Abstract
Herein, we report the systematic investigation of stereopure phosphorothioate (PS) and phosphoryl guanidine (PN) linkages on siRNA-mediated silencing. The incorporation of appropriately positioned and configured stereopure PS and PN linkages to N-acetylgalactosamine (GalNAc)-conjugated siRNAs based on multiple targets (Ttr and HSD17B13) increased potency and durability of mRNA silencing in mouse hepatocytes in vivo compared with reference molecules based on clinically proven formats. The observation that the same modification pattern had beneficial effects on unrelated transcripts suggests that it may be generalizable. The effect of stereopure PN modification on silencing is modulated by 2′-ribose modifications in the vicinity, particularly on the nucleoside 3′ to the linkage. These benefits corresponded with both an increase in thermal instability at the 5′-end of the antisense strand and improved Argonaute 2 (Ago2) loading. Application of one of our most effective designs to generate a GalNAc-siRNA targeting human HSD17B13 led to ∼80% silencing that persisted for at least 14 weeks after administration of a single 3 mg/kg subcutaneous dose in transgenic mice. The judicious use of stereopure PN linkages improved the silencing profile of GalNAc-siRNAs without disrupting endogenous RNA interference pathways and without elevating serum biomarkers for liver dysfunction, suggesting they may be suitable for therapeutic application.
INTRODUCTION
In 1998, the RNA interference (RNAi) pathway, or the mechanism by which short double-stranded RNAs lead to the degradation of specific mRNAs was first discovered (1), and soon thereafter, confirmation that this gene silencing pathway is conserved in mammalian cells was reported (2). To leverage this endogenous mechanism for therapeutic use, partially or fully chemically modified double-stranded RNAs, called siRNAs, with increased metabolic stability compared with natural RNA are needed (3). The first siRNA approved for therapeutic use, patisiran (https://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-targeted-rna-based-therapy-treat-rare-disease), is partially chemically modified with 2′-O-methyl (2′-OMe) ribose modifications and 2′-deoxythymidine dinucleotide overhangs on the 3′-ends (4). Subsequently approved molecules, including givosiran, inclisiran, lumasiran, and vutrisiran, feature conjugation to N-Acetylgalactosamine (GalNAc) to enable hepatocyte delivery (5), as well as 2′-deoxyfluoro (2′-F) ribose and phosphorothioate (PS) backbone modifications to increase metabolic stability (6,7). 2′-F and 2′-OMe ribose modifications are commonly used in siRNAs, as they favor a C3'-endo sugar conformation, which is RNA-like, and provides stability to the duplex and is compatible with Ago2 loading (8). Incorporation of PS modifications at the termini, in combination with ribose modifications, protects against degradation by exonucleases (9). Importantly, siRNAs featuring modifications that enhance stability also exhibit improved tissue exposure, Ago2 loading and catalysis (6,7,10).
The advent of the PS backbone helped to launch oligonucleotides as therapeutics across mechanisms (10). A consequence of PS modification, however, is the creation of a chiral center, where the molecule can adopt either an Rp or Sp configuration. During traditional synthesis, this property results in the production of stereorandom mixtures containing 2n diastereomers where n is the number of PS-modified positions (10,11). As the configuration of these linkages conveys distinct pharmacology to the resulting isomers (12,13), multiple groups have invested in methods to synthesize oligonucleotides with control over chirality of PS modifications (11,14–27). These efforts have enabled more detailed insight into how control over this chemistry can impact physicochemical and biological properties of these molecules. For example, Rp PS linkages form more stable duplexes with a target RNA, whereas Sp PS linkages tend to have higher metabolic stability (11,12,27). The activities of key endogenous enzymes, such as RNase H, are impacted by backbone stereochemistry (11,28,29). The same is true for TLR9, an innate immune receptor (12).
We were among the first to apply control over PS stereochemistry to RNAi when we investigated the application of stereopure PS linkages in the 5′- and 3′-termini of siRNAs targeting Proprotein convertase subtilisin/kexin type 9 (PCSK9) and identified stereochemistry-dependent activity differences between molecules (https://patents.google.com/patent/WO2014012081A2/en?oq=WO2014012081). More recently, others have generated stereorandom mixtures of fully PS-modified siRNAs with biased compositions of Rp or Sp configured PS linkages, with siRNAs having higher Rp PS content exhibiting greater silencing activity in cultured cells than those with higher Sp PS content (30). Others have generated and evaluated stereopure GalNAc-siRNAs based on a standard siRNA design, which incorporate six PS modified linkages—two at both termini of the antisense strand and two at 5′-terminus of the sense strand—and found that only a handful of the 64 possible isomers are active in mice (31). In a related study, siRNAs with a restricted PS modification pattern—containing a PS linkage at both termini of the antisense strand and a single PS linkage at the 5′-end of the sense strand were evaluated, and stereochemistry of the PS linkage was found to impact silencing activity, with a 5′-Rp PS and 3′-Sp PS in the antisense strand enhancing Ago2 loading and metabolic stability (32).
In this work, we set out to further explore the impact of stereopure PS linkages on activity and to test whether our recently reported phosphoryl guanidine (PN) backbone can enhance the activity, tissue distribution and durability of siRNAs as observed with other oligonucleotide modalities (23–25). To begin, we interrogated siRNA formats with established clinical relevance (6,7) to determine whether control over PS stereochemistry and application of stereopure PN chemistry can enhance silencing. During these interrogations, we explored the impact of various chemical and stereochemical modifications on silencing by evaluating hundreds of stereopure siRNAs. Because we interrogated siRNAs with a more complex PS backbone modification pattern—with six PS modifications instead of three—this work expands our understanding of how to configure chiral PS linkages to improve silencing activity compared with prior reports (32). We confirm that the introduction of appropriately positioned stereopure PN linkages further improves the potency and durability of silencing in a manner dependent upon nearby 2′-ribose modifications, particularly 3′ to the PN linkage. This improved silencing activity is due at least in part to increased Ago2 loading, with molecular modeling providing additional insight into why PN linkages have position- and stereo-specific effects. We confirm that the enhanced silencing activity does not disrupt endogenous RNAi pathways, and that the design principles developed herein are applicable to multiple targets.
MATERIALS AND METHODS
Oligonucleotide synthesis and preparation
Chemically modified stereopure oligonucleotides were synthesized, purified, and characterized as described previously with minor modifications (23,25). Stereorandom oligonucleotides were synthesized using standard phosphoramidite methodologies. Tri-antennary GalNAc conjugation was accomplished using a solution-phase or solid-phase approach as previously described (25). The predicted and measured mass of all oligonucleotides are shown (Supplementary Dataset 1).
To prepare duplexes, equimolar solutions of single strands in water were combined, vortexed and then lyophilized overnight. The resultant solid was reconstituted in 1× PBS. The concentration of the duplex solution was measured by A260, using extinction coefficients calculated from the extinction coefficients of the component single strands and the hypochromicity factor (33). For samples assayed in vivo, assessment of the percentage of remaining single strands in duplex solution were performed by UPLC.
In vitro experiments
Primary mouse or human hepatocytes (Cat. No. MSCP10 or HMCPTS, respectively, Life Technologies) were plated into collagen I coated 96-well plate in 108 μl of medium containing ∼10 × 103 cells following manufacturer's instruction. Gymnotic treatments of siRNAs were performed by adding 12 μl of siRNAs at the desired concentrations directly to cultured hepatocytes. Cells were incubated at 37°C for 48 h, and total RNA was extracted and purified using SV96 Total RNA Isolation kit (Promega, Cat. No. Z3505), and eluted with 100 μl of nuclease-free H2O. cDNA was produced with 5 μl of RNA sample in a total volume of 20 μl using High-Capacity cDNA Reverse Transcription kit (ThermoFisher Cat. No. 4368813) following manufacturer's instructions, and then 1:1 diluted with nuclease-free H2O. 4 μl of cDNA product was mixed with normalizer (Hprt and SFRS9 for mouse and human primary hepatocyte, respectively) and target probes in 1 μl, as well as 5 μl of iQ Multiplex Powermix (Bio-Rad, Cat. No. 1725849). Real time PCR was performed in a CFX System (BioRad). In vitro siRNA treatments were performed in triplicate, and real time PCR was performed in duplicate for each cDNA sample. To obtain the ΔΔCt for relative fold change, raw data from siRNA treatment experiments were normalized to mock control with no siRNA treatment. Primers for RT-PCR are shown in Supplementary Table S1.
Thermal denaturation (Tm)
Equimolar amounts of sense and antisense strands were combined and annealed in 0.1× PBS (pH 7.2) to obtain a final concentration of 1 μM of each strand (3 ml). UV absorbance at 254 nm was recorded at intervals of 30 s as the temperature was raised from 15°C to 95°C at a rate of +0.5°C per min, using a Cary Series UV–Vis spectrophotometer (Agilent Technologies). Absorbance was plotted against the temperature, and Tm values were calculated by taking the first derivative of each curve.
In vivo experiments
All animal experiments were performed at Biomedical Research Models, Ins. Dba Biomere (Worcester, MA, USA) under an approved protocol and in compliance with Biomere's Institutional Animal Care and Use Committee guidelines for care and use of animals. Mice were on a 12 h light-dark cycle. Food (lab diet 5001) and water were available ad libitum. Housing rooms were maintained at 20–26°C and relative humidity was 30–70%.
C57BL/6J mice (Stock No.: 000664) were obtained from Jackson Laboratory (Bar Harbour, ME) and utilized for all Ttr mouse studies. Homo sapiens Hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13) transgenic mice were generated by Biocytogen (Wakefield, MA). A targeting vector containing human HSD17B13 transcript (NM_178135) was incorporated into the ROSA26 locus of C57BL/6J mice using Bioctyogen's proprietary CRISPR/Cas9-based extreme genome editing system. Zygote microinjection was performed and integration of targeting allele in founder animals was confirmed by PCR and subsequent sequencing. PCR positive founder animals were bred with wild-type animals to generate F1 heterozygous mice. F1 heterozygous mice were confirmed positive by PCR. Southern blot analysis on PCR positive F1 mice confirmed PCR results and identified no random insertion sites. A breeding colony was established to generate animals for study. Male heterozygous mice between 9–17 weeks of age at time of dosing received subcutaneous administrations of test article and were necropsied at doses and times, respectively, as noted in individual figures.
Blood collections were performed via submandibular puncture at pre-dose and weekly and via cardiac puncture at the time of necropsy. Approximately 100–150 ul of blood was collected at pre-bleed and weekly and >500 ul at necropsy. Blood was aliquoted into serum separators, spun down at 9000 RPM for 10 min, serum was removed and stored at −80°C until processing. At the time of necropsy, animals were perfused with phosphate buffered saline and livers were dissected. Four to five 4 mm biopsy punches were taken from the right medial lobe, placed in 12-well dishes and frozen on dry ice. The left lateral lobe was also collected, wrapped in foil and frozen on dry ice. All samples were stored at -80°C until processing.
The whole liver was pulverized into powder and kept frozen. To isolate the RNA for gene level quantification for each animal, an aliquot of liver tissue powder was transferred to a well containing a stainless-steel bead (Qiagen, Cat. No. 69989) on a 96-well plate (Greiner BIO-ONE, Cat. No. 780201). 500 μl of Triazol (ThermoFisher Scientific, Cat. No. 15596018) was added to each well, and homogenization was performed at 25 Hz/sec for 3 min on Tissue Lyser II (Qiagen). After adding 100 μl of bromochloropropane (ThermoFisher Scientific, Cat. No. 106862500) to each well, the plate was shaken vigorously and then centrifuged at 4000 g for 5 min. The upper-level supernatant was transferred to the binding plate from SV96 Total RNA Isolation kit (Promega) for RNA extraction following manufacturer's instruction. cDNA preparation and real time PCR were the same as described for in vitro experiments.
Clinical chemistry
Serum samples were analyzed at Charles River Laboratories (CRL Shrewsbury, MA) using clinically validated assays on AU640 instrument. The following parameters were analyzed: ALT, AST, ALP, albumin and total protein. Serum mouse TTR protein levels was evaluated ELISA (Crystal Chem, Cat. No. 80657) following manufacturer's instructions.
Oligonucleotide (antisense strand) quantification by hybridization ELISA
The following probes were used to selectively quantify the antisense strand RNA by hybridization ELISA (Integrated DNA Technologies): mTTR-20167-DET-REV, / 5BioTEG/+A+A + A + A + C AG + T GT + T; mTTR-20167-CAP-R,+C+T + TGC + TCT + A + T + A + A/3AmMC6T/; HSD-42383G-CAP, / 5AmMC12/A + A + TGG + T + A + TC + A + AA; HSD-42383G-DET, A + CC + TCA + TG + TCA/3BioTEG/.
The maleic anhydride-activated 96-well plates (Pierce, Cat. No. 15110) were coated with 50 μl of capture probe at 500 nM in 2.5% NaHCO3 (Gibco, Cat. No. 25080-094) for 2 h at 37°C. The plate was then washed three times with PBS + 0.1% Tween-20 (PBST) and blocked with 5% fat-free milk-PBST at 37°C for 1 h. Payload oligonucleotide (single stranded antisense strand) was serially diluted into tissue matrix. This standard, together with original samples, were diluted in lysis buffer so that the oligonucleotide amount in all samples was < 200 ng/ml. Twenty microliters of diluted sample was mixed with 180 μl of 333 nM detection probe diluted in PBST and denatured (65°C for 10 min, 95°C for 15 min and hold at 4°C). Fifty microliters of the denatured samples were distributed in blocked ELISA plates (in duplicate) and incubated overnight at 4°C. After three washes with PBST, 50 μl of 1:2000 streptavidin-AP (Southern Biotech, Cat. No. 7100-04) in PBST were added, and the mixture incubated at room temperature for 1 h. After extensive washes with PBST on a plate washer, 100 μl of AttoPhos (Promega, S1000) was added, incubated at room temperature in the dark for 10 min and read on the plate reader (Molecular Device, M5; excitation wavelength of 435 nm; emission, 555 nm). The concentration of antisense strand in each sample were calculated according to standard curve by four-parameter regression.
