Summary
Many aspects of health and disease are modeled using the abstraction of a “pathway”—a set of protein or other subcellular activities with specified functional linkages between them. This metaphor is a paradigmatic case of a deterministic, mechanistic framework that focuses biomedical intervention strategies on altering the members of this network or the up-/down-regulation links between them—rewiring the molecular hardware. However, protein pathways and transcriptional networks exhibit interesting and unexpected capabilities such as trainability (memory) and information processing in a context-sensitive manner. Specifically, they may be amenable to manipulation via their history of stimuli (equivalent to experiences in behavioral science). If true, this would enable a new class of biomedical interventions that target aspects of the dynamic physiological “software” implemented by pathways and gene-regulatory networks. Here, we briefly review clinical and laboratory data that show how high-level cognitive inputs and mechanistic pathway modulation interact to determine outcomes in vivo. Further, we propose an expanded view of pathways from the perspective of basal cognition and argue that a broader understanding of pathways and how they process contextual information across scales will catalyze progress in many areas of physiology and neurobiology. We argue that this fuller understanding of the functionality and tractability of pathways must go beyond a focus on the mechanistic details of protein and drug structure to encompass their physiological history as well as their embedding within higher levels of organization in the organism, with numerous implications for data science addressing health and disease. Exploiting tools and concepts from behavioral and cognitive sciences to explore a proto-cognitive metaphor for the pathways underlying health and disease is more than a philosophical stance on biochemical processes; at stake is a new roadmap for overcoming the limitations of today’s pharmacological strategies and for inferring future therapeutic interventions for a wide range of disease states.
Keywords: pathway, drug, network, disease, learning, memory, training
The bigger picture
Disorders ranging across drug addiction, injury, and cancer have proven difficult to definitively repair by focusing on the molecular hardware inside cells. Drug design and genomic editing face fundamental limitations of context, complexity, and cellular resistance. Fortunately, computer science and behavioral science are beginning to point the way to a transformative regenerative medicine in which pharmaceutical efforts focused on molecules will be complemented by top-down approaches that exploit the collective intelligence of cells and the native control mechanisms that establish form and function.
Powerful methods of controlling complex body systems include taking advantage of their newly discovered cognitive properties: memory, problem solving, and reprogrammability. Emerging advances in placebo research, non-neural bioelectric networks, and the diverse intelligence of cells, tissues, and organs suggest that the medicine of the future may look more like a kind of somatic psychiatry than chemistry or genetics. Therapeutic interventions will communicate and behavior-shape body processes, exploiting the software of life for novel solutions to disease, injury, and aging.
Biomedical approaches that target cellular pathways as hardwired mechanisms face significant limitations with safety and efficacy. Here, we provide a framework for recognizing and exploiting the proto-cognitive aspects of molecular networks and cell behavior. This enables top-down control of complex system-level functions through the use of tools and concepts adapted from cybernetics and behavioral neuroscience. Through recognizing the learning capacities of cellular pathways and their sensitivity to high-level cognitive context, biomedical problems ranging across addiction, pharmaceutical resistance, and regeneration after injury can be addressed.
Introduction
Understanding health and disease, and the discovery of biomedical interventions for complex system-level disorders, requires a model of the underlying mechanisms regulating physiological, developmental, and neurobiological processes. One of the most powerful formalisms has been the notion of a “pathway” (Figure 1): a well-defined topological network specifying some number of biochemical activities and the functional interactions between them.1 Implicit in this metaphor is the expectation that necessary information for prediction and control is local—specified by the connectome and states of the pathway itself. This view of causation meshes well with mechanistic approaches to the life sciences, identifying a deterministic, tractable set of dynamics at a single level of organization and de-emphasizing the complexity of history, context, and larger-scale systems within which the network is embedded.2,3,4,5,6 It guides approaches in data science both with respect to which kinds of data are gathered and used for modeling and the kinds of models that analyze these data for correlations and causal relationships.
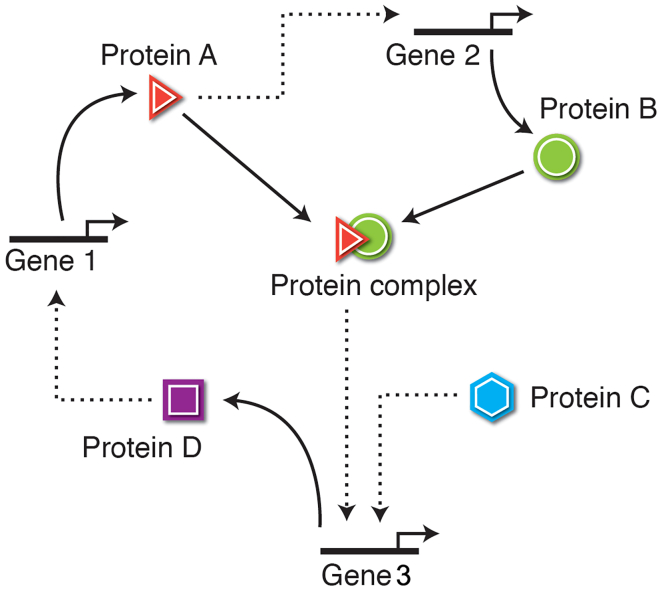
Figure 1.
A schematic of a generic GRN/pathway paradigm
A standard conceptual formalism with which to understand biological outcomes is that of a “pathway” including protein interaction and gene-regulatory networks. It describes the relationships between nodes and the strength and direction of their interaction (repressive, dashed lines, or activating, solid lines). Loops and self-loops are common. The dynamics of the system as a whole (its “behavior”) are taken to be accurately described by the topology of the connections and parameters in this circumscribed way—in other words, it draws a causal boundary around a given set of molecular players and focuses attention on their interactions. It provides a fundamentally mechanistic, local, single-level organization perspective. Image courtesy of Jeremy Guay.
This class of models also implies a specific strategy for intervention: adding new nodes or removing nodes, and manipulating the connections between them (Figure 2A), often via gene therapy or other transgenic technology, protein design, or drugs that alter the connections between nodes.7,8,9 Despite many successes, this strategy faces important limitations: the complexity of pathways makes it difficult to infer which intervention at the molecular level will give rise to desired system-level states (health) while avoiding undesirable side effects10—the so-called “inverse problem.”11 This approach to modeling healthy and diseased function is extremely popular today, leading to the near-universal assumption among both academia and the biotech industry that all control must be exerted at the hardware (molecular medicine) level. However, it is widely acknowledged that despite the ever-increasing deluge of omics data and a mature set of computational tools for understanding dynamical systems, there is immense unmet medical need,12,13,14 suggesting that additional research perspectives are needed.
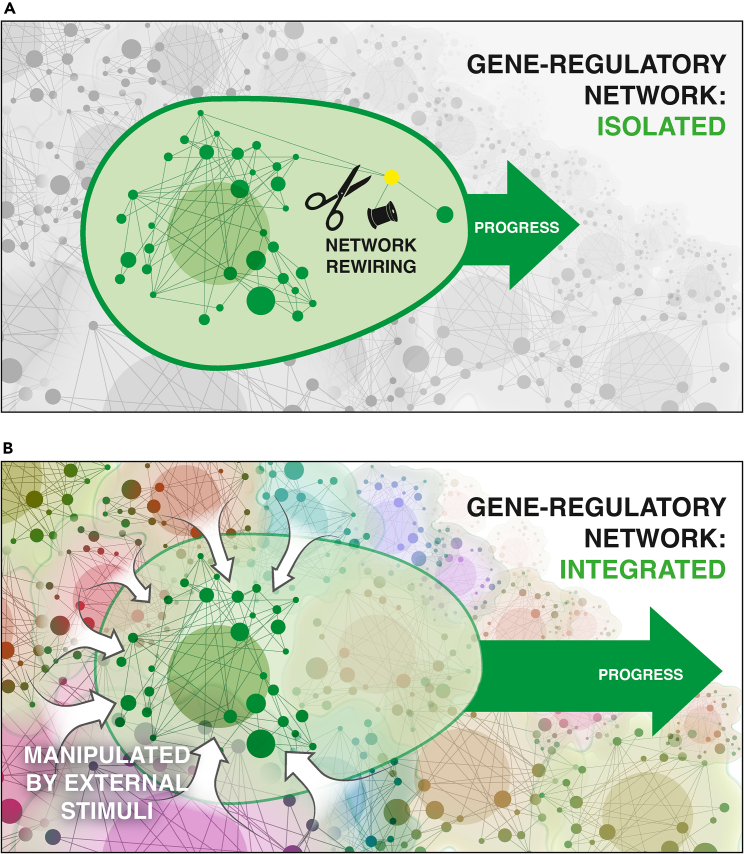
Figure 2.
Strategies for regenerative medicine
Two fundamental broad approaches toward the control of biological function in biomedicine. The first (A) focuses on the structure of a specific pathway and seeks to enhance functionality or repair a disease state via hardware rewiring—adding or deleting nodes or changing relationships between nodes. In practical terms, this means gene therapy (e.g., CRISPR) to modify the pathway at the lowest level of control. The second, elaborated in this perspective (B), sees function as the interplay of a pathway machine with its context, using multi-scale relationships with other components of the body to pursue anatomical or physiological goal states. This leads to the specific and testable proposal that optimal control over system-level phenotypes can be exerted using techniques from a discipline ideally suited for control of fundamentally multi-level phenomena: behavioral neuroscience. Image used with the permission of Jeremy Guay.
One example of a highly successful research roadmap is in computer science, which moved from physical rewiring of hardware in the 1940s and 1950s to programming at increasingly higher levels of abstraction. The remarkable state of information technology today is due in large part to commitment to the idea that some classes of devices can be controlled in software—by providing inputs that take advantage of the computational architecture and its built-in capabilities.15,16,17 An understanding of the software-hardware axis and the different strategies that can be brought to bear on reprogrammable systems enables highly efficient control. If this strategy could be applied in biomedicine, we could avoid the intractable inverse problem of deriving low-level interventions for large-scale physiological or anatomical outcomes.
However, in many ways, biomedicine is still in the phase that information technology was 70 years ago. Exciting advances are being made on ever-finer levels of the hardware of life—e.g., CRISPR, single-molecule controls, protein design—and the pharma sector focuses discovery efforts on interventions at the molecular level. Despite this torrent of resources, a wide range of complex neurological problems, physiological states, and anatomical disorders (birth defects, injury, and cancer) remain unaddressed, and side effects of efficacious new drugs are often severe, highly variable, and hard to predict.18,19,20,21 Achieving regenerative medicine that effectively moves the living system to a stable health state, as opposed to targeting symptoms without fixing the underlying dynamics, requires new ways to manipulate pathways. Is it possible that biological hardware has properties in common with reprogrammable devices and neural “software” systems that are reprogrammable in situ—that learn from experiences and offer powerful top-down control strategies? Could concepts and tools from the computational science of behavior22,23,24,25,26 complement current molecular mechanistic perspectives and provide a new way to exploit the allostatic and homeorhetic27,28 properties of living systems (Figure 2B)?
Computer metaphors are not meant to reduce the remarkable capacities of biology to the level of simple machines. On the contrary, we argue for expanding and exploiting a view of living tissue as a multi-scale agential material,29,30,31,32 amplifying its inherent capacities. Developing high-level “programming languages” that emphasize salient, context-specific information processing among biological components can complement current, largely reductionist, paradigms that focus primarily on the molecular level. As Marr famously argued in the context of vision,33 mechanism is but one important level of analysis in multi-scale control problems such as somatic health and cognition.
Such “programming” approaches are entering neuroscience and biomedicine of the brain. The emerging field of computational psychiatry links pharmacology and neuroanatomy with computational models focused on cellular information processing and perceptual control.22,23,24,34,35 While such cognitive features are readily imagined in the context of the CNS, it is increasingly recognized that many aspects of brain function are evolutionary pivots of ancient problem-solving capacities still present in most cells; thus, the insights of neuroscience extend far beyond classical neurons.36,37 Indeed, recent work has begun to emphasize the learning and memory capacities of ubiquitous and highly conserved molecular signaling machinery.38
Here, we suggest a new roadmap for medical interventions: exploiting the deep symmetries between pathway models and neural network functions,32,39,40,41 along with the ancient evolutionary origins of learning and memory mechanisms.36,42,43,44 Evolution’s use of a modular multi-scale architecture30 provides numerous computational competencies in cells and tissues that could be harnessed for prediction and control. Taking this proto-cognitive, multi-scale perspective on cellular signaling invites use of concepts and tools from behavioral and computational neuroscience, including cybernetics and basal cognition. We first review concepts from behavior science that help explain important clinical and physiological/cell-biological phenomena with respect to pathways and the drugs that have been used to control their functionality. We then discuss work showing new capabilities revealed with this approach and suggest a roadmap for future experiments to further explore the invariants across these fields. Importantly, use of a “cognitive lens”45,46 to understand and control events outside of brains goes beyond a philosophical stance: facilitating the use of powerful concepts from other fields may help overcome limitations of today’s discovery efforts, offering predictions that can be evaluated empirically.
Tolerance and sensitization: The ubiquitous atoms of complex behavior
We recently proposed the idea of competency in navigating problem spaces as a central invariant unifying behavioral science, cell biology, and physiology.37 On this view, evolution adapted some of the same strategies across metabolic, transcriptional, physiological, and anatomical spaces that feature so prominently in the study of behavior in familiar 3D space. Thus, we have been exploring the applications of concepts from behavioral science to understand aspects of the control of form and function.31,32,47 The most basic aspects of proto-cognitive function, seen even in unicellular systems,42 are habituation and sensitization. These simplest kinds of memory (ability to alter future behavior in light of past experience) tie the temporal dynamics of pathway activity to questions in behavioral science, and we next discuss some molecular mechanisms by which these proto-cognitive effects are implemented.
Tolerance and reverse tolerance (sensitization) are, in their most general forms, a decrease or increase, respectively, in the magnitude of reaction to a repeated stimulus.48 The many types of tolerance include behavioral, pharmacokinetic, and pharmacodynamic,49 while sensitization can be behavioral or pharmacokinetic. Both have a large impact on human health. Tolerance to alcohol or other drugs can lead to increased consumption, dependency, and ultimately addiction,50,51 key factors in the current public health crisis of overdose deaths.52,53 Sensitization also plays a part in these deaths because it can amplify incentive and behavioral effects, such as drug seeking, while tolerance is decreasing the molecular effects.54,55
A crucial type of behavioral tolerance develops in a context-specific way: the environmental cues presented at the time of drug administration trigger the behavioral tolerance to the drug’s effect. Context-specific tolerance has been observed for a wide range of drugs including alcohol, ketamine, and morphine.56,57,58 The mechanism behind context-specific behavioral tolerance in ketamine use has been explained neurochemically by changes in dopamine release in different parts of the brain, specifically an increase in the infralimbic cortex and a decrease in the nucleus accumbens shell.56 Thus, in some cases, the surrounding context is a key input to the control circuit and must be taken into account in designing interventions.
