Abstract
Food Microbial contamination is one of the most serious problems. A large percentage of food-borne illnesses are caused by food-borne pathogens, and diarrheal agents comprise more than half of the overall prevalence of food-borne illnesses in the globe, and more commonly in developing countries. This study aimed to identify the most-common foodborne organisms from foods in Khartoum state by PCR.
A total of 207 food samples (raw milk, fresh cheese, yogurt, fish, sausage, mortadella, and eggs) were collected. DNA was extracted from food samples by guanidine chloride protocol, and then species-specific primers were used to identify Escherichia coli O157: H7, Listeria monocytogenes, Salmonella spp., Vibrio cholerae, V. parahaemolyticus, and Staphylococcus aureus. Out of 207 samples, five (2.41%) were positive for L. monocytogenes, one (0.48%) was positive for S. aureus, and one (0.48%) was positive for both Vibrio cholerae and Vibrio parahaemolyticus. From 91 fresh cheese samples, 2 (2.19%) were positive for L. monocytogenes, and one (1.1%) sample was positive for two different foodborne pathogens (V. cholerae and V. parahaemolyticus). Out of 43 Cow's milk samples, three (7%) samples were positive for L. monocytogenes, and out of 4 sausage samples, one (25 %) was positive for S. aureus. Our study revealed the presence of L. monocytogenes and V. cholera in raw milk and fresh cheese samples. Their presence is considered a potential problem and needs intensive hygiene efforts and standard safety measures before, during, and after food processing operations.
Keywords: Foodborne illness, Escherichia coli O157: H7, Listeria monocytogenes, Salmonella spp., Vibrio cholerae, Vibrio parahaemolyticus
1. Introduction
Millions of people worldwide suffer from diseases transmitted through contaminated food and water (KAFERstEIN et al., 2019). Transmission of foodborne pathogens causes annually about 600 million cases, and 420 000 deaths, 30% of deaths occur in children less than five years old (Jaffee et al., 2018). In the past two to three decades, there is an increase in the percentage of foodborne diseases, with reports of serious outbreaks transmitted through contaminated foods and water such as cholera, listeriosis, and salmonellosis (Heredia and García, 2018). It is estimated that annually there are about four million cases of cholera in the world, which causes 21,000 to 143,000 deaths annually (Salcedo, 2018). Unlike most bacteria, L. monocytogenes can survive and multiply in low temperatures and can withstand acidity and salinity, which enables them to contaminate many types of foods (Swaminathan and Gerner-Smidt, 2007, de Noordhout et al., 2014). E. coli O157: H7 is considered one of the most dangerous serotypes of E. coli, in America alone, it causes 73,000 illnesses, more than two thousand hospital stays, and 60 deaths annually, which costs the American economy 405 million dollars a year (Lim et al., 2010). Salmonella species are considered one of the most important microbes that cause foodborne infections in the world and cause a group of diseases. The most important types are typhoid, paratyphoid, bacteremia, and gastroenteritis (Ibrahim et al., 2013). Annually 500,000 Salmonella-related death reported worldwide (Deng et al., 2003). In Sudan, there is a continuous occurrence of foodborne-related outbreaks such as cholera (Shanan et al., 2011), the last cholera outbreakone was reported recently in December 2019 (Moskvitina et al., 2020).
Sudan is one of the African countries suffering from a substantial internal conflict that has weakened the country’s health system and affected the country's overall stability (Sulieman et al., 2020). Recently Khartoum and different parts of Sudan suffered from a flash flood (Elsafi, 2021), and during and after the floods different outbreaks were recorded (Ibrahim and Saeed, 2020, Abdelnabi et al., 2022). contaminated water among the main sources of food contamination (Abdelnabi et al., 2022). Bad hygiene and contamination of slaughterhouses were reported in Khartoum, in which a high percentage of E. coli, S. aureus, and Salmonella spp. were reported (Salih, 2020). A high prevalence of Salmonella species among food handlers was reported by Ahmed et al. (Ahmed et al., 2019), they reported out of 387 healthy food handlers 17 (4.4%) were positive for Salmonella species. Different outbreaks of cholera in Sudan were characterized as “watery diarrhea” recently the Sudanese minister of health declared a cholera outbreak in 2019 (Kunna, 2020). Diarrhea is considered one of the leading causes of hospital admission and death in Sudan, especially for those under 5 years old children (Elmanssury et al., 2022), contaminated food and water are the major cause of diarrhea.