Tissue lysate preparation for assessing Ago2 loading
Tissue lysate was prepared according to prior reports (34,35). Liver punches equivalent to 25 mg were excised from the liver of individual treated animals and flash-frozen at time of harvest. The frozen tissues were homogenized in 500 μl ice cold freshly prepared lysis buffer (50 mM Tris–HCl at pH 7.5, 200 mM NaCl, 0.5% Triton X-100, 2 mM EDTA, 1 mg/ml heparin) supplemented with Pierce protease inhibitor tablet (ThermoFisher Scientific; Cat. No. PIA32953) at 1:10 dilution. The homogenization was performed in 2 ml deep well plates with 5 mm stainless steel beads (Qiagen Cat. No. 69989) using a tissue homogenizer (Qiagen Tissue Lyser II) at 30 Hz for 3 min. After processing, lysates were transferred to clean labeled 1.5 ml Eppendorf tubes and centrifuged at 21 000 g for 45 min at 4°C. The supernatants were aliquoted, and total protein concentrations were determined using Pierce® BCA Protein Assay Kit microplate procedure.
Immunoprecipitation and small RNA quantification
Immunoprecipitation and small RNA quantification were performed according to prior reports (34,35). For immunoprecipitation in 96-well plate format (Corning Cat. No. 3903), 200 ng of antibody was absorbed to 50 μl of magnetic Protein G Dynabeads (Invitrogen Cat. No. 10004D) during a 2-h incubation at 4°C in 100 μl of lysis buffer on a shaker. The beads were washed twice with 100 μl lysis buffer to remove unbound antibody. For Ago2 immunoprecipitation and control immunoprecipitation, anti-mouse Ago2 monoclonal antibody (Wako Cat. No. 01822021) and mouse IgG antibody (P3.6.2.8.1 eBioscience Cat. No. 16-4714-82) were used respectively as capture antibodies. 100 μl of lysate (300 μg of total protein) was incubated with the antibody-bound beads overnight at 4°C with mixing. The beads were washed five times with lysis buffer, followed by a washing with a buffer containing 50 mM Tris–HCl at pH 7.5, 200 mM NaCl, and 0.2% Triton X-100. The small RNAs coimmunoprecipitated with the beads were extracted by incubation with Cell Lysis buffer (cells-cDNA II Kit) by incubating at RT for 2 min.
Custom Taqman small RNA assay reagents for mouse Ttr and human HSD17B13 siRNA antisense strands and Taqman miRNA assays were obtained from ThermoFisher Scientific. 5 μl of Cell Lysis buffer eluents were used for quantification using stem-loop reverse transcription, followed by real-time qPCR. Levels of siRNA antisense strands were normalized to those of respective coimmunoprecipitated miR-122.
Structural models and molecular mechanics minimizations
Crystal structure of the human Argonaute-2 in complex with a micro-RNA (PDB code: 4F3T) was retrieved from the Protein Data Bank (www.rcsb.org). The Protein Preparation Wizard in Schrödinger Maestro (version 12.9.123, installed in a Valence VWS-1690441-SMD workstation with CentOS 7) was used to prepare the downloaded structure. PN backbone modifications were incorporated into the RNA (placed optimally to minimize steric clashes with neighboring side chains) using the structure editing option in Maestro. The RNA-protein complex model with PN modifications was energy-minimized using the OPLS4 force field as implemented in Maestro Protein Preparation Wizard using an RMSD Convergence criteria of 0.3 Å on heavy atoms. All drawings were generated using the Maestro software.
Statistics
Statistical analyses were run via the R computing environment (v 3.6.0) (https://www.r-project.org/) and the KNIME (Konstanz Information Miner) platform (v 4.5.0) (36). For all statistical comparisons, assumptions of equal variance and normality were tested using Levene's tests across all experimental factors and Shapiro-Wilk tests of model residuals, respectively (R package rstatix, v 0.7.0.999) (https://rpkgs.datanovia.com/rstatix/). For multi-factor comparisons of non-longitudinal data, Type III Two-way ANOVAs were run to investigate all possible interactions between factors (R package car, v 3.0–7) (37). In the event of unequal variance (Levene's test P < 0.05) in non-longitudinal data, Welch's one-way ANOVAs were run in place of typical one-way ANOVAs, while two-way ANOVAs were White-adjusted to allow for unequal variance. For longitudinal data, linear mixed effects models controlling for random subject intercept effects were fit to the data using the lmerTest R package (38). To compare groups in each experiment, two-tailed pairwise and Dunnett post hoc tests were extracted from the corresponding regression model via the multcomp R package (v 1.4–13) (39). Post-hoc P values were Bonferroni-corrected for multiple hypotheses. For non-longitudinal experiments featuring unequal variance, robust HC3 covariance estimation was implemented in post-hoc comparisons to allow heteroscedasticity (R package sandwich, v 2.5–1) (40–42). For the fully factored design of stereopure sense 5′ and 3′ chiral configurations and stereopure antisense chiral configurations in the reference format 1 (Figure 1B), ETA2 (η2) values were calculated from the full-interaction ANOVA model to determine proportion of variance explained by each variable. A follow-up regression model utilizing all features and interactions with high statistical significance (P < 0.0001) was utilized to generate interaction and main effects plots.
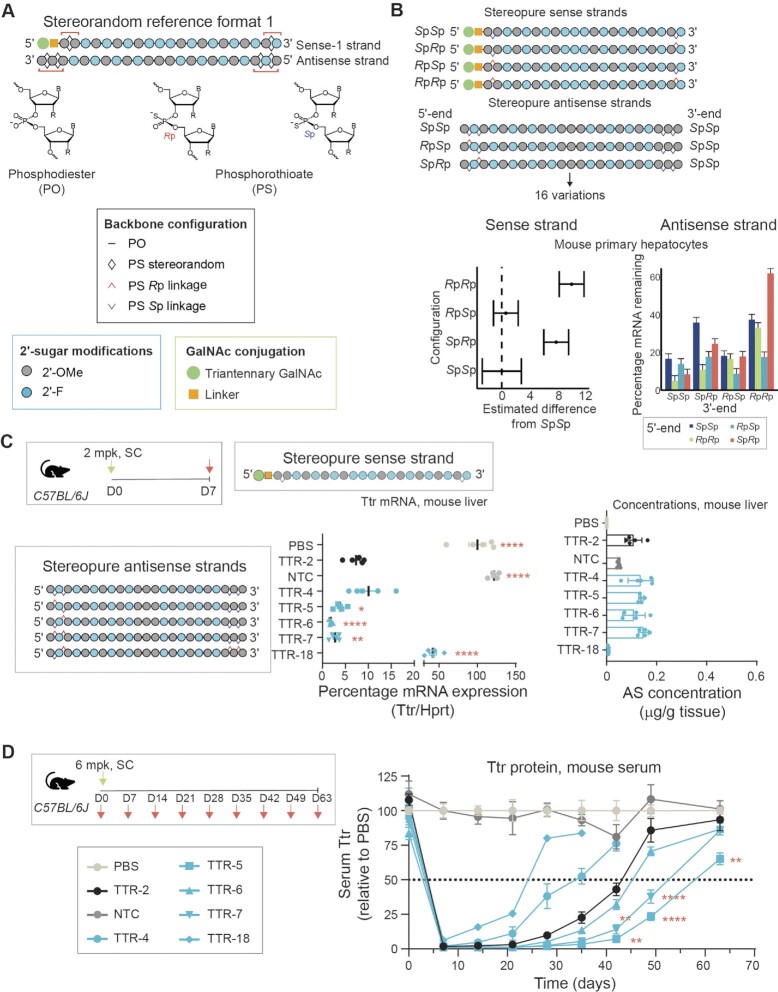
Figure 1.
Application of chiral control over PS modification to siRNA. (A) General design of siRNAs showing stereorandom reference format-1 (30) with GalNAc-modified sense strand (5’-end), with 2’-F and 2’-OMe ribose modifications and terminal stereorandom PS modification on each end (red brackets). Antisense (AS) strands have two terminal stereorandom PS modifications on each end (red brackets). Chemical structures for phosphodiester (PO) and phosphorothioate (PS) backbones are also shown along with legend for chemical modifications. (B) Application of PS stereochemistry to sense and antisense strands based on stereorandom reference format 1 in mouse primary hepatocytes. We generated 4 stereopure sense strands and 16 stereopure antisense strands with all possible combinations of Rp and Sp in the PS-modified positions shown in A. Forest plot showing summary of silencing data on 64 siRNAs (4 sense × 16 antisense) highlighting impact of PS stereochemistry in sense strand (bottom left) with SpSp configuration showing the highest mean activity independent of AS strand. All other configurations are shown with respect to activity of SpSp. Interaction between PS stereochemistry in 5'- and 3'-ends of the AS strand is shown (bottom right). See also Supplementary Table S1 and Supplementary Figure S1. (C) Ttr mRNA expression 1-week post 2 mg/kg dose. The same stereopure sense strand (shown) was used for all indicated stereopure antisense strands. Stereopure antisense strands are shown to the left of the graphs. TTR-2 is stereorandom reference format-1 and, NTC is stereorandom non-targeting control based on same format. Lines represent mean mRNA expression, and each point represents a different mouse. Stats: Welch's one-way ANOVA with Dunnett post hoc test with comparisons to TTR-2 * P < 0.05, ** P < 0.01, **** P < 0.0001. Right graph shows concentration of antisense strand detected in liver at same time point. Data are mean ± sem. Stats as described for mRNA graphic. (D) Serum Ttr protein expression after a single 6 mg/kg dose on day 0 for the same siRNAs shown in panel C. Data are presented as mean serum levels ± sem, n = 5 per group per time point. Stats: Linear mixed effects ANOVA with post-hoc test with comparisons to TTR-2 ** P < 0.01, **** P < 0.0001.
RNA-sequencing analysis (RNA-seq)
The following protocol was used for mouse Ttr ‘off-target’ experiment: QuantSeq 3′-mRNA-Seq library preparation kit (Lexogen GmbH, Vienna, Austria) was used to prepare the library according to manufacturer's protocol. Briefly, mRNA was primed by oligo(dT), second strand synthesis was initiated by random priming. After magnetic bead-based purification, the library was bar-coded by PCR, multiplexed, purified, then sequenced on NovaSeq SP chip with 100 cycle run (R1 is 100 bp + 12 bp R2).
Reads were trimmed using cutadapt v1.15. Reads were trimmed for both adapter sequencing as well as poly-A sequences. Trimmed reads were then mapped to mouse genome assembly GRCm38 (mm10). Mapping was done with hisat2 v2.1.0 using default settings. Alignment files were then processed using Rsubread v2.0.1 using featureCounts to extract counts per gene. For counting reads with multimapping features the feature only the annotation with the largest overlap is counted. The minimum overlap of bases between read and feature was set to 1. Finally, DESeq2 v1.26 was used to determine differential gene expression between treatment groups. Differentially expressed genes were considered to have different levels of expression if gene expression has <0.05 adjusted P value.
RESULTS
Chiral control over PS backbone prolongs silencing activity for siRNAs with enhanced stability
To evaluate the impact of chiral control over PS-backbone modification, we evaluated a GalNAc-siRNA design (Figure 1A) that was based on a format reported to have enhanced stability (6) referred to herein as the stereorandom reference format 1. This format contains six total PS modifications, with two PS modifications positioned at the 5′- and 3′-ends of the antisense strand, one PS modification on each end of the sense strand, and a 5′-GalNAc conjugate on the sense strand. We evaluated in vitro silencing activity of 66 siRNAs, representing all combinations of 16 stereopure antisense with 4 stereopure sense strands plus 2 controls (stereorandom reference siRNA called TTR-2 and non-targeting control, NTC) (Supplementary Table S2, Figure 1B) targeting a previously published sequence in the mouse Transthyretin gene (Ttr) (6) in mouse primary hepatocytes (Supplementary Figure S1). We considered the impact of PS stereochemistry on the sense and antisense strands independently. The impact of chirality of the PS linkage on the sense strand was small overall (η2 < 3%, 4-way ANOVA), indicating that the sense strand explained less than 3% of the variance in the observed mRNA levels. An Sp PS configuration on both ends of the sense strand yielded the most activity and was significantly more active than sense strands with 5′-Sp, 3′-Rp or 5′-Rp, 3′-Rp (Figure 1B, left panel: P < 0.0001, main effects regression model with post-hoc test). For the antisense strand, an SpSp PS configuration on the 3′-end drove most of the beneficial effects on activity (η2 = 15.19%, 4-way ANOVA), but we also observed an interaction between modifications on the two ends (η2 = 8.3%, 4-way ANOVA). For example, with the most active SpSp configuration on the 3′-end, the siRNA with an RpRp on the 5′-end was the most active silencer (95% mean silencing), whereas with RpSp on the 3′-end, the siRNA with an RpSp on the 5′-end was the most active (97% mean silencing) (Figure 1B, right panel). These findings are generally consistent with but more comprehensive than prior reports that evaluated a single stereopure PS linkage at the termini of the antisense strand (31,32).
