Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron subvariants have seriously attacked the antibody barrier established by natural infection and/or vaccination, especially the recently emerged BQ.1.1 and XBB.1. However, crucial mechanisms underlying the virus escape and the broad neutralization remain elusive. Here, we present a panoramic analysis of broadly neutralizing activity and binding epitopes of 75 monoclonal antibodies isolated from prototype inactivated vaccinees. Nearly all neutralizing antibodies (nAbs) partly or totally lose their neutralization against BQ.1.1 and XBB.1. We report a broad nAb, VacBB-551, that effectively neutralizes all tested subvariants including BA.2.75, BQ.1.1, and XBB.1. We determine the cryoelectron microscopy (cryo-EM) structure of VacBB-551 complexed with the BA.2 spike and perform detailed functional verification to reveal the molecular basis of N460K and F486V/S mutations mediating the partial escape of BA.2.75, BQ.1.1, and XBB.1 from the neutralization of VacBB-551. Overall, BQ.1.1 and XBB.1 raised the alarm over SARS-CoV-2 evolution with unprecedented antibody evasion from broad nAbs elicited by prototype vaccination.
Keywords: SARS-CoV-2, Omicron subvariant, BQ.1.1, XBB.1, broadly neutralizing activity, structure analysis
Graphical abstract
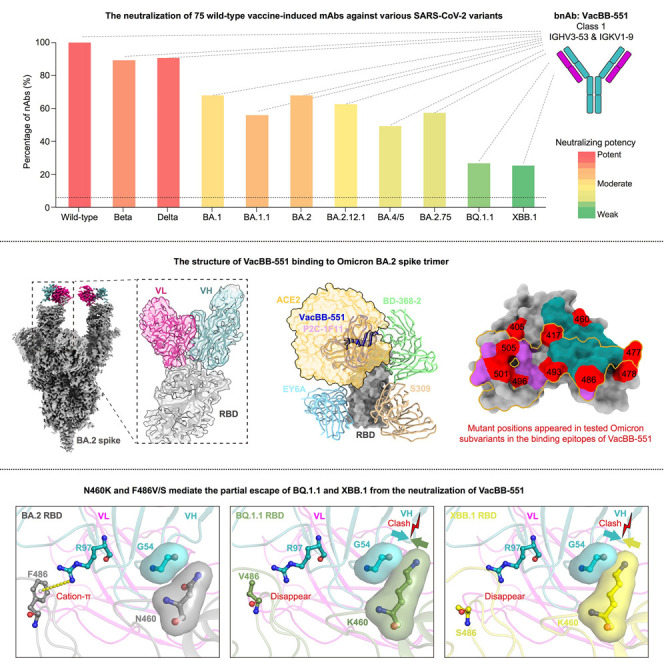
Ju et al. show that BQ.1.1 and XBB.1 partly or totally escape nearly all tested monoclonal nAbs elicited by triple prototype inactivated vaccination. Structure analysis and functional verification reveal that N460K and F486V/S contribute to the increased neutralization resistances of BQ.1.1 and XBB.1 to VacBB-551-like bnAbs.
Introduction
Since the first report of the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2021 in South Africa, it has evolved into numerous subvariants including BA.1.1, BA.2, BA.2.12.1, BA.4, BA.5, BA.2.75, etc.1 , 2 , 3 , 4 , 5 Moreover, newly emerged variants are often accompanied by striking antibody evasion from existing broadly neutralizing antibodies (bnAbs), mainly induced by prototype SARS-CoV-2 infection and vaccination.6 , 7 , 8 , 9 , 10 Recently, two Omicron subvariants, BQ.1.1 and XBB.1, caused wide public concern. BQ.1.1 was derived from BA.5 carrying three additional mutations (R346T, K444T, and N460K).11 XBB.1 (XBB+G252V) was a recombinant virus between two Omicron subvariants, BA.2.75 and BJ.1.11 Both two variants displayed unprecedented antibody escape abilities from human nAbs elicited by various vaccine immunizations and breakthrough infections,12 , 13 , 14 , 15 however, largely focusing on evaluating their neutralization susceptibilities to circulating plasma polyclonal antibodies and early isolated monoclonal Abs (mAbs). Our group have been concerned with uncovering reasons behind virus escape and maintained broad neutralization by isolating and characterizing mAbs,16 , 17 , 18 helping answer why and to what extent variants escape from the preexisting nAbs, as well as what kinds of nAbs retain the broad neutralization. Meanwhile, analyzing the neutralizing antibody response at the mAb level is also very important to evaluate the effect of multiple vaccines and guide the development and adjustment of current vaccination strategies. We now report the findings of how BQ.1.1 and XBB.1 escape SARS-CoV-2 inactivated vaccination-induced mAbs and reveal the underlying structural basis and molecular mechanism.
Results
Monoclonal nAbs isolated from triple inactivated vaccinees
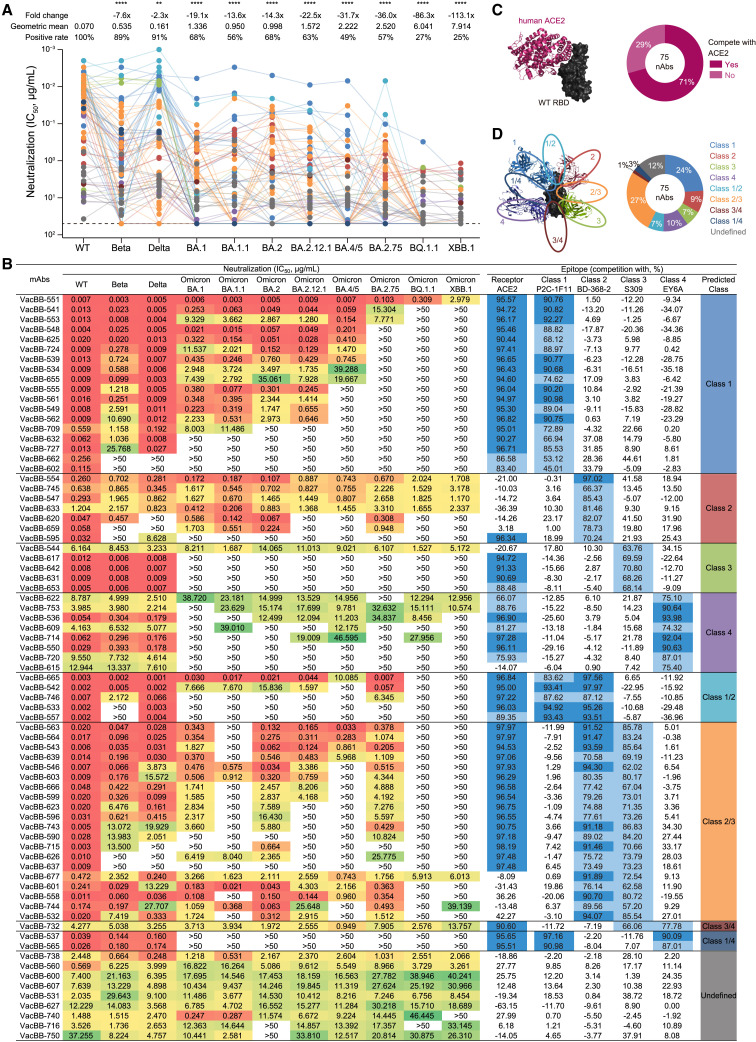
In this study, we sorted immunoglobulin G (IgG) memory B cells (MBCs) for binding to the prototype receptor-binding domain (wild type; WT-RBD) from 5 individuals at week 2 after receiving three doses of SARS-CoV-2 inactivated vaccines19 and performed single B cell PCR, gene sequencing, and antibody expression to isolate and characterize mAbs using our previous established methods.16 , 20 , 21 , 22 , 23 We obtained a total of 170 heavy- and light-chain paired antibody sequences. Due to a strong clonal expansion, some mAbs had identical sequences at the amino acid level. We expressed and purified 121 mAbs with distinct amino acid sequences and measured their neutralizing activities against WT SARS-CoV-2 pseudovirus, 63% of which showed effective neutralization with a geometric mean 50% inhibitory concentration (IC50) of 0.075 μg/mL (Figure S1). This positive rate of anti-RBD monoclonal nAbs was similar to that isolated from convalescent (58%, 52/89) and mRNA vaccine-immunized (65%, 82/127) individuals.24 , 25 Although VacBB-552 could effectively neutralize WT SARS-CoV-2 (IC50 = 14.020 μg/mL), its yield was very low in the antibody expression, which was excluded at subsequent analysis. To further determine the neutralizing breadth of the rest of 75 nAbs whose amino acid sequences were summarized in Table S1, we performed a series of neutralization assays against Beta, Delta, and Omicron subvariants including BA.1, BA.1.1 (BA.1+R346K), BA.2, BA.2.12.1, BA.4/5 (BA.4 and BA.5 sharing identical spike sequences), BA.2.75, BQ.1.1, and XBB.1 (Figure S2).
