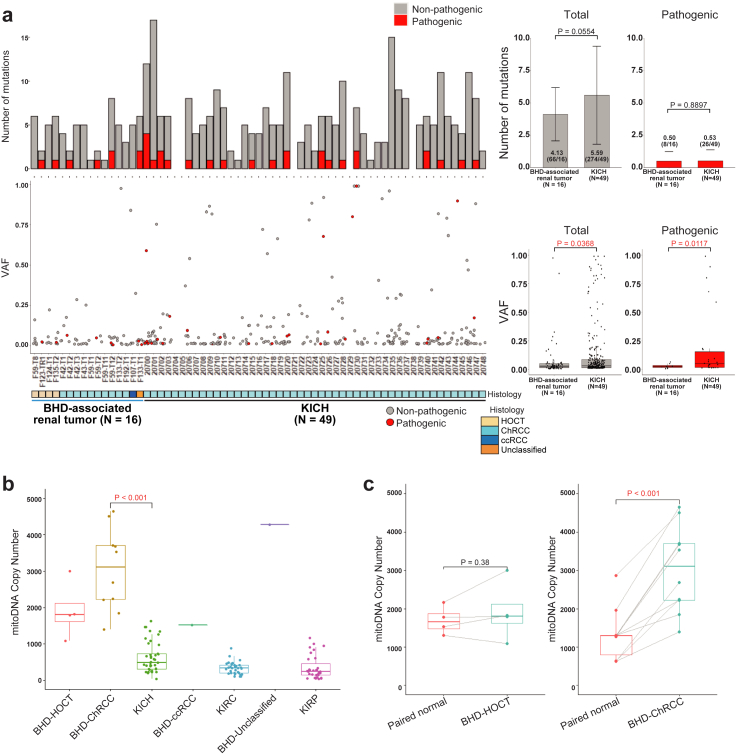
A notable proportion (22.2%) of sporadic ChRCCs harbor non-silent mtDNA variants without any variants in nuclear-encoded kidney cancer-associated genes, whereas BHD-associated renal tumours harbor no disruptive alterations in mtDNA, suggesting that non-silent mtDNA variants in sporadic ChRCCs may be drivers of tumourigenesis of sporadic ChRCCs whereas mitochondrial function is most likely retained in BHD-associated renal tumours.12,29,35 Consistent with these results, heteroplasmic level or variant allele frequencies (VAFs) of total and pathogenic variants in mtDNA of BHD-associated renal tumours were significantly lower than those of sporadic ChRCCs (p = 0.0368 and 0.0117, respectively) (Fig. 4a) (Supplementary Dataset 6). mtDNA copy numbers were higher in BHD-associated renal tumours compared to sporadic ChRCCs and variable depending on each histology (Fig. 4b and c) (Supplementary Fig. S4).
Fig. 4.
BHD-associated renal tumours harbour higher mtDNA copy number with fewer variants compared to sporadic ChRCCs. (a) Bar plots of number of mutations (left upper panel) and scatter plots of variant allele frequencies (VAF) (left lower panel) in mtDNA from sixteen BHD-associated renal tumours and forty-nine sporadic ChRCCs in TCGA cohort (KICH) are shown. Bar graphs show average number of variants shown in left upper panel (right upper panel). Box plots show average VAF shown in left lower panel (right lower panel). Welch's two sample t-test was used. (b) Box plot shows the mtDNA copy number in BHD-associated renal tumours and various sporadic renal tumours. BHD-Clear cell: BHD-associated clear cell renal cell carcinoma. BHD-Unclassified: BHD-associated unclassified renal cell carcinoma. KICH: sporadic ChRCCs in TCGA cohort. KIRC: sporadic clear cell renal cell carcinomas in TCGA cohort. KIRP: sporadic papillary renal cell carcinomas in TCGA cohort. Welch's two sample t-test was used. (c) Box plots show mitochondrial DNA copy number in each histological type of BHD-associated renal tumours and their adjacent normal kidneys (Paired normal). Welch's two sample t-test was used.
