Abstract
Systemic lupus erythematosus (SLE) is a multifactorial autoimmune disease. The sex hormones estrogen and testosterone may have an influence on the production of antibodies. In addition, the gut microbiota also shows an effect on the onset and progression of SLE. Hence, the molecular interplay between sex hormones in terms of gender difference, gut microbiota and SLE is being clarified day after day. The aim of this review is to investigate the dynamic relationship of the gut microbiota with sex hormones in systemic lupus erythematosus taking into account the bacterial strains shown to be affected, effects of antibiotics and other factors that affect the gut microbiome, which itself strongly affects the pathogenesis of SLE.
Keywords: systemic lupus erythematosus, sex hormones, gut microbiota, autoimmunity, molecular mimicry
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease including both complex and multifactorial components. Systemic lupus erythematosus involves an interplay of environmental factors, genetic factors, sex hormones and factors relating to a deficient immune mechanism and is characterized by the production of antibodies which target the cell nucleus. The etiology remains unknown, but there are key clinical manifestations which are indicative of SLE, including inflammation, vasculitis, vasculopathy and deposition of immune complexes [1].
Sex hormones are also known as gonadocorticoids and include estrogen, progesterone and androgens. Their mechanism of action occurs via a nuclear receptor mediated signaling cascade. Sex hormones play a major role in sexual differentiation, but also exert an important effect on the immune system.
The influence that is implemented on the immune system results in sexual dimorphism. Females tend to produce “more vigorous cellular and more vigorous humoral immune reactions” with a higher resistance to particular infections and “suffer a higher incidence of autoimmune diseases” [2].
Both estrogen and testosterone may have an influence on the production of antibodies/autoantibodies. Testosterone has been shown to decrease the production of antibodies, therefore having an immunosuppressive effect, whereas estrogen increases the production of antibodies, which is immunoenhancing [2].
This enhanced capability of immune reactivity in females applies an effective resistance to infections, but on the contrary also enhances susceptibility to auto-immunity and immune pathogenic effects [3]. Estrogen impairs the process of negative selection of high affinity autoreactive B cells and modulates the Th2 response [3].
Additionally to its effect on B cells, estrogen also plays a role in the induction of T cell homing by increasing the homing marker CCR5 [3]. On the other hand, testosterone modulates the Th1 response and stimulates CD8 cells, while down-regulating several inflammatory factors and up-regulating anti-inflammatory factors.
On the other hand, residing in the human body we have bacteria, viruses and other microbes which comprise the human microbiome. The effect these microbes have on human physiology in both diseased and non-diseased states is enormous, and in healthy adults over 1,000 species of bacteria may be identified [4].
Research concerning the human gut microbiome has exploded in recent years with new information regarding its role in metabolism, immune defense and behavior surfacing [5]. There are several factors which influence the microbiome of the gut, including aging, environmental factors, diet, stress and exposure to certain medications [5].
For these reasons, the interplay between all 3 aspects is being clarified day after day and it is thus our interest to investigate the dynamic relationship of gut microbiota with sex hormones in SLE.
Sex hormones and gut microbiota
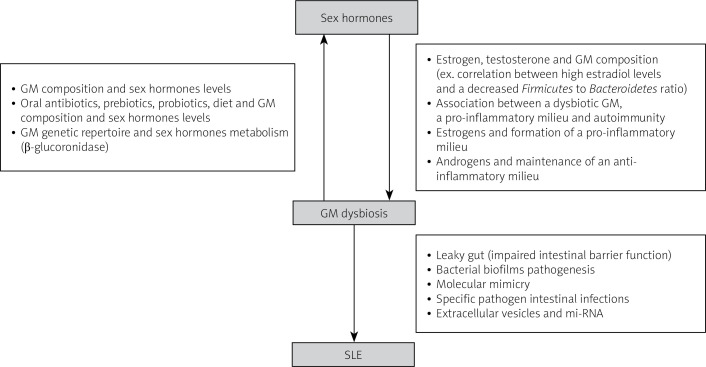
There is a bidirectional interaction between sex hormones and gut microbiota that contributes to sex-based variations in disease pathology, particularly in SLE (Fig. 1).