SARS-CoV-2 variants exhibited varying degrees of antibody evasion
As shown in Figure 1 A, most nAbs still effectively neutralized Beta and Delta variants (89% and 91%), but their geometric mean IC50s (0.535 and 0.161 μg/mL) were higher than that against WT (0.070 μg/mL). Consistent with previous studies,6 , 7 , 8 , 18 , 26 , 27 the Omicron BA.1 carrying 15 mutations in the RBD reduced or abolished the neutralization of majority of nAbs, whose positive rate was 68% and geometric mean IC50 was 1.336 μg/mL. More seriously, BA.1.1 carrying an additional R346K mutation further abolished the neutralization of some nAbs, reducing the positive rate down to 56%. The Omicron BA.2 variant shared 13 mutant sites in the RBD with BA.1, and the remaining 3 substitutions were unique.1 , 2 Perhaps for these differences, the antigenic property of BA.2 had changed and affected its antibody escape ability. Just as we observed here, BA.2 also showed a marked resistance to most of the nAbs with a geometric mean IC50 of 0.998 μg/mL. Interestingly, some inactive or severely impaired nAbs for BA.1 regained effective or potent neutralizing activities against BA.2, such as VacBB-724, VacBB-715, VacBB-744, etc. (Figure 1B), highlighting their different antibody evasion properties.
Figure 1.
Neutralizing activity and binding epitope of 75 monoclonal nAbs isolated from prototype inactivated vaccinees
(A and B) The neutralizing activity (IC50) was measured based on the SARS-CoV-2 pseudovirus-neutralization assay, whose cutoff value was set as 50 μg/mL (summarized in A and detailed in B). The data are means of at least two independent experiments. The positive rate, geometric mean IC50, fold change, and significance of difference are labeled on the top. “-” represents decreased neutralization. The statistical significance was performed using two-tailed paired Wilcoxon test. ∗∗∗∗p < 0.0001; ∗∗p < 0.01. The neutralizing potency is represented by a heatmap. Red: high, yellow: moderate, and green: weak.
(B–D) Competition ELISA was performed to predict the binding epitope of 75 nAbs (detailed in B and summarized in C and D). Human ACE2 and four representative mAbs of four classes (class 1: P2C-1F11, class 2: BD-368-2, class 3: S309, and class 4: EY6A) were used as the competitor for binding to the WT RBD. The data are means of at least two independent experiments. Dark blue: high competition (>90%), light blue: moderate competition (45%–90%), and none: weak or no competition (<45%). “Undefined” means that mAbs do not compete with any tested references (P2C-1F11, BD-368-2, S309, and EY6A).
The classifications of binding epitopes of mAbs are indicated in different colors, which are identical in (A), (B), and (D). See also Figures S1 and S2 and Table S1.
With the continuous mutation of SARS-CoV-2, the Omicron variant has evolved into more subvariants including BA.2.12.1, BA.4/5, and BA.2.75.4 , 28 Based on a head-to-head comparison, neutralization profiles of most of these 75 nAbs against the BA.2.12.1 were similar to BA.1, BA.1.1, and BA.2, showing largely reduced neutralization (63%, 1.572 μg/mL) compared with WT, Beta, and Delta (Figure 1A). BA.4/5 and BA.2.75 showed more serious resistance to the neutralization of 75 nAbs, with lower positive rates (49% and 57%) and relatively weaker potencies (2.222 and 2.520 μg/mL). Despite this, several bnAbs still maintained potent neutralization against BA.4/5 and/or BA.2.75, such as VacBB-551, VacBB-541, and VacBB-665 (Figure 1B). By contrast, BQ.1.1 and XBB.1 exhibited the most serious antibody escape from these 75 nAbs elicited by prototype vaccination, less than 30% of which maintained effectively neutralizing activities with geometric mean IC50s of 6.041 and 7.914 μg/mL, respectively (Figure 1A). Throughout the whole regions of RBDs of Omicron subvariants, the mutations appeared in or near the epitopes recognized by multiple classes of RBD-specific nAbs, which should be strongly related to their antibody escape abilities. Therefore, the detailed analysis for binding epitopes of these 75 nAbs is urgently needed.
Binding epitopes and neutralizing breadths
Previous structural studies reveal that the RBD in spike of SARS-CoV-2 has an up or down conformation, and the receptor-binding site (RBS) on the RBD is exposed only when the RBD is in an up conformation, mediating the virus binding to the receptor (angiotensin-converting enzyme 2 [ACE2]) and entry to the target cells.29 , 30 , 31 , 32 Direct competition with ACE2 for binding to the RBD is one of important neutralization mechanisms of anti-RBD nAbs, blocking engagement between the viral spike and cell receptor.23 , 33 , 34 , 35 , 36 Therefore, we first measured the potential competitions with human ACE2 of 75 nAbs using our previously established assay.17 , 18 As shown in Figures 1B and 1C, 71% of nAbs (53/75) exhibited obvious competitions with ACE2, suggesting that the remainders might bind to the region away from the RBS on the RBD and utilize other mechanisms to prevent the virus infection, just like S309, EY6A, etc.37 , 38 , 39 , 40 To identify more detailed epitopes recognized by these 75 nAbs, we further measured their competitions with four representative nAbs of classes 1 to 4 (P2C-1F11, BD-368-2, S309, and EY6A, respectively), which were classified by the competition with ACE2 and recognized the RBD conformation (up or down).16 , 17 , 18 , 41 27% of nAbs (20/75) belonged to class 2/3, recognizing an epitope between class 2 and 3 nAbs, followed by class 1 nAbs occupying 24% (Figures 1B and 1D). The percentages of other classes of nAbs ranged from 1% to 10%, also including some undefined nAbs (12%), which did not complete with any tested representative nAbs, such as VacBB-738, VacBB-560, etc. (Figure 1B).
Then, we made a comprehensive analysis between antibody-binding epitopes and broad neutralization against BQ.1.1 and XBB.1 (Figure 1B). All identified 18 nAbs of class 1 directly competed with ACE2, showing potent neutralizing activities against WT, yet were easily abolished by BQ.1.1 and XBB.1. Only VacBB-551 could neutralize all tested SARS-CoV-2 variants, despite some reduction in neutralizing against BQ.1.1 and XBB.1. A portion of bnAbs from classes 2 and 3 also maintained neutralization against BQ.1.1 and XBB.1, yet with relatively moderate potencies. The local conformation changes of the S371-S373-S375 loop in the RBD mainly affected the binding of class 4 nAbs,26 which displayed weak or no neutralization against various Omicron subvariants. Nearly all bnAbs of classes 1/2, 2/3, 3/4, and 1/4 (93%, 26/28) totally lost their neutralizing activities against BQ.1.1 and XBB.1. Only VacBB-677 and VacBB-732 could neutralize all tested SARS-CoV-2 variants with moderate potencies. A class of bnAbs, such as VacBB-738 and VacBB-560, did not compete with ACE2 and four tested representative mAbs (P2C-1F11, BD-368-2, S309, and EY6A) but exhibited good broad spectrum for neutralizing all tested SARS-CoV-2 variants including BQ.1.1 and XBB.1. Collectively, VacBB-551 was the first-best bnAb among 75 isolated nAbs derived from multiple classes in this study. Therefore, VacBB-551 was selected to further explore the broadly neutralizing mechanism and the reason for reduced neutralizing potency.
