Abstract
A DNA cleavage reagent, specifically tethered to residue 581 of the Escherichia coli RNA polymerase ς70 subunit, has been used to investigate the location of ς70 region 4 in different complexes at the galp1 promoter and the effect of the cyclic AMP receptor protein. The positions of DNA cleavage by the reagent are not affected by the cyclic AMP receptor protein. We conclude that transcription activation at the galp1 promoter by the cyclic AMP receptor protein does not involve major conformation changes in or repositioning of ς70 region 4.
Many bacterial transcription activators bind to DNA sites that overlap the −35 region of target promoters and interact directly with holo-RNA polymerase (RNAP). In most cases, these activators have specific interactions with region 4 of the principal RNAP ς subunit, ς70, that appear both to recruit RNAP to the promoter DNA and to accelerate the transition from the closed to the open complex (4, 8, 16; see also reference 11). Although it is well known that a major function of ς70 region 4 is to contact promoter −35 elements, little is known about activator-ς70 interactions (7). For example, does the activator-ς70 interaction merely recruit ς70 region 4 to the target promoter or does it reposition region 4? To investigate this point, we have exploited previous work in which ς70 region 4 was specifically labeled with a DNA cleavage reagent. DNA cleavage by this reagent was then used to investigate the position of region 4 of ς70 in different RNAP-promoter complexes.
Previous publications have described the cloning of the rpoD gene (encoding ς70) that had been mutated to remove all three cysteine codons and the subsequent introduction of a cysteine codon at position 581 in ς70 region 4 (13, 14). We previously overexpressed and purified [Cys581] ς70, showed that it was functional, and described how Cys581 could be specifically tagged with the DNA cleavage agent p-bromoacetamidobenzyl-EDTA-Fe (FeBABE). It was shown previously that RNAP, reconstituted with FeBABE-tagged [Cys581] ς70, could form open complexes at different activator-independent promoters (3, 13). Activation of the tagged FeBABE reagent by addition of sodium ascorbate leads to cleavage of specific bases in the −35 region of the promoter DNA.
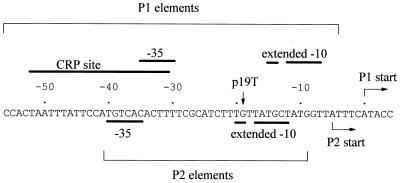
In this work, we have studied the interactions of RNAP containing FeBABE-tagged [Cys581] ς70 with the Escherichia coli gal operon promoter region, which contains two overlapping promoters, P1 and P2, that are regulated by the cyclic AMP receptor protein (CRP) (Fig. 1). It is well established that, in the absence of CRP, RNAP initiates transcripts at P2. CRP binds to a DNA target that overlaps the −35 element of both promoters and switches RNAP from P2 to P1 by repressing P2 and activating P1 (12). Here we have exploited the galp19T mutation that totally inactivates P2 (Fig. 1). In previous studies with galp19T, it was shown that P1 can function in the absence of CRP but that transcription initiation at P1 is stimulated by CRP (2, 9, 10). Thus, the use of galp1 allows us to study the position of ς70 region 4 in open complexes at the same promoter either with or without CRP. In this work, we used previously established protocols to purify wild-type and mutant CRP derivatives (6, 15) and DNA fragments carrying gal promoter region sequences that were labeled on either strand (1). Since very similar results were obtained with fragments labeled on either strand, Fig. 2 and 3 show experiments solely with fragments labeled on the template strand.
FIG. 1.
Base sequence of the wild-type E. coli gal operon regulatory region, showing the two overlapping promoters P1 and P2. The sequence is numbered with the P1 transcript start as +1. Above the sequence are shown the various P1 elements: the 22-bp DNA site for CRP and the −35 and extended −10 elements. Below the sequence are shown the P2 −35 and extended −10 element and the P2 start point. The location of the p19T substitution, which completely inactivates P2, is indicated. With the gal regulatory region carrying the p19T substitution, RNAP initiates transcription at P1 in both the absence and the presence of CRP.
FIG. 2.