In pharmacokinetic tolerance, repeated exposure increases drug metabolism or transport, reducing exposure to the active form of the drug.59 Ethanol, nicotine, tyrosine kinase inhibitors, anesthetics, and anti-epileptics can all increase the level of cytochrome P450 enzymes that are responsible for breaking them down. Drug metabolites can both drive transcription of those enzymes and stabilize them post-translationally by preventing their degradation.60,61,62 Many anti-epileptics also elicit pharmacokinetic tolerance by increasing reactive oxygen species (ROS) levels and thereby the expression of multi-drug transporters such as P-glycoprotein at the blood-brain barrier (BBB).63,64,65,66
Pharmacodynamic tolerance occurs when repeated exposure to a drug affects how the drug itself interacts with the body. Anti-epileptic drugs (AEDs) can lose their effectiveness over prolonged use due to different pharmacodynamic mechanisms.49 AEDs that specifically target and activate the γ-aminobutyric acid type A receptor (GABAA) and therefore increase cellular Cl− influx, making neurons less likely to fire, can down-regulate receptor expression at the cell surface by increasing its endocytosis in an agonist-dependent manner.67 Nicotine tolerance depends on an increase in nicotinic cholinergic receptors (nAChRs) over time, accompanied by nicotine-mediated desensitization of those receptors.68 Anti-depressants, specifically selective serotonin reuptake inhibitors (SSRIs) like fluoxetine (Prozac), can also elicit tolerance by desensitization of the 5-HT1A receptor.69,70 Even diuretics, used to treat hypertension brought on by high sodium concentrations, can show pharmacodynamic tolerance over time due to activation of the renin-angiotensin-aldosterone system and sympathetic nervous system activation resulting from increased loss of sodium and water.71,72 All of these are examples of how the molecular machinery, most of it highly conserved throughout the body, can underlie simple forms of memory in pathways guiding key biological processes. Memory of this kind presents an important limitation on the use of pharmaceuticals.
Understanding that interoceptive early-drug onset cues (DOCs) create an association with larger drug effects and therefore lead to tolerance or behavioral sensitization can inform addiction treatment protocols. These may not only work on extinguishing environmental cues but also on interoceptive cues that can be elicited by the administration of very small quantities of the drug, a method that may lead to less relapse in a non-abstinence environment.73 Tolerance to opioid analgesics is known to depend on the intrinsic efficacy of the opioid.74 However, this dependence of tolerance on intrinsic efficacy is removed when intermittent dosing is used instead of continuous dosing, which could mean that a more thorough exploration of non-traditional dosing regimens will result in decreased tolerance.74,75
Resistance (drug habituation)
Acquired resistance to repeated stimuli is a kind of memory frequently studied in behavior science as habituation. Recent advances in targeted cancer therapies, especially tyrosine kinase inhibitors, are promising but can suffer from drug resistance—either present initially or that develops due to tumor cell population changes brought on by selection of genetic mutations, alternative splicing, alternative/compensatory signaling pathways, or epigenetic changes.76 Pharmacokinetic and pharmacodynamic mechanisms take part in cancer resistance due to genetic differences in drug-metabolizing enzymes, transporters, or receptor affinities.
The tyrosine kinase inhibitor imatinib treats chronic myeloid leukemia (CML), which is caused by an oncogenic non-receptor tyrosine kinase (BCR-ABL1) that constitutively activates cell proliferation and survival without regulation by normal extracellular signals.77,78 Resistance to imatinib arises primarily (50%–90%) from mutations to the ABL kinase domain or by overproduction of BCR-ABL1 by genomic amplification (10%).79 Recurrence of cancer upon cessation of imatinib treatment may be due to differential resistance to imatinib by hematopoietic stem cells but not hematopoietic progenitor cells, due to their expression of the P-glycoprotein drug transporter. This could also explain why genetic resistance can develop over time.80 Imatinib is also used to treat gastrointestinal stromal tumors (GISTs), which similarly exhibit mutations that cause both initial and acquired resistance.81 However, unlike for CML, no genetic amplifications were observed in people with acquired resistance,81 suggesting dynamic physiological habituation mechanisms.
Drug conditioning: Proto-cognitive aspects of physiological signaling
Environmental cues involved in certain types of tolerance and sensitization show how powerful context is in eliciting responses from the body. This conditioned tolerance is similar to Pavlovian conditioning in that it results in a decrease in alcohol-induced hypothermia in an alcohol-paired environment when alcohol is administered but also in a small hyperthermic response in the paired environment without administration.82,83,84 The same Pavlovian conditioning is observed in conditioned tolerance to alcohol-induced tachycardia since unpaired environments did not show the same level of conditioned tolerance as paired environments.85 Newlan proposed that conditioned responses can counter the drug’s effect, resulting in tolerance to a depressant like alcohol or in behavioral sensitization to a stimulant like cocaine, but that both responses are due to increased stimulation of the reward centers.82
The relationship between environmental context and tolerance can have deadly consequences. Siegel posits that a large majority of heroin overdose deaths are not due to the quantity of the heroin administered but occur in cases where the environmental context normally paired with the heroin administration was not present, resulting in a loss of conditioned tolerance.86 In support of this theory, Siegel shows that rats given the same increasing dosages of heroin had differential responses to an even higher dose of heroin depending on whether the higher dose was paired with the same environmental cue associated with the previous dosages or an environmental cue associated with dextrose injections: those injected in the heroin-paired environment were significantly more likely to survive those given a high dose.87 It is becoming increasingly clear that environmental cues have a strong impact on conditioning for drugs.88,89,90,91,92,93
The significance of place conditioning94,95,96 on drug use is this: to understand and predict the action of a particular pharmacological agent, it is not sufficient to know the details of the pathway of its actions—the environmental context of its administration is a key input. Our understanding of what functionally defines “a pathway” cannot only be a high-resolution focus on the local chemical interactions. It must broaden to include events distant in time (past experiences of the patient) and in space (effects in the brain and nervous system even if the drug-relevant tissue is elsewhere in the body). This realization modifies our understanding of what it means for a physiological process to be controlled by a regulatory molecular pathway. The “light cone” of events that impinge on the process is much wider and encompasses more scales of organization than are described by the currently standard arrow diagram for representing disease pathways. Beyond context setting for drugs of abuse, this suggests a novel roadmap: computational modeling will increasingly include inputs such as past cognitive and physiological events, and therapeutics will seek to modify the functional outcome of pharmacological interventions using other stimuli (perhaps delivered by virtual reality platforms).
For example, interventions in the environmental cue (conditioned stimulus) pathways involved in tolerance could be used to disrupt the Pavlovian conditioning required to develop them.97 Partial reinforcement does this by periodically presenting the environmental cues without the drug (unconditioned stimulus) and has been shown to slow the development of tolerance to morphine in rats.88 Latent reinforcement can also slow the development of tolerance by presenting the environmental cue without the drug many times before the drug is ever given. An excellent example of this was shown by a study that administered an immunostimulatory synthetic polynucleotide (poly I:C) to mice paired with a complex environmental cue. Natural killer (NK) cell activity decreased over repeated stimulation, showing tolerance; however, when the complex environmental cue was given repeatedly before the first poly (I:C) administration, tolerance did not occur.89 That same study also showed that after tolerance was established, repeated exposure to the environmental cue without administration of poly (I:C) significantly diminished tolerance, a perfect example of Pavlovian extinction.89 Another example of reducing tolerance using methods derived from Pavlovian conditioning is found in Fanselow’s study using explicitly unpaired delivery, where rats that had grown conditionally tolerant to the locomotor-inhibitory effects of morphine lost their tolerance when morphine was administered in an unpaired environment.90 Interestingly, a change in temperature in the paired environment is sufficient to explicitly unpair the administration of alcohol, resulting in loss of tolerance.84 In agreement with Pavlov’s98 theory of tolerance, Siegel also showed that a novel extraneous stimulus, e.g., strobing light, delivered to already conditionally tolerant rats right after alcohol administration, attenuated tolerance to the hypothermic effect of alcohol.91 New studies examining drug-drug conditioning show that pairing one drug with another in a specific environmental context can cause the administration of one drug alone to cause the effect of the second as long as it is administered in the same environment.99 Drug-drug conditioning could therefore be used to amplify the effects of single drugs or give them new effects.99
Pavlovian conditioning that involves a contextual environmental cue paired with drug administration is an example of delay conditioning and is mediated by the cerebellum92 and other brain regions.100,101 Another form of Pavlovian conditioning, called trace conditioning, is mediated by the hippocampus and forebrain and occurs when a short time interval separates termination of cue presentation and onset of drug presentation.92 These two types of Pavlovian conditioning correspond to declarative memory and procedural memory, respectively, and Carey proposes that delayed conditioning of the environmental cues to drug administration elevates the environmental cue’s salience, allowing trace conditioning to occur by repeatedly activating the limbic system, followed by an activation of the frontal lobes and hippocampus to shift goal-directed behaviors toward the drug.92 This agrees with Robinson and Berridge’s view of the incentive-sensitization theory of drug addiction.55,102 Methods of disrupting trace conditioning so that the environmental cues are linked instead to an anti-drug state may be a way of using Pavlovian conditioning to combat addiction.92,93 O’Brian et al. further theorized that withdrawal may also be a conditioned response arising from the “drug opposite” conditioned responses that occur when the right environmental cues are triggered.93
Possible methods of pairing an anti-drug state with environmental cues can be inferred from studies examining the mechanism of the conditioned response. A study of fetal rats showed that pairing dynorphin A, an endogenous κ-opioid receptor agonist, with an artificial nipple induced a κ receptor-dependent conditioned response (reduction of face wiping), similar to the natural unconditioned stimulus of their first drink of milk.103 In other mechanistic studies, researchers are looking at how the actin cytoskeleton shuttles ion channels and neurotransmitter receptors to the synapse and regulates receptor channel opening during memory formation and consolidation.104 In a study of the effect of conditioning on cytoskeletal protein function in the sea slug Hermissenda crassicornis, a one-trial pairing of a light stimulus with serotonin delivery to the isolated circumesophageal nervous system resulted in phosphorylation of Ser-122 of Csp24, an actin-binding protein that contributes to the decrease of an A-type transient potassium current necessary for enhanced excitability.105 Phosphorylated Csp24 binds to 14-3-3 protein, potentially affecting cytoskeletal dynamics.106 Understanding the extent to which the CNS is involved in conditioning, and the mechanisms involved in that process, would allow development of targeted therapies to de-couple environmental stimuli from drug administration. This would have profound implications for drug addiction therapy but also for clinical tolerance to a wide range of therapeutic agents.
The influence of psychogenic factors on various tissue behaviors is just beginning to be explored. Dykman and Gantt studied the degree to which blood pressure could be influenced by the CNS in dogs and found that pairing a tone with a shock would cause an increase in blood pressure and that the increase in blood pressure could subsequently be induced by the tone alone.107 They also showed that pairing different tones with different shock intensities resulted in different levels of hypertension induced by the tones alone.107 Hypertension resulting from the presentation of tone occurred rapidly and took much longer to disappear than the conditioned movements the shocks created, a perfect example of schizokinesis. Gantt and others over the years found that skin resistance, respiration, heart rate, motor responses, heat regulation, spleen contraction, kidney secretions, pancreas secretions, and stomach secretions could all be conditioned.107 Similarly, the immune and endocrine systems can be conditioned, and the type of paired environmental cue had a significant effect on the strength of the conditioning.108,109,110,111 Further explorations are necessary to discover what other internal biological processes can be conditioned.
Toward training cell signaling pathways to regulate health and disease
The profound possibilities of these kinds of training for cell regulation are only now beginning to be realized. A broad class of systems, from molecular networks112 to physiological networks in somatic organs,113,114 exhibit plasticity and history-based remodeling of stable dynamical states. Could gene-regulatory networks (GRNs) likewise exhibit history dependence that could help understand variability of cellular responses and be exploited to control their function by modulating the temporal sequence of inputs? This is a different approach from existing conceptions of memory as changes at the epigenetic and protein levels,115,116,117,118 focusing on memory as a shift between stable attractors in the dynamical state portrait of a physiological circuit. Several studies have proposed memory phenomena in network models.41,119,120,121,122,123,124,125,126,127,128 We recently analyzed a set of GRN models129,130 to discover several predicted classes of learning, including associative conditioning. We found that many biological (but not random) networks can stably store multiple memories, are highly resistant to noise, and can perform simple computational tasks such as counting discrete stimulus events.130 Most importantly, the memories are readily controlled by specific stimuli, which can be discovered by a computational process,130 without rewiring network structure or altering promoter connection weights. While these remain to be tested in vivo, the implications are potentially very significant. For example, drugs that are too strong to be used in patients for extended periods could be paired with neutral drugs, giving rise to Pavlovian conditioning of the pathway, after which the neutral drug could be administered alone, causing the same response for some period of time. Interestingly, we found that random GRN models do not exhibit the trainability properties seen in biological networks, suggesting that this capacity is evolutionarily important and likely to be prevalent.
Beyond habituation and sensitization: More complex cell behavioral competencies revealed by morphogenetic robustness?
This demonstration that GRNs, previously assumed to be purely mechanical systems, support several kinds of learning41,128,129,131 highlights an important part of a roadmap for future biomedicine: the need to examine regulatory mechanisms for computational competency as standard practice. In addition, Csermely et al. described a number of adaptive molecular mechanisms in individual non-neuronal cells as examples of generalized Hebbian learning in signaling networks.38,132,133,134 They also proposed that this demonstration of cellular learning opens new possibilities in drug design by providing new targets for intervention, as well as new models for artificial intelligence. Here, we expand the analysis of cell signaling topologically, conceptually, and temporally, going beyond Hebbian learning and beyond the level of the single cell to model cell signaling in the context of past experience and of tissue, organ, organism, and environment (Figure 2). We propose that a proto-cognitive metaphor for cellular signaling provides a framework and toolkit for experimenters to ascertain the behavioral competencies of tissues, organs, and the regulatory machinery of a wide range of unconventional substrates. This is already beginning to be worked out for synthetic biology and bioengineered life forms.135 What other capacities could plausibly be found in biomedically relevant processes?