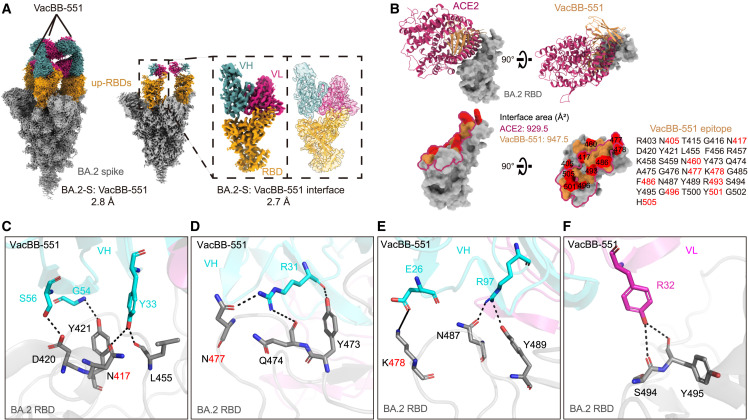
Structural basis of VacBB-551 broadly neutralizing SARS-CoV-2 variants
To define the structural basis of broad neutralization of VacBB-551, its cryoelectron microscopy (cryo-EM) structure in complex with the Omicron BA.2 spike trimer (BA.2-S) was determined at the overall resolution of 2.8 Å (Figures 2 A and S3; Table S2), showing that VacBB-551 bound to RBDs in the up conformation. Then, we performed localized refinement and further obtained the interface of BA.2-S: VacBB-551 at a resolution of 2.7 Å. Consistent with the above result of a competition ELISA (Figure 1B), VacBB-551 belonged to class 1 nAbs, causing an obvious clash with ACE2 for binding to the RBD (Figure 2B). VacBB-551 has a similar binding footprint with ACE2 on the BA.2 RBD, whose interface areas are 947.5 and 929.5 Å2, respectively. A total of 30 paratope residues of VacBB-551 interact with 31 epitope residues on the BA.2 RBD. Among the residues mentioned above, 10 mutant positions appearing in all tested Omicron subvariants were located in the epitopes recognized by VacBB-551. To further explore broad neutralization mechanisms of VacBB-551 against various SARS-CoV-2 variants and explain how several mutations located in epitopes affect the neutralizing activities of VacBB-551, we performed a more detailed analysis of interactions between VacBB-551 and RBD (Figures 2C–2F and S4; Table S3). VacBB-551 attachment utilizes 6 paratope residues of heavy chain (E26, R31, Y33, G54, S56, and R97) and 1 residue of light chain (R32) to form 11 potential hydrogen bonds and 1 salt bridge around N417, D420, Y421, L455, Y473, Q474, N477, K478, N487, Y489, S494, and Y495 epitope residues on the BA.2 RBD. Although three substitutions appearing at 417, 477, and 478 positions on the RBD, Y33, R31, and E26 of VacBB-551 still form 3 strong interactions (2 hydrogen bonds and 1 salt bridge) with mutated K417N, S477N, and T478K, revealing its probable broad neutralization mechanism against various SARS-CoV-2 variants.
Figure 2.
Cryo-EM structure basis of VacBB-551 binding to Omicron BA.2 RBD
(A) The cryo-EM density map of VacBB-551 in complex with Omicron BA.2 spike trimer. Spike is shown in gray and RBDs in orange. Heavy chain of VacBB-551 is shown in dark cyan and light chain in deep pink. Atomic model of RBD-Fab interacting region is shown as a cartoon, fitted in the corresponding transparency density map.
(B) Structure and binding footprint of VacBB-551 on BA.2 RBD. ACE2 is shown in medium violet red and VacBB-551 in orange. Epitopes recognized by VacBB-551 are listed, and mutant positions that appeared in tested Omicron subvariants are highlighted in red. The interface area of the ACE2-BA.2 RBD was calculated based on a published structure (PDB: 7XB0).
(C–F) Interactions of heavy chain (C–E) and light chain (F) of VacBB-551 with BA.2 RBD. Heavy chain is shown in cyan and light chain in magenta. Interface area (Å2) and potential hydrogen bond (black dotted line) or salt bridge (black solid line) were calculated by PISA v.1.52 (https://www.ebi.ac.uk/pdbe/pisa).
See also Figures S3 and S4 and Tables S2 and S3.
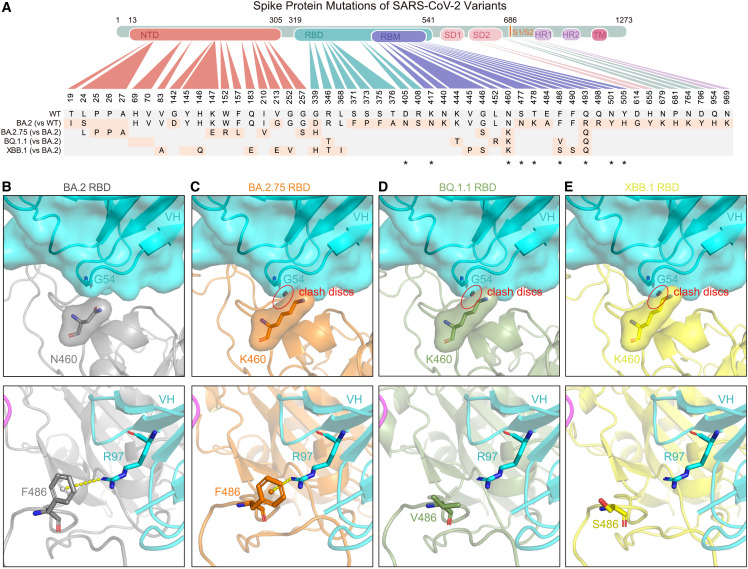
Sequence and structural analysis of variants carrying N460K and F486V/S
VacBB-551 maintained overall high neutralization potencies against all tested SARS-CoV-2 variants including Omicron subvariants except for BA.2.75, BQ.1.1, and XBB.1, whose IC50 values were decreased to some extent compared with those against BA.2 (0.103, 0.309, and 2.979 vs. 0.005 μg/mL, respectively) (Figure 1B). Sequence alignment of RBDs from all tested variants revealed that the N460K mutation that appeared in BA.2.75 was located in the interface of VacBB-551 (Figures 3 A and S5). To explain the slight reduction of VacBB-551, further structural analysis was performed around N460K. An atomic model for the BA.2.75 RBD was predicted by AlphaFold242 and aligned to the structure of BA.2-S: VacBB-551, with a root-mean-square deviation (RMSD) of 0.807 over 156 atoms. As shown in Figures 3B and 3C, the lysine at position 460 (N460K) might have a slight clash with the loop formed by G54-S56 at the heavy chain of VacBB-551,5 causing the impaired neutralization activity but not abolishing it completely.
Figure 3.
Sequence alignment and structural analysis of Omicron BA.2.75, BQ.1.1, and XBB.1 subvariants
(A) Key mutations in the spike protein of Omicron BA.2, BA.2.75, BQ.1.1, and XBB.1. “∗” represents mutations appeared in the binding epitopes of VacBB-551.
(B–E) Local region of N460 and F486 of BA.2 RBD (B), K460 and F486 of BA.2.75 RBD (C), K460 and V486 of BQ.1.1 RBD (D), and K460 and S486 of XBB.1 RBD (E) with G54 and R97 of VacBB-551 heavy chain, respectively. The structures of BA.2.75, BQ.1.1, and XBB.1 RBDs were predicted by AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb). The clash discs were displayed using the “Show bumps” plugin (https://pymolwiki.org/index.php/Show_bumps) in Pymol (C–E). The pseudoatom of F486 is shown as nb_sphere and the cation-π interaction is shown in a yellow dashed line (B and C). N460, K460, F486, V486, S486, G54, and R97 residues were shown as sticks. The VacBB-551 heavy chain was shown as cartoon/surface colored by cyan. RBDs of BA.2, BA.2.75, BQ.1.1, and XBB.1 are shown in gray, orange, smudge, and yellow, respectively.
