Abstract
Linker H1 histones play an important role in animal and human pathogenesis, but their function in plant immunity is poorly understood. Here, we analyzed mutants of the three canonical variants of Arabidopsis H1 histones, namely H1.1, H1.2 and H1.3. We observed that double h1.1h1.2 and triple h1.1h1.2h1.3 (3h1) mutants were resistant to Pseudomonas syringae and Botrytis cinerea infections. Transcriptome analysis of 3h1 mutant plants showed H1s play a key role in regulating the expression of early and late defense genes upon pathogen challenge. Moreover, 3h1 mutant plants showed enhanced production of reactive oxygen species and activation of mitogen activated protein kinases upon pathogen-associated molecular pattern (PAMP) treatment. However, 3h1 mutant plants were insensitive to priming with flg22, a well-known bacterial PAMP which induces enhanced resistance in WT plants. The defective defense response in 3h1 upon priming was correlated with altered DNA methylation and reduced global H3K56ac levels. Our data place H1 as a molecular gatekeeper in governing dynamic changes in the chromatin landscape of defense genes during plant pathogen interaction.
INTRODUCTION
Plants face constant changes in their homeostasis, so they have evolved complex mechanisms to deal with external abiotic and biotic stresses. Plants can sense microbes by membrane-localized pattern recognition receptors (PRRs) of conserved pathogen/microbe-associated molecular patterns (PAMPs/MAMPs), thereby triggering PAMP/MAMP triggered immunity (PTI/MTI) (1). After recognition, downstream signaling responses are triggered, which include production of reactive oxygen species (ROS), activation of mitogen-activated protein kinases (MAPKs), and activation of defense genes (2). PTI also changes the plant defense hormone profiles (like salicylic acid and jasmonic acid) to optimize the immune response toward pathogens (3). Successful pathogens deliver effector proteins into the plant cells to overcome PTI, causing disease in susceptible plants called effector-triggered susceptibility (ETS). However, resistant plants recognize effectors via intracellular nucleotide-binding domain leucine-rich repeat (NLR) receptors to mount effector triggered immunity (ETI) (4).
In plants, a previous stressful experience modulates the cellular machinery to confer a robust response upon a recurring stress; a phenomenon called priming (5). Priming of plants can efficiently increase the tolerance and survival of plants to different stresses. One of the well-characterized PAMPs is a 22-amino-acid long epitope of Pseudomonas flagellin (flg22), which is recognized by the FLS2 receptor (6). Pretreatment with flg22 induces defense priming in Arabidopsis enhancing plant resistance to subsequent challenge with pathogenic Pseudomonas syringae (7).
The execution of early and late immune responses upon pathogen recognition is fundamentally modulated by optimal gene expression (8,9). Chromatin changes and nucleosome dynamics fine-tune gene expression in stress conditions. Besides core nucleosomal histones, linker histones are major constituents of eukaryotic chromatin (10). In eukaryotes, linker histone H1 has a conserved tripartite structure consisting of (i) a short and flexible N-terminal tail, (ii) a dyad binding central globular domain (GH1) and (iii) a structurally disordered lysine-rich C-terminal tail (11). Linker histone H1 binds to the nucleosome to facilitate chromatin folding usually into higher order structures (12). In addition to regulating basic biological processes like DNA replication, chromosome segregation, and DNA repair, H1 regulates gene expression by modulating RNA polymerase II accessibility to chromatin (12–14).
H1 deposition is part of the crosstalk with the epigenetic landscape, notably DNA methylation and histone H3 methylation (15–17). H1 mutants are difficult to study owing to the high functional redundancy between H1 variants. Deletion of H1 in mouse and Drosophila is lethal but not in other organisms like Tetrahymena, yeast, fungi and worms (12,18). Arabidopsis has three canonical variants of H1 namely, H1.1, H1.2 and stress-regulated H1.3 (19). Interestingly, the h1.1h1.2h1.3 triple mutant (3h1) is viable with no significant developmental defects (17). In Arabidopsis the triple H1 mutant (3h1) was generated by crossing h1.1h1.2 with h1.3 (17,20). In Arabidopsis root and shoot tissues, H1.1 and H1.2 are ubiquitously expressed during vegetative development. At the genome level, H1 is distributed in heterochromatin and euchromatin showing enrichment at 3′ and 5′ ends of transposable elements (TEs) (21,22). Over gene bodies, H1 presence is usually anticorrelated to transcription and H3K4me3 levels (21). Detailed microscopic imaging reveled that the euchromatin in 3h1 nuclei shows reduced homogeneity with spatially distributed chromatin nanodomains as compared to a highly regular distribution in wild-type chromatin (17). The heterochromatic organization was also disrupted in the 3h1 mutant (17). All DNA methylation contexts in Arabidopsis (i.e. CG, CHG, CHH), are regulated by H1 mainly in heterochromatin but also in euchromatin (21,23–25). H1 possibly creates a structural barrier that restricts RNA-dependent DNA methylation to euchromatic regions and requires DDM1-mediated accessibility to DNA methyltransferases MET1, CMT1 and CMT2 for maintenance of heterochromatic DNA methylation (22,23,26).
