Abstract
The complete nucleotide sequence of Tn10 has been determined. The dinucleotide signature and percent G+C of the sequence had no discontinuities, indicating that Tn10 constitutes a homogeneous unit. The new sequence contained three new open reading frames corresponding to a glutamate permease, repressors of heavy metal resistance operons, and a hypothetical protein in Bacillus subtilis. The glutamate permease was fully functional when expressed, but Tn10 did not protect Escherichia coli from the toxic effects of various metals.
Tn10 is a composite transposon in which the genes for tetracycline resistance are flanked by inverted repeats of IS10 elements. The present isolate of Tn10 originated from the enteric bacterium Shigella flexneri, where it was discovered on a drug resistance factor related to the F episome in Escherichia coli. According to Sharpe et al. (15), this factor was first isolated by Nakaya et al. (11) as NR1 and later referred to as R222 by Watanabe and Fukasawa (19) and as R100 by Sugino and Hirota (16).
Electron microscopy of the heteroduplex formed by self annealing of single-stranded R100 DNA gave an early indication of the unusual structure of Tn10 (15, 17). A single-stranded DNA loop of 6.4 kb was seen to emanate from a double-stranded “stalk” of 1.4 kb (15). Of course, the stalk, or stem, was formed by base pairing of the inverted repeats of IS10, and the loop represented the unique sequences which contain the tetracycline resistance genes.
Tn10 is one of the most thoroughly studied transposons (8, 9), but until now there was available no accurate DNA sequence for almost 50% of Tn10. In the loop material, there was only about 500 bp of somewhat inaccurate sequence to the left of tetR, and almost half of IS10-Left remained unsequenced. In the newly sequenced region, there are three new open reading frames (ORFs) and an additional difference between the transposase genes of IS10-Left and IS10-Right.
We note that during the preparation of this article, the complete sequence of R100 was deposited in GenBank under the accession number AP000342.
Sequence determination.
The complete nucleotide sequence of Tn10 was determined using a primer walking strategy (Fig. 1). The gap between tetR and IS10-Left has been closed with almost 3.5 kb of new sequence. The regions previously in the database were resequenced and 18 errors were corrected.
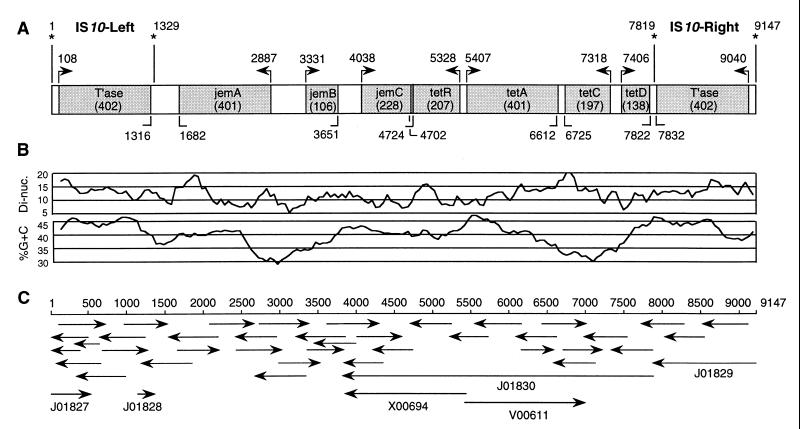
FIG. 1.
Sequencing strategy and map of Tn10. (A) The sequence of Tn10 was used to construct an ORF map which shows the name of each ORF and the predicted number of amino acids. The direction and location of predicted translational start sites are indicated above the map. The locations of predicted stop codons are indicated below the map. The locations of the ends of the flanking IS elements are indicated by asterisks. Analysis of DNA sequences was carried out using web-based resources at the National Center for Biotechnology Information: ORF Finder, BLAST, GenBank, and SwissProt. Additional searches were performed using OmniBlast at the Sanger Centre. (B) The percent G+C and the percent difference in dinucleotide signature were plotted for Tn10 using a circular permutation of the sequence and a window size of 500 bp by N. J. Saunders, J. S. Mirsky, and S. Jarvis at The Institute of Molecular Medicine, University of Oxford. (C) Tn10 was sequenced using a primer walking strategy with plasmids pNK81, pNK82, and pNK83 (4) as templates. The location, direction, and length of sequence data acquired from each sequencing reaction are indicated by arrows below the scale bar. Regions of Tn10 sequenced previously are indicated by arrows accompanied by GenBank accession numbers. Oligonucleotide primers were from Sigma Genosys, and fluorescent sequencing was carried out by the Department of Biochemistry DNA Sequencing Unit using machines from Applied Biosystems.