To understand the impact of 3′-end PS stereochemistry on the antisense strand and silencing activity, we modelled the oligonucleotide-human Argonaute-2 (AGO2) interaction using existing crystal structures of AGO2 in complex with a mixed population of RNAs (41) or with micro-RNA-20a (42) (Supplementary Figure S2). In the models, Rp PS substitutions in the two most 3′ linkages (P21 and P22) appear less favorable than Sp PS substitutions. For P21, the Rp PS substitution is predicted to disrupt a hydrogen (H) bond between a nonbridging oxygen in the PO linkage and the Arg315 sidechain, where an Sp PS substitution is predicted to preserve this interaction. At the P22 linkage, although the Rp PS substitution preserves key interactions between nonbridging oxygens and amino acid side chains, the Sp PS substitution is predicted to both preserve key interactions and create a new electrostatic interaction, which would explain why the Sp PS modification is favored over both the Rp PS and PO linkages (Supplementary Figure S2).
We evaluated whether the impact of PS stereochemistry observed in primary mouse hepatocytes translated to wild-type mice in vivo by assessing some of the most promising stereopure configurations. The modification pattern for TTR-2 and NTC is as shown in Figure 1A. For all stereopure siRNAs, the sense strand is the same, incorporating an Sp PS configuration at both ends, and the antisense strands are shown (Figure 1C). Most antisense strands tested contain a fixed 3′-SpSp configuration and a variable 5′-end configuration (TTR-4—TTR-7). We also tested a stereopure siRNA that was weakly active in vitro (TTR-18, which contains a 3′-RpRp configuration) (Figure 1C). In mice injected subcutaneously with PBS or 2 mg/kg of NTC GalNAc-siRNA, we observed little to no silencing activity 1-week post-dose as expected (Figure 1C, left panel). The stereorandom GalNAc-siRNA control TTR-2 yielded 7.3% Ttr mean mRNA expression, and three of the five stereopure configurations outperformed stereorandom (mean Ttr mRNA expression: TTR-5 3.8%; TTR-6 1.8%; TTR-7 2.7%), with TTR-5 (P < 0.05, Dunnett post-hoc test), TTR-6 (P < 0.0001), and TTR-7 (P < 0.01) exhibiting significantly more silencing than TTR-2. The improved silencing activity for these stereopure siRNAs was unlikely due to an exposure benefit, as the mean concentrations of these antisense strands in liver was comparable to TTR-2 (Figure 1C, right panel). Low concentrations of TTR-18 in liver were well-correlated with observed activity for this siRNA. Observations on mRNA expression at 1 week were generally consistent with Ttr serum protein levels observed over longer duration after mice were treated with 6 mg/kg GalNAc-siRNA (Figure 1D). PBS and NTC had a negligible impact on serum Ttr. Stereorandom GalNAc-siRNA TTR-2 led to ∼98% decrease in Ttr protein by day 7 and recovered to levels comparable to PBS- or NTC-treated controls by ∼49 days post-treatment. TTR-18 and TTR-4 performed worse than TTR-2, with Ttr serum protein levels reaching levels significantly higher than TTR-2 from day 28 onwards (p < 0.01, post-hoc test from Linear mixed effects model). TTR-5, TTR-6 and TTR-7 performed at least as well as TTR-2, with TTR-5 showing significantly more serum Ttr protein reduction than TTR-2 at time points from day 42 onwards (P < 0.01), and TTR-7 showing significantly more Ttr protein reduction at days 42 and 49 (P < 0.0001). TTR-5, the siRNA with the most durable effect, differs from the other stereopure siRNAs only at the 5′-end of the antisense strand (5′-RpSp), indicating that this configuration yields the best potency and durability profile. These data also indicate the impact of PS stereochemistry on the 5′-end of the antisense strand is more evident in vivo than in vitro.
Application of PN backbone chemistry to siRNA
PN-modified backbones (Figure 2A) have recently been reported to enhance the properties of oligonucleotides acting via various mechanisms, including silencing with RNase H, splice switching, and RNA base editing with ADAR, in preclinical studies (23–25). We investigated whether application of a stereopure PN backbone could also enhance the activity of siRNAs. For this evaluation, we first assessed the impact of single Sp or Rp PN linkages in siRNAs designed based on TTR-5 and TTR-6, which differ from each other only in the configuration of the 5′-PS linkages (TTR-5: 5′-RpSp PS; TTR-6: 5′-SpRp PS).
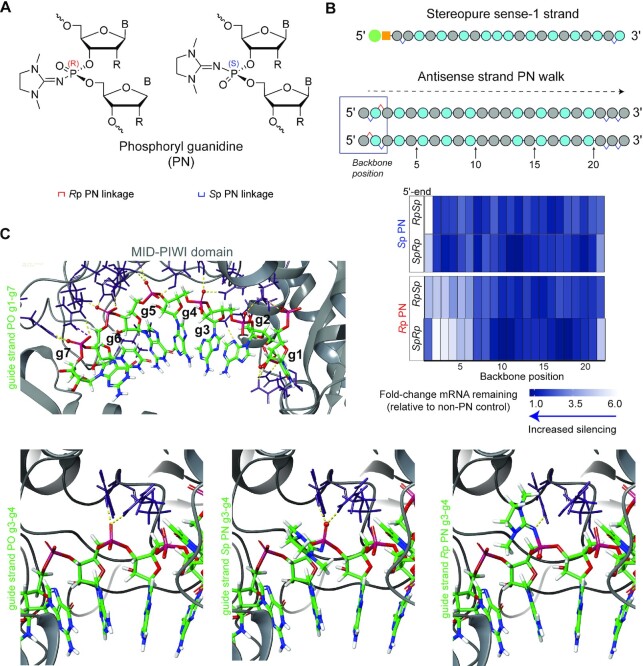
Figure 2.
Application of stereopure PN chemistry to siRNA and interrogation of the sense strand in vitro. (A) Chemical structures for phosphoryl-guanidine (PN) backbone with legend depicting how these stereopure backbone modifications are denoted in subsequent panels. (B) Heat map depicting percentage of mouse Ttr mRNA remaining relative to stereopure reference oligonucleotide with no PN backbone modifications in primary mouse hepatocytes treated with stereopure siRNAs composed of the depicted stereopure sense-1 strand and one of 88 antisense strands (Supplementary Table S2). The antisense strands were generated by walking either an Rp PN linkage or an Sp PN linkage through the backbone while fixing two Sp linkages on the 3’-end with an RpSp or an SpRp configuration on the 5’-end. Increasingly darker shades of blue correspond to increasing silencing; decreasing blue (increasing white) corresponds with decreasing silencing. (C) Models of stereopure siRNA interaction with Ago2 based on (55,56). View of antisense seed region and interactions with Ago2 for fully PO backbone (top left) and zoomed in views of backbone at position 3 in seed of antisense strand with PO linkage (lower left), Sp PN linkage (lower middle), or Rp PN linkage (lower right).
To assess this question, we generated 88 antisense strands that were duplexed with the same stereopure sense strand used in Figure 1 (sense-1, Figure 2B, Supplementary Table S3) to evaluate the impact of Sp and Rp PN linkages at every position of the backbone in silencing experiments performed in primary mouse hepatocytes. Two groups of PN-containing siRNAs were generated and benchmarked compared to TTR-5 and TTR-6, respectively, depending on the PS configuration of the 5′-end. The benchmark siRNAs TTR-5 and TTR-6 had comparable activity in this assay, with 500 and 1500 pM of both siRNAs resulting in 80% and 89% Ttr mRNA reduction, respectively (Supplementary Figure S3A). Data for the 88 siRNAs are summarized in the heat map shown in Figure 2B. In general, we found that the impact of PN linkages was not substantially impacted by the PS configuration of the 5′-end, with Sp PN linkages at most positions featuring mRNA expression changes close to 1, indicating similar expression compared with controls that lack PN modifications. Notably, the negative impact of Rp PN linkages seemed most profound in the seed region (nucleotides 2–8 of the antisense strand), as Rp PN linkages in the seed region exhibited higher levels of mRNA expression as compared with controls lacking PN modifications. (Figure 2B).
To understand the difference in activity between the Sp and Rp PN linkages, we once again modelled these substitutions using existing crystal structures of AGO2 (41,42) (Figure 2C, Supplementary Figure S4). In the AGO2-miRNA-20a structure, most interactions between AGO2 and the seed region are mediated through the phosphate backbone (42), suggesting that the orientation of chemical modifications to the backbone could impact these interactions. In the reference structure, R(Arg)792 and Y(Tyr)790 interact with a non-bridging oxygen of the PO backbone between nucleotides 3 and 4 (P3–P4) (Figure 2C, left upper and lower panels). In the model showing replacement of this backbone position with an Sp PN linkage, these H bonds with the non-bridging oxygen are preserved, and the PN modification is positioned away from the protein (Figure 2C, lower middle). With an Rp PN linkage modelled in this position, these H bonds are disrupted, and the guanidine moiety projects into AGO2, creating steric interference (Figure 2C, lower right). These observations are consistent with decreased silencing activity observed with siRNAs containing Rp PN substitutions in backbone position 3 of the antisense strand (Figure 2B). We observed similar disruption of AGO2-RNA interactions with Rp PN substitutions throughout the seed region except for the linkage between nucleotides 7 and 8 (P7) (Supplementary Figure S4), which is consistent with diminished activity observed for siRNAs with Rp PN substitutions throughout the seed region, suggesting the Rp PN backbone modification in the seed region may cause steric hindrance.
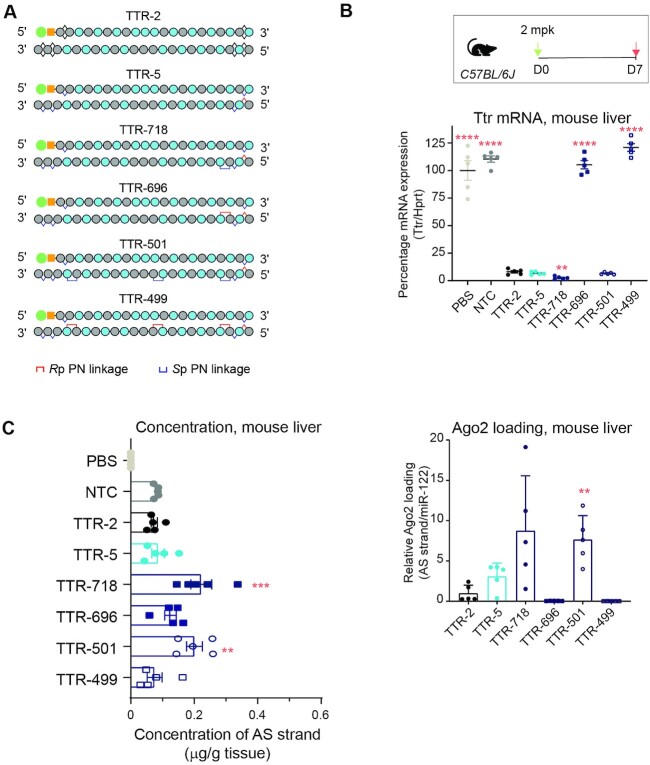
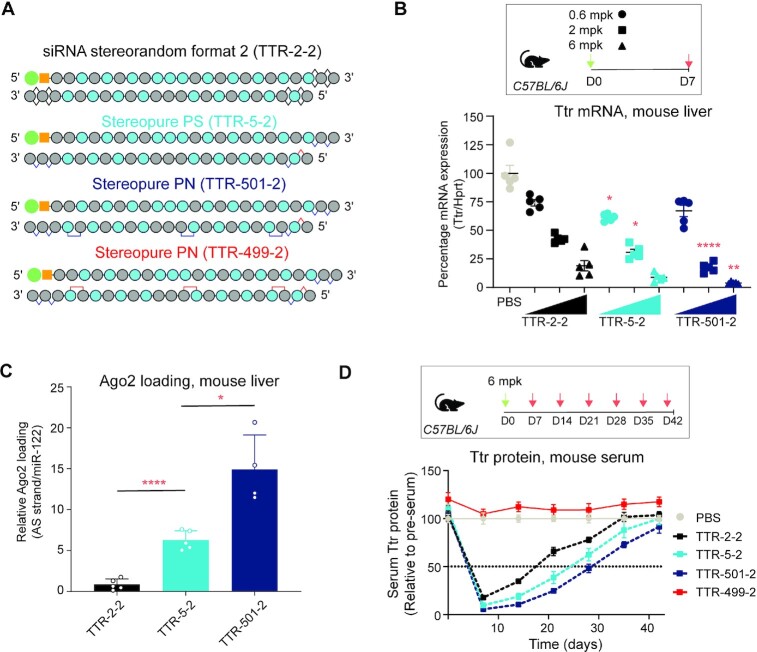
To further explore the impact of PN linkages on silencing activity, we evaluated whether varying the number of PN linkages affected silencing activity in vitro in primary mouse hepatocytes (Supplementary Figure S3B). We tested siRNAs based on TTR-5 (with 5′-RpSp PS configuration), and we added a variable number of Sp PN linkages, which generally supported more silencing activity than Rp PN linkages (Figure 2B). These siRNAs (TTR-718, TTR-500, TTR-501, TTR-549 and TTR-553) incorporated Sp PN linkages at backbone positions 3, 10, 19, 8 and 16. The inclusion of up to three Sp PN linkages in TTR-501 maintained in vitro potency comparable to TTR-5 (Supplementary Figure S3B). With the addition of four or five PN linkages (in TTR-549 and TTR-553), silencing activity declined, indicating that three PN linkages are sufficient.