Previous work, using MNase seq and FRAP of 3h1 suggested that loss of H1 causes partial loss of structural differentiation of the chromatin states (17). Also, a recent study demonstrates that the double h1.1h1.2 mutant has minor but significant increase in accessibility of H3K27me3 marked gene body regions (27). The role of the stress inducible H1.3 variant in regulating abiotic stress has been previously described (21). H1.3 expression was strongly induced by a combination of drought and low light stresses, while with h1.3 mutant plants failing to mount a typical adaptive response to drought stress (21). In addition, H1 variants collectively fine-tune developmental aspects like dormancy, lateral root formation, stomata and root hair density and flowering in Arabidopsis (17). In this study, we demonstrate that Arabidopsis H1 knockout mutants have elevated basal immune levels and are resistant against a bacterial and a fungal pathogen. However, 3h1 mutant plants are compromised in flg22 triggered priming. The altered DNA methylation and reduced H3K56ac profiles in 3h1 after flg22-treatment provide strong evidence of the molecular mechanism of the priming deficient phenotype of 3h1 mutant.
MATERIALS AND METHODS
Plant material and growth conditions
The experiments were performed by using Arabidopsis (Arabidopsis thaliana) lines in the Col-0 ecotype background unless stated otherwise. We obtained the single, double and h1.1h1.2h1.3 (3h1) triple mutant from the Baroux lab (University of Zürich, Switzerland) as described previously (17). Plants were grown on soil (jiffy pots, http://www.jiffypot.com/), in plant growth chambers (Percival Scientific) under short-day conditions (8 h light/ 16 h dark) at 22°C.
Pathogen infection assays
P. syringae tomato pv. DC3000 (Pst DC3000) strain was grown on King B agar plates with 50 μg/ml rifampicin at 28°C. Bacteria were adjusted to 106 cfu/ml in 10 mM MgCl2 and inoculated to four-week-old plants by either syringe or spray infection. Bacteria were released from three leaf discs (4mm) in 500 μl of 10 mM MgCl2 with 0.01% Silwet 77 by incubating at 28°C at 1000 rpm for 1 h. Serial dilutions were plated on LB medium with appropriated antibiotic. After incubation at 28°C, bacterial colonies were counted. The experiment was repeated three times with similar results.
Botrytis cinerea infection was performed as previously described (28). Briefly, 4-week-old plants were inoculated with Botrytis cinerea (strain: B05.10) by placing a 5 μl droplet of a spore suspension (5 × 105 spores/ml) on each rosette leaf (three fully expanded leaves per plant). Trays were covered by a transparent plastic lid to maintain high humidity. Lesion diameter was measured after 2 days of infection using ImageJ analysis tool.
ROS burst assays
ROS burst was determined by the luminol-based assay described before with modifications (29). Leaf discs were incubated overnight in a white 96-well plate in water to reduce the wounding effect. Next day, the water was replaced by 100 μl of reaction solution containing 50 μM of luminol (Sigma) and 10 μg/ml of horseradish peroxidase (Sigma) supplemented with 1 μM of flg22. The measurement was conducted immediately with a luminometer (GloMax, Promega) for a period of 40 min with a 1 min reading interval between readings. The measurements are shown as means of RLU (relative light units). The experiments were repeated three times with similar results.
MAPK activation assays
Adult plants or seedlings treated with 1 μM of flg22 were harvested at the indicated time points. Proteins were extracted using extraction buffer. The frozen seedlings were homogenized in 100 μL of extraction buffer (150 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 2 mM EGTA, 5% glycerol, 10 mM DTT and 1× Pierce Protease and Phosphatase tablet (thermo#A32959)). Twenty micrograms of protein were separated on a 10% polyacrylamide gel and transferred on a PVDF membrane. After overnight blocking with TBST–5% milk, the membrane was washed 3 times with TBST. Immunoblot analysis was performed using anti-phospho-p44/42 MAPK (1:5000; Cell Signaling Technology) for 2 h as primary antibody and peroxidase-conjugated goat anti-rabbit antibody (1:10 000; Promega) for 1 h with 5 times 10 min washes in-between the incubations. The experiment was repeated twice with similar results.