Tn10 is 9,147 bp long with a G+C content of 40%. There are nine substantial ORFs with homology to entries in sequence databases. Of the nine, three were previously unknown, and these have been designated jemA, jemB, and jemC. The database entries X00694 and J01830 (Fig. 1C) contain the entire sequence of jemC, which was not recognized previously because of two sequencing errors which resulted in frame shifts.
Properties of the sequence.
The sequence of Tn10 was analyzed by plotting the percent G+C content and the percent difference of the dinucleotide signature (Fig. 1B). These parameters are sensitive to the “foreignness” of DNA sequences, and sudden discontinuities may indicate junctions at which DNAs from different sources have been joined by recombination (7). For Tn10, the percent difference in dinucleotide signature had limited variation, being confined almost exclusively within the 5 to 15% range (Fig. 1B). The G+C content of Tn10 was also confined to a narrow range, and it therefore appears that Tn10 does not contain any regions composed of DNA from widely divergent sources. The regions of Tn10 with the highest percent G+C were the flanking IS10 elements, which had an average of 44% G+C. Within the limited range, the fluctuation of percent G+C and dinucleotide signature was relatively smooth and did not correlate with other features of the sequence such as ORFs. Together, these observations suggest either that the constituent parts of Tn10 come from a single source or that Tn10 is old enough for the base composition and dinucleotide signature of different modules to have been homogenized.
Whatever the evolutionary age of Tn10 in its present form, the G+C content of 40% indicates that it probably did not arise in the enteric bacteria, which have a base composition much closer to 50%. This idea is supported by the sequence of plasmid R100, the source of the present isolate of Tn10, which has 52% G+C (GenBank no. AP000342). Another indication that Tn10 may not be a native component of the enteric bacteria is that the closest relatives in the sequence database of jemB and jemC are genes encoding hypothetical proteins from Bacillus subtilis, a gram-positive organism with a base composition of about 45% G+C. The implication of the difference in base composition of Tn10 and its host organism is that Tn10 may be imperfectly adapted to enteric bacteria such as E. coli and Salmonella enterica serovar Typhimurium, where its behavior and patterns of gene regulation may not accurately reflect those in the original host.
JemA is a sodium-dependent glutamate permease.
The jemA ORF codes for a predicted protein of 401 amino acids which is related to the sodium-dependent glutamate permease from E. coli and Haemophilus influenzae (2, 5). Wild-type E. coli K-12 is unable to grow on glutamate as a source of carbon, nitrogen, or energy because it is not transported across the cell membrane. We therefore tested whether the presence of pNK81, a pBR322-based derivative which carries a complete copy of Tn10, would allow E. coli K-12 strains to grow on minimal glutamate media. Strains MK416 (5) and MM294 (13) were transformed with pNK81 but failed to grow on M9 glutamate agar.
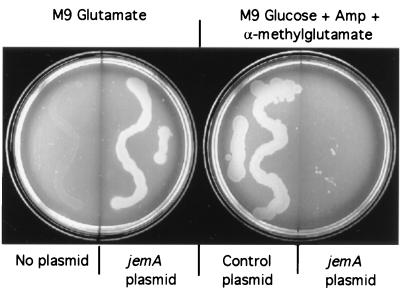
The failure of Tn10 to complement the Glt− phenotype could have been due to the inability of E. coli K-12 to express the protein. We therefore cloned jemA downstream of the ptac promoter. Expression of JemA in response to IPTG (isopropyl-β-d-thiogalactopyranoside) complemented the Glt− phenotype of MK416 (Fig. 2). Expression of JemA also rendered the cells sensitive to the toxic effects of α-methylglutamate (Fig. 2), indicating that JemA is a GltI type transporter in which glutamate transport is driven by the influx of sodium (5).