To confirm these findings generally translate in vivo in wild-type mice, we evaluated stereopure siRNAs containing one or three PN linkages in both an Sp and Rp configuration (Figure 3A). One-week after subcutaneous injection, the reference stereorandom compound (TTR-2) decreased Ttr mRNA expression by ∼95% compared with PBS or NTC treatment. Stereopure TTR-5, with no PN modifications, yielded about the same silencing activity. Stereopure siRNAs, with one (TTR-718) or three (TTR-501) Sp PN modifications, also yielded at least 95% Ttr mRNA knockdown, with TTR-718 slightly outperforming TTR-2 (P < 0.01). The stereopure siRNAs, with one (TTR-696) or three (TTR-499) Rp PN linkages, yielded nominal activity, closely matching observations in mice treated with PBS or NTC (Figure 3B). These findings are consistent with our in vitro observations and indicate that siRNAs with up to three Sp PN linkages support robust silencing activity in vivo.
Figure 3.
Optimization of number, position, and stereochemistry of PN linkages in vivo. (A) Cartoons depicting siRNAs. (B) Percentage of Ttr mRNA expression in wild-type mice 1-week post-dose after treatment with the indicated siRNAs. Horizontal bars depict mean, and individual points represent one mouse. Stats compared with TTR-2: ** P < 0.01 **** P < 0.0001, Welch's One-way ANOVA with Dunnett post-hoc test comparisons to TTR-2. (C) Concentration of antisense strand in mouse liver 1-week post-dose (left). Stats: One-way ANOVA with Dunnett post-hoc test comparisons to TTR-2 ** P < 0.01, *** P < 0.001. Relative Ago2 loading for the indicated siRNAs (right) standardized to miR-122 and normalized to TTR-2 for the indicated oligonucleotide. Data represent mean ± sd, n = 5. Stats: Welch's one-way ANOVA with Dunnett post-hoc test comparisons to TTR-2 ** P < 0.01.
To confirm predictions from our modelling experiments, we also assessed the concentration of antisense strands in the liver and the amount of antisense mouse Ago2 loading from this experiment (Figure 3C). The concentration of NTC, TTR-2 and TTR-5 were comparable (∼0.08 μg/g liver). Antisense strands with Sp PN linkages (TTR-718 and TTR-501) reached significantly higher tissue concentrations than TTR-2 (0.2 μg/g liver, Figure 3C, left; P < 0.01 Dunnett post-hoc test). TTR-5, the stereopure siRNA without PN linkages also improved Ago2 loading ∼3-fold compared with stereorandom TTR-2. Both siRNAs with Sp PN linkages (TTR-718 and TTR-501) improved Ago2 loading ∼8-fold compared with TTR-2, but only TTR-501 was statistically significant (Figure 3C, right; P < 0.01, Dunnett's post-hoc test). siRNAs with Rp PN linkages yielded nominal Ago2 loading (Figure 3C), which corresponded with their in vivo activity profile (Figure 3B). These data are consistent with our modelling, which predicted that an Rp PN linkage in the seed region would disrupt Ago2 interaction. By contrast, an Sp PN linkage in the seed region supports siRNA silencing activity that is at least comparable to stereorandom reference and stereopure TTR-5 siRNAs, with enhanced exposure in the liver and enhanced Ago2 loading.
Design of the sense strand impacts silencing activity
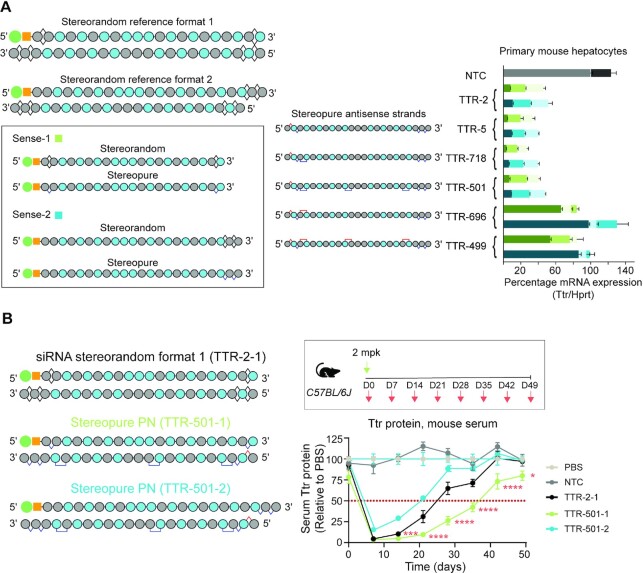
We next set out to determine whether the configuration of the sense strand impacts silencing activity. We designed siRNAs with formats based on Stereorandom reference format 1 and Stereorandom reference format 2 (Figure 4A). For siRNAs based on format 1, we used the sense-1 configuration, with 5′-GalNAc, terminal PS linkages on each end of a 21mer. In this format, the 3′-end of the sense strand aligns with the 5′-end of the antisense strand. For siRNAs based on format 2, we used the sense-2 configuration, with 5′-GalNAc, a pair of PS linkages on the 3′-end of a 23mer. In this format, the 3′-end of the sense strand overhangs the 5′-end of the antisense strand by two nucleotides. We tested a series of siRNAs incorporating the same antisense strands with either sense-1 or sense-2 formats (e.g. TTR-5–1, TTR-5–2) in primary mouse hepatocytes (Figure 4A). For all siRNAs tested and at all concentrations, the siRNAs incorporating sense-1 showed slightly more silencing activity in vitro in primary mouse hepatocytes than those with sense-2 configuration (Figure 4A). The trend was observed whether the siRNAs were stereorandom or stereopure. To confirm these findings translated in vivo, we evaluated stereopure TTR-501–1 and TTR-501–2 in wild-type mice. TTR-501–1 outperformed stereopure TTR-501–2, yielding lower levels of Ttr serum protein at all time points evaluated, reaching a statistically significant difference by day 14 (Figure 4B) (mean Ttr levels TTR-501–1: 5%, TTR-501–2: 29%, P < 0.001 linear mixed effects ANOVA with post-hoc test). A statistically significant difference between silencing observed with the two siRNAs was preserved from day 14 through the end of the study, with TTR-501–1 emerging as the more effective configuration. These data indicate that the sense strand configuration can impact silencing, with sense strand 1 outperforming sense strand 2.
Figure 4.
Evaluation of alternate sense strand formats. (A) Cartoons illustrating stereorandom siRNA reference formats 1 and 2, showing double-stranded configurations. Alternative sense strand configurations (Sense-1 and Sense-2) are shown (in box) for stereorandom and stereopure designs. Assessment of sense strand impact on siRNA activity, with siRNAs based on stereorandom reference formats (TTR-2 and NTC) or composed of the illustrated stereopure antisense strand with either stereopure sense-1 or stereopure sense-2. Silencing activity is shown for increasing concentrations (150, 500, 1500 pM) of the indicated siRNAs in primary mouse hepatocytes with the same series of antisense strands (shown to left of graph). Data are shown as mean ± sd, n = 3 per concentration per siRNA. (B) Percentage Ttr serum protein in mice over time after treatment with the indicated siRNA. The reference compound (TTR-2–1) is in the stereorandom format-1. TTR-501 is tested in siRNAs containing sense-1 (green) or sense-2 (light blue) configurations. Data are presented as mean ± sd, n = 5 per group. Stats: * P < 0.05, *** P < 0.001, **** P < 0.0001 linear mixed effects ANOVA with post-hoc test comparing TTR-501–1 to TTR-501–2.
To further explore whether PN chemistry could improve the activity of siRNAs based on stereorandom reference format 2 (TTR-2–2) (Figure 5A). Stereopure siRNAs were designed in the same general format (Figure 5A). After 1 week of treatment, all siRNAs led to dose-dependent decreases in Ttr mRNA expression in the mouse liver (Figure 5B). TTR-5–2, the stereopure non-PN containing siRNA, decreased Ttr mRNA (69% decrease) significantly more than TTR-2–2 at the 2 mg/kg dose (57% decrease; P < 0.05, 2-way ANOVA with post-hoc test). TTR-501–2, the siRNA containing Sp PN linkages, decreased Ttr mRNA significantly more than TTR-2–2 at the 2 mg/kg and 6 mg/kg doses (2 mg/kg: TTR-2–2 57%, TTR-501–2 83%; 6 mg/kg: TTR-2–2 81%, TTR-501–2 96%; P < 0.01, 2-way ANOVA with post-hoc test). Thus, the inclusion of three Sp PN linkages in TTR-501–2 significantly improved the potency of these siRNAs in vivo in mouse liver.
Figure 5.
Application of stereopure PN chemistry and interrogation of the sense strand in vivo. (A) Cartoons of siRNAs evaluated in panels B–D, which contain sense-2 strand configuration and the indicated antisense strands. (B) Percentage of Ttr liver mRNA expression in mice 1-week after treatment with the indicated siRNA. Dosing regimen is shown, with green arrow indicating time of treatment and red arrow indicating time of tissue collection. Horizontal bar is mean, and each dot represents a different mouse, n = 5 per group. Stats: Two-way ANOVA with post-hoc comparison to stereorandom siRNA at the same dose. * P < 0.05, ** P < 0.01, **** P < 0.0001. (C). Relative Ago2 loading of antisense strand (AS) standardized to miR-122 and normalized to TTR-2–2 in livers treated with 2 mg/kg siRNAs as illustrated in panel B. Data are presented as mean ± sd, n = 5 per group. Stats: Welch's one-way ANOVA with post-hoc comparisons * P < 0.05, **** P < 0.0001. (D) Percentage Ttr serum protein in mice over time after treatment with 6 mg/kg of the indicated siRNA. Dosing regimen is shown, with green arrow indicating time of treatment and red arrow indicating time of serum collection. A pre-dose serum sample was also taken for each animal. Data are presented as mean ± sd, n = 5 per group. Stats: linear mixed effects ANOVA with post-hoc tests.
To understand the reason for this increased potency, we evaluated loading into Ago2 for the middle dose and concentration of antisense strand in the liver at all doses (Figure 5C, Supplementary Figure S5). At 2 mg/kg, TTR-5–2 yielded ∼7-fold more Ago2 loading of the antisense strand than TTR-2–2 (P < 0.0001, Welch's 1-way ANOVA with post-hoc test). At the same dose, TTR-501–2 yielded ∼17-fold more Ago2 loading than TTR-2–2 and ∼2.5-fold more than TTR-5–2 (P < 0.05. Welch's 1-way ANOVA with post-hoc test). At most concentrations, TTR-2–2, TTR-5–2, and TTR-501–2 accumulated to comparable concentrations in the liver, except at the highest dose where TTR-501–2 reached concentrations ∼3-fold higher than the other antisense strands (Supplementary Figure S5). These data suggest potency improvements for stereopure TTR-5–2 and PN-containing TTR-501–2 over stereorandom TTR-2–2 are likely driven by improvements in Ago2 loading.
We next evaluated the durability of these siRNAs in vivo. For this experiment, we also included siRNA TTR-499–2, which contains Rp PN linkages at the same position as the Sp PN linkages in TTR-501–2 to confirm the impact of PN chirality persists over time. In this experiment, mice received a single 6 mg/kg dose of siRNA and serum Ttr protein was evaluated for over a month. TTR-2–2 the reference compound led to maximum silencing (83%) around 1-week post-dose, and silencing decreased over time until day ∼35 when serum Ttr levels no longer differed significantly from those in mice treated with PBS (Figure 5D). TTR-5–2 and TTR-501–2 followed the same general pattern as TTR-2–2, with greater maximal silencing (TTR-5–2 91%; TTR-501–2 95%) that was significantly lower than that observed for TTR-2–2 through day 21 for TTR-5–2 (P < 0.01, linear mixed effect ANOVA with post-hoc test) and at all time points between days 14 and 35 for TTR-501–2 (P < 0.01, linear mixed effects ANOVA with post-hoc test). By contrast, TTR-499–2 (with Rp PN linkages) showed an activity profile comparable to PBS. These data confirm that control over PS stereochemistry can improve the silencing profile for siRNAs and demonstrate that the inclusion of Sp but not Rp PN modifications in the antisense strand provide additional activity benefits due to improved Ago2 loading.
Application of stereochemistry and PN backbone chemistry to another siRNA format
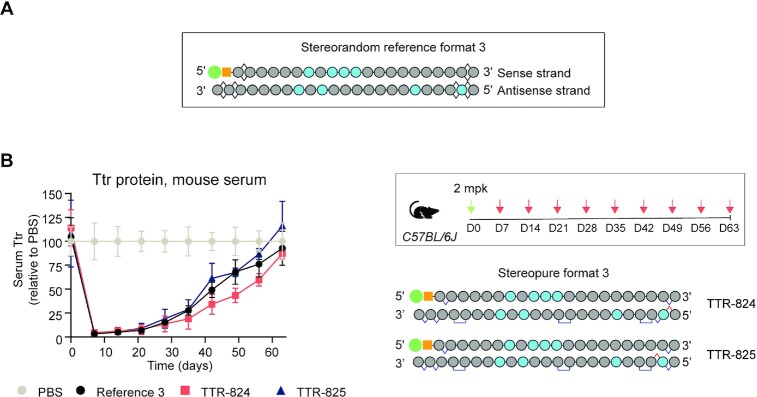
The design of siRNAs in the stereorandom reference format was improved by decreasing the amount of 2′-F-modifications (7) This newer enhanced stability format is shown in Figure 6A (Stereorandom reference format 3). To determine whether control over stereochemistry and PN backbone chemistry have a beneficial impact on reference format 3, we applied them using the same general design strategy that we employed in Figure 5, placing Sp PN linkages at backbone positions 3, 10 and 19 of the antisense strands, with RpSp (TTR-824) or SpRp (TTR-825) PS linkages on the 5′-ends. The sense strand incorporated an Sp PS linkage at each end (Figure 6B). We evaluated these stereopure siRNAs compared to Reference 3 in wild-type mice. After a 2 mg/kg dose, Reference 3 led to a mean 97% reduction in Ttr serum protein ∼1-week post-dose. Ttr serum proteins remained below 50% of those in PBS-treated mice through ∼43 days and recovered to match those in PBS-treated mice by 63-days post dose. TTR-824 and TTR-825 followed this same general trend, suggesting that this configuration of stereochemistry and PN backbone chemistry did not significantly improve siRNA-mediated silencing compared with Reference 3.