Hormone measurements
The extraction of phytohormones and camalexin was performed as described (30). The compounds were quantified by HPLC-ESI-SRM in a Thermo Fisher TQS-Altis Triple Quadrupole Mass Spectrometer coupled to a Thermo Scientific Vanquish MD HPLC system. The chromatographic separation was carried out in a UPLC column (Agilent Eclipse Plus C18, RRHD, 1.8 um, 2.1 × 50 mm), and the compounds were eluted using water (A) and acetonitrile (B) as mobile phase at 0.6 mL/min and in a gradient elution mode as following: 10% B for 0.5 min, 10–55% of B at 4.5 min, 55–100% B at 4.7 min, 100% until 6.0 min, 100–10% B at 6.1% and 10% until 8 min. The column was kept at 55°C. The statistical significance from three replicates was evaluated by ANOVA followed by Tukey Test (Tukey HSD).
Callose deposition
Callose deposition was conducted as described by Wang et al. (31) with modifications. Briefly, 10-day-old WT and 3h1 mutant plants were treated with 1 μM of flg22. After 24 h, seedlings from each genotype were destained in ethanol:acetic acid solution (3:1) for 3–4 h. Samples were washed with 150 mM K2HPO4 for 30 min. Cleared leaves were stained with 0.01% aniline blue (Sigma) in 150 mM K2HPO4 (pH 9.5) overnight in dark with constant shaking. Callose deposits were visualized under a DAPI filter using a fluorescence microscope. Callose deposits were counted using ImageJ software.
RNA extraction and real-time RT-PCR analysis
Total RNAs were extracted from 4-week-old adult plants using NucleoSpin RNA Plant (MACHEREY-NAGEL) kit, according to the manufacturer's instructions. First strand cDNA was synthesized from 1μg of total RNAs using SuperScript First-Strand Synthesis System for RT-PCR according to the manual instructions. Then 2 μl of 10 times diluted cDNA was used for CFX96 Touch Real-Time PCR Detection System (Bio-Rad) for Syber green based quantitative PCR analysis. The specificity of amplification products was determined by melting curves. Tubulin was used as internal control for signal normalization. The relative expression level of the selected genes was calculated based on ΔΔCt method. The primers used are listed in Supplementary Table S2.
RNA sequencing and analysis
RNA-seq was performed from mRNA libraries with 1μg of total plant RNA using TrueSeq standard mRNA Library Prep kit (Illumina). Pooled libraries were sequenced using Illumina HiSeq 4000 platform. RNA-seq generated 1538 million raw read pairs from 36 libraries (including three time points; 0, 6 and 24 h in methylation data). Adapters, primers, and low-quality bases were removed from the ends of raw reads using Trimmomatic v0.38 (32). The resulting trimmed reads were mapped to the TAIR10 genome using Tophat2 v2.1.1 (33). Read counts were generated for all samples from corresponding bam files using BEDTools v2.29.0 (34). DESeq2 (35) was run with read counts to identify DEGs between several conditions (comparison description: refer to RNA-seq excel file) with FDR ≤0.01. All the heatmaps generated for RNA-seq were plotted using the R package pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html). Clustering of genes based on their expression within conditions was performed in basic R language using hierarchical and k -means clustering. Functional enrichment of DEGs was carried out with AgriGO (36) using default settings. GO terms with P ≤ 0.05 were considered significant, and the occurrence of at least five times in the background set was additionally required for DEGs.
Whole genome bisulfite sequencing and analysis
DNAs from plant samples were extracted by NucleoSpin Plant II kit from Macherey-Nagel. DNA samples were processed by Zymo Research (USA) for WGBS using Methyl-MaxiSeq® Library Preparation procedure. Briefly, 250 ng of starting input genomic DNA was digested with 1 unit of Zymo Research dsDNA ShearaseTM Plus (Cat#: E2018-50). The fragments produced were end-blunted and 3’-terminal-A extended, then purified using DNA Clean & ConcentratorTM-5 (Cat#: D4003). The A-tailed fragments were ligated to pre-annealed adapters containing 5’-methyl-cytosine instead of cytosine according to Illumina's specified guidelines. Adaptor-ligated fragments ≥ 50 bp in size were recovered using the DNA Clean & ConcentratorTM-5 (Cat#: D4003). The fragments were then bisulfite-treated using the EZ DNA Methylation-LightningTM Kit (Cat#: D5030). PCR was performed with Illumina indices and the resulting products were purified with DNA Clean & ConcentratorTM-5 (Cat#: D4003). Size and concentration of the fragments were confirmed on the Agilent 2200 TapeStation. Libraries were sequenced on an Illumina NovaSeq instrument.