Fig. 1.
Summary of the proposed interaction between sex hormones and gut microbiota in the pathogenesis of systemic lupus erythematosus.
The feedback loop between gut microbiota and sex hormones affects autoimmunity by triggering either a tolerogenic or inflammatory effect. Investigating specific bacteria with pro-inflammatory or protective roles is needed to better understand these interactions [6].
Several studies have shown that the diversity of gut microbiota is influenced by various factors, including sex differences [7]. Certain bacterial strains, such as Erwinia, Allobaculum, and Anaeroplasma, were found to be more frequent in male mice than in female mice [8].
Testosterone levels in female mice were shown to increase when fecal microbiota was transferred from male to female mice. Hormonal replacement and gonadectomy can substantially alter the gut microbiota composition [9–12].
Major differences in gut microbiota composition between males and females are first seen following puberty onset and can be reversed after male castration. A correlation was observed between hyperandrogenemia and gut microbiota in mice with polycystic ovary syndrome [13–15].
Progesterone was shown to increase the growth of Prevotella intermedius and oral Bacteroides species in a cell-level study performed in the 1980s [16].
Estrogens
The level of estrogens is associated with specific gut microbiota (GM) composition, as shown in several studies on healthy men and women [17, 18]. For instance, an increase in estrogen metabolites correlates with a decrease in the Firmicutes to Bacteroidetes ratio and an increase in the Ruminococcaceae family in healthy women [17].
A direct correlation was found between high estradiol levels in healthy women and a decrease in the number of Firmicutes, and increase in the number of Bacteroidetes, and hence an overall decrease in the Firmicutes to Bacteroidetes ratio [19]
Moreover, there is a link between GM diversity and estrogen levels [20], with a strong association between alpha diversity and total estrogens in healthy men, but not in premenopausal women [18].
In women with polycystic ovary syndrome (PCOS), a correlation was observed between hyperandrogenemia and GM. These findings suggest that sex hormones play a role in shaping the GM and may contribute to sex-based variations in disease pathology.
Several studies performed in healthy women and men led to the finding that there is an association between the level of estrogen and specific GM composition [12, 17–21].
A positive correlation was found between the Subdoligranulum, Sutterella, Agathobacter and Collinsella genera and (E2) estradiol levels in obese women with PCOS [21]. However, a negative correlation was seen with Ruminococcus gnavus and Escherichia-Shigella genera [21].
Testosterone
A study conducted by Insenser et al. [20] showed a correlation between the abundance of the Raoultella genus and the ratio of total testosterone, free testosterone and free testosterone to free estrogen. Also, during the analysis of sex and sex hormones and their effect on GM genera, it turned out that Paraprevotella had an inverse association with estrogen and the strongest positive association with testosterone [20].
Sloan et al. [22] found a positive association between certain bacterial genera such as Escherichia/Shigella, Streptococcus, Alistipes, Rothia, Peptostreptococcus, Bacteroides, Blautia, Para bacteroides and testosterone levels in women with and without PCOS.
Also, an inverse correlation was found between certain genera such as Alistipes, Coprococcus, Akkermansia, Ruminococcus and Collinsella and testosterone levels [23].
Furthermore, a positive association was found between testosterone and the Prevotella genus in nonobese women with PCOS and obese women with PCOS [24]. However, a negative association was found between Eubacterium hallii group, Ruminiclostridium, Subdoligranulum, Lachnospiraceae, Ruminococcaceae, Tyzzerella and testosterone [24]. Moreover, a study was conducted on healthy men and a statistically significant association was found between the diversity of GM and testosterone levels [19].
Another study showed a significant association between the free testosterone to free estradiol ratio and alpha diversity (Shannon and Chaol indexes), and this correlation was also seen with the total testosterone level [20].
In addition, a negative association was found between diversity (PD and richness indexes) and testosterone levels when combining women with PCOS and healthy women [25].