The following are novel examples of somatic information processing that can be addressed via behavioral neuroscience perspectives. We focus on a just few examples of unique aspects that complement Csermely’s work38,132,133,134 and other recent dynamical systems perspectives on molecular networks.38,132,133,134,136,137,138,139,140 William James defined intelligence as the capacity to reach the same goal by different means.141 Our previous work provides evidence for this capacity at the level of tissue morphogenesis, where groups of cells coordinately navigate morphospace—the space of all possible anatomical trajectories—to produce appropriate anatomies.37 One example of robust problem solving in morphospace31 is craniofacial remodeling in Xenopus, a system often used to model birth defects.142 Specifically, tadpoles must rearrange their facial features to metamorphose into frogs. We found that this is not a hardwired process where each organ blindly moves a certain distance in a certain direction. Instead, tadpoles with scrambled faces (e.g., eyes on top of the head, jaws off to the side, etc.) still make largely normal frogs because the organs keep moving until they reach the correct target morphology.143 This extreme example is related to regulative development (e.g., mammalian embryos cut into pieces give rise to normal monozygotic twins) and reveals the plasticity and problem-solving capacity of processes that underlie development, repair, and cancer suppression. To harness such capacities for repair and cancer normalization,144,145 future work will characterize the parameters that cell collectives measure and the pattern-homeostatic mechanisms that enable error detection relative to a stored setpoint. Bioelectric mechanisms for setpoint storage are already beginning to be understood (see below), as are the attractor-selector mechanisms that provide metastability.146,147
A complementary example demonstrates problem-solving capabilities across both physiological and transcriptional space. In planarian flatworms exposed to the non-specific potassium channel blocker barium, neurons die of cytotoxicity due to excessive de-polarization, resulting in rapid degradation of the head. Remarkably, when kept in barium, they soon regenerate new, barium-resistant, heads.148 They do this by changing the transcriptional levels of just a handful of genes. Because planaria never experience barium in the wild, there has been no selection pressure to evolve a response to barium poisoning. This indicates that when confronted by a new physiological stressor, these cells manage to navigate a path in a very high-dimensional transcriptional space to up- and down-regulate exactly the right genes to solve their problem. This is a highly efficient process: because these cells turn over slowly, there is no time for a bacteria-like population-level search over all possible transcriptional actions and a survival of a small number of cells. This is therefore an example of drug resistance that is not merely the result of one-shot learning but also requires problem solving that can deal with both novelty and cross-modal (physiology → transcriptional) action. It is currently unknown how planarian cells solve this problem, but it is clear that this is the kind of computational search capacity (of successfully mapping stressors to transcriptional responses) that could be artificially reproduced, or harnessed directly, for biomedical purposes to help address the perennial problem of which genes to target to achieve a desired response to a disease state.
Bioelectricity: A roadmap for a biomedicine of reprogrammable tissues
The proto-cognitive functions (memory, context sensitivity, etc.) of cell signaling pathways are not unique to chemical signaling networks but are also implemented by bioelectrical networks. It has long been known that networks of neural cells exhibit learning, plasticity, and other information-processing capabilities that enable a very powerful form of functional control: behavior shaping (training). Bioelectric signaling in the brain binds a large number of individual cells into a larger-scale emergent agent with specific cognitive properties that do not belong to any of the cells individually. The exciting emerging field of developmental bioelectricity emphasizes that these capacities are not unique to the CNS but are evolutionary modifications of an ancient bioelectric communication system that earlier used the same strategies to navigate a different problem space—morphospace.37
Configuring the body in morphospace is a critical task throughout the lifespan; disorders manifest as birth defects or as failure to regenerate after damage, cancer, and aging.32 Much of the information processing that enables distributed groups of cells to ascertain current anatomy and execute changes to bring it closer to a species-specific target morphology is carried out by bioelectrical communication across tissues (Figure 3) using the same highly conserved molecular components used by neurons: ion channels and electrical synapses.152 Specific endogenous bioelectric dynamics (via slowly changing patterns of resting potential) are required for the formation of organs such as the wing,153,154 face,155,156,157 eye,158 brain,159 and heart160,161 and also function in control of single-cell parameters such as stem cell differentiation,162,163 cancer suppression,164,165 and organ-level size control.166,167,168
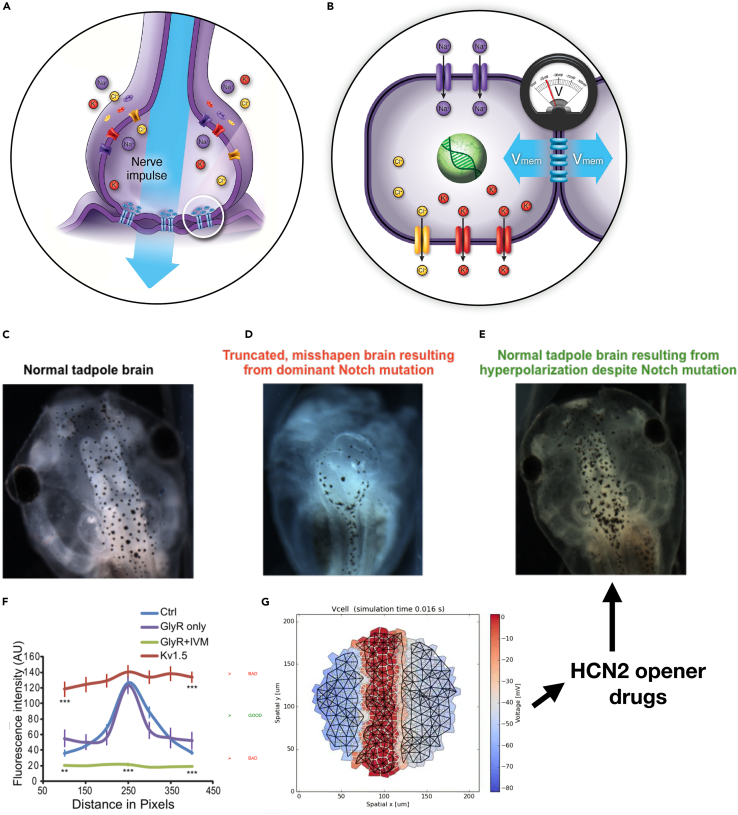
Figure 3.
Bioelectric pathways
(A) The hardware that enables complex computations in neuronal networks consists of ion channel proteins that determine cells’ electrical state and gap junctions (electrical synapses) that selectively propagate this state to their neighbors. (B) This hardware is evolutionarily ancient and ubiquitous, enabling similar (albeit slower) dynamics in non-neuronal cells. Taking advantage of this bioelectrical interface to the circuits that make anatomical decisions has significant biomedical implications. For example, normal brain development in the tadpole model (C) requires a specific bioelectrical prepattern in the nascent anterior ectoderm. Disrupting this pattern with a range of teratogens or via mutations of the NOTCH protein results in severe brain defects and a loss of learning behaviors (D). Remarkably, correct brain structure, gene expression, and learning rates can be reinstated (E) when the correct bell-curve-shaped bioelectric prepattern (F) is enforced according to a computational model that suggests specific drugs that target ion channels to enforce the correct bioelectric prepattern (G).149 (F) shows the effect of various manipulations on embryonic bioelectric prepattern: control (blue), microinjection with voltage-gated potassium channel (Kv1.5, red), microinjection with glycine-gated chloride channel (GlyR, purple), and microinjection with GlyR plus ivermectin treatment (IVM; green). (A) and (B) are courtesy of Jeremy Guay and are taken with permission from Levin.30 (C)–(E) are courtesy of Vaibhav Pai. (F) is taken with permission from Pai et al.150 (G) is courtesy of Alexis Pietak.151.
The foundations for therapeutic tissue reprogramming have been laid by showing that functional interventions into bioelectric circuit function can induce development of complete organs such as eyes158 and regeneration of tails169 and limbs170 and can abrogate cancer phenotypes in amphibians in vivo171,172,173 and in human in vitro systems.174 They can also dictate species-specific structural variations in anatomical features.175 Moreover, in a set of recent studies,149,150,159,160,176 a computational platform for simulating endogenous bioelectrical control mechanisms identified human-approved ion channel-targeting drugs that could repair drastic brain defects in a tadpole model by enforcing the appropriate bioelectric prepattern despite the action of powerful teratogens. The most remarkable aspect of these is that heart, gut, and brain defects induced by a mutation of the important neurogenesis gene NOTCH could be repaired by transient exposure to pharmacological electroceuticals (specifically HCN2-opener drugs) without repairing the mutation.150,160 This reinforces the message of the examples cited above. Experience-dependent changes to pathway function can alter outcome without gene therapy or genomic editing, demonstrating that some biological hardware problems are fixable “in software”—without rewiring the molecular machinery or directly repairing the defect.
Bioelectric signaling is also relevant to the “disease of geometry” known as cancer.177 Cells are normally bound to large-scale morphogenetic goals (such as organ building or maintenance) by a collection of electrochemical cues, creating a cellular network that maintains anatomical homeostasis,178,179 or homeorhesis, as pointed out by Waddington, since, over the long term, what is maintained are trajectories, not fixed states.28 Disruptions in this system can induce cancer phenotypes,164,165 even in the absence of mutation or DNA damage, because the society of the body is a dynamical type of order that must be actively maintained.180,181,182 Fundamentally, when cells dissociate from tight informational links with their neighbors, they revert back to a unicellular ancient state, effectively seceding from the multi-cellular collective and treating it as simply external environment. The result is resurgence of behaviors appropriate to the unicellular state, i.e., overproliferation and metastasis. This cognition-inspired perspective on cancer as fundamentally a disorder of setting informational boundaries between the cellular self and the outside world178,183 has already given rise to effective treatments that reverse and prevent cancer in amphibia171 and are now being tested in human tissues.174
The important insight emerging from this field184,185,186 is that the bioelectric signals are not just another set of microstates that could be targeted to force specific cell-level outcomes. Instead, bioelectric events are highly modular computations that implement tissue- or organ-level morphogenetic decisions, inducing complex structures. Evolution exposes a powerful control interface on tissues—ion channels that enable morphogenetic outcomes to be reprogrammed by transient experiences, just as in neuroscience, using, for example, optogenetic pulses187,188 or brief drug exposure.189 Taking advantage of the modular, homeostatic, learning, and perhaps even more complex capacities of bioelectric circuits is an excellent complement to similar approaches targeting biochemical pathways. A huge pool of ion channel-targeting drugs, many already approved for patient use in various indications, could be used to exploit the wider notion of control mechanisms as active, computational, and proto-cognitive matter, moving beyond limiting views of pathways as simple machines.
Top-down control, multi-scale information flows, and clinical medicine
Words and drugs have the same mechanisms of action. – Fabrizio Benedetti190
The data on psychological effects in medicine provide support for cross-scale computation and integration of stimuli.191,192,193,194,195 For example, the “white coat” effect is now being studied with respect to changes in gene expression induced by the very high-level stimulus of social hierarchy-induced stress.196 Similarly, it is now becoming clear that the relationship between therapist and patient is a key input into drug efficacy.197,198 Such cross-level information transfer is also manifest in biofeedback, a set of techniques that allow conscious control of autonomic body states.199,200,201,202 Biofeedback is now being studied in non-human systems such as closed-loop control of hybrots,203 organoids, and other contexts204,205,206 that will facilitate the understanding of cross-level control modalities.
Studies of placebo/nocebo effects191,194,195,207,208,209 also reflect this multi-scale integration of inputs. For example, Benedetti’s work shows that the analgesic/anxiolytic effect of potent drugs is remarkably reduced if the person is not aware that they were administered.191,192,193,194,195 Remarkably, open-label studies show that placebos work even when the patient is aware of what they are getting.210,211 All of this has important implications for predictive coding and Bayesian brain hypotheses.212 Crucially, this phenomenon is useful not only at the level of the patient but also needs to be explored and exploited in cells and tissues. For example, the association of a powerful stimulus to a neutral one (whether in tissues or in GRN models129) is essentially a minimal placebo model, where the neutral stimulus exerts effects that are not explained by a direct consideration of its mechanism of action. The efficacy is borne by a much wider context established by the learning and information-processing capacities within and across levels of organization from subcellular networks to whole-body organ systems.
Conclusion: Future medicine from data to interventions
Decades of research with pharmacological and recreational drugs have revealed interesting aspects of cellular regulatory pathways that implement basic properties of cognition, including tolerance and sensitization. Studies of morphogenesis and of psychological regulation of clinical outcomes hint that non-neuronal cell signaling can exhibit even more complex proto-cognitive behaviors, integrating computation across scales to support top-down, context-sensitive control and reprogrammability at levels other than that of molecular properties. The data suggest several main points.
-
•
Simple forms of learning, such as habituation and conditioning, exist outside the brain. Hints of more advanced capacities, such as solving novel problems in navigation of transcriptional spaces,148 physiological spaces,47 and morphospace31,213 suggest that such capacities of neural networks in unconventional substrates can provide exciting targets for biomedical intervention.
-
•
While a major problem for the development of effective therapeutics, tolerance is not an inherently negative capacity. Indeed, its failure can result in overdose.86,97,214,215,216,217,218 The challenge is to develop therapeutic approaches that manipulate these homeostatic properties in efficient ways.
-
•
Behavioral neuroscience is uniquely and rigorously comfortable with multi-scale approaches to these kinds of complex control problems. In particular, the disciplines of behavior science, control theory/cybernetics, and computational neurobiology provide rich toolkits of concepts, formalisms, and techniques that can be applied to somatic signaling systems (Figure 4).32 Examples include exploiting generic principles of behavior shaping to control phenotypes in synthetic biology and bioengineering,135 using concepts of perceptual control theory to understand instructive information in bioelectrical networks,47 and using the frameworks of active inference to understand and manipulate collective decision-making in regenerative and developmental morphogenesis.219,220,221
-
•
The remarkable capacities of pathways in biomedical contexts are very likely scaled-up aspects of extremely ancient properties of our unicellular ancestors, including multi-modal integration of signals, learning, and complex decision-making.42,46,222,223,224,225,226 The study of basal cognition and diverse intelligence is poised to strongly impact biomedicine by revealing the fundamental evolutionary roots of homeostasis, homeorhesis, and similar processes by minimal biochemical, biomechanical, and bioelectrical agents. Early efforts in regenerative medicine160,172,173,176,227 indicate the promise of this approach.
-
•
Conversely, analyses of cell biological and clinical data are revealing how a collection of molecules in a network can work together as a special kind of dynamical system that can process a history of experiences (inputs, stimuli) and gain associative or other kinds of memories that belong to no individual molecule or molecular link but to the system as a whole.38
-
•
Pathways and other clinical targets can no longer be considered self-contained dynamical systems with a small set of local interactions and mechanical behavior; optimal prediction and control of their functionality requires probing their proto-cognitive capacities and functional connections with higher levels of organism function, such as environmental cues and cognitive attitudes.