See also Figure S5.
By contrast, the potency (IC50) of VacBB-551 against BQ.1.1 or XBB.1 was largely weaker than that against BA.2 (0.309 or 2.979 vs. 0.005 μg/mL) (Figure 1B), which might be mediated by N460K in combination with F486V/F486S substitutions at the footprint region between VacBB-551 and BA.2 RBD (Figures 3A and S5). RBD structures of BQ.1.1 and XBB.1 were predicted by AlphaFold2.42 RBD-BQ.1.1 and RBD-XBB.1 were used to align to BA.2-S: VacBB-551 with RMSDs of 0.843 over 158 atoms and 0.895 over 163 atoms. The clash was also observed between K460 on the RBD of BQ.1.1 or XBB.1 and G54 of the VacBB-551 heavy chain (Figures 3D and 3E). By checking the residues around F486 with VacBB-551, a cation-π interaction43 was found between the aromatic ring of F486 on the BA.2 RBD and R97 of the VacBB-551 heavy chain (Figure 3B). Mutations of F486V on BQ.1.1 and F486S on XBB.1 cause the loss of this cation-π interaction (Figures 3D and 3E), partly explaining the neutralization decline of VacBB-551 against BQ.1.1 and XBB.1. These results raised concerns about the potential escape risks of VacBB-551-like bnAbs by future SARS-CoV-2 variants, especially those carrying mutations at N460 and F486 positions.
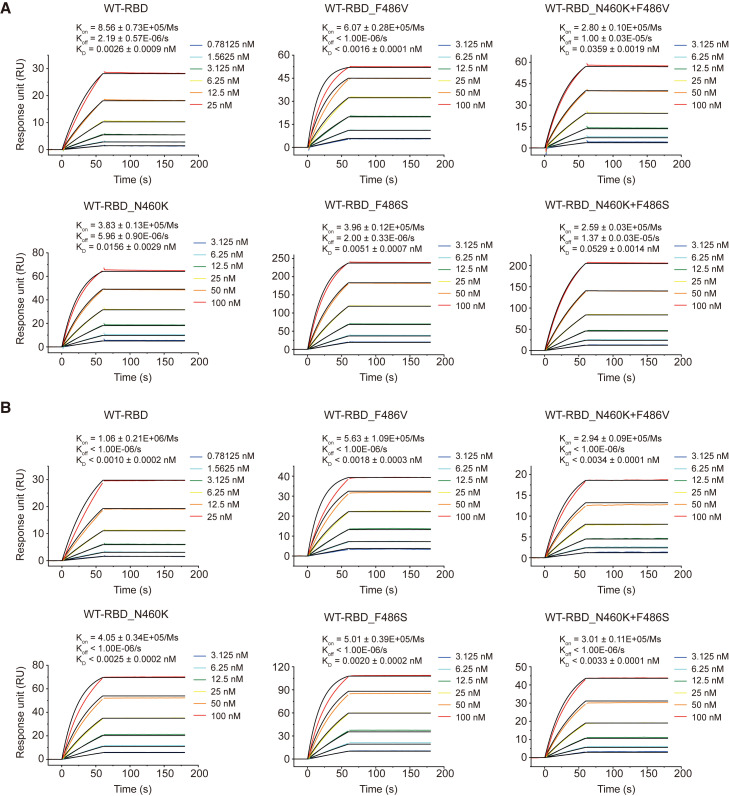
VacBB-551 maintained relatively high binding affinities to mutated RBDs based on the WT
As shown in Figure 1B, despite varying degrees of reduction in the neutralization of VacBB-551 against several mutated pseudoviruses, it still effectively neutralized all tested variants. To explore the underlying mechanism of broad neutralization, we constructed, expressed, and purified WT and a series of mutated RBD proteins bearing N460K, F486V, F486S, N460K+F486V, or N460K+F486S, respectively. Meanwhile, we also prepared the fragment of antigen binding (Fab) of VacBB-551 to measure the binding affinities to RBDs in parallel comparison with its IgG forms by surface plasmon resonance (SPR). As shown in Figures 4A and 4B, despite slight changes, both Fab-form and IgG-form VacBB-551 bound to WT and mutated RBDs with high affinities down to a picomolar level ranging from 0.0010 to 0.0529 nM. These results suggested that single-point or double-point mutations at N460 and F486 on the WT RBD did not greatly reduce the binding affinity of VacBB-551 whether in monovalent Fab form or bivalent IgG form.
Figure 4.
Binding affinity of Fab-form and IgG-form VacBB-551 to WT and mutated RBD proteins
SPR analysis of Fab-form VacBB-551 (A) and IgG-form VacBB-551 (B) binding to WT, N460K, F486V, F486S, N460K+F486V, or N460K+F486S RBD proteins, respectively. The dissociation constant (KD), association rate constant (Kon), and dissociation rate constant (Koff) are calculated from three independent experiments and represented in mean values ±standard deviation (SD). One representative curve is presented here.
Additional N460 and F486 mutations based on Omicron variants mediated them escaping from the neutralization of VacBB-551
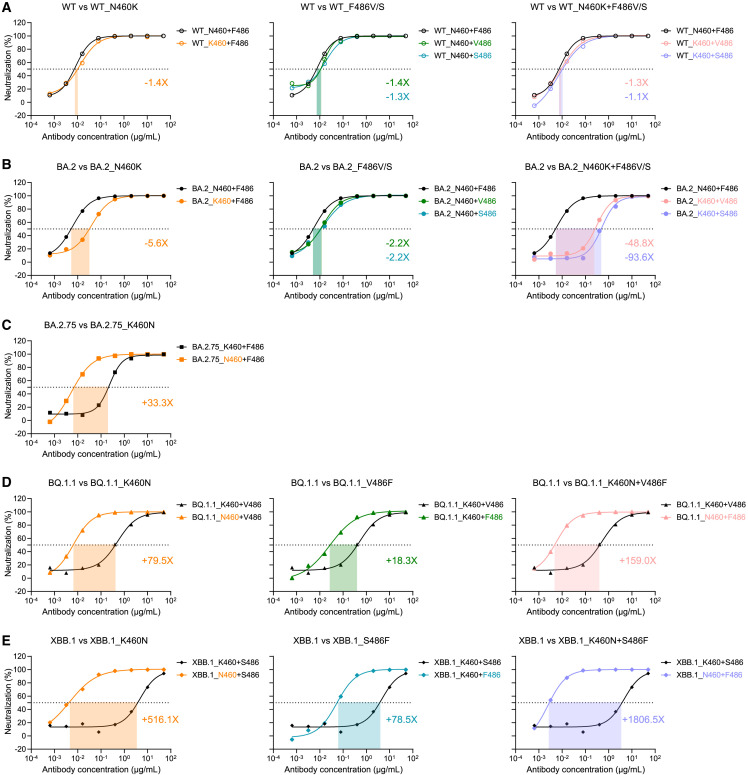
To validate the influence of N460K and F486V/S mutations, we constructed a series of mutated pseudoviruses based on WT, BA.2, BA.2.75, BQ.1.1, and XBB.1, respectively, and then measured their neutralization susceptibilities to VacBB-551. Consistent with the binding affinity results (Figure 4), VacBB-551 still neutralized WT_N460K, WT_F486V/S, and WT_N460K+F486V/S with a similar potency to that against WT (Figure 5 A). By contrast, a single-substitution N460K directly caused a 5.6-fold decline of neutralization of VacBB-551 against BA.2 (Figure 5B). A relatively slighter influence of F486V or F486S was observed on the neutralization of VacBB-551. Moreover, the combination of N460K and F486V/S largely enhanced the neutralization resistances of BA.2 to VacBB-551 by 48.8- to 93.6-fold. Conversely, we also reversed these mutations back to the original residuals and evaluated their neutralization susceptibilities to VacBB-551 (Figures 5C–5E). The reverse mutation K460N could largely increase the neutralization of VacBB-551 against BA.2.75, BQ.1.1, and XBB.1 by 33.3-, 79.5-, and 516.1-fold, respectively. The V486F and S486F reverse mutations showed similar trends with K460N in enhancing the susceptibility of BQ.1.1 and XBB.1. Not surprisingly, VacBB-551 could neutralize both BQ.1.1_K460N+V486F and XBB.1_K460N+S486F with similar potencies to that against WT, BA.2, or other susceptible variants. Collectively, these results demonstrated that the additional mutations on N460 and F486 residuals indeed influenced the neutralization of SARS-CoV-2 Omicron subvariants by the bnAb VacBB-551.
