Abstract
Buchnera aphidicola, the prokaryotic endosymbiont of aphids, complements dietary deficiencies with the synthesis and provision of several essential amino acids. We have cloned and sequenced a region of the genome of B. aphidicola isolated from Acyrthosiphon pisum which includes the two-domain aroQ/pheA gene. This gene encodes the bifunctional chorismate mutase-prephenate dehydratase protein, which plays a central role in l-phenylalanine biosynthesis. Two changes involved in the overproduction of this amino acid have been detected. First, the absence of an attenuator region suggests a constitutive expression of this gene. Second, the regulatory domain of the Buchnera prephenate dehydratase shows changes in the ESRP sequence, which is involved in the allosteric binding of phenylalanine and is strongly conserved in prephenate dehydratase proteins from practically all known organisms. These changes suggest the desensitization of the enzyme to inhibition by phenylalanine and would permit the bacterial endosymbiont to overproduce phenylalanine.
Endosymbiosis is one of the main factors that facilitated the diversification of the major insect groups and their adaptation to a wide variety of ecological niches that would otherwise have been inadequate (8). Aphids are strict phloem-feeders that maintain an endosymbiotic association with Buchnera aphidicola, a member of the class Proteobacteria (1). The association is obligate for both partners, and it is commonly accepted that the main role of endosymbionts is the provision of essential nutrients to the aphids. However, definitive evidence is rare, and only the provision of the amino acids tryptophan and leucine through the translocation of their biosynthetic genes to plasmids is well documented (2, 6). Phenylalanine seems to be overproduced by the endosymbiont, since lower levels are found in antibiotic-treated aphids (aposymbiotic aphids) than in symbiotic aphids (7). In bacteria, the main phenylalanine biosynthetic pathway starts with chorismate, which is converted to prephenate by the enzyme chorismate mutase (CM; EC 5.4.99.5). This compound is converted to phenylpyruvate by prephenate dehydratase (PDT; EC 4.2.1.51) and later transaminated to phenylalanine (9).
The evolution of the genes encoding CM (aroQ) and PDT (pheA) in prokaryotic and eukaryotic lineages comprises several duplication and fusion events between them and with other genes. Two of the three major divisions of gram-negative bacteria possess a multienzyme protein (CM/PDT) with the CM and PDT activities. The gene encoding this protein, although frequently denoted pheA, should be named aroQ/pheA in order to show the existence of the two domains (3).
In Escherichia coli, one of the closest free-living relatives of Buchnera, the biosynthesis of phenylalanine is subjected to gene and feedback enzyme regulation. A feedback inhibition of both CM and PDT activities by phenylalanine has been described (9). This amino acid has been proposed to bind an unknown site in the C-terminal part of the protein where the PDT domain is located (3, 12).
In this work, we have compared the Buchnera aroQ/pheA gene and its encoded protein with those from other organisms, especially E. coli, searching for changes that show the adaptation of Buchnera to endosymbiosis.
Cloning of the Buchnera (Acyrthosiphon pisum) aroQ/pheA gene and flanking regions.
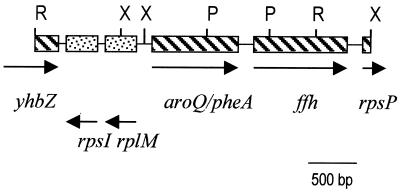
Based on the sequence of the aroQ/pheA gene from enteric bacteria, we designed two degenerate primers in the PDT domain of the gene (PheAd1, 5′-ATCCTCARCCNTTYCARC-3′; and PheAd2, 5′-GTAGAACATYTCYTCCCA-3′). Using total DNA isolated from the aphid A. pisum, we amplified by PCR an expected 400-bp fragment which was isolated and sequenced. It was highly similar to pheA genes from enteric bacteria, but with a high A+T content (>70%) typical of Buchnera genes. A Southern blot with Buchnera total DNA helped us to make a restriction map of the region of the bacterial chromosome where the pheA gene was placed (Fig. 1) and identified EcoRI and XbaI as restriction enzymes suitable for the cloning of the complete gene with an inverse PCR strategy. This experiment yielded fragments of around 3.5 and 3.0 kb for EcoRI and XbaI, respectively.
FIG. 1.
Genetic map of the 4.4-kb fragment containing the aroQ/pheA gene from Buchnera. The positions of the EcoRI (R), PstI (P), and XbaI (X) sites are indicated. Arrows show transcription directions.
Structure of the pheA genomic region.
The DNA region included in the overlapping EcoRI and XbaI fragments was sequenced. Its 4,371 bp contained four complete genes: rpsI, encoding the small subunit ribosomal protein S9; rplM, encoding the large subunit ribosomal protein L13; the aroQ/pheA gene, encoding the bifunctional CM/PDT protein; and the gene ffh, encoding the signal recognition particle protein (also called “fifty-four homolog”). In addition, two incomplete genes were present at the ends: the yhbZ gene, encoding a hypothetical GTP-binding protein; and the rpsP gene, encoding the small subunit ribosomal protein S16 (Fig. 1). Proteins encoded by the genes described above showed the highest similarity to proteins from E. coli in all cases, except for the CM/PDT protein, which more closely resembled that of Erwinia herbicola.