BHD-associated renal tumours and sporadic ChRCC may arise from different normal nephron cells
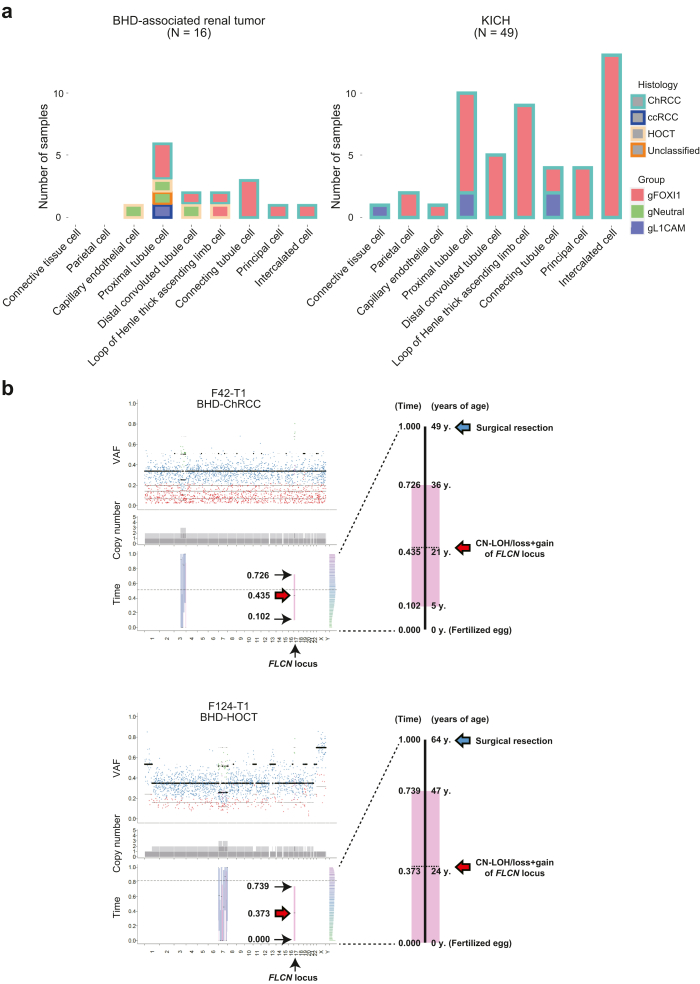
A recent machine learning algorithm integrating bulk or single cell-level ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) data of normal cell types with somatic point mutation frequency derived from the WGS data of tumours enables us to predict putative cancer cell-of-origin (COO), based on a hypothesis that the somatic variants might preferentially accumulate outside of the open chromatin regions of the cancer cell of origin.36 In this study, we applied the COOBoostR algorithm to predict a putative cancer COO of BHD-associated renal tumours and sporadic ChRCCs.26 Interestingly, the COOBoostR predicted that BHD-associated renal tumours and sporadic ChRCCs may arise from a variety of normal nephron cells. The most frequently predicted cell-of-origin was proximal tubules for BHD-associated renal tumours and intercalated cells for sporadic ChRCCs, implying the possibility that these two tumour types might arise from different cells of origin (Fig. 5a) (Supplementary Dataset 7). Of note, none of L1CAM expressing tumours in BHD-associated renal tumours and sporadic ChRCCs might arise from intercalated cells, suggesting that cell-of-origin may determine tumour characteristics of these tumours.
Fig. 5.
Bioinformatic algorithms depict natural history of BHD-associated renal tumourigenesis. (a) The bar plots show putative cell of origin of BHD-associated renal tumours (left panel) and sporadic ChRCCs in TCGA cohort (KICH) (right panel) predicted by COOBoost algorithm. Groups of gFOXI1, gNeutral and gL1CAM correspond to those in Fig. 2d and e. (b) The plots show the results of MutationTimeR algorithm. The dot plots show variant allele frequencies (VAF) of point mutations; purple dot indicates clonal mutation occurred at late stage, green dot indicates clonal mutation occurred at early stage, blue dot indicates clonal mutation occurred at unknown stage, red dot indicates subclonal mutation (left upper panels in each case). The stacked barplots show copy number of each allele; dark and light grey bars indicate major and minor allele, respectively (left middle panels in each case). The box plots with median values and 95% confidence intervals indicate predicted mutation timings of primary and secondary gains; blue bar indicates mono-allelic gains (N:1), pink bar indicates copy neutral (CN)-LOH/loss + gain (N:0) and green bar indicates bi-allelic gains (N:2) (left lower panels in each case). The histograms show magnified timelines of left lower panels in each case (right panels in each case).