There is a lack regarding the prevalence and source of the common born food pathogens in foods, so this study aimed to detect the presence of the most common foodborne pathogens from foods (raw milk, fresh cheese, yogurt, fish, sausage, mortadella, and eggs) collected in Khartoum state by using multiplex PCR.
2. Materials and methods
2.1. Samples collection and processing
This cross-sectional study was carried out in the Khartoum state (Khartoum- Bahri- Omdurman) during the period from May to June 2018. The permission to do this study was obtained from the Khartoum state ministry of health. Different food samples were collected as follows: Fresh cheese samples (n = 91) were collected from containers used regularly for selling and storage of fresh cheese at room temperature in shops. Milk samples (n = 54) were milked directly by herders into sterile containers from cows and goats (the cows' samples were from the same farm). Eggs (n = 29), fish (n = 20), locally prepared yogurt (n = 7), sausage (n = 4), and mortadella (n = 2) samples were collected from different markets and shops in Khartoum state (Khartoum, Bahri, and Omdurman). All samples were collected using sterile gloves in clean, sterile, and labeled containers, then immediately transferred to the laboratory for further processing. Samples not processed immediately were kept in −20 °C.
Ethical approval for sample collection was obtained from the University Medical Science & Technology ethical committee and the Khartoum state ministry of health.
2.2. DNA extraction
We used the guanidine chloride procedure for DNA extraction as described previously (Sabeel et al., 2017). Five grams of fresh cheese, fish, sausage, and mortadella samples, were used for DNA extraction. Sterile Pasteur pipettes are used to transfer five ml of milk, egg, and yogurt samples into sterile disposable falcon tubes (15 ml). Furthermore, 20 μl of proteinase K, 350 μl of ammonium acetate, 2 ml of cell lysis buffer, and 1 ml of guanidine chloride were mixed. The mixture was mixed and incubated overnight (at 37℃). A vortex was used to thoroughly combine the tube content after 2 ml of pre-chilled chloroform had been added. The mixture was then centrifuged for 10 min (at 6000 RPM). The DNA was precipitated by the addition of 8 ml of cold ethanol to each falcon tube while gently mixing after the supernatant was transferred into a new 15 ml falcon tube. After being centrifuged for 10 min at 6000 RPM, the pellets were rinsed with 4 ml of 70% ethanol and centrifuged for 10 min, then incubated at −20 °C overnight. The tubes were centrifuged and were blotted on filter paper, and allowed to air dry. Once the drying process was complete, 100 μl of DW was added, and the DNA elution process was completed and then stored at 4 °C for a short period until used for PCR.
2.3. Multiplex PCR
A set of primers were used to detect the Escherichia coli O157: H7 verocytotoxin stx gene, the Listeria. monocytogenes hemolysin hly gene, the Salmonella spp. invasion invA gene, the Vibrio cholerae toxin ctx gene, the Vibrio parahaemolyticus thermolabile hemolysin tlh gene, the S. aureus thermostable nuclease (nuc) gene (Lei et al., 2008) (Table 1).
Table 1.
Primer sequences and expected size of PCR-amplified gene targets of six species of foodborne pathogens (Lei et al., 2008).
| Pathogen | Primer name | DNA sequence (5ʼ to 3ʼ) | Amplicons size (bp) |
|---|---|---|---|
| V. cholerae |
ctxAB _F ctxAB _R |
TGAAATAAAGCAGTCAGGTG GGTATTCTGCACACAAATCAG |
777 |
| E. coli |
stx_F stx_R |
TGGGTTTTTCTTCGGTATCC CCAGTTCAGAGTGAGGTCCA |
632 |
| Salmonella spp. |
invA_F invA_R |
TACTAACAGTGCTCGTTTAC ATAAACTTCATCGCACCGTCA |
570 |
| V. parahaemolyticus |
tlh_F tlh_R |
CGGATTATGCAGAAGCACTG ACTTTCTAGCATTTTCTCTGC | 444 |
| S. aureus |
nuc_F nuc_R |
GCGATTGATGGTGATACGGTT AGCCAAGCCTTGACGAACTAAAGC |
270 |
| L. monocytogenes |
hly_F hly_R |
GCATCTGCATTCAATAAAGA TGTCACTGCATCTCCGTGGT |
174 |
The Maxime PCR Pre-Mix kit was used to perform multiplex PCR in a 25 μl volume (iNtRON Biotechnology, Seongnam, Korea). The premix was dissolved in 16 μl of Distilled Water and transferred into a 0.2 ml PCR tube. For each tube, 0.6 μl of each primer and 2 μl of DNA were added.