Figure 5.
Functional verification of N460 and F486 mutations influencing the neutralization susceptibility of SARS-CoV-2 variants to VacBB-551
The neutralization of VacBB-551 against the WT-related (A), BA.2-related (B), BA.2.75-related (C), BQ.1.1-related (D), and XBB.1-related (E) mutated pseudoviruses. Fold changes of neutralization (IC50) are calculated on mean values of two independent experiments. One representative curve is presented here. “-” represents decreased neutralization, and “+” represents increased neutralization.
Discussion
In this study, we isolated 75 monoclonal nAbs from individuals who received three doses of prototype inactivated vaccines and comprehensively evaluated their broad neutralization against a series of SARS-CoV-2 variants, including Beta, Delta, BA.1, BA.1.1, BA.2, BA.2.12.1, BA.4/5, BA.2.75, BQ.1.1, and XBB.1. We also analyzed the relationship between virus escape features and antibody-binding epitopes, indicating that nearly all potent nAbs totally lost their neutralizing activities against BQ.1.1 and XBB.1, consistent with previous studies on polyclonal plasma and mAbs, as well as some antibody drugs used in clinical.11 , 12 , 44 By contrast, some moderate (class 2) or even weak (undefined) nAbs binding to epitopes away from the RBS maintained effective neutralization against all tested SARS-CoV-2 variants. As a whole, the neutralization pattern of 75 mAbs elicited by triple inactivated vaccination displayed a mosaic feature against these concerned variants. Combining different epitope-targeted mAbs into an antibody cocktail is still the optimal strategy to fight against SARS-CoV-2 evolution and escape.
A typical class 1 bnAb, VacBB-551, could neutralize all tested SARS-CoV-2 variants with a high potency (geometric mean IC50 = 0.018 μg/mL), although BA.2.75, BQ.1.1, and XBB.1 escaped its neutralization to a certain extent. Genetic analysis showed that the heavy chain and light chain of VacBB-551 utilized the IGHV3-53 and IGKV1-9 germline genes with 6.67% and 4.55% of somatic hypermutations, respectively, highlighting once again the key role of IGHV3-53 public bnAbs against SARS-CoV-2 variants.45 , 46 , 47 The cryo-EM structure analysis of VacBB-551 in complex with the BA.2 spike trimer revealed its broad neutralization mechanism of tolerating a certain degree of mutation at the binding epitopes, such as K417N, S477N, and T478K.
Sequence alignment and structure modeling partly explained the reason for the varying reduced neutralizing activities of VacBB-551 against BA.2.75, BQ.1.1, and XBB.1. However, the SPR analysis and neutralization assay indicated that single-point mutations (N460K, F486V, and F486S) and double-point mutations (N460K+F486V and N460K+F486S) had very little impact on the binding and neutralizing activity of WT SARS-CoV-2 by VacBB-551. By contrast, based on BA.2, BA.2.75, BQ.1.1, and XBB.1, additional mutations that appeared in the 460 and 486 positions indeed contributed to their changed neutralization susceptibilities to VacBB-551. Greaney and colleagues have been mapped that class 1 nAbs might be escaped by mutations to N460 and F486 sites using a yeast-display system.48 Here, we performed further functional verification of N460K and F486V/S mutations causing the enhanced neutralization resistances of SARS-CoV-2 Omicron subvariants to VacBB-551-like bnAbs. Nowadays, the directed modification of mAbs has been widely used in the improvement of neutralizing activities. The potencies of DRVIA7 and VRC07, two HIV-1-specific bnAbs, were markedly increased by deleting two N-terminal amino acids along with the introduction of some other modifications in the light chain.20 , 49 Our previous study also stated that a key F27I mutation in the heavy chain contributed to the enhanced neutralization of P2C-1F11-like antibodies against SARS-CoV-2.46 Considering that BQ.1.1 and XBB.1 have largely reduced the neutralization of VacBB-551 and many available identified bnAbs,11 , 15 structure-guided antibody engineering needs to be performed to improve or even rescue their neutralizing activities in the following research.
Limitations of the study
In this study, we performed the detailed analysis of N460K and F486V/S on the RBD influencing the neutralization susceptibility of BA.2.75, BQ.1.1, and XBB.1 to VacBB-551. However, we did not exclude the potential synergistic effects of other mutations appearing in these SARS-CoV-2 variants. Moreover, we did not analyze and validate why BA.2.75, BQ.1.1, and XBB.1 totally escaped from the neutralization of other bnAbs. More mechanism analyses need to be performed in the future. In addition, we did not isolate and characterize mAbs recognizing other regions of spike including the N-terminal domain (NTD) and S2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| P2C-1F11 | Ge et al.35 | PDB code 7CDI |
| BD-368-2 | Du et al.50 | PDB code 7CHH |
| S309 | Pinto et al.37 | PDB code 6WPS |
| EY6A | Zhou et al.39 | PDB code 6ZCZ |
| VRC01 | Zhou et al.51 | PDB code 4LST; RRID: AB_2491019 |
| Bacterial and virus strains | ||
| E. coli DH5a | Takara | Cat# 9057 |
| SARS-CoV-2 pseudovirus/wild-type (WT) | Ju et al.18 | N/A |
| SARS-CoV-2 pseudovirus/Beta | Ju et al.18 | N/A |
| SARS-CoV-2 pseudovirus/Delta | Ju et al.18 | N/A |
| SARS-CoV-2 pseudovirus/Omicron BA.1 | Ju et al.18 | N/A |
| SARS-CoV-2 pseudovirus/Omicron BA.1.1 | This paper | N/A |
| SARS-CoV-2 pseudovirus/Omicron BA.2 | Guo et al.52 | N/A |
| SARS-CoV-2 pseudovirus/Omicron BA.2.12.1 | Guo et al.52 | N/A |
| SARS-CoV-2 pseudovirus/Omicron BA.4/5 | Guo et al.52 | N/A |
| SARS-CoV-2 pseudovirus/Omicron BA.2.75 | Guo et al.52 | N/A |
| SARS-CoV-2 pseudovirus/Omicron BQ.1.1 | This paper | N/A |
| SARS-CoV-2 pseudovirus/Omicron XBB.1 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle Medium | Gibco | Cat# 11965–092 |
| Fetal bovine serum | Gibco | Cat# 10099–141C |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat# 15140163 |
| HEPES (1M) Buffer Solution | Gibco | Cat# 15630–080 |
| Opti-MEM Reduced Serum Medium | Gibco | Cat# 51985034 |
| FreeStyle 293 expression medium | Gibco | Cat# 12338026 |
| Polyethylenimines (PEIs) 25K | PolySciences | Cat# 23966 |
| Ampicillin | Amresco | Cat# 69-52-3 |
| DEAE-Dextran hydrochloride | Sigma Aldrich | Cat# D9885-10G |
| Acetate 4.5 | Cytiva | Cat# BR100350 |
| Acetate 5.0 | Cytiva | Cat# BR100351 |
| HBS-EP+ Buffer 10× | Cytiva | Cat# BR100669 |
| Glycine 1.5 | Cytiva | Cat# BR100354 |
| Glycine 2.0 | Cytiva | Cat# BR100355 |
| BeyoGold His-tag Purification Resin | Beyotime | Cat# P2233-100mL |