The analysis of WGBS was performed as described by Cokus et al. (37). Briefly, sequencing of the 12 libraries (4 conditions, 3 biological replicates each) resulted in 991 million read pairs. Adapters were trimmed from the raw sequences using Trimmomatic v0.38. Subsequently, trimmed reads were mapped to the TAIR10 genome using Bowtie2 v2.2.5 (38), and methylation calls were performed using Bismark v0.22.3 (39). Four filters were used to reduce false positives. First, we removed the reads with three or more consecutive CHH methylations as described earlier (37). Second, with k methylated reads mapping, the probability of it occurring through sequencing error (that is, unmethylated position appearing as methylated) was modeled using a binomial distribution B(n, p), where n is the coverage (methylated + unmethylated reads) and p is the probability of sequencing error (set to 0.01). We kept positions with k methylated reads if P(X ≥ k) < 0.05 (post-FDR correction). Third, retained methylated positions had to have ≥1 methylated read in all three biological replicates of at least one growth condition. Finally, the median coverage of retained positions across all 12 samples had to be ≥10. After calling significant methylation in each replicate separately, we merged the replicates from the corresponding condition by taking the mean for each methylation context. This yields a final average bedgraph file for each of the four conditions which have been used for further analyses. All the average line plots of methylation around and within the genes have been plotted by deepTools (40). All boxplots for methylation have been generated using the R package ggplot2. The snapshots used in the analyses were taken from the integrated genome browser IGB v9.8.1.
Assignment of genomic context to methylated cytosines
On the basis of the gene annotation of the TAIR10 (GFF3 file) and the positional coordinates of the methylated cytosines produced by Bismark, we annotated every methylated cytosine based on the genomic context using bedtools intersect, which is a common genome analysis toolkit. Based on the GFF3 file we included whether the methylated position resides in a genic or intergenic region, and the distances to the 5′ and 3′ ends of each genomic feature (gene/intergenic region/exon/intron).
PCA and correlation matrices
Median methylation levels of methylated genes and log2FPKM of expressed genes were shifted to be zero-centered and analyzed by PCA using the prcomp function in R. Using the same data, we calculated correlation matrices (Pearson correlation coefficient) and clustered samples with hclust implemented in R using complete linkage and Euclidean distance.
Western blotting of histones
Nuclei were isolated from ground powder as already described (41). 5× SDS Loading dye was directly added to the nuclei and boiled at 85°C for 10 min and later loaded on 15% SDS-PAGE gel. Later, western blotting was performed as described in MAPK activation assays section.
DNaseI accessibility PCR
DNaseI (Promega) treatment was given to isolated nuclei for 5 min at 37°C and reaction was stopped by adding EDTA to the tubes. Later DNA was isolated from the samples and accessibility PCR assay performed as already described (42).
RESULTS
Linker histone H1 regulates plant immunity against pathogen infection
In an effort to understand the role of linker histone proteins in plant immunity, we analyzed Arabidopsis H1 mutants for developmental and pathogen phenotypes. The H1 mutants used in the study were either knockouts of single, double or all three isoforms of H1. As described previously (17), all H1 mutants were viable and grew well with no visible morphological defects in pots (Figure 1A). We tested 4-week-old single mutants (h1.2 and h1.3), double mutants (h1.1h1.2, h1.1h1.3 and h1.2h1.3) and the triple mutant h1.1h1.2h1.3 (called 3h1 from now onwards) against virulent Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). We observed that h1.1h1.2 and 3h1 are significantly resistant to Pst DC3000 infection while single h1s, double h.1.1h1.3 and h1.2h1.3 mutants allow proliferation of Pst DC3000 to a similar extent as found in wild type (WT) plants (Figure 1B). These observations suggest that two main isoforms H1.1 and H1.2 together are important in modulating plant immunity. As 3h1 showed the strongest resistance phenotype, we carried out the rest of the analysis using the triple 3h1 mutant. When challenged with the fungal pathogen Botrytis cinerea, 3h1 plants efficiently restricted fungal infection compared with WT plants as depicted by the smaller lesion size (Figure 1C).
Figure 1.
Linker histone mutant 3h1 is resistant to pathogen infection. (A) Morphology of 4-week-old wild-type (WT), single (h1.2, h1.3), double (h1.1h1.2, h1.1h1.3, h1.2h1.3) and triple (h1.1h1.2h1.3 or 3h1) mutants of Arabidopsis H1 grown on pots. (B) Quantification of bacterial colonies in different h1 mutants after spray inoculation with Pseudomonas syringae pv DC3000 (Pst DC3000) at 3 and 72 hours post infection (hpi). (C) Quantification of fungal infection as reflected by the lesion size in the WT and 3h1 after 48 h of Botrytis cinerea drop inoculation on 4-week-old plants. The data is represented as mean ± SEM where * and *** indicate P < 0.05 and 0.001 compared to WT, as determined by ANOVA significance test.