2.4. The protocol used for the amplification of the genes
A DNA thermal cycler was used to carry out each PCR reaction (K960 Healforce, China). The template DNA was initially denatured at 95 °C for 3 min, and the Taq polymerase was then activated. Then, 35 times each of the following PCR temperature cycling settings were used: primer annealing for 90 s at 55°Celsius, DNA extension for 90 s at 72°Celsius, and denaturation for 60 s at 95°Celsius. After target amplification, the reaction warmed and was held at 4 °C while the incompletely synthesized DNA underwent one final extension at 72 °C for 10 min. Instead of template DNA, a negative reaction control mixture including only sterile distilled water was utilized (Lei et al., 2008).
2.5. Detection of PCR-amplified products
Using the electrophoresis apparatus, the amplified PCR products were separated at 100 V for 30 min in a 1.5% (wt/vol) agarose gel containing ethidium bromide. In order to identify the individual amplified products, bands were compared with 100 bp of standard ladders (INTRON biotechnology, Korea) using a UV transilluminator (UVitec-UK).
3. Results
As shown in Table 2, the frequencies of collected (2 0 7) samples were as follows; 43 (20.8%) cow's milk, 11 (5.3%) goat's milk, 91 (44%) fresh cheese, 29 (14%) of egg, 7 (3.4%) yogurt, 20 (9.7%) fish, 4 (1.9%) sausage and 2 (1%) mortadella. Samples were collected from localities in Khartoum state as shown in Fig. 1.
Table 2.
Distribution and frequencies of collected samples according to Khartoum state localities.
| Samples | Locality |
|||
|---|---|---|---|---|
| Bahri | Khartoum | Omdurman | Total | |
| Fresh cheese | 16 (59.3%) | 27 (41%) | 48 (41.7)% | 91(44%) |
| Egg | 6 (22.2%) | 8 (12%) | 15 (13%) | 29 (14%) |
| Fish | 3 (11%) | 12 (19%) | 5 (4.3%) | 20 (9.7%) |
| Cow's Milk | 1(3.7%) | 9 (14%) | 33 (28.7%) | 43(20.8%) |
| Goat's Milk | 0 | 0 | 11(9.6%) | 11(5.3%) |
| Mortadella | 0 | 2(3%) | 0 | 2 (1%) |
| Sausage | 1(3.7%) | 2 (3%) | 1 (0.9%) | 4 (1.9%) |
| Yogurt | 0 | 5 (8%) | 2 (1.7%) | 7 (3.4%) |
| Total | 27 (100%) | 65(100%) | 115(100%) | 207(100%) |
Fig. 1.
Frequency of collected samples according to localities in Khartoum state.
Among all samples, eight foodborne pathogens were detected in seven samples (3.4%): five samples (2.4%) were positive for L. monocytogenes, one (0.48%) was positive for both V. cholerae, and V. parahaemolyticus and one (0.48%) was positive for S. aureus. From 91 fresh cheese samples, two (2.2%) were positive for L. monocytogenes, and one (1.1%) was positive for both V. cholerae and V. parahaemolyticus (Fig. 2). Out of 43 Cow's milk samples, three (7.0%) samples were positive for Listeria monocytogenes (Fig. 3), and out of 4 sausage samples, one (25.0%) was positive for S. aureus (Fig. 2), more details are in Table 3 and Table 4.
Fig. 2.
Multiplex PCR amplification on 1.5% agarose gel electrophoresis. Lane M is a DNA ladder: MW 100–1500 bp. Lane 7 shows a typical band size of (777 bp &444 bp) corresponding to the molecular size of ctx and tlh genes of V. cholerae, and V. parahaemolyticus. Lanes 1, 2, 3, 4, and 5 are negative samples.
Fig. 3.