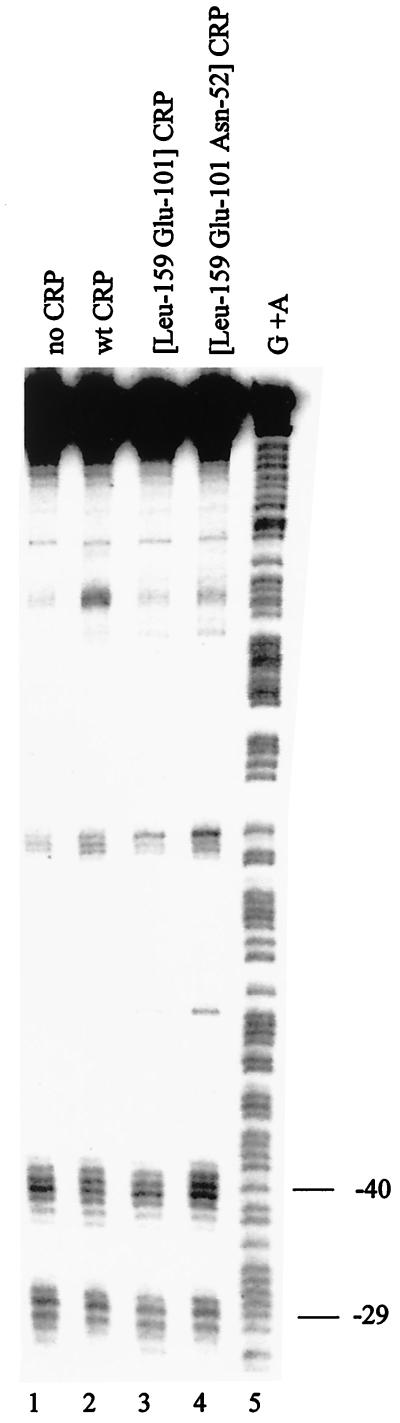
DNA cleavage of galp19T by RNAP containing FeBABE-tagged [Cys581] ς70. Purified wild-type (wt) CRP (150 nM) or mutant derivatives were mixed, as indicated, with 32P-end-labeled AatII-HindIII DNA fragments (0.4 nM, labeled on the template strand) carrying the gal promoter region with the p19T mutation in a reaction volume of 35 μl [20 mM HEPES (pH 8.0), 5 mM MgCl2, 200 μM cyclic AMP, 50 mM potassium glutamate, 50 μg of bovine serum albumin per ml, 5 μg of poly(dI-dC)]. RNAP holoenzyme (30 nM) reconstituted with FeBABE-tagged [Cys-581] ς70 was then added and incubated at 37°C for 20 min. Complexes were challenged with heparin (200 μM for 5 min), and DNA cleavage was initiated by the addition of sodium ascorbate (2 mM) followed by incubation at 37°C for 20 min. Modified DNA was extracted with phenol-chloroform and analyzed on a 6% polyacrylamide sequencing gel, which was calibrated with Maxam-Gilbert G+A sequence ladders and processed and scanned using a Molecular Dynamics PhosphorImager (full protocols are given in reference 3). Bands due to DNA cleavage near positions −40 and −29 upstream of the galp1 transcription start point are indicated. Lane 1, no CRP; lane 2, wild-type CRP; lane 3, [Leu159 Glu101] CRP; lane 4, [Leu159 Glu101 Asn52] CRP; lane 5, G+A sequencing ladder.
FIG. 3.