-
•
We suggest exploring scale-free228 dynamics that may be conserved between proto-cognition and evolution.229,230 For example, molecular resistance on coevolutionary timescales, such as the ecotoxinology of venoms,231,232 may offer interesting parallels to organism-scale resistance effects. Likewise, exciting work hints at relevant coevolutionary dynamics driven by stress233,234,235 and cellular adaptation to novel conditions.236,237
-
•
This perspective has key implications for data science on biological control. It suggests we need to widen the data mined around any biological endpoint to include other regions of the body and other levels of organization (i.e., psychological in addition to molecular pathway data). This will require novel data structures to handle diverse information at different levels of organization, integrating advanced data mining techniques across modalities from physiological measurements to behavioral ones. In addition, unsupervised artificial intelligence (AI) approaches will be necessary to identify factors and dynamics that are as yet unknown and to infer interventions via integrated multi-scale models that exploit innate, multi-scale competencies from cell networks to organ systems.
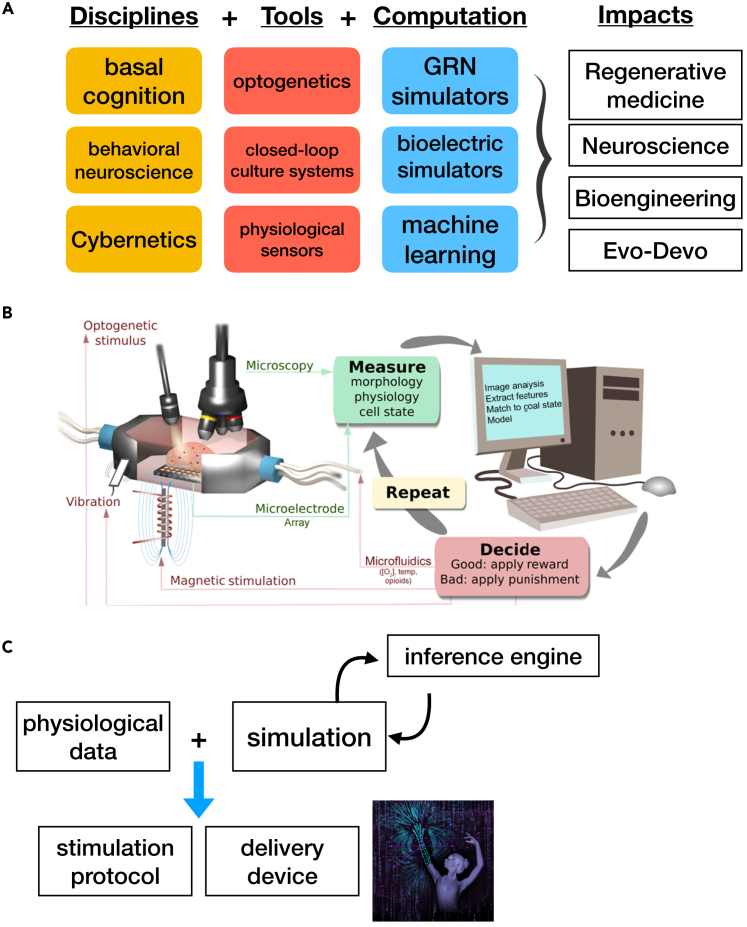
Figure 4.
Conceptualization of key features of the emerging field of multi-scale biomedicine
(A) Schematic illustration of a multi-disciplinary field, suggesting impacts that insights from several diverse disciplines, specific tools, and computational approaches will have in biomedicine, neuroscience, synthetic bioengineering, and basic evolutionary developmental biology.
(B) A schematic of a generic closed-loop system in which an ex vivo cultured tissue or organ is being trained by rewards and punishments for specific morphogenetic, transcriptional, or physiological outcomes. Such systems can be used in basic research to identify the proto-cognitive capacities of cells and tissues or in regenerative/bioengineering contexts to achieve desired complex endpoints.
(C) A schematic of the deployment pipeline for medical intervention in which computational/AI algorithms (inference engine), fed with physiomic data and operating over an in silico simulation environment, can suggest stimulation protocols with specific drugs or other modalities, delivered by novel devices such as wearable bioreactors and other stimulation technology.
(B) is by Alexis Pietak. Image in (C) is by Jeremy Guay of Peregrine Creative.
Here, we have extended the concept of “pathways” along 3 dimensions: space, time, and level of organization. Data indicate that for cell signaling, history matters, and large-scale (non-local) inputs matter. Place modulation and placebo/association studies discussed above, as well as conditioned immunosuppression,238,239,240 reveal that an exclusive molecular pathway view of drug action is highly incomplete. It is now clear that information crosses levels: changes of gene expression and cytokines by intentional meditation,241,242,243,244,245,246,247 psychoneuroimmunology,248,249,250 somatic and transcriptional memories of experience of stress/trauma,251,252,253,254 and hypnodermatology255 all show that high-level mental states change cell-level phenotypes. One of the most interesting implications of these data is the interactions across levels of organization: for example, the efficacy of a drug on cell physiology is in part a function of the patient’s environment and their psychological relationship with a therapist. Thus, it forms a model system for the holy grail of neuroscience: how do higher psychological levels arise and relate to the dynamical circuits of behavior and the mechanisms of ion channels and neurotransmitter signaling below that? How does the mind (at whatever level of sophistication) relate to the matter of the body that implements it?
Thus, the use of reagents to manipulate disease pathways is likely way more complex than currently appreciated by biopharma efforts in mechanistic screening, drug-protein interaction predictions, and machine learning focused on pathway models. The existence of complex behaviors at all scales, and multi-scale interactions, means that it is hard to control the details. The good news is that we should not have to control the details. In vivo, the modular architectures (much like behavioral repertoires) are already good at dealing with complexity, noise, and uncertainty. We can therefore focus on developing behavior-shaping protocols that exploit these high-level properties. Early examples of such protocols in preclinical models include a 1-day exposure to a trigger drug blend that can kickstart 18 months of leg regeneration in a frog model189 and a 1-h exposure to a specific ionophore electroceutical that can induce regeneration of a tail (with spinal cord).169
We propose a research roadmap (Figure 4) in which we view health and disease pathways as multi-scale, predictive computational agents that offer interfaces for prediction and control at multiple scales. Given the recent explosion of work on nervous systems as predictive agents in active inference formalisms,256,257,258 all the tools now exist (Figure 4A) to begin formulating sophisticated models of pharmacological signals and physiological/behavioral environment states that can predict which drug metabolism or receptor pathway needs to be modulated for therapeutic effects and to test those predictions. Moving even further upstream, it should be possible to train tissues and organs for long-term changes in fundamental dynamics by appropriate positive and negative reinforcement. Indeed, many of the advances from the cognitive and behavioral sciences,22,23,24,34,35 including ones in which the efficacy of pharmaceutical therapeutics is highly dependent on high-level features such as “therapeutic alliance” and not only on molecular structure,197,259 are ideally poised to be translated into a kind of “somatic psychiatry” that maximizes efficacy by interventions at multiple scales.208
The non-neural bioelectricity framework,47,220,260 together with perspectives from the diverse intelligence and computer science communities,42,261,262 are revealing a biomedical roadmap that is not stuck at the molecular level any more than modern cognitive science is exclusively focused on synaptic machinery. This will not only drive the design of much more effective interventions for regenerative medicine but will also advance neuroscience (by solving key problems in simpler, evolutionarily more basal, contexts), enable better control of complex morphogenesis for bioengineered synthetic constructs (such as novel biorobotics), and even the field of evolutionary developmental biology (by improving our understand of the relationship between the genetically specified hardware and the phenotypes that result from the physiological software that drives biological robustness and competency). Transformative impacts are also predicted for evolutionary developmental biology, basal cognition, and synthetic bioengineering by harnessing clinical data to spur novel ways to understand universal dynamics operating across scales.228 We believe the future of biomedicine will look a lot more like behavior shaping than molecular engineering,29,144,145 with all the benefits of high-level communication and control that the intelligence of the body enables.
Acknowledgments
We thank Douglas Brash, Karl Friston, Adam Goldstein, Anna Kane, Michael Hufford, Eric Kuelker, Eric Lagasse, Giovanni Pezzulo, Alex Schmidt, and Richard Watson for helpful discussions; Randy Ellis and Cody Rasmussen-Ivey for very insightful comments on a draft of the paper; Susan Lewis for editorial assistance with the manuscript; and two referees for their helpful suggestions. We gratefully acknowledge funding from Templeton World Charity Foundation (TWCF0606) and the John Templeton Foundation (62212), as well as sponsored research agreements to Tufts University from Astonishing labs and Morphoceuticals.
Declaration of interests
M.L. is a scientific cofounder of a company called Astonishing Labs, which includes computational approaches to information processing by multi-scale pathways as one of its long-term goals. He is also a scientific cofounder of a company called Morphoceuticals, which includes bioelectric approaches to regeneration as one of its long-term goals.
Biography
About the author
Michael Levin earned undergraduate degrees in computer science and biology. He received his PhD in genetics from Harvard and post-doctoral training in cell biology at Harvard School of Medicine. His group focuses on the evolution and scaling of diverse intelligence: proto-cognitive competencies in cells, organs, and swarms in transcriptional, physiological, anatomical, and behavioral problem spaces. He has developed tools for reading and writing information in somatic bioelectric networks that guide growth and form. Using machine learning and developmental biophysics, he has applied the tools of behavioral science toward applications in birth defects, regeneration, cancer remodeling, and the bioengineering of novel synthetic life forms.
References
- 1.Emmert-Streib F., Dehmer M., Haibe-Kains B. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Front. Cell Dev. Biol. 2014;2:38. doi: 10.3389/fcell.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brash D.E. Rethinking Causation for Data-intensive Biology: constraints, Cancellations, and Quantized Organisms: causality in complex organisms is sculpted by constraints rather than instigators, with outcomes perhaps better described by quantized patterns than rectilinear pathways. Bioessays. 2020;42:e1900135. doi: 10.1002/bies.201900135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noble D. A theory of biological relativity: no privileged level of causation. Interface Focus. 2012;2:55–64. doi: 10.1098/Rsfs.2011.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bizzarri M., Brash D.E., Briscoe J., Grieneisen V.A., Stern C.D., Levin M. A call for a better understanding of causation in cell biology. Nat. Rev. Mol. Cell Biol. 2019;20:261–262. doi: 10.1038/s41580-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 5.Simeoni C., Dinicola S., Cucina A., Mascia C., Bizzarri M. Systems biology approach and mathematical modeling for analyzing phase-space switch during epithelial-mesenchymal transition. Methods Mol. Biol. 2018;1702:95–123. doi: 10.1007/978-1-4939-7456-6_7. [DOI] [PubMed] [Google Scholar]
- 6.Bizzarri M., Palombo A., Cucina A. Theoretical aspects of systems biology. Prog. Biophys. Mol. Biol. 2013;112:33–43. doi: 10.1016/j.pbiomolbio.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 7.Das S.K., Menezes M.E., Bhatia S., Wang X.Y., Emdad L., Sarkar D., Fisher P.B. Gene therapies for cancer: strategies, challenges and successes. J. Cell. Physiol. 2015;230:259–271. doi: 10.1002/jcp.24791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paananen J., Fortino V. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2020;21:1937–1953. doi: 10.1093/bib/bbz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrov D.S. Therapeutic proteins. Methods Mol. Biol. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamb A., Wee S., Lengauer C. Why is cancer drug discovery so difficult? Nat. Rev. Drug Discov. 2007;6:115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- 11.Lobo D., Solano M., Bubenik G.A., Levin M. A linear-encoding model explains the variability of the target morphology in regeneration. J. R. Soc. Interface. 2014;11:20130918. doi: 10.1098/rsif.2013.0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scannell J.W., Bosley J. When quality beats quantity: decision theory, drug discovery, and the reproducibility crisis. PLoS One. 2016;11:e0147215. doi: 10.1371/journal.pone.0147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scannell J.W., Bosley J., Hickman J.A., Dawson G.R., Truebel H., Ferreira G.S., Richards D., Treherne J.M. Predictive validity in drug discovery: what it is, why it matters and how to improve it. Nat. Rev. Drug Discov. 2022;21:915–931. doi: 10.1038/s41573-022-00552-x. [DOI] [PubMed] [Google Scholar]
- 14.Seyhan A.A. Lost in translation: the valley of death across preclinical and clinical divide – identification of problems and overcoming obstacles. Transl. Med. Commun. 2019;4:18. doi: 10.1186/s41231-019-0050-7. [DOI] [Google Scholar]
- 15.Akl S.G. Information and computation: the essence of it all. Int. J. Unconv. Comput. 2017;13:187–194. [Google Scholar]
- 16.Hopfield J.J. Physics, computation, and why biology looks so different. J. Theor. Biol. 1994;171:53–60. doi: 10.1006/jtbi.1994.1211. [DOI] [Google Scholar]
- 17.Ellis G., Drossel B. How downwards causation occurs in digital computers. Found. Phys. 2019;49:1253–1277. doi: 10.1007/s10701-019-00307-6. [DOI] [Google Scholar]
- 18.Rachwalski M., Khonsari R.H., Paternoster G. Current approaches in the development of molecular and pharmacological therapies in craniosynostosis utilizing animal models. Mol. Syndromol. 2019;10:115–123. doi: 10.1159/000493535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galgano M., Toshkezi G., Qiu X., Russell T., Chin L., Zhao L.R. Traumatic brain injury: current treatment strategies and future endeavors. Cell Transplant. 2017;26:1118–1130. doi: 10.1177/0963689717714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeClerc S., Easley D. Pharmacological therapies for autism spectrum disorder: a review. Pharm. Therapeut. 2015;40:389–397. [PMC free article] [PubMed] [Google Scholar]
- 21.Pokhriyal R., Hariprasad R., Kumar L., Hariprasad G. Chemotherapy resistance in advanced ovarian cancer patients. Biomark. Cancer. 2019;11 doi: 10.1177/1179299X19860815. 1179299X19860815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friston K.J., Stephan K.E., Montague R., Dolan R.J. Computational psychiatry: the brain as a phantastic organ. Lancet Psychiatr. 2014;1:148–158. doi: 10.1016/S2215-0366(14)70275-5. [DOI] [PubMed] [Google Scholar]