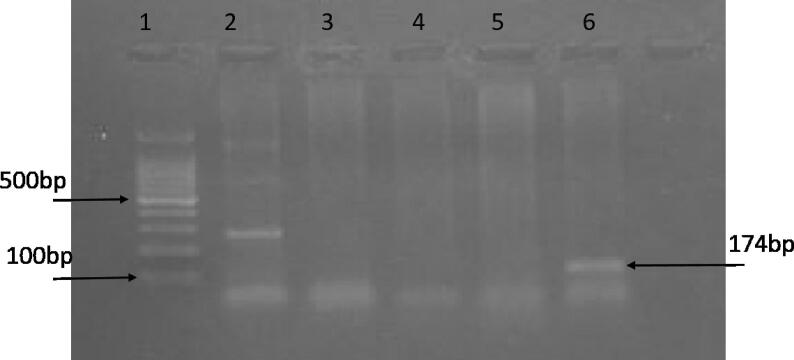
Multiplex PCR amplification on 1.5% agarose gel electrophoresis. Lane 1 is a DNA ladder: MW 100–1500 bp. Lane 2 shows a typical band size of (270 bp) corresponding to the molecular size of nuc gene of S. aureus. Lanes 3, 4, and 5 are negative samples. Lane 6 shows a typical band size of (174 bp) corresponding to the molecular size of hly gene of L. monocytogenes.
Table 3.
Frequencies of detected foodborne pathogens on different foods samples.
| sample | Foodborne pathogens |
||||
|---|---|---|---|---|---|
| L. monocytogenes | S. aureus | V. cholerae | V. parahaemolyticus | Total | |
| Fresh cheese (n = 91) | 2 (2.2%) | 0 | 1(1.1%) | 1(1.1%) | 4(4.4%) |
| Egg (n = 29) | 0 | 0 | 0 | 0 | 0 |
| Fish (n = 20) | 0 | 0 | 0 | 0 | 0 |
| Cow's Milk (n = 43) | 3 (7 %) | 0 | 0 | 0 | 3 (7%) |
| Goat's Milk (n = 11) | 0 | 0 | 0 | 0 | 0 |
| Mortedella (n = 2) | 0 | 0 | 0 | 0 | 0 |
| Sausage (n = 4) | 0 | 1 (25%) | 0 | 0 | 1 (25%) |
| Yogurt (n = 7) | 0 | 0 | 0 | 0 | 0 |
| Total (n = 207) | 5 (2.4%) | 1 (0.48%) | 1 (0.48%) | 1 (0.48%) | 8 (3.9%) |
Table 4.
Frequencies of positive samples according to storage type.
| Storage | Results |
Total | ||||
|---|---|---|---|---|---|---|
| -ve | Lm | S. aureus |
V. cholerae |
V. haemolyticus | ||
| Cooled | 21 | 1 | 0 | 0 | 0 | 22 |
| 95.5% | 4.5% | 0.0% | 0.0% | 0.0% | 100.0% | |
| Freeze | 8 | 0 | 1 | 0 | 0 | 9 |
| 88.9% | 0.0% | 11.1% | 0.0% | 0.0% | 100.0% | |
| Fresh | 35 | 3 | 0 | 0 | 0 | 38 |
| 92.1% | 7.9% | 0.0% | 0.0% | 0.0% | 100.0% | |
| RT | 136 | 1 | 0 | 1 | 1 | 138 |
| 97.9% | 0.7% | 0.0% | 0.7% | 0.7% | 100.0% | |
| Total | 200 | 5 | 1 | 1 | 1 | 207 |
| 96.1% | 2.4% | 0.5% | 0.5% | 0.5% | 100.0% | |
Abbreviations: RT, room temperature; Lm, Listeria monocytogenes; -ve, negative.
The detected organisms were from the Omdurman locality (n = 4): Three were L. monocytogenes from fresh milk, V. cholerae, and V. parahaemolyticus detected in one fresh cheese sample. In the Khartoum locality, three samples were positive: one S. aureus was detected in one sausage sample, and L. monocytogenes were detected in two fresh cheese samples.
According to storage type, out of 22 samples that were stored in the refrigerator (cooled) 1 (4.5%) sample was positive for Listeria monocytogenes, out of 9 samples that were stored in a freezer 1 sample (11.1%) was positive with S. aureus, out of 38 fresh samples 3 (7.9%) samples were positive with Listeria monocytogenes, out of 138 samples which were stored at room temperature 1(0.7%) sample was positive for S. aureus and 1 (0.7%) sample was positive with two organisms which were Vibrio cholera and Vibrio parahaemolyticus as described in details in supplementary material Table S1.