Second hit mutations of the FLCN gene may occur in early third decade of life in BHD patients
Recent bioinformatics methodology allows us to predict the timing of the occurrence of a large duplication from the proportion of point variants in that duplicated region.28 We applied the MutationTimeR pipeline to our WGS data and predicted variant acquisition timing. In our WGS study, two samples of F42-T1 (BHD-associated ChRCC) and F124-T1 (BHD-associated HOCT) harboured a 17p copy neutral loss of heterozygosity (CN-LOH)/loss + gain, where FLCN gene was located. Therefore, we applied the MutationTimeR pipeline to these cases and found that a second hit mutation in the remaining FLCN gene may have occurred at median age of 21 years (ranging from 5 to 36 years of age) in F42-T1 patient and median age of 24 years (ranging from 0 to 47 years of age) in F124-T1 patient (Fig. 5b) (Supplementary Fig. S5).
Discussion
Here, we present comparative analyses of BHD-associated renal tumours and sporadic renal tumours, which exhibited molecular similarities as well as differences between these histologically similar renal tumours (Table 3). WGS analysis revealed that very few genes were commonly mutated in BHD-associated renal tumours except for FLCN and no common structural variants were observed. Therefore, FLCN alteration appears to be a relatively strong tumour driver given the fact that FLCN-deficiency leads to kidney cell proliferation in both in-vivo and in-vitro models, whereas mutation of other kidney cancer-associated genes including VHL and fumarate hydratase (FH) in genetically manipulated murine models only develop renal cysts.6,9,37, 38, 39, 40 Taken together, BHD-associated renal tumourigenesis may be largely attributed to the exclusive loss of FLCN as the main tumour driver.
Table 3.
Molecular differences between BHD-associated renal tumours and sporadic ChRCCs.
| BHD-associated renal tumours | Sporadic ChRCCs | |
|---|---|---|
| Expressions | ||
| Upregulated pathways (GSEA) | Oxydative phosphorylation, Glutathione metabolism | Cancer-related pathways, Cell cycle, DNA replication |
| Heterogeneity | Intra-tumour heterogeneity conffered by mutually exclusive expressions of L1CAM and FOXI1 | Inter-tumour heterogeneity conffered by mutually exclusive expressions of L1CAM and FOXI1 |
| Nuclear DNA | ||
| Driver gene mutation | FLCN | TP53, PTEN |
| Copy number alteration (CNA) | Low | Copy loss of chr 1, 2, 6, 10, 13 and 17 |
| Tumour mutation burden (TMB) | Low | High |
| Mutational signature | SBS1, SBS5, SBS40 | SBS1, SBS5, SBS40 |
| Cell of origin (most frequent) | Proximal tubule cell | Intercalated cell of collecting duct |
| Timing of second hit alteration | Early twenties | – |
| Mitochondrial DNA | ||
| Variant allele frequency (VAF) | Low | High |
| Copy number | High | Low |
In this study, we identified inter-tumour heterogeneity of sporadic ChRCCs, characterized by mutually exclusive expressions of L1CAM and FOXI1. FOXI1 is an important transcription factor for differentiation of intercalated cells of the collecting duct and, importantly, Notch signalling regulates differentiation of ureteric bud cells either into L1CAM-positive principal cells of the collecting duct when Notch signalling is active, or into FOXI1-positive intercalated cells of the collecting duct when Notch signalling is inactive.18 Notably, defective Notch signalling drives renal cyst formation and yields intratumour heterogeneity.41,42 Our single-cell transcriptomic analysis of BHD-associated HOCT tumours revealed that expressions of Notch genes as well as those of Notch downstream genes including HES and HEY family members were increased in the cluster of L1CAM expressing cells, suggesting that Notch signalling may associate with transcriptomic intratumour heterogeneity (tITH) of BHD-associated renal tumours. However, in sporadic ChRCCs, although expressions of genes in the Notch signalling-mediated network, i.e., ARNT, PARP1, RB1 and STAT3, exhibited positive correlation with L1CAM expression and inverse correlation with FOXI1 expression, expressions of HES1, HES5, HES7, HEY1 and HEY2, direct downstream genes of Notch signalling, were not correlated with L1CAM or FOXI1 expression, suggesting that Notch signalling may be dysregulated in sporadic ChRCCs (Supplementary Fig. S6).