Sex hormones, gut microbiota dynamics and molecular interactions
The genetic repertoire of the gut microbiota that are capable of metabolizing estrogens is called the estrobolome [26]. Estrogens are metabolized from their conjugate forms to their deconjugated forms via microbial secreted β-glucuronidase [27], then, by binding to estrogen α and β receptors, estrogens and phytoestrogens exert their effect and activate downstream intracellular signaling pathways, hence affecting gene expression [28].
The administration of oral antibiotics led to the increase of conjugated estrogen excretion lowering urinary estrogens [29–31]. Also, many in vitro and in vivo animal model studies show the importance of the estrobolome in the metabolism of estrogens [32–34].
Several studies have shown that probiotics and diet were able to alter β-glucuronidase activity, hence showing the importance of taking the diet and probiotics into account in further studies discussing the relationship between sex hormones and GM [35, 36].
Limited information is available regarding the molecular pathways and dynamics between GM and testosterone. The deconjugation of glucuronidated testosterone and dihydrotestosterone into their active/deconjugated form that will enter the bloodstream is facilitated by bacterial β-glucuronidase [37, 38].
Furthermore, in vitro conversion of glucocorticosteroids in the bile into androgens by Clostridium scindens may affect systemic and local testosterone levels in vivo [39, 40].
A dysbiotic GM can lead to a pro-inflammatory mucosal milieu which may lead to autoimmunity. As we previously mentioned, estrogen favors the formation of a pro-inflammatory milieu but androgens help maintain an anti-inflammatory milieu. Thus, diet and probiotics can alter the GM composition and diversity and can change the disease outcome, specifically in males [6].
Gut microbiota and systemic lupus erythematosus
Recently, a link between the gut microbiota and SLE has been proposed [41]. A breakthrough was reported Hevia et al. [42], showing that patients with SLE have restricted GM in comparison with twenty healthy counterparts.
Patients with SLE have an altered gut microbiota composition and microenvironment (differing from the GM of healthy individuals), including high presence of Actinobacteria, Bacteroidetes, and Proteobacteria and lower levels of Firmicutes in comparison with healthy controls [43].
It is important to note that SLE disease activity index is inversely associated with Firmicutes; hence lupus progression can be delayed by Firmicutes presence [44]. In addition, SLE patients have a decrease in the diversity and richness of GM in comparison with healthy controls [45, 46].
In studies done on mice, it was found that levels of Lachnospiraceae increased and Lactobacilli decreased in the GM of Murphy Roths large mice homozygous for the lymphoproliferation spontaneous mutation FASlpr (MRL/lpr) (more severe in females). Also, the intestinal colonization of Lactobacillaceae was negatively associated with SLE activity in mice and lupus progression is strongly correlated with Lachnospiraceae in MRL/lpr mice [47, 48].
Pathways linking GM dysbiosis and SLE pathogenesis include leaky gut (impaired intestinal barrier function), imbalance of the immune system, molecular mimicry, biofilms, sex hormones and others.
Leaky gut hypothesis
The intestinal mucosa has an intestinal barrier that protects it from bacteria, viruses, fungi, toxins, foreign and food antigens [49]. Calcium-containing protein from immune cells such as macrophages and neutrophils is a biomarker for damaged intestinal mucosa (impaired function). Stool samples from SLE patients contained high levels of calprotectin, hence revealing impaired intestinal barrier function [50, 51].
Moreover, many sera soluble markers such as lipopolysaccharides, CD14, and alpha1-acid glycoprotein were high in SLE patients, hence revealing the presence of intestinal bacterial translocation [46]. A study was conducted by Thim-Uam et al. [52] whereby a leaky gut was induced in lupus mice following administration of dextran sulfate solution.
The result was progression and aggravation of SLE disease in the mice because endotoxins and other chemicals were translocated out through the leaky gut, thereby promoting apoptosis [52].
In addition, anti-dsDNA autoantibodies and immune complex deposition were promoted by the leaky gut, leading to aggravation of the disease [53]. Also, after the administration of probiotics, antibiotics or diet change, the gut barrier and lupus were highly ameliorated in lupus mice [54, 55]. Hence, these previous studies confirm that damage to the intestinal barrier can lead or aggravated SLE.