- 23.Corlett P.R., Fletcher P.C. Computational psychiatry: a Rosetta Stone linking the brain to mental illness. Lancet Psychiatr. 2014;1:399–402. doi: 10.1016/S2215-0366(14)70298-6. [DOI] [PubMed] [Google Scholar]
- 24.Montague P.R., Dolan R.J., Friston K.J., Dayan P. Computational psychiatry. Trends Cogn. Sci. 2012;16:72–80. doi: 10.1016/j.tics.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramstead M.J., Sakthivadivel D.A., Heins C., Koudahl M., Millidge B., Da Costa L., Klein B., Friston K.J. On Bayesian mechanics: a physics of and by beliefs. arXiv. 2022 doi: 10.48550/arXiv.2205.11543. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friston K. A free energy principle for a particular physics. arXiv. 2019 doi: 10.48550/arXiv.1906.10184. Preprint at. [DOI] [Google Scholar]
- 27.McEwen B.S. Stress, adaptation, and disease. Allostasis and allostatic load. Ann. N. Y. Acad. Sci. 1998;840:33–44. doi: 10.1111/j.1749-6632.1998.tb09546.x. [DOI] [PubMed] [Google Scholar]
- 28.Matsushita Y., Kaneko K. Homeorhesis in Waddington's landscape by epigenetic feedback regulation. Phys. Rev. Res. 2020;2:023083. doi: 10.1103/PhysRevResearch.2.023083. [DOI] [Google Scholar]
- 29.Davies J., Levin M. Synthetic morphology with agential materials. Nature Reviews Bioengineering. 2023;1:46–59. doi: 10.1038/s44222-022-00001-9. [DOI] [Google Scholar]
- 30.Levin M. Technological approach to mind everywhere: an experimentally-grounded framework for understanding diverse bodies and minds. Front. Syst. Neurosci. 2022;16:768201. doi: 10.3389/fnsys.2022.768201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pezzulo G., Levin M. Top-down models in biology: explanation and control of complex living systems above the molecular level. J. R. Soc. Interface. 2016;13:20160555. doi: 10.1098/rsif.2016.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pezzulo G., Levin M. Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integr. Biol. 2015;7:1487–1517. doi: 10.1039/c5ib00221d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marr D. W.H. Freeman; 1982. Vision : A Computational Investigation into the Human Representation and Processing of Visual Information. [Google Scholar]
- 34.Adams R.A., Huys Q.J.M., Roiser J.P. Computational Psychiatry: towards a mathematically informed understanding of mental illness. J. Neurol. Neurosurg. Psychiatry. 2016;87:53–63. doi: 10.1136/jnnp-2015-310737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X.J., Krystal J.H. Computational psychiatry. Neuron. 2014;84:638–654. doi: 10.1016/j.neuron.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fields C., Bischof J., Levin M. Morphological coordination: a common ancestral function unifying neural and non-neural signaling. Physiology. 2020;35:16–30. doi: 10.1152/physiol.00027.2019. [DOI] [PubMed] [Google Scholar]
- 37.Fields C., Levin M. Competency in navigating arbitrary spaces as an invariant for analyzing cognition in diverse embodiments. Entropy. 2022;24:819. doi: 10.3390/e24060819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Csermely P., Kunsic N., Mendik P., Kerestély M., Faragó T., Veres D.V., Tompa P. Learning of signaling networks: molecular mechanisms. Trends Biochem. Sci. 2020;45:284–294. doi: 10.1016/j.tibs.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Vohradský J. Neural model of the genetic network. J. Biol. Chem. 2001;276:36168–36173. doi: 10.1074/jbc.M104391200. [DOI] [PubMed] [Google Scholar]
- 40.Vohradský J. Neural network model of gene expression. FASEB J. 2001;15:846–854. doi: 10.1096/fj.00-0361com. [DOI] [PubMed] [Google Scholar]
- 41.Watson R.A., Buckley C.L., Mills R., Davies A. 2010. Associative Memory in Gene Regulation Networks; pp. 194–201. held in Odense. [Google Scholar]
- 42.Baluška F., Levin M. On having No head: cognition throughout biological systems. Front. Psychol. 2016;7:902. doi: 10.3389/fpsyg.2016.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keijzer F., van Duijn M., Lyon P. What nervous systems do: early evolution, input-output, and the skin brain thesis. Adapt. Behav. 2013;21:67–85. doi: 10.1177/1059712312465330. [DOI] [Google Scholar]
- 44.Prindle A., Liu J., Asally M., Ly S., Garcia-Ojalvo J., Süel G.M. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manicka S., Levin M. The Cognitive Lens: a primer on conceptual tools for analysing information processing in developmental and regenerative morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374:20180369. doi: 10.1098/rstb.2018.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyon P. The biogenic approach to cognition. Cogn. Process. 2006;7:11–29. doi: 10.1007/s10339-005-0016-8. [DOI] [PubMed] [Google Scholar]
- 47.Pezzulo G., LaPalme J., Durant F., Levin M. Bistability of somatic pattern memories: stochastic outcomes in bioelectric circuits underlying regeneration. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376:20190765. doi: 10.1098/rstb.2019.0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart J., Badiani A. Tolerance and sensitization to the behavioral effects of drugs. Behav. Pharmacol. 1993;4:289–312. [PubMed] [Google Scholar]
- 49.Löscher W., Schmidt D. Experimental and clinical evidence for loss of effect (tolerance) during prolonged treatment with antiepileptic drugs. Epilepsia. 2006;47:1253–1284. doi: 10.1111/j.1528-1167.2006.00607.x. [DOI] [PubMed] [Google Scholar]
- 50.Scholz H., Franz M., Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penderson C.A. In: International Review of Neurobiology. T.T.E., editor. Academic Press; 2017. Oxytocin, tolerance, and the dark side of addiction; pp. 239–274. [DOI] [PubMed] [Google Scholar]
- 52.Esser M.B., Sherk A., Liu Y., Naimi T.S., Stockwell T., Stahre M., Kanny D., Landen M., Saitz R., Brewer R.D. Deaths and years of potential life lost from excessive alcohol use - United States, 2011-2015. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1428–1433. doi: 10.15585/mmwr.mm6939a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattson C.L., Tanz L.J., Quinn K., Kariisa M., Patel P., Davis N.L. Trends and geographic patterns in drug and synthetic opioid overdose deaths - United States, 2013-2019. MMWR Morb. Mortal. Wkly. Rep. 2021;70:202–207. doi: 10.15585/mmwr.mm7006a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steketee J.D., Kalivas P.W. Drug wanting: behavioral sensitization and relapse to drug-seeking behavior. Pharmacol. Rev. 2011;63:348–365. doi: 10.1124/pr.109.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berridge K.C., Robinson T.E. Liking, wanting, and the incentive-sensitization theory of addiction. Am. Psychol. 2016;71:670–679. doi: 10.1037/amp0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva-Cardoso G.K., Nobre M.J. Context-specific tolerance and pharmacological changes in the infralimbic cortex-nucleus accumbens shell pathway evoked by ketamine. Neurochem. Res. 2021;46:1686–1700. doi: 10.1007/s11064-021-03300-6. [DOI] [PubMed] [Google Scholar]
- 57.White A.M., Roberts D.C., Best P.J. Context-specific tolerance to the ataxic effects of alcohol. Pharmacol. Biochem. Behav. 2002;72:107–110. doi: 10.1016/s0091-3057(01)00731-6. [DOI] [PubMed] [Google Scholar]
- 58.Siegel S., Ellsworth D.W. Pavlovian conditioning and death from apparent overdose of medically prescribed morphine: a case report. Bull. Psychon. Soc. 2013;24:278–280. doi: 10.3758/bf03330140. [DOI] [Google Scholar]
- 59.Dumas E.O., Pollack G.M. Opioid tolerance development: a pharmacokinetic/pharmacodynamic perspective. AAPS J. 2008;10:537–551. doi: 10.1208/s12248-008-9056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.García-Suástegui W.A., Ramos-Chávez L.A., Rubio-Osornio M., Calvillo-Velasco M., Atzin-Méndez J.A., Guevara J., Silva-Adaya D. The role of CYP2E1 in the drug metabolism or bioactivation in the brain. Oxid. Med. Cell. Longev. 2017;2017:4680732. doi: 10.1155/2017/4680732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hakkola J., Hukkanen J., Turpeinen M., Pelkonen O. Inhibition and induction of CYP enzymes in humans: an update. Arch. Toxicol. 2020;94:3671–3722. doi: 10.1007/s00204-020-02936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okamoto M., Rao S.N., Reyes J., Rifkind A.B. Recovery from dispositional and pharmacodynamic tolerance after chronic pentobarbital treatment. J. Pharmacol. Exp. Ther. 1985;235:26–31. [PubMed] [Google Scholar]
- 63.Löscher W., Potschka H. Role of multidrug transporters in pharmacoresistance to antiepileptic drugs. J. Pharmacol. Exp. Ther. 2002;301:7–14. doi: 10.1124/jpet.301.1.7. [DOI] [PubMed] [Google Scholar]
- 64.Tang F., Hartz A.M.S., Bauer B. Drug-resistant epilepsy: multiple hypotheses, few answers. Front. Neurol. 2017;8:301. doi: 10.3389/fneur.2017.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grewal G.K., Kukal S., Kanojia N., Saso L., Kukreti S., Kukreti R. Effect of oxidative stress on ABC transporters: contribution to epilepsy pharmacoresistance. Molecules. 2017;22:365. doi: 10.3390/molecules22030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sisodiya S.M., Lin W.R., Harding B.N., Squier M.V., Thom M. Drug resistance in epilepsy: expression of drug resistance proteins in common causes of refractory epilepsy. Brain. 2002;125:22–31. doi: 10.1093/brain/awf002. [DOI] [PubMed] [Google Scholar]
- 67.Chaumont S., André C., Perrais D., Boué-Grabot E., Taly A., Garret M. Agonist-dependent endocytosis of gamma-aminobutyric acid type A (GABAA) receptors revealed by a gamma2(R43Q) epilepsy mutation. J. Biol. Chem. 2013;288:28254–28265. doi: 10.1074/jbc.M113.470807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benowitz N.L. Pharmacology of nicotine: addiction, smoking-induced disease, and therapeutics. Annu. Rev. Pharmacol. Toxicol. 2009;49:57–71. doi: 10.1146/annurev.pharmtox.48.113006.094742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dawson L.A., Nguyen H.Q., Smith D.L., Schechter L.E. Effect of chronic fluoxetine and WAY-100635 treatment on serotonergic neurotransmission in the frontal cortex. J. Psychopharmacol. 2002;16:145–152. doi: 10.1177/026988110201600205. [DOI] [PubMed] [Google Scholar]
- 70.McGrath P.J., Stewart J.W., Quitkin F.M., Chen Y., Alpert J.E., Nierenberg A.A., Fava M., Cheng J., Petkova E. Predictors of relapse in a prospective study of fluoxetine treatment of major depression. Am. J. Psychiatry. 2006;163:1542–1548. doi: 10.1176/ajp.2006.163.9.1542. [DOI] [PubMed] [Google Scholar]
- 71.Burnier M., Brunner H.R. Neurohormonal consequences of diuretics in different cardiovascular syndromes. Eur. Heart J. 1992;13(Suppl G):28–33. doi: 10.1093/eurheartj/13.suppl_g.28. [DOI] [PubMed] [Google Scholar]
- 72.Wakelkamp M., Alván G., Gabrielsson J., Paintaud G. Pharmacodynamic modeling of furosemide tolerance after multiple intravenous administration. Clin. Pharmacol. Ther. 1996;60:75–88. doi: 10.1016/s0009-9236(96)90170-8. [DOI] [PubMed] [Google Scholar]
- 73.Sokolowska M., Siegel S., Kim J.A. Intraadministration associations: conditional hyperalgesia elicited by morphine onset cues. J. Exp. Psychol. Anim. Behav. Process. 2002;28:309–320. doi: 10.1037/0097-7403.28.3.309. [DOI] [PubMed] [Google Scholar]
- 74.Duttaroy A., Yoburn B.C. The effect of intrinsic efficacy on opioid tolerance. Anesthesiology. 1995;82:1226–1236. doi: 10.1097/00000542-199505000-00018. [DOI] [PubMed] [Google Scholar]
- 75.Ibrahim K., Donyai P. Drug holidays from ADHD medication: international experience over the past four decades. J. Atten. Disord. 2015;19:551–568. doi: 10.1177/1087054714548035. [DOI] [PubMed] [Google Scholar]
- 76.Rosenzweig S.A. Acquired resistance to drugs targeting tyrosine kinases. Adv. Cancer Res. 2018;138:71–98. doi: 10.1016/bs.acr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siveen K.S., Prabhu K.S., Achkar I.W., Kuttikrishnan S., Shyam S., Khan A.Q., Merhi M., Dermime S., Uddin S. Role of non receptor tyrosine kinases in hematological malignances and its targeting by natural products. Mol. Cancer. 2018;17:31. doi: 10.1186/s12943-018-0788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alves R., Gonçalves A.C., Rutella S., Almeida A.M., De Las Rivas J., Trougakos I.P., Sarmento Ribeiro A.B. Resistance to tyrosine kinase inhibitors in chronic myeloid leukemia-from molecular mechanisms to clinical relevance. Cancers. 2021;13:4820. doi: 10.3390/cancers13194820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah N.P. Loss of response to imatinib: mechanisms and management. Hematology. Am. Soc. Hematol. Educ. Program. 2005;2005:183–187. doi: 10.1182/asheducation-2005.1.183. [DOI] [PubMed] [Google Scholar]
- 80.Michor F. Quantitative approaches to analyzing imatinib-treated chronic myeloid leukemia. Trends Pharmacol. Sci. 2007;28:197–199. doi: 10.1016/j.tips.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 81.Antonescu C.R., Besmer P., Guo T., Arkun K., Hom G., Koryotowski B., Leversha M.A., Jeffrey P.D., Desantis D., Singer S., et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin. Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 82.Newlin D.B. A comparison of drug conditioning and craving for alcohol and cocaine. Recent Dev. Alcohol. 1992;10:147–164. doi: 10.1007/978-1-4899-1648-8_8. [DOI] [PubMed] [Google Scholar]
- 83.Crowell C.R., Hinson R.E., Siegel S. The role of conditional drug responses in tolerance to the hypothermic effects of ethanol. Psychopharmacology (Berl) 1981;73:51–54. doi: 10.1007/BF00431101. [DOI] [PubMed] [Google Scholar]
- 84.Cunningham C.L., Crabbe J.C., Rigter H. Pavlovian conditioning of drug-induced changes in body temperature. Pharmacol. Ther. 1983;23:365–391. doi: 10.1016/0163-7258(83)90019-0. [DOI] [PubMed] [Google Scholar]
- 85.Dafters R., Anderson G. Conditioned tolerance to the tachycardia effect of ethanol in humans. Psychopharmacology (Berl) 1982;78:365–367. doi: 10.1007/BF00433743. [DOI] [PubMed] [Google Scholar]