Figure 6.
Application of PN chemistry to stereorandom reference format-3 featuring reduced 2'-F content. (A) Schematic representation of Stereorandom reference format 3, designed based on (7). (B) Application of PN chemistry to reference format 3 in mice. Relative serum Ttr protein levels detected in mice treated with the indicated siRNA molecules. Data are presented as mean ± sem, n = 5 per group.
Optimization of PN backbone placement with 2′-ribose chemistry shows silencing benefit
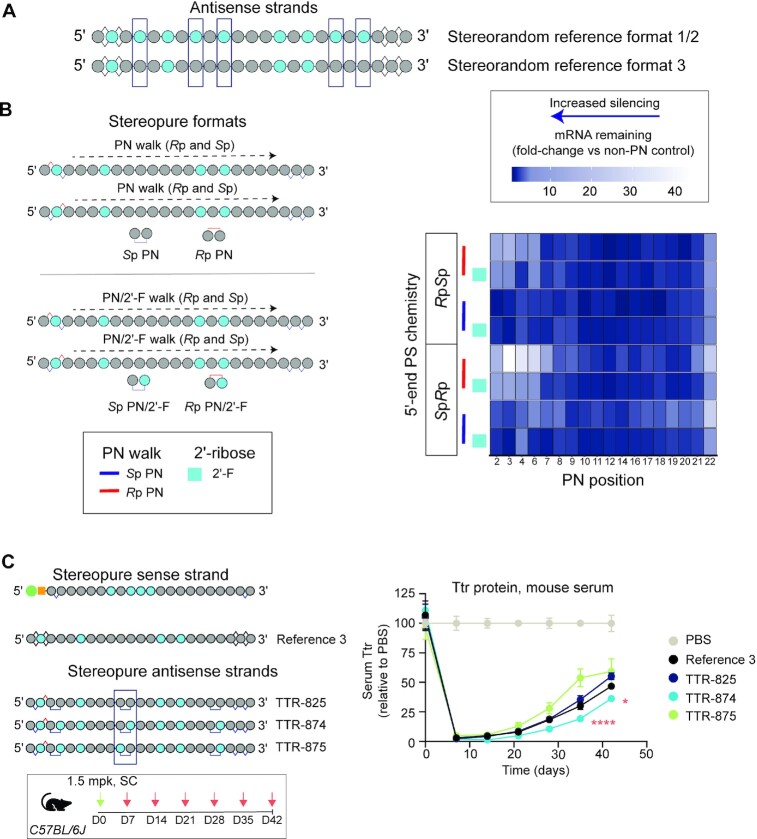
Given the impact that PN chemistry had on silencing with the initial siRNA formats, we were surprised that there appeared to be minimal benefit with reference format 3. We suspected that the lack of effect related to non-optimal placement of the linkages. To evaluate this possibility, we considered other changes in the format that could negatively impact the effect of PN chemistry that may need to be optimized in parallel. Compared with the reference formats 1 and 2, reference 3 had both fewer 2′-F modifications and more 2′-OMe modifications (Figure 7A). To investigate whether there was an interaction between 2′-ribose modifications and PN chemistry, we performed another structure-activity relationship analysis by creating 90 antisense strands that evaluate a single Rp PN or Sp PN linkage at every position in a stereopure antisense strand based on reference 3 (Figure 7B, Supplementary Figure S6A, Supplementary Table S4). In parallel, we also assessed another series of 90 stereopure antisense strands to evaluate a single Rp PN or Sp PN linkage in combination with a 2′-F modification on the nucleoside on the 3′-side of the linkage (Figure 7B, Supplementary Figure S6A, Supplementary Table S5). Within each series, we also assessed whether there was an interaction between the PN linkage and PS stereochemistry on the 5′-end of the antisense strand by generating the series with either a 5′-RpSp or a 5′-SpRp PS configuration (Figure 7B, Supplementary Figure S6A, Supplementary Tables S4 and S5). We assessed the in vitro silencing activity of the 180 antisense strands described above as siRNAs by combining them with the stereopure sense strand shown in Figure 7B. These experiments were performed at high (750 pM) and low concentrations (250 pM), and they reinforced some observations from our prior experiments including the notion that Rp PN linkages in the seed region of the antisense strand negatively impact silencing activity, as demonstrated by the high expression of mRNA with Rp PN linkages at positions 2–4 and 6 with respect to non-PN controls. In addition, we found a 2′-F or 2′-OMe modification 3′ of the PN linkage impacted silencing activity, suggesting that there may be an interaction between the PN linkage and the sugar modification 3′ of the linkage.
Figure 7.
Optimization of 2′-sugar chemistry pattern for PN linkages. (A) Schematic representation of stereorandom reference formats 1–3, highlighting differences in the 2'-ribose modification patterns on the antisense strands. (B) Optimization of PN chemistry and 2'-ribose modifications for the antisense strand in primary mouse hepatocytes. Schematics on the left illustrate ‘PN walk’ where 750 pM of siRNA containing antisense strand with Sp PN or Rp PN linkage was tested at each position of the oligonucleotide backbone (top), or an Sp PN with 2'-F or Rp PN with 2'-F was tested (bottom). All antisense strands are shown in Supplementary Tables S4 and S5. Low concentration (250 pM) data shown in Supplementary Figure S6A. Heat map (right) depicts Relative Ttr mRNA remaining for the stereopure siRNAs compared with TTR-871 or TTR-872 controls (750 pM), which lack PN linkages. Increasingly darker shades of blue correspond to increasing silencing; decreasing blue (increasing white) corresponds with decreasing silencing. The stereopure sense stand shown in panel C was used. The 5′-end PS pattern is indicated, as is PN chirality (red bars, Rp; blue bars, Sp), and sugar chemistry 3′ to the PN linkage (light blue box, 2′-F). (C) Relative serum Ttr protein levels detected in mice treated with the indicated siRNA molecules (left) after a single 1.5 mg/kg injection. Data are presented as mean ± sem, n = 5 per group. Stats *P < 0.01, **** P < 0.0001 linear mixed-effects ANOVA with post-hoc test comparing TTR-874 to TTR-875 at the same time points.
To explore this further, we assessed four siRNAs in wild-type mice including, the stereorandom Reference-3; TTR-825, which—similar to Reference 3—contains a 2′-OMe modification on both sides of the Sp PN linkage at backbone position 10; TTR-874, which contains a 2′-F modification 3′ to the Sp PN linkage; and TTR-875, which contains a 2′-F 5′ to the central Sp PN linkage (Figure 7C). Reference 3 and TTR-825, which had the same 2′-modification, exhibited a similar silencing profile over the 6-week experiment with a maximum ∼98% silencing of Ttr serum protein expression observed after 1 week, with levels recovering to ∼50% of PBS-treated levels by the end of the experiment. TTR-874 performed slightly better than Reference 3, with Ttr serum protein expression levels recovering to ∼35% of PBS-treated mice by the end of the experiment. TTR-875 performed slightly worse than Reference 3, with Ttr serum protein levels recovering to ∼60%. The differences between TTR-874 and TTR-875 were statistically significant for the final two weeks (P < 0.05, Linear mixed effects ANOVA with post-hoc test). Since TTR-875 and TTR-874 differ only in the 2′-ribose modification positioned 3′ of the Sp PN linkage at position 10, these data suggest there is an interaction between the PN linkage and the chemistry of the ribose sugar 3′ to the linkage, with 2′-F supporting more silencing activity in conjunction with the PN linkage than 2′-OMe.
To determine the number of PN linkages that yield the most activity, we evaluated the silencing activity of a small number of stereopure siRNAs compared with stereorandom Reference 3 in primary mouse hepatocytes (Supplementary Figure S6B). We first compared TTR-825, with three Sp PN linkages to TTR-932, TTR-933, and TTR-934, which each contain only two of the three Sp PN linkages from TTR-825. TTR-934 (IC50 = 149 pM) is the most active of these three siRNAs and is also slightly more active than TTR-825 (IC50 = 271 pM). We next evaluated siRNAs containing one or the other of the two Sp PN linkages in TTR-934 (TTR-936 and TTR-937, Supplementary Figure S6B). Although potent, neither of these siRNAs were quite as potent as TTR-934. Thus, we opted to advance the antisense design from TTR-934, with two instead of three PN linkages, into in vivo experiments.
PN chemistry increases Ago2 loading without disrupting endogenous RNAi pathways
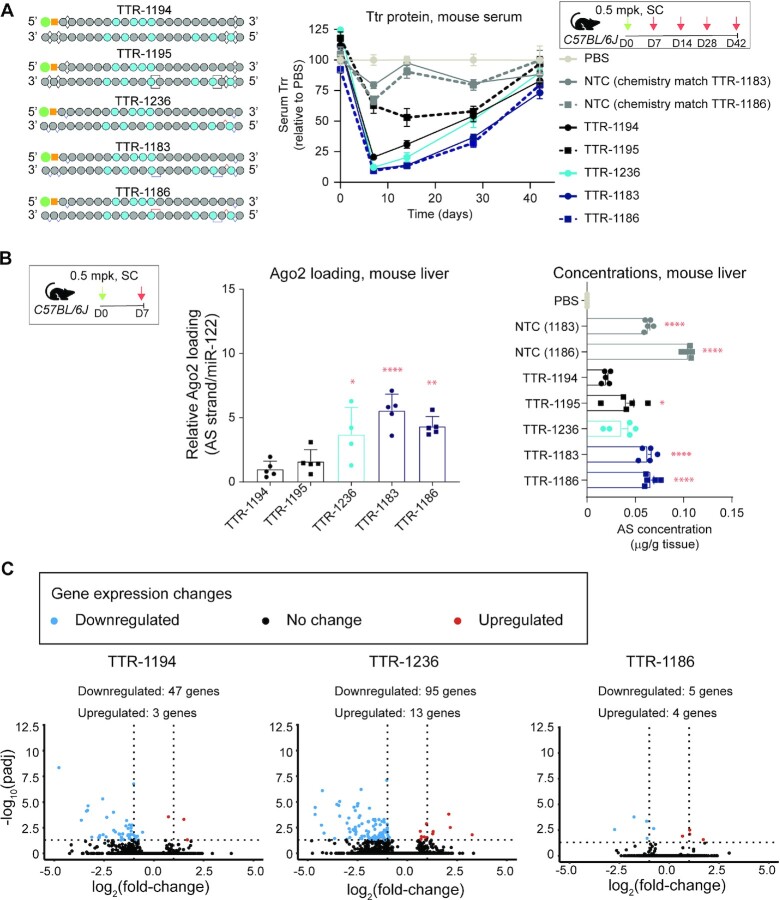
We evaluated several siRNAs designed to confirm a benefit of PN (TTR-1236) and to identify the optimal configuration for the two PN linkages (TTR-1183 and TTR-1186) and compared them to stereorandom TTR-1194, a new stereorandom reference compound based on Figure 7, and TTR-1195, a PN version of TTR-1194 containing two stereorandom PN linkages (Figure 8A). For this experiment, wild-type mice were treated with a low 0.5 mg/kg subcutaneous dose of siRNA and followed for 6 weeks. TTR-1194, the stereorandom molecule, yielded maximal ∼80% reduction in Ttr serum protein levels ∼1-week post dose, with serum protein levels rising to ∼20% of PBS-treated controls by the end of the experiment. TTR-1195 showed notably less activity, leading to a maximal 40% reduction in Ttr serum protein levels ∼1-week post dose. The stereopure siRNA TTR-1236, which lacks PN chemistry, was more potent than TTR-1194, yielding ∼90% reduction in Ttr serum protein levels ∼1-week post-dose and a similar recovery profile. The stereopure PN-containing siRNAs TTR-1183 and TTR-1186 both showed slightly more silencing than TTR-1194, yielding ∼90% reduction in Ttr serum protein levels ∼1-week post-dose. TTR-1183 and TTR-1186 maintained Ttr protein levels below those observed with TTR-1194 until 6-weeks post-dose, achieving significance at 2–3-weeks post dose (P < 0.05, linear mixed effects ANOVA with post-hoc test) (Figure 8A). Together, these data support several important points. First, controlling stereochemistry of the PS linkages in this siRNA format increases potency. Including stereorandom PN linkages conveys no silencing benefit and likely curbs potency. Finally, stereopure siRNAs containing two appropriately positioned and configured PN linkages yield more potent and durable silencing than TTR-1194.
Figure 8.
Optimized siRNA format 3 shows improved silencing activity. (A) Schematic representation of siRNAs tested in the study (left). Dosing regimen and Ttr serum protein levels in mice treated with 0.5 mg/kg of the indicated siRNA. Serum Ttr levels (compared to PBS) over time for the indicated siRNAs. Data are presented as mean ± sd, n = 5 per group per time point. (B) Antisense strand Ago2 loading standardized to miR-122 and normalized to TTR-1194 for the indicated oligonucleotide at 1-week post-dose (left). Concentration of antisense strand in liver 1-week post-dose. Data are presented as mean ± sem. Stats: One-way ANOVA with post-hoc test comparisons to TTR-1194. P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (C) Gene expression changes detected based on RNA-seq analysis of primary mouse hepatocytes treated with 2 μM of the indicated siRNA compared with PBS. Log2 fold-change in expression is plotted with respect to the P value. Dotted lines indicate statistical significance: the horizontal line denotes P = 0.01, and the vertical lines denote >1-fold-change in expression, with decreased expression to the left and increased expression to the right.