No foodborne pathogens were found in Bahri and Khartoum localities, while in the Omdurman locality three foodborne pathogens were found in different types of samples, Listeria monocytogenes were found in 3 fresh milk samples, V. cholerae and V. parahaemolyticus were found in 1 cheese sample. While in the Jabal Awleya locality two foodborne pathogens were found in different types of samples, S. aureus found in 1 sausage sample and Listeria monocytogenes found in 2 cheese samples.
4. Discussion
Foodborne infections are a serious problem affecting public health worldwide (Odeyemi, 2016). A large percentage of food-borne illnesses are caused by food-borne pathogens, and diarrheal agents comprise more than half of the overall prevalence of food-borne illnesses in the globe, the developing countries are more affected (Donkor, 2020). In this study 207 food samples were investigated for the presence of the commonly reported foodborne pathogens; 5 samples (2.4%) were positive for L. monocytogenes, one sample (0.48%) was positive for V. cholerae and V. parahaemolyticus, and one sample (0.48%) was positive for S. aureus.
L. monocytogenes is a bacterium that can infect humans and animals and can cause an outbreak of listeriosis (Hunt et al., 2012). L. monocytogenes was positive in three milk samples, and two fresh cheese samples, the presence of this organism in milk samples could be a result of these cows being infected with listeriosis or from contaminated udders. This hypothesis is supported by that all positive samples were collected from the same herd. Several reports from different regions of the world have confirmed the presence of L. monocytogenes in milk (Hunt et al., 2012, Olaimat et al., 2018, Moosavy et al., 2014). In Sudan, L. monocytogenes was detected previously in broiler chicken also (Alsheikh et al., 2012). The presence of L. monocytogenes in fresh cheese may be a result of the fact that fresh cheese is traditionally prepared from fresh milk that could be contaminated with these bacteria, or the contamination happened during the preparation of fresh cheese (Moosavy et al., 2014). The presence of L. monocytogenes in fresh cheese is more dangerous than in milk because fresh cheese is used directly, while milk goes through a heating process before use.
One fresh cheese sample was positive for both V. cholerae and V. parahaemolyticus. High temperatures will accelerate the growth of Vibrio cholera species (Asadgol et al., 2019), and raw foods will serve as good conditions for the growth and dissemination of these species. Although fermented foods could hinder cholera progression here we reported the presence of V. cholera DNA, which may be a part of a dead organism originating from a contaminated fresh milk sample (Mao et al., 2018). Before a short time of sample collection (in 2019), an outbreak of V. cholera hit Sudan mainly in Khartoum (hit African, 2019), this could be a reason for the presence of this species in our sample.
Primers targeting the species-specific S. aureus nuc gene which encodes for the extracellular thermostable nuclease protein (TNase) proved to be a useful tool for the molecular identification of S. aureus isolates (Javid et al., 2018). The presence of Staphylococcal species in food is an indicator of food contamination, meat suitable for human use must be Staphylococcus-free (Khalid et al., 2019). Sausages are uncooked meat products and the chance to get contaminated during preparation and processing is high (Hassabo et al., 2012) (Khalid et al., 2019). In this study, S. aureus was detected in ¼ (25%) of the manufactured sausage samples, the obtained result is lower than the result obtained by Nagwa et al. (Nagwa, 2015), in which they found 50% of sausage samples collected from Khartoum State were positive for S. aureus. While our finding is higher than Khalid et al. (Khalid et al., 2019) who zero reported S. aureus in investigated sausage samples from Khartoum State.
5. Limitations
Culturing of microorganisms was don’t performed due lack of resources at the time of the study, we depend only on PCR to screen a large number of different samples which required different types of culture media. The presence S. aureus is only screened by targeting the housekeeping gene (nuc), additional primers targeting the enterotoxin will differentiate the commensals from toxin strains.