Comparative analysis of aroQ/pheA genes of Buchnera and E. coli.
The expression of the aroQ/pheA genes in E. coli and other enteric bacteria is controlled solely by an attenuation system, which includes the sequences containing the stem-loop structures and a leader region encoding a 15-residue phenylalanine-rich leader peptide (11). The analysis of the 5′ region of the Buchnera gene did not show any sequence resembling such an attenuator; hence, adaptation of Buchnera to endosymbiosis has probably produced the loss of gene regulation and the change to a constitutive expression to allow for overproduction of phenylalanine.
Comparison of Buchnera CM/PDT protein with CM/PDT or PDT proteins from other organisms.
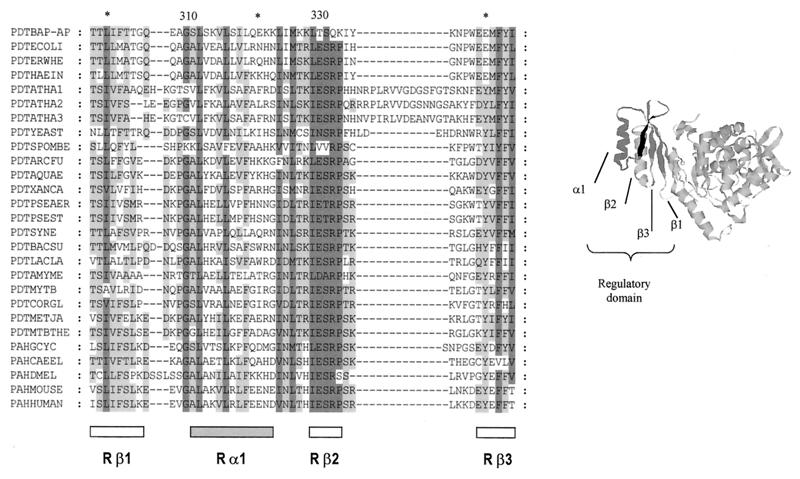
The alignment of CM/PDT from Buchnera and several other species showed that some parts of the amino acid sequence were well conserved. However, an important feature of Buchnera protein was the lack of conservation of the four-residue sequence (ESRP) located in the regulatory part of the PDT domain (Fig. 2). The homologous residues in Buchnera were TSQK (residues 329 to 332). When the alignment was extended to monofunctional and trifunctional PDT proteins, the importance of this region was reinforced, since the last two amino acids were conserved in all available sequences, and the first two were conserved in practically all of them. This suggests that the Buchnera enzyme could have changed some of its regulatory properties to adapt to the endosymbiotic way of life. Based on the following three arguments, we propose that these changes have produced the desensitization of the enzyme to the inhibitory effect of Phe. Our hypothesis implies that the ESRP sequence is part of the allosteric site of the enzyme. First, in E. coli, this sequence is placed in the vicinity of W338, and fluorescent assays have shown that the allosteric binding of Phe takes place close to this amino acid (12). Second, when the regulatory PDT domain was probed against the protein database, a significant similarity was found with the regulatory domain of metazoan aromatic amino acid hydroxylases. In at least two proteins of this family, rat and human phenylalanine hydroxylases (PAHs), it is known that Phe binds the regulatory domain, producing a conformational change that activates the protein (4). The presence of the ESRP motif in these proteins (Fig. 2) points to the involvement of this sequence in Phe binding. Besides, the crystal structure of a dimeric rat PAH, which includes the regulatory domain, has been recently reported (5). These authors raise the possibility that the regulatory Phe binding site was located near the interface between the regulatory and catalytic domains in the vicinity of the β-strand Rβ2 (Fig. 2). Third, it has been shown recently (10) that E. coli CM/PDT proteins with a change in either E329A, S330A, or R331A produced to different extents a strong increase in the concentration of Phe required to produce a 50% enzyme inhibition and a decrease in Phe binding capacity (less than 10% of the level of wild-type protein).
FIG. 2.