Recent bioinformatics methodology allows us to predict the putative timing of a large duplication from the proportion of point variants in that duplicated region.28 According to this methodology, chromosome 3p loss with concurrent chromosome 5q gain may occur during childhood to adolescence and the remaining VHL allele may be lost after this event in sporadic clear cell renal cell carcinoma.43 On the other hand, chromosome 3p loss with concurrent chromosome 5q gain may occur during childhood to adolescence in VHL-associated kidney cancer, suggesting that the earlier biallelic loss of the VHL gene may result in earlier development of kidney cancer compared to sporadic clear cell renal cell carcinoma.43 Our timing prediction analysis has shown that second hit alterations of FLCN may occur during the BHD patients’ early twenties, further supporting the concept that the earlier biallelic loss of causative genes in inherited renal tumours may lead to earlier tumour development compared to sporadic renal tumour.
So far, we have experienced only one BHD-associated oncocytoma and hence, the frequency of oncocytoma in BHD-associated renal tumours in our cohort is relatively low compared to that in other cohorts.16,44 Although a genotype–phenotype correlation has not been investigated in BHD syndrome, it is urgent to interrogate the elements that possibly affect histological variations across the cohorts.
The findings of this study, which delineate the molecular similarities as well as differences between BHD-associated renal tumours and sporadic renal tumours, further our understanding of the tumourigenesis of these renal tumours. These findings provide a foundation for the management of these tumours as well as for the development of targeted therapeutics approaches for BHD-associated tumours and sporadic renal tumours.
Contributors
Hisashi Hasumi had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Jikuya, Schmidt, Linehan, Yao, Nakagawa, Hasumi. Acquisition of samples: Jikuya, Tatenuma, Komeya, H. Ito, Y. Ito, Muraoka, Hamanoue, Makiyama, Hasumi. Acquisition of data: Jikuya, Maejima, An, Ju, Kim, Imoto, Nakagawa. Analysis and interpretation of data: Jikuya, Johnson, An, Ju, Lee, Ha, Song, Kim, Okawa, Sasagawa, Kanazashi, Fujita, Imoto, Mitome, Ohtake, Noguchi, Kawaura, Iribe, Aomori, Furuya, Kato, Fujii, Tamura, Baba, Suda, Kodama, Shuch, Nakagawa, Hasumi. Drafting of the manuscript: Jikuya, Ricketts, Schmidt, Linehan, Nakagawa, Hasumi. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: Jikuya, Johnson, Maejima, An, Ju, Lee. Obtaining funding: Komeya, H. Ito, Y. Ito, Makiyama, Yao, Nakagawa, Hasumi. Administrative, technical, or material support: Jikuya, Hasumi. Supervision: Nakagawa, Hasumi. Jikuya and Hasumi have verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
All of sequencing data were deposited in National Bioscience Database Center (NBDC) under the accession number JGAS000564, https://humandbs.biosciencedbc.jp/en/hum0277-v2.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
We thank Dr. Noboru Nakaigawa, Dr. Keiichi Kondo, Dr. Narihiko Hayashi, Dr. Kimito Osaka and Dr. Yumiko Yokomizo in Department of Urology, Yokohama City University for surgical resection of renal tumours and Dr. Akira Nishiyama and Dr. Koichi Murakami in Department of Immunology, Yokohama City University for their excellent works in scRNA-seq analysis.
Authors were supported by JSPS KAKENHI Grant Number as following; H.Hasumi. by 19K09694, I.K. by 19K16563, Y.Ito. by 19K18591, H.I. by 20K18121 and 19K23781, M.K. by 21H03068, M.Furuya. by 20K07395, M.Y. by 19K09717, S.F. by 19K07769, H.N. by 18H04049 and M.B. by 18H02938. This work was also supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104596.
Contributor Information
Hidewaki Nakagawa, Email: hidewaki@riken.jp.