Bacterial biofilm pathogenesis
A biofilm is a membrane that wraps bacteria and protects them from immune recognition and allows bacteria to develop drug resistance [56–58]. Salmonella typhimurium amyloid Curly fibers can bind to DNA in bacteria, hence forming biofilms and contributing to SLE pathogenesis [59].
After intraperitoneal injection of curli-DNA complexes in normal mice, antinuclear antibody (ANA) and anti-dsDNA autoantibodies were formed. Also, curli-DNA complexes cause proliferation of B and T cells and inflammatory monocytes and autoantibody production in lupus mice [59].
Molecular mimicry
When a particular structure of a microorganism is similar to a self-structure of the host, an autoimmune response can occur, leading to damage of host tissue. This is known as molecular mimicry [60]. Hence specific bacteria able to undergo molecular mimicry can lead SLE patients to form cross-reactive autoantibodies.
In a study conducted by Zhang and Reichlin [61], a bacterial antigen of Burkholderia and another transcriptional regulatory peptide were able to bind to dsDNA antibodies in SLE patients’ sera; this leads to the conclusion that the synthesis of anti-dsDNA antibodies in SLE patients is correlated with Burkholderia molecular mimicry.
A study showed that the molecular mimicry of environmental microbes or commensals enhances autoanti-body production in SLE, driven by HLA-DR restriction and T cells [62]. Furthermore, another study showed that mycobacterial cell wall glycolipids can bind to the dsDNA autoantibodies that are derived from SLE patients and mice [63].
An important factor in SLE pathogenesis is bacterial molecular mimicry. Stimulation of autoantibody synthesis can be through molecular mimicry. Also B and T cells are involved in the bacterial molecular mimicry process; however, limited information exists at present [64].
Specific intestinal pathogens
Many reports have associated the onset and progression of SLE and intestinal infections with specific pathogens. More studies are investigating the dynamics and pathogenesis of these specific pathogens.
One of these specific pathogens is Ruminococcus gnavus, which may be able to affect SLE disease progression. A study found an increase in sIgA-coated R. gnavus in stool samples from SLE patients, and the proliferation of the bacteria was directly proportional to the disease activity.
Also, an increase in the calprotectin levels in stool samples and in the lipopolysaccharides in sera was found in SLE patients, due to disrupted function of the intestinal barrier. Furthermore, this leaky gut leads to molecular mimicry and hence the synthesis of anti-dsDNA autoantibodies, hence aggravating SLE [46].
Another specific intestinal bacterial pathogen is Enterococcus gallinarum. The latter organism was found to have an important role in SLE pathogenesis. This bacterium damages the intestinal barrier and promotes Tfh (T follicular helper) and Th17 cell proliferation in F1 lupus mice. Subsequently, it translocates to the liver via the route of mesenteric lymph nodes and mesenteric veins. Also, it is able to enhance autoimmunity by triggering overexpression of ERV gp70 in the liver [51].
In addition, E. gallinarum was detected in liver biopsies of patients with autoimmune hepatitis and SLE, although it was not detected in non-autoimmune hepatitis patients or healthy controls. After the administration of a vaccine against this bacterium, the level of autoanti-bodies was reduced and bacterial translocation was inhibited [51]. This leads to the conclusion that therapy directed against these specific pathogens can suppress the autoimmune reaction.
Extracellular vesicle and miRNA
In a study, mice that were deficient in in IECs miRNA had GM dysbiosis and worsening of colitis, that improved after transplantation with fecal EV-derived miRNA from wild type mice [65]. This led to the conclusion that EV-derived miRNAs are able to control the GM composition and improve the progression of intestinal inflammation.
In addition, a recent study showed that the exosome miR181a relieves colitis by promoting GM homeostasis, decreasing the secretion of the pro-inflammatory factors and normal intestinal barrier function [66].
In summary, the EV-derived miRNA in the intestines can decrease or inhibit the progression of lupus by ameliorating GM homeostasis and intestinal barrier function.