Alignment of the homologous region of the regulatory domains from PDT and PAH proteins. α-Helix and β-strand positions correspond to the information obtained from the crystal structure of rat PAH (5) (shown to the right). The sequences used in this alignment were as follows (sources and accession numbers are given in parentheses): PDTBAP-AP (B. aphidicola [A. pisum], AJ239043), PDTECOLI (E. coli, P07022), PDTERWHE (E. herbicola, Q02286), PDTHAEIN (Haemophilus influenzae, P43900), PDTATHA1 (Arabidopsis thaliana, O22241), PDTATHA2 (A. thaliana, AAD30242), PDTATHA3 (A. thaliana, AAC73018), PDTYEAST (Saccharomyces cerevisiae, P32452), PDTSPOMBE (Schizosaccharomyces pombe, O14361), PDTARCFU (Archaeoglobus fulgidus, O30012), PDTAQUAE (Aquifex aeolicus, O67085), PDTXANCA (Xanthomonas campestris, O87954), PDTPSEAER (P. aeruginosa, Pseudomonas Genome Project), PDTPSEST (P. stutzeri, P27603), PDTSYNE (Synechocystis sp., P72808), PDTBACSU (Bacillus subtilis, P21203), PDTLACLA (Lactococcus lactis, P43909), PDTAMYME (Amycolatopsis methanolica, Q44104), PDTMYTB (Mycobacterium tuberculosis, P96240), PDTCORGL (Corynebacterium glutamicum, P10341), PDTMETJA (Methanococcus jannaschii, Q58054), PDTMTBTHE (Methanobacterium thermoautotrophicum, 027288), PAHGCYC (Geodia cydonium, Y16353), PAHCAEEL (Caenorhabditis elegans, CAA91286), PAHDMEL (Drosophila melanogaster, Q27599), PAHMOUSE (Mus musculus, P16331), PAHHUMAN (Homo sapiens, P00439). The numbers at the top indicate the residue position in Buchnera.
All of these arguments support the hypothesis that Buchnera PDT is not inhibited by phenylalanine, at least to the same extent as in Buchnera's free-living relatives, due to the changes in its regulatory domain described above. Since Buchnera proteins are refractory to be active in E. coli systems (P. Baumann, personal communication), demonstration of this hypothesis might be best achieved by performing inhibition tests with genetically engineered E. coli proteins with amino acid changes mimicking those observed in Buchnera, which are currently under way in our laboratory.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the GenBank/EMBL database under accession no. AJ239043.
Acknowledgments
We thank A. Latorre and B. Sabater for helpful comments and suggestions. We are indebted to the Servei de Bioinformàtica and the Servei de Seqüenciació de ADN i proteïnes (S.C.S.I.E., Universitat de València) for computer and technical support.
This work was supported by grant PB96-0793 C04-01 from Dirección General de Enseñanza Superior (Spain).
REFERENCES
- 1.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 2.Bracho A M, Martinez-Torres D, Moya A, Latorre A. Discovery and molecular characterization of a plasmid localized in Buchnera sp. bacterial endosymbiont of the aphid Rhopalosiphum padi. J Mol Evol. 1995;41:67–73. doi: 10.1007/BF00174042. [DOI] [PubMed] [Google Scholar]
- 3.Gu W, Williams D S, Aldrich H C, Xie G, Gabriel D W, Jensen R A. The AroQ and PheA domains of the bifunctional P-protein from Xanthomonas campestris in a context of genomic comparison. Microb Comp Genomics. 1997;2:141–158. doi: 10.1089/omi.1.1997.2.141. [DOI] [PubMed] [Google Scholar]
- 4.Hufton S E, Jennings I G, Cotton R G H. Structure and function of the aromatic amino acid hydroxylases. Biochem J. 1995;311:353–366. doi: 10.1042/bj3110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobe B, Jennings I G, House C M, Michell B J, Goodwill K E, Santarsiero B D, Stevens R C, Cotton R G H, Kemp B E. Structural basis of autoregulation of phenylalanine hydroxylase. Nat Struct Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- 6.Lai C-Y, Baumann L, Baumann P. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA. 1994;91:3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liadouze I, Febvay G, Guillaud J, Bonnot G. Effect of diet on the free amino acid pools of symbiotic and aposymbiotic pea aphids Acyrthosiphon pisum. J Insect Physiol. 1995;41:33–40. [Google Scholar]
- 8.Moran N A, Telang A. Bacteriocyte associated symbionts of insects. Bioscience. 1998;48:295–304. [Google Scholar]
- 9.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 458–484. [Google Scholar]
- 10.Pohnert G, Zhang S, Husain A, Wilson D B, Ganem B. Regulation of phenylalanine biosynthesis. Studies on the mechanism of phenylalanine binding and feedback inhibition in the Escherichia coli P-protein. Biochemistry. 1999;38:12212–12217. doi: 10.1021/bi991134w. [DOI] [PubMed] [Google Scholar]
- 11.Xia T, Zhao G, Jensen R A. The pheA/tyrA/aroF region from Erwinia herbicola: an emerging comparative basis for analysis of gene organization and regulation in enteric bacteria. J Mol Evol. 1993;36:107–120. doi: 10.1007/BF00166246. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Pohnert G, Kongsaeree P, Wilson D B, Clardy J, Ganem B. Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J Biol Chem. 1998;273:6248–6253. doi: 10.1074/jbc.273.11.6248. [DOI] [PubMed] [Google Scholar]