Hisashi Hasumi, Email: hasumi@yokohama-cu.ac.jp.
Appendix A. Supplementary data
References
- 1.Hasumi H., Baba M., Hasumi Y., Furuya M., Yao M. Birt-Hogg-Dube syndrome: clinical and molecular aspects of recently identified kidney cancer syndrome. Int J Urol. 2016;23(3):204–210. doi: 10.1111/iju.13015. [DOI] [PubMed] [Google Scholar]
- 2.Hasumi H., Yao M. Hereditary kidney cancer syndromes: genetic disorders driven by alterations in metabolism and epigenome regulation. Cancer Sci. 2018;109(3):581–586. doi: 10.1111/cas.13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasumi Y., Baba M., Ajima R., et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumour development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci U S A. 2009;106(44):18722–18727. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linehan W.M., Schmidt L.S., Crooks D.R., et al. The metabolic basis of kidney cancer. Cancer Discov. 2019;9(8):1006–1021. doi: 10.1158/2159-8290.CD-18-1354. [DOI] [PubMed] [Google Scholar]
- 5.Hasumi H., Baba M., Hasumi Y., et al. Regulation of mitochondrial oxidative metabolism by tumour suppressor FLCN. J Natl Cancer Inst. 2012;104(22):1750–1764. doi: 10.1093/jnci/djs418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasumi H., Baba M., Hasumi Y., et al. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumour suppression in cooperation with Flcn. Proc Natl Acad Sci U S A. 2015;112(13):E1624–E1631. doi: 10.1073/pnas.1419502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasumi H., Baba M., Hong S.B., et al. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415(1–2):60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasumi Y., Baba M., Hasumi H., et al. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet. 2014;23(21):5706–5719. doi: 10.1093/hmg/ddu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasumi H., Hasumi Y., Baba M., et al. H255Y and K508R missense mutations in tumour suppressor folliculin (FLCN) promote kidney cell proliferation. Hum Mol Genet. 2017;26(2):354–366. doi: 10.1093/hmg/ddw392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda N., Furuya M., Nagashima Y., et al. Intratumoural peripheral small papillary tufts: a diagnostic clue of renal tumours associated with Birt-Hogg-Dube syndrome. Ann Diagn Pathol. 2014;18(3):171–176. doi: 10.1016/j.anndiagpath.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Furuya M., Hong S.B., Tanaka R., et al. Distinctive expression patterns of glycoprotein non-metastatic B and folliculin in renal tumours in patients with Birt-Hogg-Dube syndrome. Cancer Sci. 2015;106(3):315–323. doi: 10.1111/cas.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis C.F., Ricketts C.J., Wang M., et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26(3):319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linehan W.M., Ricketts C.J. The metabolic basis of kidney cancer. Semin Cancer Biol. 2013;23(1):46–55. doi: 10.1016/j.semcancer.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt L.S., Marston Linehan W. FLCN: the causative gene for Birt-Hogg-Dube syndrome. Gene. 2018;15(640):28–42. doi: 10.1016/j.gene.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuya M., Yao M., Tanaka R., et al. Genetic, epidemiologic and clinicopathologic studies of Japanese Asian patients with Birt-Hogg-Dube syndrome. Clin Genet. 2016;90(5):403–412. doi: 10.1111/cge.12807. [DOI] [PubMed] [Google Scholar]
- 16.Hasumi H., Furuya M., Tatsuno K., et al. BHD-associated kidney cancer exhibits unique molecular characteristics and a wide variety of variants in chromatin remodeling genes. Hum Mol Genet. 2018;27(15):2712–2724. doi: 10.1093/hmg/ddy181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jikuya R., Murakami K., Nishiyama A., et al. Single-cell transcriptomes underscore genetically distinct tumour characteristics and microenvironment for hereditary kidney cancers. iScience. 2022;25(6) doi: 10.1016/j.isci.2022.104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barone S., Zahedi K., Brooks M., et al. Kidney intercalated cells and the transcription factor FOXi1 drive cystogenesis in tuberous sclerosis complex. Proc Natl Acad Sci U S A. 2021;118(6) doi: 10.1073/pnas.2020190118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinf. 2018;19(1):534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson T.A., Maekawa S., Fujita M., et al. Genomic features of renal cell carcinoma developed during end-stage renal disease and dialysis. Hum Mol Genet. 2022 doi: 10.1093/hmg/ddac180. [DOI] [PubMed] [Google Scholar]