- 86.Siegel S. Pavlovian conditioning and drug overdose: when tolerance fails. Addiction Res. Theor. 2001;9:503–513. doi: 10.3109/16066350109141767. [DOI] [Google Scholar]
- 87.Siegel S., Hinson R.E., Krank M.D., McCully J. Heroin "overdose" death: contribution of drug-associated environmental cues. Science. 1982;216:436–437. doi: 10.1126/science.7200260. [DOI] [PubMed] [Google Scholar]
- 88.Krank M.D., Hinson R.E., Siegel S. Effect of partial reinforcement on tolerance to morphine-induced analgesia and weight loss in the rat. Behav. Neurosci. 1984;98:72–78. doi: 10.1037//0735-7044.98.1.72. [DOI] [PubMed] [Google Scholar]
- 89.Dyck D.G., Greenberg A.H., Osachuk T.A. Tolerance to drug-induced (poly I:C) natural killer cell activation: congruence with a Pavlovian conditioning model. J. Exp. Psychol. Anim. Behav. Process. 1986;12:25–31. doi: 10.1037/0097-7403.12.1.25. [DOI] [PubMed] [Google Scholar]
- 90.Fanselow M.S., German C. Explicitly unpaired delivery of morphine and the test situation: extinction and retardation of tolerance to the suppressing effects of morphine on locomotor activity. Behav. Neural. Biol. 1982;35:231–241. doi: 10.1016/s0163-1047(82)90665-3. [DOI] [PubMed] [Google Scholar]
- 91.Siegel S., Sdao-Jarvie K. Attenuation of ethanol tolerance by a novel stimulus. Psychopharmacology (Berl) 1986;88:258–261. doi: 10.1007/BF00652251. [DOI] [PubMed] [Google Scholar]
- 92.Carey R.J., Carrera M.P., Damianopoulos E.N. A new proposal for drug conditioning with implications for drug addiction: the Pavlovian two-step from delay to trace conditioning. Behav. Brain Res. 2014;275:150–156. doi: 10.1016/j.bbr.2014.08.053. [DOI] [PubMed] [Google Scholar]
- 93.O'Brien C., Childress A., McLellan A., Ehrman R., Ternes J. Progress in understanding the conditioning aspects of drug dependence. NIDA Res. Monogr. 1988;81:395–404. [PubMed] [Google Scholar]
- 94.Ramoz L., Lodi S., Bhatt P., Reitz A.B., Tallarida C., Tallarida R.J., Raffa R.B., Rawls S.M. Mephedrone ("bath salt") pharmacology: insights from invertebrates. Neuroscience. 2012;208:79–84. doi: 10.1016/j.neuroscience.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rawls S.M., Patil T., Tallarida C.S., Baron S., Kim M., Song K., Ward S., Raffa R.B. Nicotine behavioral pharmacology: clues from planarians. Drug Alcohol Depend. 2011;118:274–279. doi: 10.1016/j.drugalcdep.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spyraki C., Nomikos G. On the role of serotonin in drug reward: studies with the place conditioning procedure. Pol. J. Pharmacol. Pharm. 1991;43:221–229. [PubMed] [Google Scholar]
- 97.MacRae J.R., Scoles M.T., Siegel S. The contribution of Pavlovian conditioning to drug tolerance and dependence. Br. J. Addict. 1987;82:371–380. doi: 10.1111/j.1360-0443.1987.tb01493.x. [DOI] [PubMed] [Google Scholar]
- 98.Pavlov I.P. Application of the results of our animal experiments to man. Dtsch. Gesundheitsw. 1953;8:32–40. [PubMed] [Google Scholar]
- 99.Sparkman N.L., Li M. Drug-drug conditioning between citalopram and haloperidol or olanzapine in a conditioned avoidance response model: implications for polypharmacy in schizophrenia. Behav. Pharmacol. 2012;23:658–668. doi: 10.1097/FBP.0b013e328358590d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng D.T., Disterhoft J.F., Power J.M., Ellis D.A., Desmond J.E. Neural substrates underlying human delay and trace eyeblink conditioning. Proc. Natl. Acad. Sci. USA. 2008;105:8108–8113. doi: 10.1073/pnas.0800374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ewald H., Glotzbach-Schoon E., Gerdes A.B.M., Andreatta M., Müller M., Mühlberger A., Pauli P. Delay and trace fear conditioning in a complex virtual learning environment-neural substrates of extinction. Front. Hum. Neurosci. 2014;8:323. doi: 10.3389/fnhum.2014.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robinson T.E., Berridge K.C. The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Res. Brain Res. Rev. 1993;18:247–291. doi: 10.1016/0165-0173(93)90013-. [DOI] [PubMed] [Google Scholar]
- 103.Varlinskaya E.I., Petrov E.S., Smotherman W.P. Classical conditioning in the fetal rat: reinforcing properties of dynorphin A (1-13) Behav. Neurosci. 1996;110:154–167. doi: 10.1037//0735-7044.110.1.154. [DOI] [PubMed] [Google Scholar]
- 104.Shaw J.E., Koleske A.J. Functional interactions of ion channels with the actin cytoskeleton: does coupling to dynamic actin regulate NMDA receptors? J. Physiol. 2021;599:431–441. doi: 10.1113/JP278702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crow T., Xue-Bian J.J. One-trial in vitro conditioning of hermissenda regulates phosphorylation of ser-122 of csp24. Ann. N. Y. Acad. Sci. 2007;1112:189–200. doi: 10.1196/annals.1415.012. [DOI] [PubMed] [Google Scholar]
- 106.Crow T., Xue-Bian J.J., Neary J.T. 14-3-3 proteins interact with the beta-thymosin repeat protein Csp24. Neurosci. Lett. 2007;424:6–9. doi: 10.1016/j.neulet.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dykman R.A., Gantt W.H. Experimental psychogenic hypertension: blood pressure changes conditioned to painful stimuli (schizokinesis) Integr. Physiol. Behav. Sci. 1997;32:272–287. doi: 10.1007/BF02688625. [DOI] [PubMed] [Google Scholar]
- 108.Tekampe J., van Middendorp H., Meeuwis S.H., van Leusden J.W.R., Pacheco-López G., Hermus A.R.M.M., Evers A.W.M. Conditioning immune and endocrine parameters in humans: a systematic review. Psychother. Psychosom. 2017;86:99–107. doi: 10.1159/000449470. [DOI] [PubMed] [Google Scholar]
- 109.Tekampe J., van Middendorp H., Sweep F.C.G.J., Roerink S.H.P.P., Hermus A.R.M.M., Evers A.W.M. Human pharmacological conditioning of the immune and endocrine system: challenges and opportunities. Int. Rev. Neurobiol. 2018;138:61–80. doi: 10.1016/bs.irn.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 110.Skvortsova A., Veldhuijzen D.S., Kloosterman I.E.M., Meijer O.C., van Middendorp H., Pacheco-Lopez G., Evers A.W.M. Conditioned hormonal responses: a systematic review in animals and humans. Front. Neuroendocrinol. 2019;52:206–218. doi: 10.1016/j.yfrne.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 111.Tekampe J., van Middendorp H., Sweep F.C.G.J., Roerink S.H.P.P., Hermus A.R.M.M., Evers A.W.M. Conditioning cortisol in humans: design and pilot study of a randomized controlled trial. Pilot Feasibility Stud. 2019;5:9. doi: 10.1186/s40814-018-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Szabó Á., Vattay G., Kondor D. A cell signaling model as a trainable neural nanonetwork. Nano Commun. Netw. 2012;3:57–64. [Google Scholar]
- 113.Turner C.H., Robling A.G., Duncan R.L., Burr D.B. Do bone cells behave like a neuronal network? Calcif. Tissue Int. 2002;70:435–442. doi: 10.1007/s00223-001-1024-z. [DOI] [PubMed] [Google Scholar]
- 114.Goel P., Mehta A. Learning theories reveal loss of pancreatic electrical connectivity in diabetes as an adaptive response. PLoS One. 2013;8:e70366. doi: 10.1371/journal.pone.0070366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nashun B., Hill P.W.S., Hajkova P. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 2015;34:1296–1308. doi: 10.15252/embj.201490649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quintin J., Cheng S.C., van der Meer J.W.M., Netea M.G. Innate immune memory: towards a better understanding of host defense mechanisms. Curr. Opin. Immunol. 2014;29:1–7. doi: 10.1016/j.coi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 117.Corre G., Stockholm D., Arnaud O., Kaneko G., Viñuelas J., Yamagata Y., Neildez-Nguyen T.M.A., Kupiec J.J., Beslon G., Gandrillon O., et al. Stochastic fluctuations and distributed control of gene expression impact cellular memory. PLoS One. 2014;9:e115574. doi: 10.1371/journal.pone.0115574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zediak V.P., Wherry E.J., Berger S.L. The contribution of epigenetic memory to immunologic memory. Curr. Opin. Genet. Dev. 2011;21:154–159. doi: 10.1016/j.gde.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 119.Watson R.A., Mills R., Buckley C.L. Global adaptation in networks of selfish components: emergent associative memory at the system scale. Artif. Life. 2011;17:147–166. doi: 10.1162/artl_a_00029. [DOI] [PubMed] [Google Scholar]
- 120.American Association for the Advancement of Science Maturing from memory. Sci. Signal. 2003;2003:tw462. [Google Scholar]
- 121.Sible J.C. Thanks for the memory. Nature. 2003;426:392–393. doi: 10.1038/426392a. [DOI] [PubMed] [Google Scholar]
- 122.Xiong W., Ferrell J.E. A positive-feedback-based bistable ‘memory module’that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 123.Levine J.H., Lin Y., Elowitz M.B. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Urrios A., Macia J., Manzoni R., Conde N., Bonforti A., de Nadal E., Posas F., Solé R. A synthetic multicellular memory device. ACS Synth. Biol. 2016;5:862–873. doi: 10.1021/acssynbio.5b00252. [DOI] [PubMed] [Google Scholar]
- 125.Macia J., Vidiella B., Solé R.V. Synthetic associative learning in engineered multicellular consortia. J. R. Soc. Interface. 2017;14:20170158. doi: 10.1098/rsif.2017.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kandel E.R., Dudai Y., Mayford M.R. The molecular and systems biology of memory. Cell. 2014;157:163–186. doi: 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 127.Ryan T.J., Roy D.S., Pignatelli M., Arons A., Tonegawa S. Engram cells retain memory under retrograde amnesia. Science. 2015;348:1007–1013. doi: 10.1126/science.aaa5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Szilágyi A., Szabó P., Santos M., Szathmáry E. Phenotypes to remember: evolutionary developmental memory capacity and robustness. PLoS Comput. Biol. 2020;16:e1008425. doi: 10.1371/journal.pcbi.1008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Biswas S., Manicka S., Hoel E., Levin M. Gene regulatory networks exhibit several kinds of memory: quantification of memory in biological and random transcriptional networks. iScience. 2021;24:102131. doi: 10.1016/j.isci.2021.102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Biswas S., Clawson W., Levin M. Learning in transcriptional network models: computational discovery of pathway-level memory and effective interventions. Int. J. Mol. Sci. 2022;24:285. doi: 10.3390/ijms24010285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fernando C.T., Liekens A.M.L., Bingle L.E.H., Beck C., Lenser T., Stekel D.J., Rowe J.E. Molecular circuits for associative learning in single-celled organisms. J. R. Soc. Interface. 2009;6:463–469. doi: 10.1098/rsif.2008.0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gyurkó D.M., Veres D.V., Módos D., Lenti K., Korcsmáros T., Csermely P. Adaptation and learning of molecular networks as a description of cancer development at the systems-level: potential use in anti-cancer therapies. Semin. Cancer Biol. 2013;23:262–269. doi: 10.1016/j.semcancer.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 133.Csermely P., Korcsmáros T., Kiss H.J.M., London G., Nussinov R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol. Ther. 2013;138:333–408. doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Csermely P., Korcsmáros T. Cancer-related networks: a help to understand, predict and change malignant transformation. Semin. Cancer Biol. 2013;23:209–212. doi: 10.1016/j.semcancer.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 135.Abramson C.I., Levin M. Behaviorist approaches to investigating memory and learning: a primer for synthetic biology and bioengineering. Commun. Integr. Biol. 2021;14:230–247. doi: 10.1080/19420889.2021.2005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Graudenzi A., Serra R., Villani M., Colacci A., Kauffman S.A. Robustness analysis of a Boolean model of gene regulatory network with memory. J. Comput. Biol. 2011;18:559–577. doi: 10.1089/cmb.2010.0224. [DOI] [PubMed] [Google Scholar]
- 137.Damiani C., Serra R., Villani M., Kauffman S.A., Colacci A. Cell-cell interaction and diversity of emergent behaviours. IET Syst. Biol. 2011;5:137–144. doi: 10.1049/iet-syb.2010.0039. [DOI] [PubMed] [Google Scholar]
- 138.Serra R., Villani M., Barbieri A., Kauffman S.A., Colacci A. On the dynamics of random Boolean networks subject to noise: attractors, ergodic sets and cell types. J. Theor. Biol. 2010;265:185–193. doi: 10.1016/j.jtbi.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 139.Damiani C., Kauffman S.A., Serra R., Villani M., Colacci A. Information transfer among coupled random boolean networks. Cell. Automata. 2010;6350:1–11. [Google Scholar]
- 140.Huang S., Ernberg I., Kauffman S. Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Semin. Cell Dev. Biol. 2009;20:869–876. doi: 10.1016/j.semcdb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.James W. H. Holt and company; 1890. The Principles of Psychology. [Google Scholar]
- 142.Dubey A., Saint-Jeannet J.P. Modeling human craniofacial disorders in Xenopus. Curr. Pathobiol. Rep. 2017;5:79–92. doi: 10.1007/s40139-017-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vandenberg L.N., Adams D.S., Levin M. Normalized shape and location of perturbed craniofacial structures in the Xenopus tadpole reveal an innate ability to achieve correct morphology. Dev. Dyn. 2012;241:863–878. doi: 10.1002/dvdy.23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mathews J., Levin M. The body electric 2.0: recent advances in developmental bioelectricity for regenerative and synthetic bioengineering. Curr. Opin. Biotechnol. 2018;52:134–144. doi: 10.1016/j.copbio.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Levin M. The wisdom of the body: future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer. Regen. Med. 2011;6:667–673. doi: 10.2217/rme.11.69. [DOI] [PubMed] [Google Scholar]
- 146.Stringer S.M., Rolls E.T., Trappenberg T.P., de Araujo I.E.T. Self-organizing continuous attractor networks and motor function. Neural Netw. 2003;16:161–182. doi: 10.1016/S0893-6080(02)00237-X. [DOI] [PubMed] [Google Scholar]