Future perspectives
As we previously discussed, GM dysbiosis is correlated with the development of SLE and its progression. Many pathways and mechanisms can disrupt the homeostasis of the GM microenvironment and lead to GM dysbiosis and aggravation of the disease, such as leaky gut, molecular mimicry, specific pathogen interactions, biofilms, sex hormones, EV-derived miRNA and others.
Since SLE is a multifactorial heterogeneous disease, the therapeutic approach towards SLE via targeting the composition and the microenvironment of the gut microbiota could be a breakthrough. Administration of oral antibiotics could possibly be a future approach; however, symbiosis in other types of bacteria and increased bacterial resistance should be taken into account and studied in more depth.
Another approach could be via prebiotics or probiotics, hence aiming to construct and remodel the GM composition diversity in a way that would help create a bacterial homeostasis and an anti-inflammatory milieu by limiting activation of the adaptive immune system and the release of inflammatory markers and cytokines.
Vaccines directed against specific intestinal pathogens could be an approach, hence aiming to reduce the intestinal barrier damage, increasing its stability and preventing the translocation of organic molecules, antigens and other toxins into the bloodstream, and hence reducing the immune activation and thus suppression of SLE.
Furthermore, this can help prevent molecular mimicry induced by antigens similar to those of the host, hence decreasing immune activation, and can reduce complexes of biofilm proteins and DNA from triggering autoantibody synthesis.
The more we understand the complex pathways between the gut microbiota, sex hormones and the immune system, the more promising will be the novel therapeutic approaches.
Conclusions
This review of SLE pathogenesis reveals that this autoimmune disease is indeed multifactorial with inter-dynamicity of gut microbiota and sex hormones playing a major role in its onset and progression. Whether estrogen or testosterone have positive or negative correlations with the diversity and composition of the gut bacterial strains, this affects the intestinal barrier’s integrity, and hence alters the local immune microenvironment.
Moreover, the remaining mechanisms and pathways that link GM dysbiosis and SLE pathogenesis are numerous, and include leaky guy, bacterial biofilm pathogenesis, molecular mimicry, specific pathogen intestinal infections and extracellular vesicles and miRNA.
Footnotes
The authors declare no conflict of interests.
References
- 1.Mok CC, Lau CS. Pathogenesis of systemic lupus erythematosus. J Clin Pathol 2003; 56: 481–490, DOI: 10.1136/jcp.56.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Hum Reprod Update 2005; 11: 411–423, DOI: 10.1093/humupd/dmi008. [DOI] [PubMed] [Google Scholar]
- 3.Taneja V. Sex Hormones determine immune response. Front Immunol 2018; 9: 1931, DOI: 10.3389/fimmu.2018.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cresci GA, Bawden E. Gut microbiome: what we do and don’t know. Nutr Clin Pract 2015; 30: 734–746, DOI: 10.1177/0884533615609899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol 2015; 31: 69–75, DOI: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez A, Luckey D, Taneja V. The gut microbiome in autoimmunity: sex matters. Clin Immunol 2015; 159: 154–162, DOI: 10.1016/j.clim.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486: 207–214, DOI: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yurkovetskiy L, Burrows M, Khan AA, et al. Gender bias in auto-immunity is influenced by microbiota. Immunity 2013; 39: 400–412, DOI: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Org E, Mehrabian M, Parks BW, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016; 7: 313–322, DOI: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He S, Li H, Yu Z, et al. The gut microbiome and sex hormone-related diseases. Front Microbiol 2021; 12: 711137, DOI: 10.3389/fmicb.2021.711137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wankhade UD, Zhong Y, Lazarenko OP, et al. Sex-specific changes in gut microbiome