3h1 mutant plants have elevated basal immune responses
To examine the effect of the loss of linker histones on basal immune responses, we performed a number of flg22-induced early and late PTI readouts in 3h1 mutant as compared to WT plants. For early PTI events, 3h1 plants produced higher levels of reactive oxygen species (ROS) than WT plants upon flg22 or chitin treatment (Figure 2A and Supplementary Figure S1A). We also observed an elevated activation of the mitogen-activated protein kinases MPK3, 4 and 6 in 3h1 mutant as compared to WT after 15 and 30 min of flg22 elicitation in both adult plants and seedlings (Figure 2B and Supplementary Figure S1B). The key plant enzymes producing ROS in responses to pathogen defense are RBOHD and RBOHF (43). We observed a higher level of both RBOHD and RBOHF levels in Pst DC3000 treated 3h1 plants (Figure 2C,D). Also, PR1 defense gene expression was elevated in 3h1 mutant while a dynamic expression of FRK1 and MYB51 was observed before and after Pst DC3000 treatment (Figure 2E and Supplementary Figure S1C, D). The defense related hormones salicylic acid (SA) and jasmonic acid (JA) are key to optimize immune outputs, we thus quantified SA and JA in 3h1 mutant plants after pathogen infection. We observed elevated levels of SA in adult 3h1 plants compared to WT when challenged with Pst DC3000, while there was a subtle but insignificant increase in JA levels (Figure 2F and Supplementary Figure S1E). Together, these results indicate that a deficiency in linker H1 affects basal immune responses in plants, possibly through H1-mediated regulation of the plant gene expression.
Figure 2.
3h1 mutant shows elevated innate immunity levels. (A) Reactive oxygen species (ROS) levels quantified over 40 min in WT and 3h1 leaf discs triggered by 1 μM flg22 treatment. (B) MAPK activation monitored with anti-pTEpY antibody indicating phosphorylation of MPK6, MPK3 and MPK4 in WT and 3h1 plants after 1 μM flg22 treatment for 15 and 30 min. CBB stain of Rubisco serves as a loading control. (C–E) Expression of key innate immunity genes RBOHD, RBOHF and PR1 after 6h of mock treatment or Pst DC3000 infection. (F) Salicylic acid (SA) quantification in WT and 3h1 plants is shown as ng/g of fresh weight. The data are shown as means ± SEMs from three replicates. Asterisk indicates a significant difference with P < 0.05.
H1 regulates the plant defense gene expression profile
In order to understand the immunity phenotype of the 3h1 mutant, we performed RNA sequencing of adult WT and 3h1 plants after challenge with Pst DC3000 for early (6 h) and late (24 h) defense responses (Figure 3A). Principal components analysis (PCA) revealed that biological replicates clustered together (Supplementary Figure S2A), indicating good reproducibility of our experiments. Additionally, the 0 to 24 hpi (hours post infection) samples were separated in the PCA plot, suggesting that gene expression changed massively as disease progressed (Supplementary Figure S2A). As 3h1 is known to have a misregulated transcriptome (17), we also identified more than 1600 genes already affected by the genotype before challenge with Pst DC3000 (Supplementary Figure S2B). More than 1000 genes were upregulated, and around 600 genes were downregulated in 3h1 in control conditions (Supplementary Figure S2B). When compared to WT, 3h1 plants displayed significant differences in transcript abundance of 924 (638 up- and 286 downregulated) and 2151 (1145 up- and 1006 downregulated) genes after 6 and 24 h of Pst DC3000 infection, respectively (Supplementary Figure S2B). Hierarchical clustering identified ten gene clusters with distinct changes in expression in response to Pst DC3000 pathogen in 3h1 mutant plants compared to WT (Figure 3B). Notably, the genes in cluster 1, which were highly upregulated in WT at 24 hpi, were repressed in 3h1 mutant plants. This cluster contained a number of defense related genes, including WRKY38 and WRKY62, which are both negative regulators of immunity (44). We confirmed the behavior of this cluster by qRT-PCR analysis of WRKY38 and WRKY62, which both showed reduced expression in 3h1 post-infection by Pst DC3000 (Figure 3C). The transcripts of cluster 5, including immunity-related defensins and cytochrome P450 family proteins, were highly upregulated in 3h1 but not in WT at 6 hpi (Figure 3B). Transcripts of cluster 6 and 7 genes showed highly elevated levels in 3h1 mutant plants at 24 hpi (Figure 3B). Whereas the GO terms of