| Imidazole | Sangon Biotech | Cat# B540146-0100 |
| L-Cysteine hydrochloride anhydrous | Sigma-Aldrich | Cat# 30120-50G |
| Papain from papaya latex lyophilized powder | Sigma-Aldrich | Cat# P4762-1G |
| Iodoacetamide | Sigma-Aldrich | Cat# I6125-25G |
| 0.5 M EDTA, pH 8.0 | Invitrogen | Cat# 15575-038 |
| SARS-CoV-2 WT RBD probe | Sino Biological Inc. | Cat# 40592-V08B |
| Human ACE2 | Sino Biological Inc. | Cat# 10108-H08H |
| SARS-CoV-2 BA.2 spike trimer | Sino Biological Inc. | Cat# 40589-V08H28 |
| SARS-CoV-2 WT RBD with His tag | Ju et al.18 | N/A |
| SARS-CoV-2 mutated RBD_N460K with His tag | This paper | N/A |
| SARS-CoV-2 mutated RBD_F486V with His tag | This paper | N/A |
| SARS-CoV-2 mutated RBD_F486S with His tag | This paper | N/A |
| SARS-CoV-2 mutated RBD_N460K + F486V with His tag | This paper | N/A |
| SARS-CoV-2 mutated RBD_N460K + F486S with His tag | This paper | N/A |
| Critical commercial assays | ||
| Bright-Lite Luciferase Assay System | Vazyme Biotech | Cat# DD1204-03 |
| Gold Hi EndoFree Plasmid Maxi Kit | CWBIO | Cat# CW2104M |
| TMB substrate | Sangon Biotech | Cat# E661007-0100 |
| Mut Express II Fast Mutagenesis Kit V2 | Vazyme Biotech | Cat# C214-01 |
| HRP Conjugation Kit | Abcam | Cat# ab102890 |
| Amine Coupling Kit | Cytiva | Cat# BR100050 |
| Deposited data | ||
| The density map of the RBD-VacBB-551 complex | This paper | EMDB: EMD-34226 |
| The atomic model of the RBD-VacBB-551 complex | This paper | PDB code 8GS9 |
| Experimental models: Cell lines | ||
| HEK-293T | ATCC | Cat# CRL-3216 |
| HEK-293T-hACE2 | YEASEN Biotech | Cat# 41107ES03 |
| HEK-293F | Gibco | Cat# R79007 |
| Oligonucleotides | ||
| Primers used in this study (Table S4) | This paper | N/A |
| Recombinant DNA | ||
| pNL4-3.Luc.R-E− | NIH AIDS Reagent Program | Cat# 3418 |
| Software and algorithms | ||
| Graphpad Prism 8.0 | GraphPad | https://www.graphpad.com |
| Biacore Evaluation software 3.0 | GE Healthcare | https://www.gehealthcare.com |
| IMGT/V-Quest | IMGT | http://www.imgt.org/IMGT_vquest/vquest |
| PyMOL | PyMOL | http://www.pymol.org |
| UCSF ChimeraX | ChimeraX | http://www.cgl.ucsf.edu/chimerax |
| Phenix | Phenix | http://www.phenix-online.org |
| Coot | Coot | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| MotionCor2 | MotionCor2 | https://emcore.ucsf.edu/ucsf-software |
| GCTF | GCTF | https://www2.mrc-lmb.cam.ac.uk/download/gctf_v1-06-and-examples |
| cryoSPARC | cryoSPARC | https://cryosparc.com |
| ColabFold v1.5.2: AlphaFold2 using MMseqs2 | AlphaFold2 | https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb |
| Molprobity | Molprobity | http://molprobity.biochem.duke.edu |
| PISA v1.52 | PISA | https://www.ebi.ac.uk/pdbe/pisa |
| Origin 2023 | OriginLab | https://www.originlab.com |
| Other | ||
| Holey Carbon Grids | Quantifoil | Cat# R1.2/1.3 Cu400 |
| Series S Sensor Chip CM5 | Cytiva | Cat# 29149603 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zheng Zhang (zhangzheng1975@aliyun.com).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Human subjects
This study was approved by the Ethics Committee of Shenzhen Third People’s Hospital, China (approval number: 2021-030). All participants had provided written informed consent for sample collection and subsequent analysis. Peripheral blood mononuclear cells (PBMCs) from individuals who received the SARS-CoV-2 inactivated vaccine (BBIBP-CorV, the Sinopharm COVID-19 vaccine, Beijing Institute of Biological Products Co., Ltd) were stored at Biobank of Shenzhen Third People’s Hospital. All participants had no history of SARS-CoV-2 infection. Flow cytometric analysis of SARS-CoV-2 WT RBD-specific MBCs have been published in our previous study.19 Here, we further performed subsequent isolation and characterization of anti-RBD mAbs from 5 individuals (BBIBP-donor 19: male, 57 years old; BBIBP-donor 21: male, 57 years old; BBIBP-donor 29: male, 23 years old; BBIBP-donor 73: female, 27 years old; BBIBP-donor 108: male, 27 years old).
Cell lines
HEK-293T cells were from ATCC. HEK-293T-hACE2 cells were from YEASEN Biotech. HEK-293F cells were from Gibco. HEK-293T and HEK-293T-hACE2 cells were cultured in Dulbecco’s modified eagle medium (DMEM, Gibco), supplemented with 10% Fetal bovine serum (FBS, Gibco), 1% penicillin-streptomycin (Gibco), and 1% HEPES (1M) buffer solution (Gibco) at 37°C with 5% CO2. HEK-293F cells were cultured in FreeStyle 293 expression medium (Gibco) at 37°C with 8% CO2 at 130 rpm.
Method details
Identification of RBD-specific mAbs
SARS-CoV-2 WT RBD-specific MBCs (CD19+CD3−CD8−CD14−CD27+IgG+RBD+)19 were sorted into the 96-well PCR plate containing cell lysis buffer and then snap-frozen on the dry ice and stored at −80°C. RT-PCR and nested PCR were performed to amplify antibody heavy- and light-chain variable genes. After sequencing (Sangon Biotech), antibody genes were analyzed by the IMGT/V-QUEST program (www.imgt.org/IMGT_vquest/vquest).16 , 20 , 21 , 22 , 23 Variable genes were synthesized and cloned into the expression vectors containing full-length heavy (IgG1) and light (kappa or lambda) chains by GenScript, respectively. Paired heavy- and light-chain plasmids were co-transfected into HEK-293F cells to express mAbs, which were purified from the culture supernatants using protein A column (GenScript).
SARS-CoV-2 pseudovirus-based neutralization assay
SARS-CoV-2 pseudoviruses were generated by co-transfection of HEK-293T cells with an env-deficient HIV-1 backbone vector (pNL4-3.Luc.R-E−) and different spike-expressing plasmids of various SARS-CoV-2 variants, respectively. Two days after transfection, culture supernatant was harvested, clarified by centrifugation, filtered, and stored at −80°C.18 , 19 , 23 , 45 , 52 To determine the neutralizing activity, mAbs were serially diluted and then incubated with an equal volume of pseudovirus at 37°C for 1 h. HEK-293T-hACE2 cells were subsequently added to 96-well plates. After a 48 h incubation, the culture medium was removed, and 100 μL of Bright-Lite Luciferase reagent (Vazyme Biotech) was added to the cells. After a 2 min incubation at RT, 90 μL of cell lysate was transferred to the 96-well white solid plates for measurements of the luminescence using the Varioskan LUX multimode microplate reader (Thermo Fisher Scientific). The 50% inhibitory concentration (IC50) was calculated using GraphPad Prism 8.0 software by log (inhibitor) vs. normalized response - Variable slope (four parameters) model. Detailed sequence information of spike proteins used in this study were listed below.
SARS-CoV-2 wild-type (WT)
Accession number: NC_045512.