composition following blueberry consumption in C57BL/6J mice. Nutrients 2019; 11: 313, DOI: 10.3390/nu11020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.d’Afflitto M, Upadhyaya A, Green A, Peiris M. Association between sex hormone levels and gut microbiota composition and diversity-a systematic review. J Clin Gastroenterol 2022; 56: 384–392, DOI: 10.1097/MCG.0000000000001676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Qi Y, Yang X, et al. Association between Polycystic ovary syndrome and gut microbiota. PLoS One 2016; 11: e0153196, DOI: 10.1371/journal.pone.0153196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley ST, Skarra DV, Rivera AJ, Thackray VG. The gut microbiome is altered in a letrozole-induced mouse model of polycystic ovary syndrome. PLoS One 2016; 11: e0146509, DOI: 10.1371/journal.pone.0146509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T, Sha L, Li Y, et al. Dietary α-linolenic acid-rich flaxseed oil exerts beneficial effects on polycystic ovary syndrome through sex steroid hormones-microbiota-inflammation axis in rats. Front Endocrinol (Lausanne) 2020; 11: 284, DOI: 10.3389/fendo.2020.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demirel KJ, Guimaraes AN, Demirel I. Effects of estradiol on the virulence traits of Porphyromonas gingivalis. Sci Rep 2022; 12: 13881, DOI: 10.1038/s41598-022-17019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuhrman BJ, Feigelson HS, Flores R, et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab 2014; 99: 4632–4640, DOI: 10.1210/jc.2014-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores R, Shi J, Fuhrman B, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med 2012; 10: 253, DOI: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin JH, Park YH, Sim Met al. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res Microbiol 2019; 170: 192–201, DOI: 10.1016/j.resmic.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Insenser M, Murri M, Del Campo R, et al. Gut microbiota and the polycystic ovary syndrome: influence of sex, sex hormones, and obesity. J Clin Endocrinol Metab 2018; 103: 2552–2562, DOI: 10.1210/jc.2017-02799. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Ni Z, Cheng W, et al. Characteristic gut microbiota and predicted metabolic functions in women with PCOS. Endocr Connect 2020; 9: 63–73, DOI: 10.1530/EC-19-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sloan TJ, Jalanka J, Major G, at al. A low FODMAP diet is associated with changes in the microbiota and reduction in breath hydrogen but not colonic volume in healthy subjects. PLoS One 2018; 13: e0201410, DOI: 10.1371/journal.pone.0201410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Zhang C, Shi Y, et al. Dysbiosis of gut microbiota associated with clinical parameters in polycystic ovary syndrome. Front Microbiol 2017; 8: 324, DOI: 10.3389/fmicb.2017.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y, Ming Q, Liang J, et al. Gut microbiota dysbiosis in polycystic ovary syndrome: association with obesity – a preliminary report. Can J Physiol Pharmacol 2020; 98: 803–809, DOI: 10.1139/cjpp-2019-0413. [DOI] [PubMed] [Google Scholar]
- 25.Torres PJ, Siakowska M, Banaszewska B, et al. Gut microbial diversity in women with polycystic ovary syndrome correlates with hyperandrogenism. J Clin Endocrinol Metab 2018; 103: 1502–151, DOI: 10.1210/jc.2017-02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Tang H, Chen P, et al. Demystifying the manipulation of host immunity, metabolism, and extraintestinal tumors by the gut microbiome. Signal Transduct Target Ther 2019; 4: 41, DOI: 10.1038/s41392-019-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega MA, Fraile-Martínez O, Naya I, et al. Type 2 diabetes mellitus associated with obesity (diabesity). the central role of gut microbiota and its translational applications. Nutrients 2020; 12: 2749, DOI: 10.3390/nu12092749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rietjens IMCM, Louisse J, Beekmann K. The potential health effects of dietary phytoestrogens. Br J Pharmacol 2017; 174: 1263–1280, DOI: 10.1111/bph.