cluster 6 genes were associated with cell wall and polysaccharide metabolism, cluster 7 genes contained a number of chromatin-related proteins, including the histone acetyltransferase HAC2 of the CBP family 2 and the histone deacetylase HDA18 of the RPD3/HDA1 superfamily (Figure 3B). By qRT-PCR, we confirmed that HDA18 and HAC2 levels strongly increased in 3h1 after Pst DC3000 infection compared to WT plants, eluding to the possible fine-tuning of gene expression by H1 (Figure 3D). Interestingly, cluster 9 genes, encoding flavonoid and anthocyanin secondary metabolite genes, were downregulated upon pathogen infection in WT but not in 3h1, which showed elevated levels of these pathways before pathogen challenge. To this end, we studied the levels of some of the key defense related secondary metabolites like Callose and Camalexin (45). We observed significantly higher callose deposition in 3h1 than in WT in control conditions (Supplementary Figure S3A, B). Also, compared to WT, higher camalexin amounts were observed in 3h1 under mock conditions, which further significantly increased after Pst DC3000 infection (Supplementary Figure S3C, D). Together, these results suggest that H1 histones contribute in orchestrating both early and late responsive plant defense genes.
Figure 3.
Dynamic transcriptome of 3h1 during pathogen infection. (A) Schematic diagram of RNA seq procedure where 4-week-old adult plants were infected with Pst DC3000 and samples harvested at 0, 6 and 24 h after infection. (B) Hierarchical clustering of Pst DC3000-infected (6 and 24 h) WT and 3h1 plants compared to control based on differentially expressed RNA transcripts from mRNA seq using DESeq2 (2.0 FC, FDR Padj < 0.01). Transcript fold-change depicted as color scale demonstrates log2 fold changes compared to mean for each transcript across genotype and condition. GO and pathway enrichment terms are depicted for the gene clusters against the background of all transcripts in the heatmap. (C, D) qRT-PCR validation of selected genes which showed altered expression pattern between WT and 3h1 before and after infection. The data are shown as means ± SEMs from three replicates. Asterisk indicates a significant difference with P < 0.05 from two-way ANOVA.
flg22 induced defense priming is attenuated in 3h1
To understand whether H1 plays a role in defense priming, we pretreated WT and 3h1 plants with flg22 for 24 hours before Pst DC3000 pathogen infection. Compared to water pre-treatment, flg22 treated WT showed reduced disease symptoms of Pst DC3000 infection (Figure 4A, B). In contrast, 3h1 plants were insensitive to flg22-induced priming as shown by the similar bacterial titers in 3h1 mutant plants with and without flg22 pre-treatment (Figure 4A, B). We observed a decrease in callose deposition in 3h1 mutant after 24 h of flg22 treatment, although callose deposition was increased in mock-treated 3h1 mutant plants (Supplementary Figure S3A, B). Also, a reduction in camalexin levels was observed in flg22 treated 3h1 plants as compared to WT before and after Pst DC3000 challenge (Supplementary Figure S3C, D). The reshaping of the transcriptome in pathogen-challenged tissues is one of the attributes of defense priming (46). To understand the compromised defense priming phenotype in 3h1, we performed transcriptome analysis of flg22 pre-treated WT and 3h1 plants challenged with Pst DC3000 at early (6 h) and late (24 h) time points (Figure 4C). PCA analysis showed the reliability of our biological replicates, and the Venn diagrams represent the numbers of up and downregulated genes after Pst DC3000 infection (Supplementary Figure S2C, D). Cluster 1 is comprised of 154 genes that get strongly upregulated in WT plants during early Pst DC3000 (6h) infection while the expression is not altered in 3h1 after infection (Figure 4D). Interestingly, GO term analysis shows strong enrichment of these genes in innate immunity and defense signaling. One of the key innate defense marker genes, WRKY29 (47), was not induced in flg22-treated 3h1 plants after Pst DC3000 infection, suggesting a possible explanation for the compromised defense priming phenotype in 3h1 mutant plants (Figure 4E). We also tested the expression pattern of other defense-related genes like WRKY31 and MAPKKK15 in Cluster 1 (Figure 4F, G and Supplementary Figure S4A). Their expression behavior was similar to WRKY29, suggesting they might orchestrate the changes in plant defense in a concerted manner.
Figure 4.