- 23.Cameron D.L., Baber J., Shale C., et al. GRIDSS2: comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biol. 2021;22(1):202. doi: 10.1186/s13059-021-02423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franch-Exposito S., Bassaganyas L., Vila-Casadesus M., et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. Elife. 2020;9 doi: 10.7554/eLife.50267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov L.B., Kim J., Haradhvala N.J., et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S., Ha K., Song W., et al. COOBoostR: an extreme gradient boosting-based tool for robust tissue or cell-of-origin prediction of tumours. Life. 2022;13(1) doi: 10.3390/life13010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muto Y., Wilson P.C., Ledru N., et al. Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat Commun. 2021;12(1):2190. doi: 10.1038/s41467-021-22368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerstung M., Jolly C., Leshchiner I., et al. The evolutionary history of 2,658 cancers. Nature. 2020;578(7793):122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y., Ju Y.S., Kim Y., et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52(3):342–352. doi: 10.1038/s41588-019-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ball M.W., Shuch B.M. Inherited kidney cancer syndromes. Curr Opin Urol. 2019;29(4):334–343. doi: 10.1097/MOU.0000000000000646. [DOI] [PubMed] [Google Scholar]
- 31.Ball M.W., An J.Y., Gomella P.T., et al. Growth rates of genetically defined renal tumours: implications for active surveillance and intervention. J Clin Oncol. 2020:JCO1902263. doi: 10.1200/JCO.19.02263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricketts C.J., Shuch B., Vocke C.D., et al. Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. J Urol. 2012;188(6):2063–2071. doi: 10.1016/j.juro.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricketts C.J., Forman J.R., Rattenberry E., et al. Tumour risks and genotype-phenotype-proteotype analysis in 358 patients with germline mutations in SDHB and SDHD. Hum Mutat. 2010;31(1):41–51. doi: 10.1002/humu.21136. [DOI] [PubMed] [Google Scholar]
- 34.Maura F., Degasperi A., Nadeu F., et al. A practical guide for mutational signature analysis in hematological malignancies. Nat Commun. 2019;10(1):2969. doi: 10.1038/s41467-019-11037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang M., Vocke C.D., Merino M.J., Schmidt L.S., Linehan W.M. Mitochondrial DNA mutations distinguish bilateral multifocal renal oncocytomas from familial Birt-Hogg-Dube tumours. Mod Pathol. 2015;28(11):1458–1469. doi: 10.1038/modpathol.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polak P., Karlic R., Koren A., et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518(7539):360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba M., Furihata M., Hong S.B., et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst. 2008;100(2):140–154. doi: 10.1093/jnci/djm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haase V.H. The VHL tumour suppressor in development and disease: functional studies in mice by conditional gene targeting. Semin Cell Dev Biol. 2005;16(4–5):564–574. doi: 10.1016/j.semcdb.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollard P.J., Spencer-Dene B., Shukla D., et al. Targeted inactivation of fh1 causes proliferative renal cyst development and activation of the hypoxia pathway. Cancer Cell. 2007;11(4):311–319. doi: 10.1016/j.ccr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Gu Y.F., Cohn S., Christie A., et al. Modeling renal cell carcinoma in mice: bap1 and Pbrm1 inactivation drive tumour grade. Cancer Discov. 2017;7(8):900–917. doi: 10.1158/2159-8290.CD-17-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Idowu J., Home T., Patel N., et al. Aberrant regulation of Notch3 signaling pathway in polycystic kidney disease. Sci Rep. 2018;8(1):3340. doi: 10.1038/s41598-018-21132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim J.S., Ibaseta A., Fischer M.M., et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545(7654):360–364. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell T.J., Turajlic S., Rowan A., et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell. 2018;173(3):611–623.e17. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruinsma F.J., Dowty J.G., Win A.K., et al. Update of penetrance estimates in Birt-Hogg-Dube syndrome. J Med Genet. 2023;60(4):317–326. doi: 10.1136/jmg-2022-109104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.