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin F, Peltonen J, Laatikainen T, et al. Excretion of progesterone metabolites and estriol in faeces from pregnant women during ampicillin administration. J Steroid Biochem 1975; 6: 1339–1346, DOI: 10.1016/0022-4731(75)90363-5. [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann P, Curtis N. The effect of antibiotics on the composition of the intestinal microbiota – a systematic review. J Infect 2019; 79: 471–489, DOI: 10.1016/j.jinf.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Adlercreutz H, Pulkkinen MO, Hämäläinen EK, Korpela JT. Studies on the role of intestinal bacteria in metabolism of synthetic and natural steroid hormones. J Steroid Biochem 1984; 20: 217–229, DOI: 10.1016/0022-4731(84)90208-5. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu K, Muranaka Y, Fujimura R, et al. Normalization of reproductive function in germfree mice following bacterial contamination. Exp Anim 1998; 47: 151–158, DOI: 10.1538/expanim.47.151. [DOI] [PubMed] [Google Scholar]
- 33.Järvenpää P, Kosunen T, Fotsis T, Adlercreutz H. In vitro metabolism of estrogens by isolated intestinal micro-organisms and by human faecal microflora. J Steroid Biochem 1980; 13: 345–349, DOI: 10.1016/0022-4731(80)90014-x. [DOI] [PubMed] [Google Scholar]
- 34.Lombardi P, Goldin B, Boutin E, Gorbach SL. Metabolism of androgens and estrogens by human fecal microorganisms. J Steroid Biochem 1978; 9: 795–801, DOI: 10.1016/0022-4731(78)90203-0. [DOI] [PubMed] [Google Scholar]
- 35.Goldin BR, Swenson L, Dwyer J, et al. Effect of diet and Lactobacillus acidophilus supplements on human fecal bacterial enzymes. J Natl Cancer Inst 1980; 64: 255–261, DOI: 10.1093/jnci/64.2.255. [DOI] [PubMed] [Google Scholar]
- 36.McIntosh FM, Maison N, Holtrop G, et al. Phylogenetic distribution of genes encoding β-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ Microbiol 2012; 14: 1876–1887, DOI: 10.1111/j.1462-2920.2012.02711.x. [DOI] [PubMed] [Google Scholar]
- 37.Colldén H, Landin A, Wallenius V, et al. The gut microbiota is a major regulator of androgen metabolism in intestinal contents. Am J Physiol Endocrinol Metab 2019; 317: E1182–E1192, DOI: 10.1152/ajpendo.00338.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li CY, Basit A, Gupta A, et al. Major glucuronide metabolites of testosterone are primarily transported by MRP2 and MRP3 in human liver, intestine and kidney. J Steroid Biochem Mol Biol 2019; 191: 105350, DOI: 10.1016/j.jsbmb.2019.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridlon JM, Ikegawa S, Alves JM, et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res 2013; 54: 2437–2449, DOI: 10.1194/jlr.M038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gare J, Kanoute A, Meda N, et al. Periodontal conditions and pathogens associated with pre-eclampsia: a scoping review. Int J Environ Res Public Health 2021; 18: 7194, DOI: 10.3390/ijerph18137194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Qing P, Yang H, et al. Gut microbiome and metabolites in systemic lupus erythematosus: link, mechanisms and intervention. Front Immunol 2021; 12: 686501, DOI: 10.3389/fimmu.2021.686501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hevia A, Milani C, López P, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 2014; 5: e01548–e01614, DOI: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Z, Shao T, Li H, et al. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog 2016; 8: 64, DOI: 10.1186/s13099-016-0146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He J, Chan T, Hong X, et al. Microbiome and metabolome analyses reveal the disruption of lipid metabolism in systemic lupus erythematosus. Front Immunol 2020; 11: 1703, DOI: 10.3389/fimmu.2020.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen BD, Jia XM, Xu JY, et al. An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol 2021; 73: 232–243, DOI: 10.1002/art.41511. [DOI] [PubMed] [Google Scholar]