- 147.Doboli S., Minai A.A., Best P.J. Latent attractors: a model for context-dependent place representations in the hippocampus. Neural Comput. 2000;12:1009–1043. doi: 10.1162/089976600300015484. [DOI] [PubMed] [Google Scholar]
- 148.Emmons-Bell M., Durant F., Tung A., Pietak A., Miller K., Kane A., Martyniuk C.J., Davidian D., Morokuma J., Levin M. Regenerative adaptation to electrochemical perturbation in planaria: a molecular analysis of physiological plasticity. iScience. 2019;22:147–165. doi: 10.1016/j.isci.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pai V.P., Pietak A., Willocq V., Ye B., Shi N.Q., Levin M. HCN2 Rescues brain defects by enforcing endogenous voltage pre-patterns. Nat. Commun. 2018;9:998. doi: 10.1038/s41467-018-03334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pai V.P., Lemire J.M., Paré J.F., Lin G., Chen Y., Levin M. Endogenous gradients of resting potential instructively pattern embryonic neural tissue via notch signaling and regulation of proliferation. J. Neurosci. 2015;35:4366–4385. doi: 10.1523/JNEUROSCI.1877-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pietak A., Levin M. Exploring instructive physiological signaling with the bioelectric tissue simulation engine (BETSE) Front. Bioeng. Biotechnol. 2016;4:55. doi: 10.3389/fbioe.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mathews J., Levin M. Gap junctional signaling in pattern regulation: physiological network connectivity instructs growth and form. Dev. Neurobiol. 2017;77:643–673. doi: 10.1002/dneu.22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dahal G.R., Rawson J., Gassaway B., Kwok B., Tong Y., Ptácek L.J., Bates E. An inwardly rectifying K+ channel is required for patterning. Development. 2012;139:3653–3664. doi: 10.1242/dev.078592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dahal G.R., Pradhan S.J., Bates E.A. Inwardly rectifying potassium channels influence Drosophila wing morphogenesis by regulating Dpp release. Development. 2017;144:2771–2783. doi: 10.1242/dev.146647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Belus M.T., Rogers M.A., Elzubeir A., Josey M., Rose S., Andreeva V., Yelick P.C., Bates E.A. Kir2.1 is important for efficient BMP signaling in mammalian face development. Dev. Biol. 2018;444(Suppl 1):S297–S307. doi: 10.1016/j.ydbio.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Adams D.S., Uzel S.G.M., Akagi J., Wlodkowic D., Andreeva V., Yelick P.C., Devitt-Lee A., Paré J.F., Levin M. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J. Physiol. 2016;594:3245–3270. doi: 10.1113/JP271930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vandenberg L.N., Morrie R.D., Adams D.S. V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev. Dyn. 2011;240:1889–1904. doi: 10.1002/dvdy.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pai V.P., Aw S., Shomrat T., Lemire J.M., Levin M. Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development. 2012;139:313–323. doi: 10.1242/dev.073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pai V.P., Lemire J.M., Chen Y., Lin G., Levin M. Local and long-range endogenous resting potential gradients antagonistically regulate apoptosis and proliferation in the embryonic CNS. Int. J. Dev. Biol. 2015;59:327–340. doi: 10.1387/ijdb.150197ml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pai V.P., Levin M. HCN2 channel-induced rescue of brain, eye, heart and gut teratogenesis caused by nicotine, ethanol and aberrant notch signalling. Wound Repair Regen. 2022;30:681–706. doi: 10.1111/wrr.13032. [DOI] [PubMed] [Google Scholar]
- 161.Pai V.P., Willocq V., Pitcairn E.J., Lemire J.M., Paré J.F., Shi N.Q., McLaughlin K.A., Levin M. HCN4 ion channel function is required for early events that regulate anatomical left-right patterning in a nodal and lefty asymmetric gene expression-independent manner. Biol. Open. 2017;6:1445–1457. doi: 10.1242/bio.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pai V., Levin M. In: Stem Cells. Calegari F., Waskow C., editors. CRC Press; 2013. Bioelectric controls of stem cell function; pp. 106–148. [Google Scholar]
- 163.Liebau S., Kleger A., Levin M., Yu S.P. Stem cells and ion channels. Stem Cells Int. 2013;2013:238635. doi: 10.1155/2013/238635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Blackiston D., Adams D.S., Lemire J.M., Lobikin M., Levin M. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis. Model. Mech. 2011;4:67–85. doi: 10.1242/dmm.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Morokuma J., Blackiston D., Adams D.S., Seebohm G., Trimmer B., Levin M. Modulation of potassium channel function confers a hyperproliferative invasive phenotype on embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:16608–16613. doi: 10.1073/pnas.0808328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yi C., Spitters T.W., Al-Far E.A.D.A., Wang S., Xiong T., Cai S., Yan X., Guan K., Wagner M., El-Armouche A., Antos C.L. A calcineurin-mediated scaling mechanism that controls a K(+)-leak channel to regulate morphogen and growth factor transcription. Elife. 2021;10:e60691. doi: 10.7554/eLife.60691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lanni J.S., Peal D., Ekstrom L., Chen H., Stanclift C., Bowen M.E., Mercado A., Gamba G., Kahle K.T., Harris M.P. Integrated K+ channel and K+Cl- cotransporter functions are required for the coordination of size and proportion during development. Dev. Biol. 2019;456:164–178. doi: 10.1016/j.ydbio.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Daane J.M., Lanni J., Rothenberg I., Seebohm G., Higdon C.W., Johnson S.L., Harris M.P. Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci. Rep. 2018;8:10391. doi: 10.1038/s41598-018-28450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tseng A.S., Beane W.S., Lemire J.M., Masi A., Levin M. Induction of vertebrate regeneration by a transient sodium current. J. Neurosci. 2010;30:13192–13200. doi: 10.1523/JNEUROSCI.3315-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tseng A., Levin M. Cracking the bioelectric code: probing endogenous ionic controls of pattern formation. Commun. Integr. Biol. 2013;6:e22595. doi: 10.4161/cib.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chernet B.T., Levin M. Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumor development in a Xenopus model. Dis. Model. Mech. 2013;6:595–607. doi: 10.1242/dmm.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chernet B.T., Fields C., Levin M. Long-range gap junctional signaling controls oncogene-mediated tumorigenesis in Xenopus laevis embryos. Front. Physiol. 2014;5:519. doi: 10.3389/fphys.2014.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chernet B.T., Levin M. Transmembrane voltage potential of somatic cells controls oncogene-mediated tumorigenesis at long-range. Oncotarget. 2014;5:3287–3306. doi: 10.18632/oncotarget.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mathews J., Kuchling F., Baez-Nieto D., Diberardinis M., Pan J.Q., Levin M. Ion Channel drugs suppress cancer phenotype in NG108-15 and U87 cells: toward novel electroceuticals for glioblastoma. Cancers. 2022;14:1499. doi: 10.3390/cancers14061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Emmons-Bell M., Durant F., Hammelman J., Bessonov N., Volpert V., Morokuma J., Pinet K., Adams D.S., Pietak A., Lobo D., Levin M. Gap junctional blockade stochastically induces different species-specific head anatomies in genetically wild-type girardia dorotocephala flatworms. Int. J. Mol. Sci. 2015;16:27865–27896. doi: 10.3390/ijms161126065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pai V.P., Cervera J., Mafe S., Willocq V., Lederer E.K., Levin M. HCN2 channel-induced rescue of brain teratogenesis via local and long-range bioelectric repair. Front. Cell. Neurosci. 2020;14:136. doi: 10.3389/fncel.2020.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chernet B., Levin M. Endogenous voltage potentials and the microenvironment: bioelectric signals that reveal, induce and normalize cancer. J. Clin. Exp. Oncol. 2013;(Suppl 1):S1-002. doi: 10.4172/2324-9110.S1-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Levin M. Bioelectrical approaches to cancer as a problem of the scaling of the cellular self. Prog. Biophys. Mol. Biol. 2021;165:102–113. doi: 10.1016/j.pbiomolbio.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 179.McMillen P., Oudin M.J., Levin M., Payne S.L. Beyond neurons: long distance communication in development and cancer. Front. Cell Dev. Biol. 2021;9:739024. doi: 10.3389/fcell.2021.739024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sonnenschein C., Soto A.M. Springer; 1999. The Society of Cells : Cancer Control of Cell Proliferation. [Google Scholar]
- 181.Tarin D. Role of the host stroma in cancer and its therapeutic significance. Cancer Metastasis Rev. 2013;32:553–566. doi: 10.1007/s10555-013-9438-4. [DOI] [PubMed] [Google Scholar]
- 182.Rubin H. Cancer as a dynamic developmental disorder. Cancer Res. 1985;45:2935–2942. [PubMed] [Google Scholar]
- 183.Moore D., Walker S.I., Levin M. Cancer as a disorder of patterning information: computational and biophysical perspectives on the cancer problem. Converg. Sci. Phys. Oncol. 2017;3:043001. [Google Scholar]
- 184.Harris M.P. Bioelectric signaling as a unique regulator of development and regeneration. Development. 2021;148:dev180794. doi: 10.1242/dev.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bates E. Ion channels in development and cancer. Annu. Rev. Cell Dev. Biol. 2015;31:231–247. doi: 10.1146/annurev-cellbio-100814-125338. [DOI] [PubMed] [Google Scholar]
- 186.Levin M. Bioelectric signaling: reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell. 2021;184:1971–1989. doi: 10.1016/j.cell.2021.02.034. [DOI] [PubMed] [Google Scholar]
- 187.Spencer Adams D., Lemire J.M., Kramer R.H., Levin M. Optogenetics in Developmental Biology: using light to control ion flux-dependent signals in Xenopus embryos. Int. J. Dev. Biol. 2014;58:851–861. doi: 10.1387/ijdb.140207ml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Adams D.S., Tseng A.S., Levin M. Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration: sparking system-level controls in vivo. Biol. Open. 2013;2:306–313. doi: 10.1242/bio.20133665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Murugan N.J., Vigran H.J., Miller K.A., Golding A., Pham Q.L., Sperry M.M., Rasmussen-Ivey C., Kane A.W., Kaplan D.L., Levin M. Acute multidrug delivery via a wearable bioreactor facilitates long-term limb regeneration and functional recovery in adult Xenopus laevis. Sci. Adv. 2022;8:eabj2164. doi: 10.1126/sciadv.abj2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Piedimonte A., Benedetti F. Words and drugs: same mechanisms of action? J. Contemp. Psychother. 2016;46:159–166. doi: 10.1007/s10879-015-9321-4. [DOI] [Google Scholar]
- 191.Evers A.W.M., Colloca L., Blease C., Annoni M., Atlas L.Y., Benedetti F., Bingel U., Büchel C., Carvalho C., Colagiuri B., et al. Implications of placebo and nocebo effects for clinical practice: expert consensus. Psychother. Psychosom. 2018;87:204–210. doi: 10.1159/000490354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lui F., Colloca L., Duzzi D., Anchisi D., Benedetti F., Porro C.A. Neural bases of conditioned placebo analgesia. Pain. 2010;151:816–824. doi: 10.1016/j.pain.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 193.Benedetti F., Lanotte M., Lopiano L., Colloca L. When words are painful: unraveling the mechanisms of the nocebo effect. Neuroscience. 2007;147:260–271. doi: 10.1016/j.neuroscience.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 194.Colloca L., Benedetti F. How prior experience shapes placebo analgesia. Pain. 2006;124:126–133. doi: 10.1016/j.pain.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 195.Colloca L., Benedetti F. Placebos and painkillers: is mind as real as matter? Nat. Rev. Neurosci. 2005;6:545–552. doi: 10.1038/nrn1705. [DOI] [PubMed] [Google Scholar]
- 196.Rimpela J.M., Niiranen T., Jula A., Porsti I.H., Tikkakoski A., Havulinna A., Lehtimaki T., Salomaa V., Kontula K.K., Hiltunen T.P. Genome-wide association study of white-coat effect in hypertensive patients. Blood Pres. 2019;28:239–249. doi: 10.1080/08037051.2019.1604066. [DOI] [PubMed] [Google Scholar]
- 197.McKay K.M., Imel Z.E., Wampold B.E. Psychiatrist effects in the psychopharmacological treatment of depression. J. Affect. Disord. 2006;92:287–290. doi: 10.1016/j.jad.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 198.Totura C.M.W., Fields S.A., Karver M.S. The role of the therapeutic relationship in psychopharmacological treatment outcomes: a meta-analytic review. Psychiatr. Serv. 2018;69:41–47. doi: 10.1176/appi.ps.201700114. [DOI] [PubMed] [Google Scholar]
- 199.Miller N.E. Biofeedback and visceral learning. Annu. Rev. Psychol. 1978;29:373–404. doi: 10.1146/annurev.ps.29.020178.002105. [DOI] [PubMed] [Google Scholar]
- 200.Keck J.O., Staniunas R.J., Coller J.A., Barrett R.C., Oster M.E., Schoetz D.J., Jr., Roberts P.L., Murray J.J., Veidenheimer M.C. Biofeedback training is useful in fecal incontinence but disappointing in constipation. Dis. Colon Rectum. 1994;37:1271–1276. doi: 10.1007/BF02257795. [DOI] [PubMed] [Google Scholar]
- 201.McKee M.G., Moravec C.S. Biofeedback in the treatment of heart failure. Cleve. Clin. J. Med. 2010;77(Suppl 3):S56–S59. doi: 10.3949/ccjm.77.s3.10. [DOI] [PubMed] [Google Scholar]
- 202.Moravec C.S., McKee M.G. Biofeedback in the treatment of heart disease. Cleve. Clin. J. Med. 2011;78(Suppl 1):S20–S23. doi: 10.3949/ccjm.78.s1.03. [DOI] [PubMed] [Google Scholar]
- 203.Potter S.M., Wagenaar D.A., Madhavan R., DeMarse T.B. Long-term bidirectional neuron interfaces for robotic control, and in vitro learning studies. Proc. Ann. Int. IEEE Eng. Med. Biol. Soc. 2003;25:3690–3693. doi: 10.1109/Iembs.2003.1280959. [DOI] [Google Scholar]
- 204.Aaser P., Knudsen M., Ramstad O.H., van de Wijdeven R., Nichele S., Sandvig I., Tufte G., Bauer U.S., Halaas Ø., Hendseth S., et al. ECAL 2017: The 14th European Conference on Artificial Life. MIT Press; 2017. Towards making a cyborg: a closed-loop reservoir-neuro system. [Google Scholar]