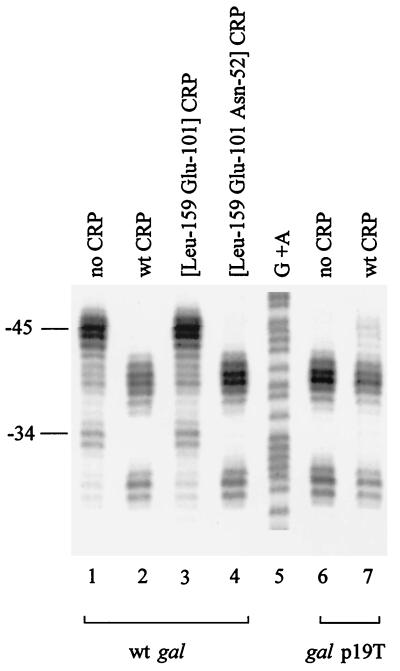
DNA cleavage of wild-type (wt) gal by RNAP containing FeBABE-tagged [Cys581] ς70. Purified wild-type CRP (150 nM) or mutant derivatives were mixed, as indicated, with 32P-end-labeled AatII-HindIII DNA fragments (0.4 nM, labeled on the template strand) carrying gal promoter DNA in a reaction volume of 35 μl, and RNAP holoenzyme (30 nM) reconstituted with FeBABE-tagged [Cys581] ς70 was then added. Samples were processed exactly as described in the legend to Fig. 2 (full protocols are given in reference 3). Bands due to DNA cleavage at different positions upstream of the galp1 transcription start point are indicated. Lane 1, wild-type gal, no CRP; lane 2, wild-type gal, wild-type CRP; lane 3, wild-type gal, [Leu159 Glu101] CRP; lane 4, wild-type gal, [Leu159 Glu101 Asn52] CRP; lane 5, G+A sequencing ladder; lane 6, galp19T, no CRP; lane 7, galp19T, wild-type CRP.
Figure 2 shows the pattern of DNA cleavage that results when open complexes were formed by RNAP containing FeBABE-tagged [Cys581] ς70 at galp1, using DNA carrying the galp19T mutation. Consistent with previous reports (3, 13), the FeBABE tag results in cleavage of promoter DNA near positions −29 and −40. Our results show that the pattern of cleavage is identical in both the absence and the presence of CRP, implying that CRP does not reposition ς70 region 4 during transcription activation. To confirm this conclusion, we exploited two CRP derivatives that are altered in their ability to activate transcription at promoters such as galp1, where the DNA site for CRP overlaps the target promoter −35 element (class II CRP-dependent promoters: reviewed in reference 5). First, [Leu159 Glu101] CRP carries the His159Leu and Lys101Glu substitutions that inactivate the two principal activating regions of CRP that interact with RNAP at class II CRP-dependent promoters (5, 15). Second, [Leu159 Glu101 Asn52] CRP carries an additional substitution, Lys52Asn, that improves the function of a third activating region that is known to interact directly with ς70 region 4 (11). Previous studies have shown that the Lys52Asn substitution in CRP greatly increases the ability of CRP to activate transcription at class II CRP-dependent promoters (11, 17). The results in Fig. 2 show that these substitutions have little or no effect on the pattern of digestion due to FeBABE-tagged [Cys581] ς70 at galp1.
As a control, the experiment with galp19T DNA shown in Fig. 2 was repeated in exactly the same conditions, using the wild-type gal regulatory region. Figure 3 shows the DNA cleavage due to the FeBABE tag when RNAP containing FeBABE-tagged [Cys581] ς70 was incubated with wild-type gal DNA either with or without different CRP derivatives. In the absence of CRP, the FeBABE tag results in cleavage of promoter DNA near positions −34 and −45, due to occupation of P2 (see Fig. 1 for the location of P2). The presence of CRP shifts the sites of DNA cleavage downstream to positions −29 and −40, as P2 is repressed and P1 is activated (this pattern of cleavage is similar to that seen with galp19T DNA). Further results in Fig. 3 show that [Leu159 Glu101 Asn52] CRP but not [Leu159 Glu101] CRP is able to shift RNAP from P2 to P1. The result with [Leu159 Glu101] CRP was unexpected and implies that, in our conditions, mere occupation of the DNA site for CRP is insufficient to shift RNAP from P2 to P1. However, since with [Leu159 Glu101 Asn52] CRP the sole functional activating region of CRP interacts with ς70 region 4, we can infer that this interaction is sufficient to shift RNAP from P2 to P1 at the wild-type gal operon regulatory region.