- 46.Azzouz D, Omarbekova A, Heguy A, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis 2019; 78: 947–956, DOI: 10.1136/annrheumdis-2018-214856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Liao X, Sparks JB, Luo XM. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol 2014; 80: 7551–7560, DOI: 10.1128/AEM.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mu Q, Tavella VJ, Kirby JL, et al. Antibiotics ameliorate lupus-like symptoms in mice. Sci Rep 2017; 7: 13675, DOI: 10.1038/s41598-017-14223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu Q, Kirby J, Reilly CM, Luo XM. Leaky gut as a danger signal for autoimmune diseases. Front Immunol 2017; 8: 598, DOI: 10.3389/fimmu.2017.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gisbert JP, Bermejo F, Pérez-Calle JL, et al. Fecal calprotectin and lactoferrin for the prediction of inflammatory bowel disease relapse. Inflamm Bowel Dis 2009; 15: 1190–1198, DOI: 10.1002/ibd.20933. [DOI] [PubMed] [Google Scholar]
- 51.Manfredo Vieira S, Hiltensperger M, Kumar V, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018; 359: 1156–1161, DOI: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thim-Uam A, Surawut S, Issara-Amphorn J, et al. Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci Rep 2020; 10: 777, DOI: 10.1038/s41598-019-57275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Wang G, Banerjee N, et al. Aberrant gut microbiome contributes to intestinal oxidative stress, barrier dysfunction, inflammation and systemic autoimmune responses in MRL/lpr mice. Front Immunol 2021; 12: 651191, DOI: 10.3389/fimmu.2021.651191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toral M, Robles-Vera I, Romero M, et al. Lactobacillus fermentum CECT5716: a novel alternative for the prevention of vascular disorders in a mouse model of systemic lupus erythematosus. FASEB J 2019; 33: 10005–10018, DOI: 10.1096/fj.201900545RR. [DOI] [PubMed] [Google Scholar]
- 55.Abdelhamid L, Cabana-Puig X, Swartwout B, et al. Retinoic acid exerts disease stage-dependent effects on pristane-induced lupus. Front Immunol 2020; 11: 408, DOI: 10.3389/fimmu.2020.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costerton JW. Introduction to biofilm. Int J Antimicrob Agents 1999; 11: 217–239, DOI: 10.1016/s0924-8579(99)00018-7. [DOI] [PubMed] [Google Scholar]
- 57.Suh JD, Ramakrishnan V, Palmer JN. Biofilms. Otolaryngol Clin North Am 2010; 43: 521–530, DOI: 10.1016/j.otc.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet 2001; 358: 135–138, DOI: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 59.Gallo PM, Rapsinski GJ, Wilson RP, et al. Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 2015; 42: 1171–1184, DOI: 10.1016/j.immuni.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blank M, Barzilai O, Shoenfeld Y. Molecular mimicry and auto-immunity. Clin Rev Allergy Immunol 2007; 32: 111–118, DOI: 10.1007/BF02686087. [DOI] [PubMed] [Google Scholar]
- 61.Zhang W, Reichlin M. A possible link between infection with burkholderia bacteria and systemic lupus erythematosus based on epitope mimicry. Clin Dev Immunol 2008; 2008: 683489, DOI: 10.1155/2008/683489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Z, Ren J, Dai C, et al. Nature of T cell epitopes in lupus antigens and HLA-DR determines autoantibody initiation and diversification. Ann Rheum Dis 2019; 78: 380–390, DOI: 10.1136/annrheumdis-2018-214125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shoenfeld Y, Zamir R, Joshua H, et al. Human monoclonal anti-DNA antibodies react as lymphocytotoxic antibodies. Eur J Immunol 1985; 15: 1024–1028, DOI: 10.1002/eji.1830151012. [DOI] [PubMed] [Google Scholar]
- 64.Pan Q, Guo F, Huang Y, et al. Gut microbiota dysbiosis in systemic lupus erythematosus: novel insights into mechanisms and promising therapeutic strategies. Front Immunol 2021; 12: 799788, DOI: 10.3389/fimmu.2021.799788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S, da Cunha AP, Rezende RM, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016; 19: 32–43, DOI: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gu L, Ren F, Fang X, et al. Exosomal microRNA-181a derived from mesenchymal stem cells improves gut microbiota composition, barrier function, and inflammatory status in an experimental colitis model. Front Med (Lausanne) 2021; 8: 660614, DOI: 10.3389/fmed.2021.660614. [DOI] [PMC free article] [PubMed] [Google Scholar]