- 205.Tessadori J., Bisio M., Martinoia S., Chiappalone M. Modular neuronal assemblies embodied in a closed-loop environment: toward future integration of brains and machines. Front. Neural Circuits. 2012;6:99. doi: 10.3389/fncir.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kagan B.J., Kitchen A.C., Tran N.T., Habibollahi F., Khajehnejad M., Parker B.J., Bhat A., Rollo B., Razi A., Friston K.J. In vitro neurons learn and exhibit sentience when embodied in a simulated game-world. Neuron. 2022;110:3952–3969.e8. doi: 10.1016/j.neuron.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Colloca L., Lopiano L., Lanotte M., Benedetti F. Overt versus covert treatment for pain, anxiety, and Parkinson's disease. Lancet Neurol. 2004;3:679–684. doi: 10.1016/S1474-4422(04)00908-1. [DOI] [PubMed] [Google Scholar]
- 208.Faria V., Gingnell M., Hoppe J.M., Hjorth O., Alaie I., Frick A., Hultberg S., Wahlstedt K., Engman J., Månsson K.N.T., et al. Do you believe it? Verbal suggestions influence the clinical and neural effects of escitalopram in social anxiety disorder: a randomized trial. EBioMedicine. 2017;24:179–188. doi: 10.1016/j.ebiom.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Beauregard M. Mind does really matter: evidence from neuroimaging studies of emotional self-regulation, psychotherapy, and placebo effect. Prog. Neurobiol. 2007;81:218–236. doi: 10.1016/j.pneurobio.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 210.Kaptchuk T.J. Open-label placebo: reflections on a research agenda. Perspect. Biol. Med. 2018;61:311–334. doi: 10.1353/pbm.2018.0045. [DOI] [PubMed] [Google Scholar]
- 211.Kaptchuk T.J., Miller F.G. Open label placebo: can honestly prescribed placebos evoke meaningful therapeutic benefits? Br. Med. J. 2018;363:k3889. doi: 10.1136/bmj.k3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kaptchuk T.J., Hemond C.C., Miller F.G. Placebos in chronic pain: evidence, theory, ethics, and use in clinical practice. Br. Med. J. 2020;370:m1668. doi: 10.1136/bmj.m1668. [DOI] [PubMed] [Google Scholar]
- 213.Levin M. In: Evolution “on Purpose” : Teleonomy in Living Systems. Corning P.A., Kauffman S.A., Noble D., Shapiro J.A., Vane-Wright R.I., Pross A., editors. MIT Press; 2023. Collective intelligence of morphogenesis as a teleonomic process; pp. 175–198. [Google Scholar]
- 214.Siegel S., Baptista M.A., Kim J.A., McDonald R.V., Weise-Kelly L. Pavlovian psychopharmacology: the associative basis of tolerance. Exp. Clin. Psychopharmacol. 2000;8:276–293. doi: 10.1037//1064-1297.8.3.276. [DOI] [PubMed] [Google Scholar]
- 215.Siegel S.F. The compensatory conditioned response to brain stimulation-induced feeding a preliminary study. Psychol. Rep. 1986;59:1244–1246. doi: 10.2466/pr0.1986.59.3.1244. [DOI] [PubMed] [Google Scholar]
- 216.Siegel S., Hinson R.E., Krank M.D. The role of predrug signals in morphine analgesic tolerance: support for a Pavlovian conditioning model of tolerance. J. Exp. Psychol. Anim. Behav. Process. 1978;4:188–196. doi: 10.1037//0097-7403.4.2.188. [DOI] [PubMed] [Google Scholar]
- 217.Siegel S. Pavlovian conditioning analysis of morphine tolerance. NIDA Res. Monogr. 1978:27–53. [PubMed] [Google Scholar]
- 218.Siegel S. Morphine analgesic tolerance: its situation specificity supports a Pavlovian conditioning model. Science. 1976;193:323–325. doi: 10.1126/science.935870. [DOI] [PubMed] [Google Scholar]
- 219.Kuchling F., Friston K., Georgiev G., Levin M. Morphogenesis as Bayesian inference: a variational approach to pattern formation and control in complex biological systems. Phys. Life Rev. 2020;33:88–108. doi: 10.1016/j.plrev.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 220.Friston K., Levin M., Sengupta B., Pezzulo G. Knowing one's place: a free-energy approach to pattern regulation. J. R. Soc. Interface. 2015;12:20141383. doi: 10.1098/rsif.2014.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Pio-Lopez L., Kuchling F., Tung A., Pezzulo G., Levin M. Active inference, morphogenesis, and computational psychiatry. Front. Comput. Neurosci. 2022;16:988977. doi: 10.3389/fncom.2022.988977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kramer B.A., Sarabia Del Castillo J., Pelkmans L. Multimodal perception links cellular state to decision-making in single cells. Science. 2022;377:642–648. doi: 10.1126/science.abf4062. [DOI] [PubMed] [Google Scholar]
- 223.Lyon P., Keijzer F., Arendt D., Levin M. Reframing cognition: getting down to biological basics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376:20190750. doi: 10.1098/rstb.2019.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Levin M., Keijzer F., Lyon P., Arendt D. Uncovering cognitive similarities and differences, conservation and innovation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376:20200458. doi: 10.1098/rstb.2020.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lyon P. Of what is “minimal cognition” the half-baked version? Adapt. Behav. 2020;28:407–424. doi: 10.1177/1059712319871360. [DOI] [Google Scholar]
- 226.Lyon P. The cognitive cell: bacterial behavior reconsidered. Front. Microbiol. 2015;6:264. doi: 10.3389/fmicb.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gumuskaya G., Srivastava P., Cooper B.G., Lesser H., Semegran B., Garnier S., Levin M. Motile living biobots self-construct from adult human somatic progenitor seed cells. bioRxiv. 2022 doi: 10.1101/2022.08.04.502707. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fields C., Levin M. Scale-free biology: integrating evolutionary and developmental thinking. Bioessays. 2020;42:e1900228. doi: 10.1002/bies.201900228. [DOI] [PubMed] [Google Scholar]
- 229.Watson R.A., Szathmáry E. How can evolution learn? Trends Ecol. Evol. 2016;31:147–157. doi: 10.1016/j.tree.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 230.Watson R.A., Mills R., Buckley C.L., Kouvaris K., Jackson A., Powers S.T., Cox C., Tudge S., Davies A., Kounios L., Power D. Evolutionary connectionism: algorithmic principles underlying the evolution of biological organisation in Evo-Devo, Evo-Eco and evolutionary transitions. Evol. Biol. 2016;43:553–581. doi: 10.1007/s11692-015-9358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Holding M.L., Drabeck D.H., Jansa S.A., Gibbs H.L. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr. Comp. Biol. 2016;56:1032–1043. doi: 10.1093/icb/icw082. [DOI] [PubMed] [Google Scholar]
- 232.Jackson T.N.W., Jouanne H., Vidal N. Snake venom in context: neglected clades and concepts. Front. Ecol. Evol. 2019;7 doi: 10.3389/fevo.2019.00332. [DOI] [Google Scholar]
- 233.Soen Y. Environmental disruption of host-microbe co-adaptation as a potential driving force in evolution. Front. Genet. 2014;5:168. doi: 10.3389/fgene.2014.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Soen Y., Knafo M., Elgart M. A principle of organization which facilitates broad Lamarckian-like adaptations by improvisation. Biol. Direct. 2015;10:68. doi: 10.1186/s13062-015-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Elgart M., Snir O., Soen Y. Stress-mediated tuning of developmental robustness and plasticity in flies. Biochim. Biophys. Acta. 2015;1849:462–466. doi: 10.1016/j.bbagrm.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 236.Freddolino P.L., Yang J., Momen-Roknabadi A., Tavazoie S. Stochastic tuning of gene expression enables cellular adaptation in the absence of pre-existing regulatory circuitry. Elife. 2018;7:e31867. doi: 10.7554/eLife.31867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Freddolino P.L., Tavazoie S. Beyond homeostasis: a predictive-dynamic framework for understanding cellular behavior. Annu. Rev. Cell Dev. Biol. 2012;28:363–384. doi: 10.1146/annurev-cellbio-092910-154129. [DOI] [PubMed] [Google Scholar]
- 238.Rogers M.P., Peteet J.R., Reich P. Conditioned immunosuppression? Am. J. Psychiatry. 1983;140:1110–1111. doi: 10.1176/ajp.140.8.1110b. [DOI] [PubMed] [Google Scholar]
- 239.Rogers M.P., Dubey D., Reich P. The influence of the psyche and the brain on immunity and disease susceptibility: a critical review. Psychosom. Med. 1979;41:147–164. doi: 10.1097/00006842-197903000-00008. [DOI] [PubMed] [Google Scholar]
- 240.Rogers M.P., Reich P., Strom T.B., Carpenter C.B. Behaviorally conditioned immunosuppression: replication of a recent study. Psychosom. Med. 1976;38:447–451. doi: 10.1097/00006842-197611000-00009. [DOI] [PubMed] [Google Scholar]
- 241.Dutcher J.M., Cole S.W., Williams A.C., Creswell J.D. Smartphone mindfulness meditation training reduces Pro-inflammatory gene expression in stressed adults: a randomized controlled trial. Brain Behav. Immun. 2022;103:171–177. doi: 10.1016/j.bbi.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 242.Dada T., Bhai N., Midha N., Shakrawal J., Kumar M., Chaurasia P., Gupta S., Angmo D., Yadav R., Dada R., Sihota R. Effect of mindfulness meditation on intraocular pressure and trabecular meshwork gene expression: a randomized controlled trial. Am. J. Ophthalmol. 2021;223:308–321. doi: 10.1016/j.ajo.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 243.Venditti S., Verdone L., Reale A., Vetriani V., Caserta M., Zampieri M. Molecules of silence: effects of meditation on gene expression and epigenetics. Front. Psychol. 2020;11:1767. doi: 10.3389/fpsyg.2020.01767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Black D.S., Christodoulou G., Cole S. Mindfulness meditation and gene expression: a hypothesis-generating framework. Curr. Opin. Psychol. 2019;28:302–306. doi: 10.1016/j.copsyc.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Dada T., Mittal D., Mohanty K., Faiq M.A., Bhat M.A., Yadav R.K., Sihota R., Sidhu T., Velpandian T., Kalaivani M., et al. Mindfulness meditation reduces intraocular pressure, lowers stress biomarkers and modulates gene expression in glaucoma: a randomized controlled trial. J. Glaucoma. 2018;27:1061–1067. doi: 10.1097/IJG.0000000000001088. [DOI] [PubMed] [Google Scholar]
- 246.Buric I., Farias M., Jong J., Mee C., Brazil I.A. What is the molecular signature of mind-body interventions? A systematic review of gene expression changes induced by meditation and related practices. Front. Immunol. 2017;8:670. doi: 10.3389/fimmu.2017.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Saatcioglu F. Regulation of gene expression by yoga, meditation and related practices: a review of recent studies. Asian J. Psychiatr. 2013;6:74–77. doi: 10.1016/j.ajp.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 248.Bauer S.M. Psychoneuroimmunology and cancer: an integrated review. J. Adv. Nurs. 1994;19:1114–1120. doi: 10.1111/j.1365-2648.1994.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 249.Vollhardt L.T. Psychoneuroimmunology: a literature review. Am. J. Orthopsychiatry. 1991;61:35–47. doi: 10.1037/h0079226. [DOI] [PubMed] [Google Scholar]
- 250.Houldin A.D., Lev E., Prystowsky M.B., Redei E., Lowery B.J. Psychoneuroimmunology: a review of literature. Holist. Nurs. Pract. 1991;5:10–21. doi: 10.1097/00004650-199107000-00004. [DOI] [PubMed] [Google Scholar]
- 251.Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E., Mansuy I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jawaid A., Roszkowski M., Mansuy I.M. Transgenerational epigenetics of traumatic stress. Prog. Mol. Biol. Transl. Sci. 2018;158:273–298. doi: 10.1016/bs.pmbts.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 253.Thumfart K.M., Jawaid A., Bright K., Flachsmann M., Mansuy I.M. Epigenetics of childhood trauma: long term sequelae and potential for treatment. Neurosci. Biobehav. Rev. 2022;132:1049–1066. doi: 10.1016/j.neubiorev.2021.10.042. [DOI] [PubMed] [Google Scholar]
- 254.Woldemichael B.T., Jawaid A., Kremer E.A., Gaur N., Krol J., Marchais A., Mansuy I.M. The microRNA cluster miR-183/96/182 contributes to long-term memory in a protein phosphatase 1-dependent manner. Nat. Commun. 2016;7:12594. doi: 10.1038/ncomms12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mason A.A. A case of congenital ichthyosiform erythrodermia of Brocq treated by hypnosis. Br. Med. J. 1952;2:422–423. doi: 10.1136/bmj.2.4781.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pezzulo G., Rigoli F., Friston K.J. Hierarchical active inference: a theory of motivated control. Trends Cogn. Sci. 2018;22:294–306. doi: 10.1016/j.tics.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kirchhoff M., Parr T., Palacios E., Friston K., Kiverstein J. The Markov blankets of life: autonomy, active inference and the free energy principle. J. R. Soc. Interface. 2018;15:20170792. doi: 10.1098/rsif.2017.0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Pezzulo G., Rigoli F., Friston K. Active Inference, homeostatic regulation and adaptive behavioural control. Prog. Neurobiol. 2015;134:17–35. doi: 10.1016/j.pneurobio.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zuroff D.C., Blatt S.J. The therapeutic relationship in the brief treatment of depression: contributions to clinical improvement and enhanced adaptive capacities. J. Consult. Clin. Psychol. 2006;74:130–140. doi: 10.1037/0022-006X.74.1.130. [DOI] [PubMed] [Google Scholar]
- 260.Pezzulo G. Disorders of morphogenesis as disorders of inference: comment on "Morphogenesis as Bayesian inference: a variational approach to pattern formation and control in complex biological systems" by Michael Levin et al. Phys. Life Rev. 2020;33:112–114. doi: 10.1016/j.plrev.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 261.Dodig-Crnkovic G. Cognition as morphological/morphogenetic embodied computation in vivo. Entropy. 2022;24:1576. doi: 10.3390/e24111576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Abrego L., Zaikin A. Integrated information as a measure of cognitive processes in coupled genetic repressilators. Entropy. 2019;21:382. doi: 10.3390/e21040382. [DOI] [PMC free article] [PubMed] [Google Scholar]