To confirm our earlier observations that the benefits of PN chemistry derive at least in part from an increase in Ago2 loading, we quantified the amount of antisense strand loaded into Ago2 as well as the amount detected in liver at 1 week, which approximates the time of maximal silencing. Compared with TTR-1194, all other siRNAs increased loading (TTR-1195 1.6-fold; TTR-1236 ∼3.7-fold; TTR-1183 ∼5.5-fold; TTR-1186 ∼4.3-fold), but only the stereopure siRNAs were statistically better (Figure 8B, p < 0.05, Dunnett post-hoc test). We also observed an increase in the concentration of antisense strands in the liver (TTR-1195 ∼2-fold; TTR-1236 ∼2.0-fold; TTR-1183 ∼3.0-fold; TTR-1186 ∼3.0-fold) with TTR-1183 and TTR-1186 being statistically significant (Figure 8B, P < 0.0001, Dunnett post-hoc test). The data suggest that for these siRNAs, stereopure PN chemistry increases both the amount of siRNA in the liver and the amount of Ago2 loading.
With increased Ago2 loading comes a risk that these siRNAs could disrupt endogenous RNAi pathways that rely on Ago2. If these siRNAs disrupt RNAi pathways, we would expect to see an increase in gene expression upon treatment, as they would diminish endogenous pools of microRNAs, which generally repress gene expression. To evaluate whether these siRNAs impact endogenous RNAi pathways, we performed whole transcriptome expression analysis using RNA sequencing (RNA-seq) on primary mouse hepatocytes treated with TTR-1194, TTR-1236, TTR-1186 siRNAs or PBS (Figure 8C, Supplementary Figure S7, Supplementary Data set 2). Compared with PBS, we observed relatively minor changes in gene expression upon treatment with stereorandom TTR-1194, including downregulation of 47 genes and upregulation of 3 genes at the highest concentration tested. The gene expression changes observed with TTR-1186, the stereopure PN-containing siRNA, were similar (Figure 8C, Supplementary Figure S7, Supplementary Dataset 2), indicating that the increased Ago2 loading activity exhibited by TTR-1186 does not disrupt endogenous RNAi pathways.
Improved silencing corresponds to lower thermal stability
To further investigate how the inclusion of PN linkages at backbone position 3 of the antisense strand impacts silencing, we assessed the thermal stability of siRNAs without these linkages and those with these linkages for both Reference format 1 and Reference format 3 configurations (Supplementary Table S6). For TTR-5, which is a format 1 stereopure siRNA that lacks PN linkages, the measured Tm was 69.6 °C. Adding a PN linkage at position 3 in either an Sp (TTR-718) or Rp configuration (TTR-696) lowered Tm by ∼1°C. For TTR-872, which is a format 3 stereopure siRNA that lacks PN linkages and has a 2′-OMe on the 3′-side of the third backbone position, the measured Tm was 64.4°C. Adding a PN linkage at position 3 in either an Sp (TTR-937) or an Rp configuration (TTR-1034) preserved or increased Tm compared with TTR-872. By contrast for TTR-1237, which is format 3 with the more favorable sugar pattern (having a 2′-F on the 3′-side of the third backbone position), the Tm decreased by ∼1°C by the addition of a PN linkage in either an Sp (TTR-1165) or Rp configuration (TTR-1147). These data demonstrate that the inclusion of PN linkages in format 1 increases thermal instability, and comparable increases in thermal instability for format 3 are observed if the PN linkage is accompanied by an adjacent 2′-F ribose modification but not a 2′-OMe modification 3′ of the linkage.
PN-containing siRNAs targeting Ttr are well-tolerated in wild-type mice following subcutaneous injections
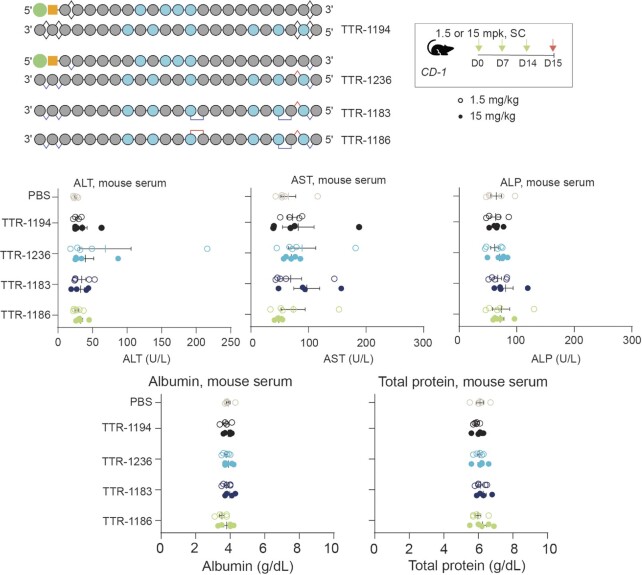
Because siRNAs with PN modifications have not been tested in vivo before, we evaluated the impact of these molecules on a series of serum biomarkers that provide insight into the health of the liver including alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP) albumin and total protein levels. For these experiments, we treated wild-type mice with three weekly low (1.5 mg/kg) or high doses (15 mg/kg) of siRNA, including Reference 3, TTR-1236, TTR-1183, and TTR-1186, or PBS, subcutaneously. Serum was evaluated one day after the final dose. None of the siRNAs had a significant impact on the liver biomarkers compared with PBS (Figure 9). These data indicate that these Ttr-targeting PN-containing siRNAs are well tolerated in wild-type mice at doses up to 15 mg/kg.
Figure 9.
PN-based RNAi format has nominal impact on liver function or serum proteins in mice and is specific in primary hepatocytes. General design for siRNAs tested and dosing regimen for evaluation of liver function in wild-type mice. Graphs show liver enzyme function for alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), albumin and total serum proteins in CD-1 mice treated with the indicated siRNA or PBS. Each symbol represents one mouse and data are presented as mean ± sd, n = 5 per group.
Application of PN chemistry to HSD17B13 silencing
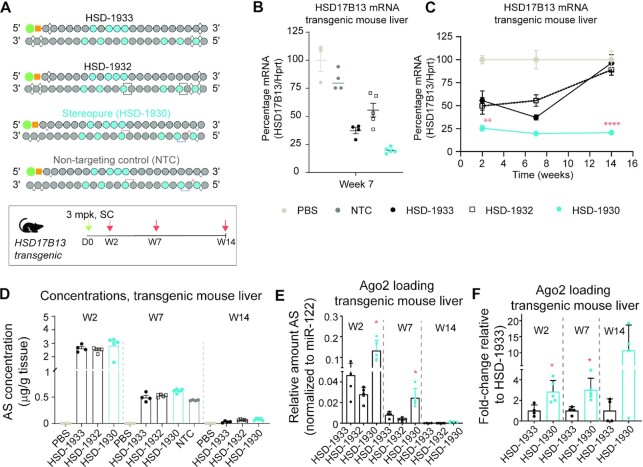
We assessed whether the design strategy for the deployment of PN chemistry with appropriately positioned 2′-ribose modifications we developed for Ttr was beneficial for another target with an unrelated sequence. For these experiments, we opted to develop siRNAs for the clinically relevant liver gene Hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13) (43). We identified a new sequence amenable to silencing via siRNAs that is comparable in potency to a previously reported sequence (https://patents.google.com/patent/WO2019183164A1/en?oq = WO±2019%2f183164) (Supplementary Figure S8A). Experiments in primary human hepatocytes confirmed that a GalNAc-siRNA with chemistry format based on one of our best-performing Ttr siRNAs (TTR-1186) that targets HSD17B13, HSD-1930, is more potent than stereorandom siRNAs designed based on HSD-1933 (HSD-1592) or with stereorandom PN linkages (HSD-1932) (Supplementary Figure S8B: IC50 HSD-1933 759 pM; HSD-1932 1836 pM; HSD-1930 557 pM).To confirm these benefits translate in vivo, we generated mice expressing a human HSD17B13 transgene and evaluated siRNA-mediated silencing of HSD17B13 mRNA expression in the liver.
We tested three GalNAc-siRNAs targeting the same sequence: one designed according to TTR-1194 (HSD-1933), HSD-1932, and HSD-1930, as well as a NTC (Figure 10A). We injected transgenic mice with a single 3 mg/kg dose of the indicated siRNA and evaluated HSD17B13 mRNA expression in the liver over time. At week 7, the control, HSD-1933, led to a mean silencing of ∼60% HSD17B13 mRNA in liver. HSD-1932 performed slightly worse (mean silencing: ∼45%), and HSD-1930 performed the best (mean silencing: ∼80%) (Figure 10B). By 14-weeks post-dose, these differences increased, with HSD17B13 mRNA levels recovering in HSD-1933-treated samples (mean silencing: ∼5%) but remaining significantly suppressed in HSD-1930-treated samples (mean silencing: ∼80%, P < 0.0001, Two-way ANOVA with post-hoc test) (Figure 10C). Once again, we evaluated the concentration of antisense strands and Ago2 loading in the mouse liver. The differences in antisense strand liver concentrations were nominal at all time points (Figure 10D). By contrast, Ago2 loading differences were substantial, with significantly more HSD-1930 than HSD-1933 found in complex with Ago2 at 2-weeks and 7-weeks post-dose (Figure 10E,F). Although Ago2 loading decreased for all antisense strands through the experiment, by the final 14-week time point, ∼10-fold more HSD-1930 was loaded onto Ago2 than HSD-1933 (Figure 10E,F; mean relative to miR-122, HSD-1933 1.03 × 10−4; HSD-1930 1.10 × 10−3), indicating that differences in silencing were likely driven, at least in part, to an Ago2-loading benefit for HSD-1930.
Figure 10.
PN-based GalNAc-siRNA format translates to an additional liver target. (A) Schematic representation of siRNAs tested in the study. Dosing regimen for transgenic HSD17B13 mice, with dosing at day 0 (D0, green arrow) and mouse sacrifice and tissue evaluation at the indicated times (orange arrows) in weeks (W) post-dose. (B, C) HSD17B13 mRNA levels in liver of transgenic mice treated with the indicated siRNA 7-weeks post dose (B) or for the entire 14-week experiment (C). Data in B are presented as scatter plots with lines denoting mean mRNA levels. Data in C are presented as mean ± sem, n = 5. Stats: Two-way ANOVA with post-hoc comparison between HSD-1933 and HSD-1930 at each time point. ** P < 0.01, **** P < 0.0001. (D) Concentration of antisense strands in livers 2-, 7- or 14-weeks post-dose for the indicated treatment. Data are presented as mean ± sd, n ≥ 3. (E) Relative Ago2 loading for antisense strands in mouse livers over time with normalization to miR-122. Data are presented as mean ± sd, n = 5. Stats: Two-way ANOVA with post-hoc comparisons between HSD-1930 and HSD-1933 at each time point. (F) Fold-change Ago2 loading for HSD-1933 compared to HSD-1930 in mouse livers over time after normalization to miR-122. Data are presented as mean ± sd, n = 5.
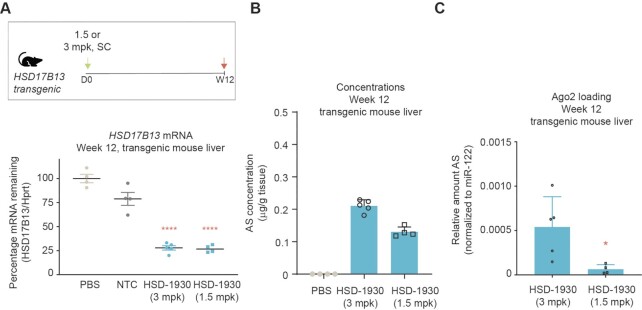
Because HSD17B13 silencing persisted at the longest timepoint after treatment with 3 mg/kg HSD-1930, we evaluated this same siRNA at a lower dose, treating HSD17B13 transgenic mice with 1.5 or 3 mg/kg of HSD-1930 and evaluating silencing 3 months later (Figure 11). At both doses, HSD-1930 led to mean HSD17B13 silencing ∼75% after 3 months (Figure 11A), suggesting that HSD-1930 has saturated the system, and a lower 1.5 mg/kg dose is sufficient to maximize silencing. In the liver, we detected comparable amounts of antisense strand 3-months post-dose at both doses (Figure 11B), and ∼8-fold more antisense strand loaded onto Ago2 from the mice treated with the higher dose (Figure 11C). Taken together, these data support the notion that HSD-1930 is a potent silencer of HSD17B13 expression in part due to an Ago2-loading benefit over comparator siRNAs.
Figure 11.
HSD17B13 silencing with low, single dose. (A) Dosing regimen for transgenic HSD17B13 mice, with dosing at day 0 (D0) and tissue evaluation at the indicated time in weeks (W) post-dose. HSD17B13 mRNA levels in liver of transgenic mice treated with the indicated siRNA at 1.5 or 3.0 mg/kg 12-weeks post-dose. Data are presented as scatter plots with lines denoting mean mRNA levels, n ≥ 4. Stats: One-way ANOVA with post-hoc test comparison to PBS ****P < 0.0001. (B) Concentration of antisense strands in livers 12-weeks post-dose for the indicated treatment. Data are presented as mean ± sd, n ≥ 4. (C) Relative Ago2 loading for antisense strands in mouse livers over time with normalization to miR-122. Data are presented as mean ± sd, n = 5. Stats: Unpaired, two-tailed t test * P < 0.05.