6. Conclusion
This study revealed the presence of dangerous foodborne pathogens such as L. monocytogenes (2.4%) and V. cholera (1.1%) in raw milk and fresh cheese samples in Khartoum state. Their presence is considered a potential problem and needs intensive hygiene efforts and standard safety measures before, during, and after processing operations. Among 43 samples of cow's fresh milk, 3 of them (7%) tested positive for Listeria monocytogenes in samples from the Omdurman locality from the same herd suggesting the endemic presence of these bacteria in the herd.
7. Authors’ contributions
HNA, RMB and EHM conceived the study. KC and HNA did the statistical analysis and drafted the manuscript. RMB did the investigations and sample collection. All authors contributed to the writing of the paper and approved it.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors acknowledge the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah for funding this work under Grant No. D-302-130-1441.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2023.103653.
Contributor Information
Hisham N. Altayb, Email: hdemmahom@kau.edu.sa.
Rania M. Badri, Email: ahmed.hassanab@sustech.edu.
Kamel Chaieb, Email: kalshaib@kau.edu.sa.
Ehssan Moglad, Email: e.moglad@psau.edu.sa.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data Availability
All data used in this study are included in the manuscript.
References
- Abdelnabi G.H., Abdelaziz S.A., Alia O., Abeer A., Hind E., Sabiel Y., Muna E., Abass A.M. Assessment of bacterial contamination in drinking water sources in Khartoum/Sudan. J. Adv. Microbiol. 2022:50–56. [Google Scholar]
- Ahmed N.E.M.A., Hassan A.N., Holi M.A.I., Galander I. Prevalence and antibiotic susceptibility of salmonella species among food handlers in Khartoum-State (Sudan) Prevalence. 2019;35 [Google Scholar]
- Alsheikh A., Mohammed G., Abdalla M. First isolation and identification of listeria monocytogenes from fresh raw dressed broiler chicken in Sudan. Res. J. Microbiol. 2012;7:319–326. [Google Scholar]
- Asadgol Z., Mohammadi H., Kermani M., Badirzadeh A., Gholami M. The effect of climate change on cholera disease: the road ahead using artificial neural network. PLoS One. 2019;14:e0224813. doi: 10.1371/journal.pone.0224813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Noordhout C.M., Devleesschauwer B., Angulo F.J., Verbeke G., Haagsma J., Kirk M., Havelaar A., Speybroeck N. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Liou S.R., Plunkett G., III, Mayhew G.F., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor E.S. Cockroaches and food-borne pathogens. Environ. Health Insights. 2020;14 doi: 10.1177/1178630220913365. 1178630220913365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmanssury A., Elnadif D., Safa A. Prevalence of diarrhea and association with socio-demographic factors among children under five in Mayo Camp-Khartoum State Sudan. Pakistan J. Medical Health Sci. 2022;16:1100. [Google Scholar]
- Elsafi, S. H. 2021. Difference in the extreme flooded years in Khartoum region using water Indices from Landsat images (1988-1994-1998-1999-2013-2020). مجلة بحوث کلية الآداب. جامعة المنوفية.
- Hassabo A., Eisa M., Ishag I., Osman S., Bushara I. Usage of antibiotic as milk preservative in the slums of Khartoum state. J. Anim. Prod. Adv. 2012;2:138–141. [Google Scholar]
- Heredia N., García S. Animals as sources of food-borne pathogens: a review. Animal nutrition. 2018;4:250–255. doi: 10.1016/j.aninu.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIT AFRICAN, F. 2019. Public health round-up. Bull World Health Organ, 97, 793-794. [DOI] [PMC free article] [PubMed]
- Hunt K., Drummond N., Murphy M., Butler F., Buckley J., Jordan K. A case of bovine raw milk contamination with Listeria monocytogenes. Ir. Vet. J. 2012;65:13. doi: 10.1186/2046-0481-65-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S., Asakir S., Idris A., Martinez-Urtaza J., Elsafi H.H. Prevalence of Salmonella species among asymptomatic food handlers in Khartoum State, Sudan. Brit. J. Biomed. Sci. 2013;70:88. doi: 10.1080/09674845.2013.11978267. [DOI] [PubMed] [Google Scholar]
- Ibrahim M.E.A., Saeed H.A. Serodetection of Hepatitis A Virus among Food-handlers in Khartoum Locality, Sudan. Microbiology. 2020 [Google Scholar]
- Havelaar, A.H., Kirk, M.D., Torgerson, P.R., Gibb, H.J., Hald, T., 2018. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. University of Southern California Los Angeles. [DOI] [PMC free article] [PubMed]
- Javid F., Taku A., Bhat M.A., Badroo G.A., Mudasir M., Sofi T.A. Molecular typing of Staphylococcus aureus based on coagulase gene. Vet. World. 2018;11:423. doi: 10.14202/vetworld.2018.423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaferstein F.K., Motarjemi Y.M., Moy G.G., Quevado F. Food safety: a worldwide public. International Food Safety Handbook: Science, International Regulation, and Control. 2019:1. [Google Scholar]
- Khalid A.M., Ali A.A., Hussain S.K., Awad F.N., Elhassan I.H., Ismaiel A.E., Basheer E.O. Microbial characteristics of meat products in Khartoum State. Int. J. Innovative Sci. Eng. Technol. 2019;6:225. [Google Scholar]
- Kunna, E. 2020. Sudan: Managing COVID-19 Pandemic During a Time of Transition.