The main conclusion from this work comes from the study with galp19T, where RNAP can form open complexes at P1 both in the absence and in the presence of CRP. Our study shows that both [Leu159 Glu101 Asn52] CRP and wild-type CRP, in conditions where they can stimulate transcription and participate in ternary CRP-RNAP-galp1 complexes, have no effect on the location of ς70 region 4. Thus, we conclude that the primary function, at this particular promoter, of the CRP-ς70 interaction is not to trigger a major conformation change or repositioning of ς70 region 4 but rather to recruit region 4 to the promoter −35 element. It will be interesting to see if this conclusion holds for other transcription activators that bind to targets that overlap promoter −35 elements and interact with ς70.
Acknowledgments
This study was funded by a project grant from the Wellcome Trust and an Anglo-Japanese Scientific Collaboration Award from the Royal Society.
REFERENCES
- 1.Belyaeva T, Bown J, Fujita N, Ishihama A, Busby S. Location of the C-terminal domain of the RNA polymerase alpha subunit in different open complexes at the E. coli galactose operon regulatory region. Nucleic Acids Res. 1996;24:2243–2251. doi: 10.1093/nar/24.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingham A, Ponnambalam S, Chan B, Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41:67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- 3.Bown J, Owens J, Meares C, Fujita N, Ishihama A, Busby S, Minchin S. Organization of open complexes at Escherichia coli promoters: location of promoter DNA sites close to region 2.5 of the ς70 subunit of RNA polymerase. J Biol Chem. 1999;274:2263–2270. doi: 10.1074/jbc.274.4.2263. [DOI] [PubMed] [Google Scholar]
- 4.Busby S, Ebright R. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–748. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Busby S, Ebright R. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 6.Ghosaini L, Brown A, Sturtevant J. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry. 1988;27:5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- 7.Gross C, Lonetto M, Losick R. Bacterial sigma factors. In: McKnight S, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 129–176. [Google Scholar]
- 8.Ishihama A. Promoter selectivity control of RNA polymerase. In: Eckstein F, Lilley D, editors. Mechanisms of transcription. Heidelberg, Germany: Springer-Verlag; 1997. pp. 53–70. [Google Scholar]
- 9.Lavigne M, Kolb A, Buc H. Transcription activation by cAMP receptor protein at the Escherichia coli galP1 promoter—crucial role for the spacing between the CRP binding site and the −10 region. Biochemistry. 1992;31:9647–9656. doi: 10.1021/bi00155a018. [DOI] [PubMed] [Google Scholar]
- 10.Lavigne M, Herbert M, Kolb A, Buc H. Upstream curved sequences influence the initiation of transcription at the Escherichia coli galactose operon. J Mol Biol. 1992;224:293–306. doi: 10.1016/0022-2836(92)90995-v. [DOI] [PubMed] [Google Scholar]
- 11.Lonetto M, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase ς70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 12.Musso R, Di Lauro R, Adhya S, de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977;12:847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- 13.Owens J, Chmura A, Murakami K, Fujita N, Ishihama A, Meares C. Mapping the promoter DNA sites proximal to conserved regions of ς70 in an Escherichia coli RNA polymerase-lacUV5 open promoter complex. Biochemistry. 1998;37:7670–7675. doi: 10.1021/bi980188n. [DOI] [PubMed] [Google Scholar]
- 14.Owens J, Miyake R, Murakami K, Chmura A, Fujita N, Ishihama A, Meares C. Mapping the ς70 subunit contact sites on Escherichia coli RNA polymerase with a ς70-conjugated chemical protease. Proc Natl Acad Sci USA. 1998;95:6021–6026. doi: 10.1073/pnas.95.11.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhodius V, West D, Webster C, Busby S, Savery N. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 1997;25:326–333. doi: 10.1093/nar/25.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodius V, Busby S. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 17.Williams R, Rhodius V, Bell A, Kolb A, Busby S. Orientation of functional activating regions in the E. coli CRP protein during transcription activation at class II promoters. Nucleic Acids Res. 1996;24:1112–1118. doi: 10.1093/nar/24.6.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]