Multiple PN modifications maintain silencing by RNAi
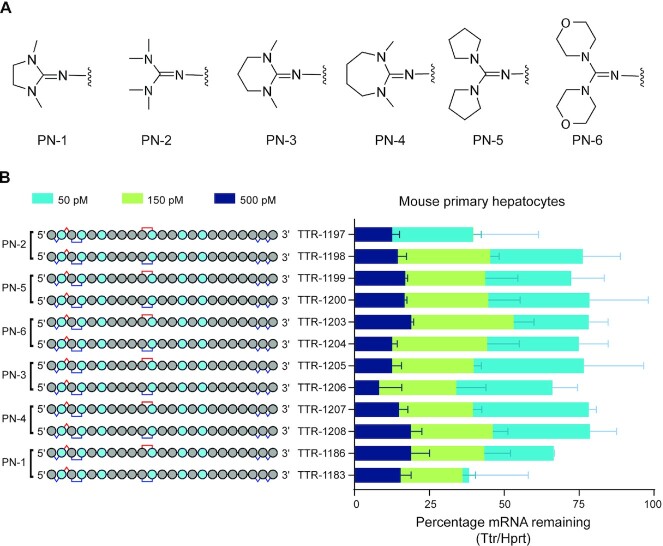
Finally, we explored whether other types of PN backbone linkages could have the same impact as the linkage used throughout this work (PN-1) on silencing by generating a series of stereopure PN-modified siRNAs based on TTR-1183 and TTR-1186. These stereopure siRNAs employ the same sense strand (shown in Figure 8), and the antisense strands have the same sequence, chemistry, and pattern of backbone modification. The only elements that changed from one antisense strand to the next was the chemical moiety deployed as the PN modification and the chiral configuration of that moiety. We explored the impact of simpler PN modification (PN-2), as well as ring size in five-(PN-1), six-(PN-3) and seven-membered rings (PN-4), and exocyclic rings (PN-5, PN-6) (Figure 12A). In cultured mouse primary hepatocytes, the siRNAs led to dose-dependent decreases in Ttr mRNA expression, with expression levels generally similar to TTR-1183 or TTR-1186 (Figure 12B). These data indicate that multiple variations of the PN-1 backbone may convey a silencing benefit when incorporated into the antisense strand of a stereopure siRNA.
Figure 12.
Assessment of multiple PN backbone chemistries. (A) Chemical structures of PN backbone variants. (B) Mean Ttr expression in mouse primary hepatocytes after gymnotic treatment with the indicated siRNA. Data are presented as mean ± sd, n = 2 per siRNA per concentration.
DISCUSSION
Impact of PS chirality
We have previously shown the importance of controlling backbone chemistry and chirality for other oligonucleotide modalities such as RNase H-mediated silencing (11,28,44), splice modulation (24), and RNA base editing with ADAR (25), and we now extend the application of these finding from our initial work on siRNA-mediated silencing (https://patents.google.com/patent/WO2014012081A2/en?oq=WO2014012081) and the subsequent work of others (31,32) to confirm control over PS chirality improves siRNA-mediated silencing. Previously published reports limited evaluation of chirality to a single PS linkage at the terminals or a small number of stereoisomers (31,32). By contrast, this report more thoroughly investigates the impact of control over PS stereochemistry by deploying high-throughput synthesis methods, which enable us to test many more iterations of stereochemistry. We demonstrate that a 3′-SpSp PS configuration in the antisense strand supports more silencing activity than a 3′-RpRp PS configuration likely due to steric interference between the Rp PS linkages and Ago2 and the creation of new electrostatic interactions between Ago2 and the Sp PS linkage at the terminal linkage (P22). We confirm that a 5′-RpSp PS in the antisense strand is favored in an siRNA format with high 2′-F content and without PN linkages. We also show that an Sp PS configuration on both ends of the sense strand is favored, which is consistent with the Sp configuration being more resistant to nucleases than Rp PS (11,12,27). We believe this work in combination with growing evidence in the literature (11,23,24,25,28,29,31,32,45–47) should put to rest questions about whether control over the chirality of PS linkages can impact the biological activity of oligonucleotides.
Chirality, position of PN backbone and interaction with 2′-ribose modifications influence silencing
In contrast to prior reports (48), we found that the application of PN chemistry to the siRNA modality enhanced both potency and durability. Enhancing the activity of the comparator siRNA formats used in this work was challenging, as these designs were optimized over decades (6,7,49,50). The enhanced potency and durability observed with stereopure GalNAc-siRNAs designed to silence HSD17B13 RNA expression that contain PN chemistry was particularly promising, with a single 1.5 mg/kg subcutaneous dose leading to ∼75% silencing for at least 3 months in mice. These advances were possible because of the general high-throughput synthesis methods we developed for generating stereopure chimeric backbones that enabled facile interrogation of hundreds of molecules. With these interrogations, we discovered PN backbone chemistry impacted silencing activity based on three factors: i) chirality; ii) position; and iii) nearby 2′-ribose modifications.
Based on our modelling and in vitro silencing assays, we believe Rp PN linkages in the seed region interfere with antisense-Ago2 interactions, whereas Sp PN linkages in the seed region preserve antisense-Ago2 interactions, with the exceptions of positions 5 and 6 (P5 and P6), where an Sp PN linkage is predicted to disrupt a H bond and where we observed decreased in vitro silencing activity compared with a PO linkage. These findings may help to explain why one configuration of the PN backbone works better than the other and why others who interrogated the impact of stereorandom PN-containing siRNAs, which would likely incorporate Rp PN linkages in the seed region, concluded that the linkage was not suitable for use in the antisense strand due to steric interference with RNAi machinery (48). Based on these findings we opted for an Sp PN linkage at position 3 (P3) in the seed region. We also found that an Sp or Rp PN linkage in the central region at position 10 (P10) of the antisense strand yielded excellent silencing activity. Finally, we also uncovered beneficial interactions between the PN backbone and adjacent sugar modifications, with 2′-F modifications in the position 3′ to the PN linkage supporting more silencing activity than a 2′-OMe modification.
Thermal instability may drive enhanced Ago2 loading
The introduction of PN linkages near the 5′-end (P3) and in the middle of the antisense strand (P10) improved the potency and durability of GalNAc-siRNAs targeting both Ttr and HSD17B13 in vitro and in vivo. That these design strategies are generalizable is supported by the observation that the same design pattern had beneficial effects on two unrelated transcripts. For the P3 position, the incorporation of PN linkages decreased the thermal stability of the siRNA duplex in accordance with observed improvements in silencing and Ago2 loading. For Reference format 3, this destabilizing effect was observed when a 2′-F modification but not a 2′-OMe modification was located 3′ to the linkage. These observations align with reports indicating that each PN substitution in a modified backbone is predicted to reduce the thermal stability by an average of 1.2 °C (48), and PN linkages adjacent to a 2′-F modification have a more reliable destabilizing effect than PN linkages alone (48).
For siRNAs targeting Ttr and HSD17B13, the PN-containing designs improved Ago2 loading compared with reference formats. Both reference formats featured optimized 2′-ribose modification patterns that enhanced metabolic stability but not Ago2 loading (6,7). The observation that PN chemistry especially with appropriately positioned 2′-F modifications enhanced Ago2 loading is consistent with the hypotheses of others (50,51) who predicted that thermal instability in the seed could increase activity by improving duplex unwinding, strand selection, and/or RISC activation. By destabilizing the 5′-end of the antisense strand, the Sp PN backbone linkage may improve the efficiency of Ago2 loading by promoting selection of the antisense strand over the sense strand in the right orientation (5′-end over 3′-end) to enable silencing. Although the Rp PN backbone linkage would also increase thermal instability of the 5′-end of the antisense strand, it is also predicted to interfere sterically with Ago2, which would undermine any benefit for Ago2 loading.
Chemical modifications to siRNAs are known to impact intracellular trafficking, and different endosomal classes are associated with distinct trafficking patterns, with some endosomes leading to distinct intracellular locations and other recycling to the plasma membrane (52). GalNAc conjugation is known to extend the durability of silencing by siRNAs by directing them to acidic intracellular compartments that can serve as long-term depots that enable continuous RNAi activity over time (53). PN backbone modification has been shown to increase the intracellular stability of single-stranded oligonucleotides taken up by muscle cells (24). Thus, PN linkages could also improve Ago2 loading by ensuring more successful intracellular trafficking of siRNAs to the rough endoplasmic reticulum (ER), where RISC is activated (54). This effect could be direct, by enhancing productive trafficking to the rough ER, or indirect, by enhancing stable release from acidic intracellular depots that enable durable silencing.
Importantly, the improved Ago2 loading observed with PN-containing siRNAs did not disrupt endogenous RNAi pathways, as off-target gene expression changes in the presence of these siRNAs were minimal in hepatocytes. This suggests there may be ample untapped capacity in endogenous RNAi pathways to further improve siRNA-mediated silencing.
Implications for oligonucleotide therapeutics
With this work, we demonstrate that the judicious use of stereopure PN backbone chemistry can improve on the activity of siRNAs inspired by designs with a clinically proven track record (50), and we add to the evidence that PN chemistry has a profound impact on the pharmacologic properties of stereopure PS-modified oligonucleotides (23,24,25). Thus, this work highlights the importance of developing methodologies to control backbone chirality, as well as developing synthetic chemistries that are compatible across backbones and ribose modifications, enabling the synthesis of chimeric stereopure molecules. Through the study of these molecules, we have discovered that the PS and PN backbones convey distinct benefits. PS linkages in combination with 2′-ribose modifications increase the metabolic stability of siRNAs (6,7). We now show that stereopure Sp PS linkages on the 3′-end of the antisense strand may create favorable interactions with Ago2 that improve loading. We also show that Sp PN linkages at P3 increase the thermal instability of siRNAs to drive Ago2-loading. Because these advantageous modifications impact silencing activity in complementary ways, they can be combined to make molecules that are superior to those containing any of these modifications alone. Although work remains to fully understand the mechanisms by which PN linkages can improve oligonucleotide activity, work across modalities promises to provide insights into properties of these molecules that are universal (i.e. independent of modality) and those that are unique (i.e. specific to the biological mechanism).
In 2021, the first PN-containing molecules advanced into clinical trials (44) (https://ir.wavelifesciences.com/news-releases/news-release-details/wave-life-sciences-announces-initiation-dosing-phase-1b2a-focus). Preliminary data from these ongoing trials suggest the pharmacologic benefits of PN chemistry may be translating, as low, single doses of investigational WVE-004 (44) showed evidence for target engagement in the central nervous system (https://ir.wavelifesciences.com/news-releases/news-release-details/wave-life-sciences-announces-positive-update-ongoing-phase-1b2a). These early clinical observations with PN-modified molecules bolster our enthusiasm for the longer-term impact of PN chemistry on RNAi. Similar to other modalities, the inclusion of PN linkages improved both the potency and durability of GalNAc-siRNAs in preclinical models. These observations in combination with evidence indicating that PN-containing siRNAs do not interfere with endogenous RNAi pathways and can be well-tolerated in the liver suggest that PN-containing GalNAc-siRNAs may also be suitable for therapeutic application.
DATA AVAILABILITY
Source data for Figures 1B-D, 2B, 3B-C, 4A-B, 5B-D, 6B, 7B-C, 8A-C, 9, 10B-E, 11A-C, 12B and Supplementary Figs. S1, S4A, S5, S6A-B and S8A-B are provided with the paper. RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE220502 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE220502).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are also grateful to Amy Donner (Wave Life Sciences) for writing, editorial and graphical support for this paper.
Contributor Information
Wei Liu, Wave Life Sciences, Cambridge, MA 02138, USA.
Naoki Iwamoto, Wave Life Sciences, Cambridge, MA 02138, USA.
Subramanian Marappan, Wave Life Sciences, Cambridge, MA 02138, USA.
Khoa Luu, Wave Life Sciences, Cambridge, MA 02138, USA.
Snehlata Tripathi, Wave Life Sciences, Cambridge, MA 02138, USA.
Erin Purcell-Estabrook, Wave Life Sciences, Cambridge, MA 02138, USA.
Juili Dilip Shelke, Wave Life Sciences, Cambridge, MA 02138, USA.
Himali Shah, Wave Life Sciences, Cambridge, MA 02138, USA.
Anthony Lamattina, Wave Life Sciences, Cambridge, MA 02138, USA.
Qianli Pan, Wave Life Sciences, Cambridge, MA 02138, USA.
Brett Schrand, Wave Life Sciences, Cambridge, MA 02138, USA.
Frank Favaloro, Wave Life Sciences, Cambridge, MA 02138, USA.
Mugdha Bedekar, Wave Life Sciences, Cambridge, MA 02138, USA.
Arindom Chatterjee, Wave Life Sciences, Cambridge, MA 02138, USA.
Jigar Desai, Wave Life Sciences, Cambridge, MA 02138, USA.
Tomomi Kawamoto, Wave Life Sciences, Cambridge, MA 02138, USA.
Genliang Lu, Wave Life Sciences, Cambridge, MA 02138, USA.
Jake Metterville, Wave Life Sciences, Cambridge, MA 02138, USA.
Milinda Samaraweera, Wave Life Sciences, Cambridge, MA 02138, USA.
Priyanka Shiva Prakasha, Wave Life Sciences, Cambridge, MA 02138, USA.
Hailin Yang, Wave Life Sciences, Cambridge, MA 02138, USA.
Yuan Yin, Wave Life Sciences, Cambridge, MA 02138, USA.
Hui Yu, Wave Life Sciences, Cambridge, MA 02138, USA.
Paloma H Giangrande, Wave Life Sciences, Cambridge, MA 02138, USA.
Michael Byrne, Wave Life Sciences, Cambridge, MA 02138, USA.
Pachamuthu Kandasamy, Wave Life Sciences, Cambridge, MA 02138, USA.
Chandra Vargeese, Wave Life Sciences, Cambridge, MA 02138, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wave Life Sciences. Funding for open access charge: Wave Life Sciences.
Conflict of interest statement. All authors are employees of Wave Life Sciences or were employees of Wave Life Sciences during completion of this work.