FIG. 2.
jemA encodes a functional glutamate transporter. E. coli K-12 gltS deletion strain MK416 (ΔgltS Kanr prototroph) (5) was plated on M9 minimal media. (Left) Plate with 0.5 mM IPTG and 0.5% glutamate as the sole source of carbon and energy. Cells were either untransformed or transformed with pRC163 expressing jemA from the ptac promoter. (Right) Plate with M9 minimal glucose agar containing 100 μg of ampicillin/ml, 0.5 mM IPTG, and 40 μg of α-methylglutamate/ml. Cells were transformed either with the control plasmid pBluescript (Stratagene) or with pRC163 expressing jemA from the ptac promoter.
jemB.
The jemB ORF codes for a predicted protein of 106 amino acids which has homology to an ORF encoding 116 amino acids located in the glnQ-ansR intragenic region of B. subtilis (SwissProt P54563). The level of identity between the proteins is only 25% but is highly significant as judged by a Monte Carlo statistical analysis (12), which gave a P value of 3.8e-13. JemB is more distantly related to another protein from Mycobacterium tuberculosis (Sanger Centre gene Rv3592) and a 166-amino-acid protein of unknown function from B. subtilis (SwissProt P38049), the gene for which is located next to a gene coding for a class A penicillin-binding protein. At present there is no known function for JemB or any of the homologs mentioned above. However, the presence of several homologs in the database, albeit hypothetical proteins, makes it likely that these are members of a family of functional proteins. Also, there is a strong predicted transcriptional start site 85 bp upstream of jemB, and it has an excellent putative ribosome-binding site of CGGAGA (see Promoter Prediction by Neural Network by Martin Reese at http://www.fruitfly.org/seq_tools/promoter.html).
jemC.
The jemC ORF overlaps tetR by 17 bp and codes for a predicted protein of 228 amino acids (Fig. 1). The first 100 amino acids of JemC are highly homologous to a family of bacterial transcriptional regulators which repress the arsenic and mercury resistance operons (6, 14). One of the most highly conserved regions between the proteins corresponds to the putative helix-turn-helix DNA-binding motif of the repressors (data not shown).
The N-terminal half of JemC is also homologous to the N-terminal half of a putative arsenate reductase (18). The arsenate reductase appears to have been assembled by the fusion of an N-terminal metal-binding repressor-like domain and a C-terminal reductase domain. By analogy with the reductase, it seems likely that the N-terminal portion of JemC constitutes a metal/DNA-binding domain and the C-terminal domain performs an as-yet-unrecognized function. JemC is also 39% identical over its entire length to the hypothetical protein YdfF from B. subtilis. This is a strong indication that both of these proteins are functional and that jemC is not simply a fusion between the remnants of two unrelated and degenerate ORFs.
The presence of Tn10 in strains of E. coli K-12 was not sufficient to provide resistance to salts of cadmium, arsenite, arsenate, mercury, cobalt, or sodium (data not shown). However, the test would not have detected less than a two-fold difference in sensitivity, and partial resistance to these metals cannot be ruled out. There is also the possibility that any resistance determinants which might be present do not function well in E. coli.
Intragenic regions.
Three intragenic regions of significant size occur in the newly sequenced half of Tn10 (Fig. 1). The region between jemA and IS10-Left is large enough to code for 118 amino acids. This region contains an ORF encoding 41 amino acids; this ORF appears to be the remnant of a gene related to the E. coli lysR transcription activator which has been interrupted by the insertion of the inside end of IS10-Left. The region between jemA and jemB shows no significant homology to any entries in the sequence database (see above).
There is another 165-amino-acid-encoding ORF at position 3,533 to 4,030 of the Tn10 sequence which overlaps the C terminus of jemB. It is homologous to hypothetical protein Cj1032 in Campylobacter jejuni. However, it is likely to be a degenerate remnant because the homology with Cj1032 is limited to the region that does not overlap jemB, which appears to be a complete protein with close relatives in the database (see above).
Distribution of Tn10/IS10 in bacterial species.