SARS-CoV-2 Beta
D80A, D251G, 242-243del, K417N, E484K, N501Y, D614G, A701V.
SARS-CoV-2 Delta
T19R, G142D, 157-158del, L452R, T478K, D614G, P681R, D950N.
SARS-CoV-2 Omicron BA.1
A67V, 69-70del, T95I, G142D, 143-145del, N211I, 212del, 215EPEins, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F.
SARS-CoV-2 Omicron BA.1.1
A67V, 69-70del, T95I, G142D, 143-145del, N211I, 212del, 215EPEins, G339D, R346K, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F.
SARS-CoV-2 Omicron BA.2
Accession number: EPI_ISL_9652748.
SARS-CoV-2 Omicron BA.2.12.1
T19I, L24S, 25-27del, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, N969K.
SARS-CoV-2 Omicron BA.4/5
Accession number: EPI_ISL_11542550.
SARS-CoV-2 Omicron BA.2.75
Accession number: EPI_ISL_13502576.
SARS-CoV-2 Omicron BQ.1.1
Accession number: EPI_ISL_14818139.
SARS-CoV-2 Omicron XBB.1
Accession number: EPI_ISL_14917761.
Some additional mutations were constructed by the site-directed mutagenesis based on above SARS-CoV-2 spike genes using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech). Detailed primer information used in this study were listed in Table S4.
Competition enzyme linked immunosorbent assay (ELISA)
SARS-CoV-2 WT RBD protein (Sino Biological) (2 μg/mL) was coated into 96-well plates at 4°C overnight. The plates were washed with PBST buffer and blocked with blocking buffer (5% skim milk and 2% bovine albumin in PBS) at RT for 1 h. Human ACE2 (Sino Biological) or four classes of SARS-CoV-2 RBD-specific mAbs (P2C-1F11,35 BD-368-2,50 S309,37 and EY6A39) coupled with HRP (Abcam)16 , 17 , 18 were mixed with an equal volume of diluted mAbs (20 μg/mL), added into the ELISA plates, and then incubated at 37°C for 1 h. The TMB substrate (Sangon Biotech) was added and incubated at RT for 20 min and the reaction was stopped by 2M H2SO4. The readout was detected at the wave length of 450nm. VRC0151 (a HIV-1-specific mAb) was used here as the negative control, meaning no competition with SARS-CoV-2-specific mAbs. The percentage of competition was calculated by the formula: (1-OD450 of tested mAb/OD450 of VRC01 control) × 100%.
Cryo-EM sample preparation and data collection
SARS-CoV-2 BA.2 spike trimer protein (Sino Biological) was concentrated to about 2 mg/mL and incubated with VacBB-551 for 30 min (1: 2 M ratio). Aliquots (3 μL) of spike-antibody complex were applied to glow-discharged holey carbon grids (Quantifoil Cu R1.2/1.3) in Vitrobot Mark IV (Thermo Scientific) at 4°C and 100% humidity. Grids were blotted for about 5 s with 0 blot force after waiting for 3 s. The flash-frozen samples were then transferred to a Titan Krios transmission electron microscope at 300 kV. Stacks were automatically collected using EPU software, with a slit width of 20 eV and a defocus range from −1.5 μm to −2.5 μm in super-resolution mode. Images for BA.2-S: VacBB-551 complex were recorded using a K3 camera with a pixel size of 0.855 Å/pixel. Exposures were performed with a total dose of 50 e−/Å2, which were fractionated into 32 frames.
Cryo-EM data processing
A total of 6153 movies stacks of BA.2-S: VacBB-551 complex were collected. MotionCor253 was used for correcting beam-induced drift. The defocus values were estimated with Gctf.54 Micrographs were imported into cryoSPARC.55 Particles were automatically picked using Blob picker with 90 Å minimum and 250 Å maximum particle diameter. The coordinates were used to extracted 4× binned particles for subsequent processing. After several rounds of Class2D, the good particles were selected and subjected to Ab-Initio Reconstruction (K = 4). Particles were further subjected to Hetero Refinement with those initial model acquired above. The coordinates for particles belonging to the desired class were used to re-extracted un-binned particles. Non-uniform Refinement was used for the whole structure without symmetry imposed. For the interaction region between RBD and antibody, particle subtraction was used and local refinement was performed to improve the quality of RBD-antibody sub-complex density maps. The resolution was estimated by the gold-standard Fourier shell correlation (FSC) cut-off value of 0.143. The local resolution of the final density map was computed in cryoSPARC. Refer to Figure S3 and Table S2 for details of data collection and processing.
Cryo-EM model building and analysis
The RBD structure of BA.2 (PDB ID: 7XB0) spike protein and predicted atomic models by AlphaFold242 of VacBB-551 were used as the initial model to fit in the density maps of BA.2-S: VacBB-551 sub-complexes using UCSF Chimera.56 The amino acids were then manually adjusted in Coot.57 The resulting coordinates were further improved through real space refinement using Phenix.58 The refinement cycle was repeated, and the quality of the final 3D atomic models were evaluated using MolProbity.59 The model refinement statistics are summarized in Table S2. The structure figures were prepared using PyMol (http://www.pymol.org) or ChimeraX.60
Preparation of fragment of antigen binding (Fab) of VacBB-551
Fab-form VacBB-551 was digested from purified IgG-form VacBB-551. IgGs were diluted to 1 mg/mL, then L-Cysteine hydrochloride (final concentration 20 mM, Sigma-Aldrich), EDTA (final concentration 20 mM, Invitrogen), and papine (final concentration 1.25 μg/mL, Sigma-Aldrich) were added and incubated at 37°C for 12–14 h. The reaction was stopped by adding 0.5 M Iodoacetamide (Sigma-Aldrich). Fabs were purified by using protein A affinity chromatography to separate the fragment crystallizable (Fcs) and redundant undigested IgGs. Unless otherwise specified, the description of VacBB-551 indicates its IgG form.
Binding affinity analysis to recombinant mutated RBD proteins by surface plasmon resonance (SPR)
SARS-CoV-2 WT RBD protein was expressed with a His tag at the C-terminus.18 Mutated RBD proteins carrying N460K, F486V, F486S, N460K + F486V, or N460K + F486S, respectively, were constructed using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech) and expressed in HEK-293F cells. After 5 days, the cell culture supernatant was collected and applied to nickel affinity column (BeyoGold His-tag Purification Resin, Beyotime). After washing with PBS containing 70 mM imidazole (Sangon Biotech), the protein was eluted with PBS containing 500 mM imidazole. The purified protein was diluted with PBS and then concentrated to remove the imidazole.
The binding assays of Fab-form VacBB-551 and IgG-form VacBB-551 to WT and mutated RBD proteins were performed using the Biacore 8K system (GE Healthcare). Specifically, one flow cell of the CM5 sensor chips were covalently coated with the RBD protein in 10 mM sodium acetate buffer (pH 5.0) for a final response unit (RU) around 250, whereas the other flow cell was left uncoated and blocked as a control. All the assays were run at a flow rate of 30 μL/min in HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween 20). Serially diluted Fab-form VacBB-551 or IgG-form VacBB-551 were injected for 60 s, respectively, and the resulting data were fit in a 1:1 binding model with Biacore Evaluation software (GE Healthcare). Every measurement was performed two times and the individual values were used to produce the mean affinity constant.
Quantification and statistical analysis
The independent experiment replicates were indicated in the figure legends. Half-maximal inhibitory concentrations (IC50) of antibodies were calculated using GraphPad Prism 8.0 software by log (inhibitor) vs. normalized response - Variable slope (four parameters) model. The values of binding affinity (KD) of antibodies were calculated using Biacore Evaluation software 3.0 by Multi-cycle kinetics/affinity model. The statistical analysis was indicated in the figure legend.