- Lei I.-F., Roffey P., Blanchard C., Gu K. Development of a multiplex PCR method for the detection of six common foodborne pathogens. J. Food Drug Anal. 2008;16 [Google Scholar]
- Lim J.Y., Yoon J.W., Hovde C.J. A brief overview of Escherichia coli O157: H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010;20:5. [PMC free article] [PubMed] [Google Scholar]
- Mao N., Cubillos-Ruiz A., Cameron D.E., Collins J.J. Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 2018;10:eaao2586. doi: 10.1126/scitranslmed.aao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavy M.-H., Esmaeili S., Mostafavi E., Amiri F.B. Isolation of Listeria monocytogenes from milks used for Iranian traditional cheese in Lighvan cheese factories. Ann. Agric. Environ. Med. 2014;21 doi: 10.5604/12321966.1129923. [DOI] [PubMed] [Google Scholar]
- Moskvitina E.A., Yanovich E.G., Kurilenko M.L., Kruglikov V.D., Titova S.V., Levchenko D.A., Vodopyanov A.S., Lopatin A.A. Cholera: Monitoring of Epidemiological Situation around the World and in Russia. Forecast For. 2020 [Google Scholar]
- Nagwa, B., Elhag, E.R.B.B., Ahmed, A.M., 2015. The Prevalence of Staphylococci in.
- Odeyemi O.A. Public health implications of microbial food safety and foodborne diseases in developing countries. Food Nutr. Res. 2016;60 doi: 10.3402/fnr.v60.29819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaimat A.N., Al-Holy M.A., Shahbaz H.M., Al‐Nabulsi A.A., Abu Ghoush M.H., Osaili T.M., Ayyash M.M., Holley R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018;17:1277–1292. doi: 10.1111/1541-4337.12387. [DOI] [PubMed] [Google Scholar]
- Sabeel S., Salih M.A., Ali M., El-Zaki S.E., Abuzeid N., Elgadi Z.A.M., Altayb H.N., Elegail A., Ibrahim N.Y., Elamin B.K. Phenotypic and genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Sudanese patients. Tuberculosis Res Treatment. 2017 doi: 10.1155/2017/8340746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo, C.M. 2018. Modification of the treatment protocol as a strategy in the control of the cholera epidemic in Haiti 2016-2017.
- Salih M.M.E. Sudan University of Science & Technology; 2020. Assessment of the Microbial Qualities of Exported Sheep and Goats’ Carcasses and the Hygiene Conditions of an Export Slaughterhouse in Khartoum State-Sudan. [Google Scholar]
- Shanan S., Abd H., Hedenström I., Saeed A., Sandström G. Detection of Vibrio cholerae and Acanthamoeba species from same natural water samples collected from different cholera endemic areas in Sudan. BMC. Res. Notes. 2011;4:109. doi: 10.1186/1756-0500-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman A.M.E., Dafallah F.E., Abdel-Rahman E.H., Alshammari N.I., Shommo S.A., Ibrahim S.E. Isolation, Identification and Characterization of Salmonella spp. from Chicken purchased at Wad Madani City, Gezira State, Sudan. Advancements Life Sci. 2020;8:98–102. [Google Scholar]
- Swaminathan B., Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
Further Reading
- Sausages at Khartoum State IJSR – Int. J. Sci. Res. 4 175-178.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are included in the manuscript.