3h1 is compromised in flg22- triggered defense priming. (A) Altered disease symptom development in mock and flg22-treated WT and 3h1 adult plants. Photographs of representative infected plants were taken after 2 days of Pst DC3000 infection. (B) Bacterial growth of WT and 3h1 plants with and without 24 h of flg22-treatment. (C) Schematic diagram of RNA seq where 4-week-old adult plants were first treated with 1 μM flg22 and after 24 h syringe infiltrated with Pst DC3000 and samples harvested at 0, 6 and 24 h after infection. (D) Heat map showing differentially expressed transcripts of Pst DC3000-infected (6 and 24 h) WT and 3h1 plants pre-treated 24 h prior with flg22. Transcript fold-change depicted as color scale demonstrates log2 fold changes compared to mean for each transcript across genotype and condition (FDR Padj < 0.01). GO pathway enrichment terms depicted for the gene clusters against the background of all transcripts in the heatmap. (E–G) qRT-PCR validation of WRKY29, WRKY31 and MAPKKK15 genes which showed altered expression pattern between WT and 3h1 before and after infection when pre-treated with flg22. The data are shown as means ± SEMs from three replicates. Asterisk indicates a significant difference with P < 0.05 from multiple comparison two-way ANOVA.
Another stark difference was observed at HAC2 and HDA18 as their expression decreased in 3h1 plants after flg22 priming (Supplementary Figure S4B, C), which is in contrast to their expression in non-primed conditions (Figure 3D). These results suggest that an altered regulation of defense priming might occur at the epigenetic level in 3h1 mutant plants.
3h1 plants exhibit dynamic DNA methylation before and after flg22 treatment
The reshaping of the epigenetic landscape leads to a transiently enhanced local immunity followed by a poorly understood defense priming mechanism (46). One of the key epigenetic modifications regulated by H1 histones is DNA methylation (22,23,25,27). To further understand the defective priming in 3h1, we analyzed genome-wide DNA methylation by whole-genome bisulfite sequencing (WGBS) in WT and 3h1 mutants pre- and post-flg22 treatment. To validate the quality of our sequencing data we analyzed the percentage of non-conversion rate by calculating mean methylation ratios of chloroplast genomes (which lacks DNA methylation) in CG, CHG and CHH contexts and observed <0.5% non-conversion in all samples, which supports that our data meet the standards of acceptable WGBS (Supplementary Table S1, Supplementary Figure S5A-F). Principal components analysis (PCA) and Pearson correlation analysis showed that the biological replicates clustered together (Supplementary Figure S6A–C, D–F) and individual methylation percentages in biological replicates again indicate good reproducibility of our experiments (Supplementary Figure S6G-I). We observed higher global CG and CHG methylation levels in 3h1 as compared to WT (Supplementary Figure S6J–L). Next, we analyzed the methylation dynamics in CG, CHG and CHH contexts for protein coding genes (PCGs) and transposable element (TE) regions. We further divided the protein coding loci into promoters (2 kb upstream) and gene body. We plotted methylation differences in CG, CHG and CHH contexts from all three individual biological replicates and analyzed the effect of flg22 treatment in Col-0 and 3h1 mutant (Supplementary Figure S7). Interestingly we observed a decrease in promoter methylation in 3h1 in CHG and CHH contexts while a subtle but insignificant increase was observed in CG context (Figure 5A, B, Supplementary Figure S7A). On the other hand, an increase in gene body CHG methylation was observed in 3h1 compared to WT (Figure 5A, B, Supplementary Figure S7B). The DNA methylation in all sequence contexts was strongly increased in 3h1 over the TE regions (Figure 5C, D, Supplementary Figure S7C). Flg22 treatment resulted in a further increase in DNA methylation in CG and CHG wile a slight decrease in CHH contexts in TE regions (Figure 5B, Supplementary Figure S7). However, a decrease in promoter and gene body methylation was observed in flg22 treated 3h1 mutant. This suggests that H1 can influence the flg22 triggered methylation changes in PCGs and TEs (Figure 5A, B, Supplementary Figure S7).
Figure 5.
Global methylation profiles are altered in 3h1 plants and after flg22-treatment. (A) Distribution of DNA methylation in all three sequence contexts in WT, WT + flg22, 3h1 and 3h1 + flg22 over protein coding genes (PCGs). Biological replicates were combined as one sample. Independent biological replicates are shown in Supplementary Figure S6G–I. (B) Boxplots of all methylation percentages shown for promoter and gene body regions for the PCGs in all three contexts. (C, D) Average distribution and boxplots of WT, WT + flg22, 3h1 and 3h1 + flg22 from all three contexts in transposable elements (TEs).