Acknowledgments
We thank the biological sample bank (BioBank) of the Shenzhen Third People’s Hospital for bio-samples and services provided. We thank all participants who received the inactivated vaccine and all the healthcare providers from the Shenzhen Third People’s Hospital for the work they have done. We thank Dr. Qingbing Zheng at Xiamen University for discussing the cryo-EM data. This study was supported by the National Key Plan for Scientific Research and Development of China (2021YFC2301900 and 2021YFC0864500); the National Science Fund for Distinguished Young Scholars (82025022); the National Natural Science Foundation of China (82002140, 92169204, 82171752, and 32100960); the Guangdong Basic and Applied Basic Research Foundation (2021B1515020034, 2023A1515011883, and 2019A1515011197); the Shenzhen Science and Technology Program (RCYX20200714114700046 and RCBS20210706092345028); the Science and Technology Innovation Committee of Shenzhen Municipality (JSGG20220226085550001, JSGG20200207155251653, and JSGG20210901145200002); and the Shenzhen Natural Science Foundation (JCYJ20190809115617365, JCYJ20200109144201725, and JCYJ20220530163405012).
Author contributions
Z.Z. and B.J. conceived and designed the study. B.J., Q.F., C.L., S.S., and M.W. performed all experiments and analyzed the data together with assistance from H.G., B.Z., and X.G. Z.Z. and B.J. wrote the manuscript, and all authors read and approved this version of the manuscript.
Declaration of interests
Patent applications have been filed on some of mAbs presented here. Z.Z., B.J., Q.F., C.L., S.S., M.W., B.Z., and X.G. are inventors.
Published: May 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112532.
Supplemental information
Footnote: a represents that identical amino acid sequences of both heavy and light chains appear more than once due to the antibody clonal expansion.
Data and code availability
The structure coordinate is deposited in the Protein Data Bank under accession code 8GS9 (BA.2-S: VacBB-551). The corresponding EM density map is deposited in the Electron Microscopy Data Bank under accession number EMD-34226 (BA.2-S: VacBB-551). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans J.P., Zeng C., Qu P., Faraone J., Zheng Y.-M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization of SARS-CoV-2 omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022;30:1093–1102. doi: 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022 doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/d41586-021-03825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/d41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M., Wu L., Zheng A., Xie Y., He Q., Rong X., Han P., Du P., Han P., Zhang Z., et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3. Immunity. 2022;55:1501–1514.e3. doi: 10.1016/j.immuni.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1421–1422. doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu P., Evans J.P., Faraone J.N., Zheng Y.M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe. 2023;31:9–17.e3. doi: 10.1016/j.chom.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurhade C., Zou J., Xia H., Liu M., Chang H.C., Ren P., Xie X., Shi P.Y. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med. 2023;29:344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 14.Miller J., Hachmann N.P., Collier A.R.Y., Lasrado N., Mazurek C.R., Patio R.C., Powers O., Surve N., Theiler J., Korber B., Barouch D.H. Substantial neutralization escape by SARS-CoV-2 omicron variants BQ.1.1 and XBB.1. N. Engl. J. Med. 2023;388:662–664. doi: 10.1056/NEJMc2214314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng L., Song S., Fan Q., Shen S., Wang H., Zhou B., Ge X., Ju B., Zhang Z. Cross-neutralization of SARS-CoV-2 Kappa and Delta variants by inactivated vaccine-elicited serum and monoclonal antibodies. Cell Discov. 2021;7:112. doi: 10.1038/s41421-021-00347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Fan Q., Zhou B., Shen Y., Zhang Y., Cheng L., Qi F., Song S., Guo Y., Yan R., et al. Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants. iScience. 2022:104431. doi: 10.1016/j.isci.2022.104431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju B., Zheng Q., Guo H., Fan Q., Li T., Song S., Sun H., Shen S., Zhou X., Xue W., et al. Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209. Cell Res. 2022;32:491–494. doi: 10.1038/s41422-022-00638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju B., Zhou B., Song S., Fan Q., Ge X., Wang H., Cheng L., Guo H., Shu D., Liu L., Zhang Z. Potent antibody immunity to SARS-CoV-2 variants elicited by a third dose of inactivated vaccine. Clin. Transl. Med. 2022;12:e732. doi: 10.1002/ctm2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong L., Ju B., Chen Y., He L., Ren L., Liu J., Hong K., Su B., Wang Z., Ozorowski G., et al. Key gp120 glycans pose roadblocks to the rapid development of VRC01-class antibodies in an HIV-1-Infected Chinese donor. Immunity. 2016;44:939–950. doi: 10.1016/j.immuni.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S., Ju B., Shapero B., Lin X., Ren L., Zhang L., Li D., Zhou Z., Feng Y., Sou C., et al. A VH1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite. Sci. Adv. 2020;6:eabb1328. doi: 10.1126/sciadv.abb1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju B., Li D., Ren L., Hou J., Hao Y., Liang H., Wang S., Zhu J., Wei M., Shao Y. Identification of a novel broadly HIV-1-neutralizing antibody from a CRF01_AE-infected Chinese donor. Emerg. Microb. Infect. 2018;7:174. doi: 10.1038/s41426-018-0175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 24.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- 27.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022;30:1527–1539.e5. doi: 10.1016/j.chom.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 33.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 34.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan R., Wang R., Ju B., Yu J., Zhang Y., Liu N., Wang J., Zhang Q., Chen P., Zhou B., et al. Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies. Cell Res. 2021;31:517–525. doi: 10.1038/s41422-021-00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 38.Fedry J., Hurdiss D.L., Wang C., Li W., Obal G., Drulyte I., Du W., Howes S.C., van Kuppeveld F.J.M., Förster F., Bosch B.J. Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11. Sci. Adv. 2021;7:eabf5632. doi: 10.1126/sciadv.abf5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 40.Lv Z., Deng Y.Q., Ye Q., Cao L., Sun C.Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dougherty D.A. The cation-pi interaction. Acc. Chem. Res. 2013;46:885–893. doi: 10.1021/ar300265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arora P., Cossmann A., Schulz S.R., Ramos G.M., Stankov M.V., Jäck H.M., Behrens G.M.N., Pöhlmann S., Hoffmann M. Neutralisation sensitivity of the SARS-CoV-2 XBB.1 lineage. Lancet Infect. Dis. 2023;23:147–148. doi: 10.1016/S1473-3099(22)00831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q., Ju B., Ge J., Chan J.F.W., Cheng L., Wang R., Huang W., Fang M., Chen P., Zhou B., et al. Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2. Nat. Commun. 2021;12:4210. doi: 10.1038/s41467-021-24514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M., Fan Q., Zhou B., Ye H., Shen S., Yu J., Cheng L., Ge X., Ju B., Zhang Z. A key F27I substitution within HCDR1 facilitates the rapid maturation of P2C-1F11-like neutralizing antibodies in a SARS-CoV-2-infected donor. Cell Rep. 2022;40:111335. doi: 10.1016/j.celrep.2022.111335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ju B., Zhang Q., Wang Z., Aw Z.Q., Chen P., Zhou B., Wang R., Ge X., Lv Q., Cheng L., et al. Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants. Nat. Immunol. 2023;24:690–699. doi: 10.1038/s41590-023-01449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudicell R.S., Kwon Y.D., Ko S.Y., Pegu A., Louder M.K., Georgiev I.S., Wu X., Zhu J., Boyington J.C., Chen X., et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I.S., Altae-Tran H.R., Chuang G.Y., Joyce M.G., et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo H., Jiang J., Shen S., Ge X., Fan Q., Zhou B., Cheng L., Ju B., Zhang Z. Additional mutations based on Omicron BA.2.75 mediate its further evasion from broadly neutralizing antibodies. iScience. 2023;26:106283. doi: 10.1016/j.isci.2023.106283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Footnote: a represents that identical amino acid sequences of both heavy and light chains appear more than once due to the antibody clonal expansion.
Data Availability Statement
The structure coordinate is deposited in the Protein Data Bank under accession code 8GS9 (BA.2-S: VacBB-551). The corresponding EM density map is deposited in the Electron Microscopy Data Bank under accession number EMD-34226 (BA.2-S: VacBB-551). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.