Tn10 has only two partial matches in the public sequence database, which includes 23 complete bacterial genomes, partial coverage of 82 bacterial genomes (work is ongoing), and 47 bacterial plasmids. The most extensive match is from the S. enterica serovar Typhi sequence at the Sanger Centre and covers the first 5,700 bases of Tn10 extending from IS10-Left into tetA. The second match from the database is from the multiantibiotic resistance locus of S. flexneri (GenBank no. G4098955), which resembles a portion of the loop region of Tn10.
Examples of IS10 are also sparsely represented in the public database. Three exact matches to IS10-Left are found in the almost-completed Salmonella serovar Typhi sequence at the Sanger Centre. One of these is associated with the truncated copy of Tn10 mentioned above. Another is flanked by the direct repeat of 9 bp characteristic of IS10/Tn10 insertions. The similarity of the repeat, GCANAGC, to the perfect consensus target hot spot, GCTNAGC (3), indicates that this insertion arrived at this location by transposition and begs the question as to the source of the transposase, since that encoded by IS10-Left is largely defective (8).
IS10 is perhaps more widespread among the enteric bacteria than the paucity of entries in the sequence database may suggest. Using DNA hybridization to survey repetitive sequences in bacteria, Matsutani (10) found 15 copies of IS10-Right in the chromosome of Enterobacter cloacae MD36, nine in Shigella sonnei HH109, and several in natural isolates of E. coli. The number of IS elements in the majority of bacterial genomes sequenced so far has proven to be limited to one or a few copies (1), and the high copy number of IS10 in MD36 is therefore unusual and remains to be explained.
Nucleotide sequence accession number.
The complete sequence of Tn10 was deposited in GenBank (accession number AF162223).
Acknowledgments
This work was funded by grants from the Wellcome Trust and The Royal Society. R.C. is a Royal Society University Research Fellow.
REFERENCES
- 1.Chalmers R, Blot M. Insertion sequences and transposons. In: Charlebois R L, editor. Organization of the prokaryotic genome. Washington, D.C.: American Society for Microbiology; 1999. pp. 151–169. [Google Scholar]
- 2.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 3.Halling S M, Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982;28:155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- 4.Halling S M, Simons R W, Way J C, Walsh R B, Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci USA. 1982;79:2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalman M, Gentry D R, Cashel M. Characterization of the Escherichia coli K12 gltS glutamate permease gene. Mol Gen Genet. 1991;225:379–386. doi: 10.1007/BF00261677. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 7.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 8.Kleckner N. Transposon Tn10. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 227–268. [Google Scholar]
- 9.Kleckner N, Chalmers R M, Kwon D K, Sakai J, Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. In: Saedler H, Gierl A, editors. Transposable elements. Vol. 204. Berlin, Germany: Springer; 1996. pp. 49–82. [DOI] [PubMed] [Google Scholar]
- 10.Matsutani S. Multiple copies of IS10 in the Enterobacter cloacae MD36 chromosome. J Bacteriol. 1991;173:7802–7809. doi: 10.1128/jb.173.24.7802-7809.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakaya R, Nakamura A, Murata Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- 12.Pearson W R. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Sedlmeier R, Altenbuchner J. Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol Gen Genet. 1992;236:76–85. doi: 10.1007/BF00279645. [DOI] [PubMed] [Google Scholar]
- 15.Sharp P A, Cohen S N, Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973;75:235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- 16.Sugino Y, Hirota Y. Conjugal fertility associated with resistance factor R in Escherichia coli. J Bacteriol. 1962;84:902–910. doi: 10.1128/jb.84.5.902-910.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tye B K, Chan R K, Botstein D. Packaging of an oversize transducing genome by Salmonella phage P22. J Mol Biol. 1974;85:485–500. doi: 10.1016/0022-2836(74)90311-8. [DOI] [PubMed] [Google Scholar]
- 18.Vlcek C, Paces V, Maltsev N, Paces J, Haselkorn R, Fonstein M. Sequence of a 189-kb segment of the chromosome of Rhodobacter capsulatus SB1003. Proc Natl Acad Sci USA. 1997;94:9384–9388. doi: 10.1073/pnas.94.17.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe T, Fukasawa T. Episome-mediated transfer of drug resistance in Enterobacteriaceae. III. Transduction of resistance factors. J Bacteriol. 1961;82:202–209. doi: 10.1128/jb.82.2.202-209.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]