REFERENCES
- 1. Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 2. Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T.. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498. [DOI] [PubMed] [Google Scholar]
- 3. Dong Y., Siegwart D.J., Anderson D.G.. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug. Deliv. Rev. 2019; 144:133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L.et al.. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018; 379:11–21. [DOI] [PubMed] [Google Scholar]
- 5. Sehgal A., Barros S., Ivanciu L., Cooley B., Qin J., Racie T., Hettinger J., Carioto M., Jiang Y., Brodsky J.et al.. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat. Med. 2015; 21:492–497. [DOI] [PubMed] [Google Scholar]
- 6. Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S.et al.. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017; 45:10969–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M.et al.. Advanced siRNA Designs Further Improve In Vivo Performance of GalNAc-siRNA Conjugates. Mol. Ther. 2018; 26:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guenther D.C., Mori S., Matsuda S., Gilbert J.A., Willoughby J.L.S., Hyde S., Bisbe A., Jiang Y., Agarwal S., Madaoui M.et al.. Role of a “Magic” methyl: 2′-deoxy-2′-α-F-2′-β-C-methyl pyrimidine nucleotides modulate RNA interference activity through synergy with 5′-phosphate mimics and mitigation of off-target effects. J Am Chem Soc. 2022; 144:14517–14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whitesell L., Geselowitz D., Chavany C., Fahmy B., Walbridge S., Alger J.R., Neckers L.M.. Stability, clearance, and disposition of intraventricularly administered oligodeoxynucleotides: implications for therapeutic application within the central nervous system. Proc. Nat. Acad. Sci. U.S.A. 1993; 90:4665–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014; 24:374–387. [DOI] [PubMed] [Google Scholar]
- 11. Iwamoto N., Butler D.C.D., Svrzikapa N., Mohapatra S., Zlatev I., Sah D.W.Y., Meena, Standley S.M., Lu G., Apponi L.H.et al.. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:845–851. [DOI] [PubMed] [Google Scholar]
- 12. Krieg A.M., Guga P., Stec W.. P-chirality-dependent immune activation by phosphorothioate CpG oligodeoxynucleotides. Oligonucleotides. 2003; 13:491–499. [DOI] [PubMed] [Google Scholar]
- 13. Koziołkiewicz M., Wójcik M., Kobylańska A., Karwowski B., Rebowska B., Guga P., Stec W.J.. Stability of stereoregular oligo(nucleoside phosphorothioate)s in human plasma: diastereoselectivity of plasma 3′-exonuclease. Antisense Nucleic Acid Drug Dev. 1997; 7:43–48. [DOI] [PubMed] [Google Scholar]
- 14. Stec W.J., Grajkowski A., Koziolkiewicz M., Uznanski B.. Novel route to oligo(deoxyribonucleoside phosphorothioates). Stereocontrolled synthesis of P-chiral oligo(deoxyribonucleoside phosphorothioates). Nucleic Acids Res. 1991; 19:5883–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stec W.J., Karwowski B., Boczkowska M., Guga P., Koziołkiewicz M., Sochacki M., Wieczorek M.W., Błaszczyk J.. Deoxyribonucleoside 3′-O-(2-Thio- and 2-Oxo-“spiro”-4,4-pentamethylene-1,3,2-oxathiaphospholane)s: Monomers for Stereocontrolled Synthesis of Oligo(deoxyribonucleoside phosphorothioate)s and Chimeric PS/PO Oligonucleotides. J. Am. Chem. Soc. 1998; 120:7156–7167. [Google Scholar]
- 16. Guo M., Yu D., Iyer R.P., Agrawal S.. Solid-phase stereoselective synthesis of 2′-O-methyl-oligoribonucleoside phosphorothioates using nucleoside bicyclic oxazaphospholidines. Bioorg. Med. Chem. Lett. 1998; 8:2539–2544. [DOI] [PubMed] [Google Scholar]
- 17. Iyer R.P., Yu D., Ho N.-H., Tan W., Agrawal S.. A novel nucleoside phosphoramidite synthon derived from 1R, 2S-ephedrine. Tetrahedron Asymmetry. 1995; 6:1051–1054. [Google Scholar]
- 18. Wilk A., Grajkowski A., Phillips L.R., Beaucage S.L.. Deoxyribonucleoside Cyclic N-Acylphosphoramidites as a New Class of Monomers for the Stereocontrolled Synthesis of Oligothymidylyl- and Oligodeoxycytidylyl- Phosphorothioates. J. Am. Chem. Soc. 2000; 122:2149–2156. [Google Scholar]
- 19. Nukaga Y., Yamada K., Ogata T., Oka N., Wada T.. Stereocontrolled solid-phase synthesis of phosphorothioate oligoribonucleotides using 2′-O-(2-cyanoethoxymethyl)-nucleoside 3′-O-oxazaphospholidine monomers. J. Org. Chem. 2012; 77:7913–7922. [DOI] [PubMed] [Google Scholar]
- 20. Oka N., Kondo T., Fujiwara S., Maizuru Y., Wada T.. Stereocontrolled synthesis of oligoribonucleoside phosphorothioates by an oxazaphospholidine approach. Org. Lett. 2009; 11:967–970. [DOI] [PubMed] [Google Scholar]
- 21. Nukaga Y., Oka N., Wada T.. Stereocontrolled solid-phase synthesis of phosphate/phosphorothioate (PO/PS) chimeric oligodeoxyribonucleotides on an automated synthesizer using an oxazaphospholidine-phosphoramidite method. J. Org. Chem. 2016; 81:2753–2762. [DOI] [PubMed] [Google Scholar]
- 22. Oka N., Yamamoto M., Sato T., Wada T.. Solid-phase synthesis of stereoregular oligodeoxyribonucleoside phosphorothioates using bicyclic oxazaphospholidine derivatives as monomer units. J Am Chem Soc. 2008; 130:16031–16037. [DOI] [PubMed] [Google Scholar]
- 23. Kandasamy P., Liu Y., Aduda V., Akare S., Alam R., Andreucci A., Boulay D., Bowman K., Byrne M., Cannon M.et al.. Impact of guanidine-containing backbone linkages on stereopure antisense oligonucleotides in the CNS. Nucleic Acids Res. 2022; 50:5401–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kandasamy P., McClorey G., Shimizu M., Kothari N., Alam R., Iwamoto N., Kumarasamy J., Bommineni G.R., Bezigian A., Chivatakarn O.et al.. Control of backbone chemistry and chirality boost oligonucleotide splice switching activity. Nucleic Acids Res. 2022; 50:5443–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Monian P., Shivalila C., Lu G., Shimizu M., Boulay D., Bussow K., Byrne M., Bezigian A., Chatterjee A., Chew D.et al.. Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat. Biotechnol. 2022; 40:1093–1102. [DOI] [PubMed] [Google Scholar]
- 26. Huang Y., Knouse K.W., Qiu S., Hao W., Padial N.M., Vantourout J.C., Zheng B., Mercer S.E., Lopez-Ogalla J., Narayan R.et al.. A P(V) platform for oligonucleotide synthesis. Science (New York, N.Y.). 2021; 373:1265–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koziolkiewicz M., Krakowiak A., Kwinkowski M., Boczkowska M., Stec W.J.. Stereodifferentiation–the effect of P chirality of oligo(nucleoside phosphorothioates) on the activity of bacterial RNase H. Nucleic Acids Res. 1995; 23:5000–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Byrne M., Vathipadiekal V., Apponi L., Iwamoto N., Kandasamy P., Longo K., Liu F., Looby R., Norwood L., Shah A.et al.. Stereochemistry enhances potency, efficacy, and durability of Malat1 antisense oligonucleotides in vitro and in vivo in multiple species. Transl. Vis. Sci. Technol. 2021; 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y., Dodart J.C., Tran H., Berkovitch S., Braun M., Byrne M., Durbin A.F., Hu X.S., Iwamoto N., Jang H.G.et al.. Variant-selective stereopure oligonucleotides protect against pathologies associated with C9orf72-repeat expansion in preclinical models. Nat Commun. 2021; 12:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jahns H., Roos M., Imig J., Baumann F., Wang Y., Gilmour R., Hall J.. Stereochemical bias introduced during RNA synthesis modulates the activity of phosphorothioate siRNAs. Nat. Commun. 2015; 6:6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakamuri S., Eltepu L., Liu D., Lam S., Meade B.R., Liu B., Dello Iacono G., Kabakibi A., Luukkonen L., Leedom T.et al.. Impact of phosphorothioate chirality on double-stranded siRNAs: a systematic evaluation of stereopure siRNA designs. Chembiochem. 2020; 21:1304–1308. [DOI] [PubMed] [Google Scholar]
- 32. Jahns H., Taneja N., Willoughby J.L.S., Akabane-Nakata M., Brown C.R., Nguyen T., Bisbe A., Matsuda S., Hettinger M., Manoharan R.M.et al.. Chirality matters: stereo-defined phosphorothioate linkages at the termini of small interfering RNAs improve pharmacology in vivo. Nucleic Acids Res. 2022; 50:1221–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tataurov A.V., You Y., Owczarzy R.. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008; 133:66–70. [DOI] [PubMed] [Google Scholar]
- 34. Pei Y., Hancock P.J., Zhang H., Bartz R., Cherrin C., Innocent N., Pomerantz C.J., Seitzer J., Koser M.L., Abrams M.T.et al.. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010; 16:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng A., Li M., Liang Y., Wang Y., Wong L., Chen C., Vlassov A.V., Magdaleno S.. Stem-loop RT-PCR quantification of siRNAs in vitro and in vivo. Oligonucleotides. 2009; 19:203–208. [DOI] [PubMed] [Google Scholar]
- 36. Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T., Ohl P., Sieb C., Thiel K., Wiswedel B.. Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R.. Data Analysis, Machine Learning and Applications. Studies in Classification, Data Analysis, and Knowledge Organization. 2008; Berlin, Heidelberg: Springer; 319–326. [Google Scholar]
- 37. Fox J., Weisberg S.. An {R} Companion to Applied Regression. 2019; 3rd edn.Thousand Oaks, CA: Sage Publications. [Google Scholar]
- 38. Kuznetsova A., Brockhoff P.B., Christensen R.H.B.. lmerTest Package: tests in linear mixed effects models. J. Stat. Softw. 2017; 82:1–26. [Google Scholar]
- 39. Hothorn T., Bretz F., Westfall P.. Simultaneous inference in general parametric models. Biom. J. Biometrische Zeitschrift. 2008; 50:346–363. [DOI] [PubMed] [Google Scholar]
- 40. Zeileis A., Köll S., Graham N.. Various versatile variances: an object-oriented implementation of clustered covariances in R. J. Stat. Softw. 2020; 95:1–36. [Google Scholar]
- 41. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science (New York, N.Y.). 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abul-Husn N.S., Cheng X., Li A.H., Xin Y., Schurmann C., Stevis P., Liu Y., Kozlitina J., Stender S., Wood G.C.et al.. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N. Engl. J. Med. 2018; 378:1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Y., Andreucci A., Iwamoto N., Yin Y., Yang H., Liu F., Bulychev A., Hu X.S., Lin X., Lamore S.et al.. Preclinical evaluation of WVE-004, aninvestigational stereopure oligonucleotide forthe treatment of C9orf72-associated ALS or FTD. Mol. Ther. Nucleic Acids. 2022; 28:558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Funder E.D., Albæk N., Moisan A., Sewing S., Koch T.. Refining LNA safety profile by controlling phosphorothioate stereochemistry. PLoS One. 2020; 15:e0232603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hansen H.F., Albaek N., Hansen B.R., Shim I., Bohr H., Koch T.. In vivo uptake of antisense oligonucleotide drugs predicted by ab initio quantum mechanical calculations. Sci. Rep. 2021; 11:6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kiełpiński Ł.J., Funder E.D., Schmidt S., Hagedorn P.H. Characterization of Escherichia coli RNase H discrimination of DNA phosphorothioate stereoisomers. Nucleic Acid Ther. 2021; 31:383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pavlova A.S., Yakovleva K.I., Epanchitseva A.V., Kupryushkin M.S., Pyshnaya I.A., Pyshnyi D.V., Ryabchikova E.I., Dovydenko I.S.. An influence of modification with phosphoryl guanidine combined with a 2′-O-methyl or 2′-fluoro group on the small-interfering-RNA effect. Int. J. Mol. Sci. 2021; 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.J.. Therapeutic siRNA: state of the art. Signal Transduct. Targeted Ther. 2020; 5:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Egli M., Schlegel M.K., Manoharan M.. Acyclic (S)-glycol nucleic acid (S-GNA) modification of siRNAs improves the safety of RNAi therapeutics while maintaining potency. RNA. 2023; 29:402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. [DOI] [PubMed] [Google Scholar]
- 52. Ly S., Navaroli D.M., Didiot M.C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D.et al.. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017; 45:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brown C.R., Gupta S., Qin J., Racie T., He G., Lentini S., Malone R., Yu M., Matsuda S., Shulga-Morskaya S.et al.. Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res. 2020; 48:11827–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stalder L., Heusermann W., Sokol L., Trojer D., Wirz J., Hean J., Fritzsche A., Aeschimann F., Pfanzagl V., Basselet P.et al.. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013; 32:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeileis A. Object-oriented computation of sandwich estimators. J. Stat. Softw. 2006; 16:1–16. [Google Scholar]
- 56. Zeileis A. Econometric computing with HC and HAC covariance matrix estimators. J. Stat. Softw. 2004; 11:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figures 1B-D, 2B, 3B-C, 4A-B, 5B-D, 6B, 7B-C, 8A-C, 9, 10B-E, 11A-C, 12B and Supplementary Figs. S1, S4A, S5, S6A-B and S8A-B are provided with the paper. RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE220502 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE220502).