Flg22 triggered differential methylation influences defense gene expression in 3h1 plants
To understand the effects of dynamic PCG methylation on gene expression, we focused on the prominent gene clusters (Cluster 1, Figure 4D) where the gene expression strongly increases at 6h of Pst DC3000 infection in flg22-treated WT plants but not in 3h1. We observed that 3h1 plants exhibited an overall decrease in average promoter methylations of these genes in all contexts (Supplementary Figure S8). Interestingly, flg22 treatment increased the overall promoter methylation levels in 3h1 with no marked changes in WT (Supplementary Figure S8). Moreover, 44 genes from Cluster 1 directly involved in plant innate defense showed a striking anti-correlation between promoter CG, CHG and CHH methylation, respectively, and repressed expression patterns in 3h1 as compared to WT after flg22 treatment (Figure 6A–C). Interestingly, no significant promoter methylation changes were observed in WT for these genes after flg22 treatment while an opposite trend in all sequence contexts was obvious in 3h1 (Figure 6D–F). This further reiterates that H1 modulates flg22-induced DNA methylation dynamics and gene expression changes in plants. As flg22 enhanced PCG DNA methylation in all contexts (CG, CHH and CHG) in 3h1 mutant plants, we speculate that the altered gene expression of WRKY29, PUB22 and Exo70H1 (Figure 4D and Supplementary Figure S4B) might be due to enhanced promoter DNA methylation at these gene loci. Accordingly, we observed increased methylation in flg22-treated 3h1 plants at these gene loci (Figure 6G–I).
Figure 6.
flg22 triggers differential methylation and gene expression in 3h1. (A–C) Heatmap of mean promoter methylation in all contexts (CG, CHG and CHH) on selected transcripts from Cluster 1 (Figure 4D) in flg22-treated WT and 3h1 samples which showed differential expression after Pst DC3000 infection. (D–F) Boxplot of average methylations in indicated contexts in the promoter (–2 kb) of these genes. p value represents Wilcoxon test. (G–I) Genome Browser snapshots showing the distribution of CH, CHG and CHH methylation in representative defense genes WRKY29, PUB22 and Exo70H1.
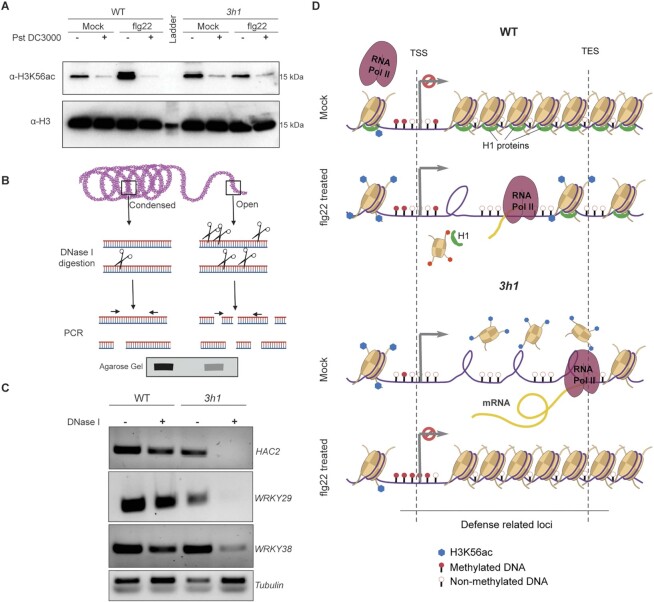
H1 histones control H3K56ac levels in plants
H1 histones control the epigenetic landscape in animals by either promoting or inhibiting various H3 histone modifications (48). Since we also observed dynamic differential regulation of HAC2 and HDA18 in 3h1 and H1 and H3K56ac can act as antagonistic regulators of nucleosome dynamics (49), we speculated that H1 might also regulate the histone epigenetic profile. To this end, we tested the global levels of two well-known active and repressive marks H3K4me3, H3K27me3, respectively, as well as H3K56ac in flg22-primed and non-primed plants after Pst DC3000 infection. flg22 or pathogen treatment did not significantly change the H3K4me3 and H3K27me3 levels between 3h1 and WT plants (Supplementary Figure S9). In contrast, although H3K56ac levels were enhanced in 3h1 plants, we observed a dramatic decrease in H3K56ac levels in flg22-treated 3h1 mutant plants (Figure 7A). In contrast, flg22 treatment increased H3K56ac in WT plants, suggesting that H3K56ac is a crucial player in flg22-triggered priming in plants. Interestingly, the H3K56ac levels decreased upon Pst DC3000 infection, suggesting a possible strategy of the pathogen to overcome plant defense. The combined regulation of DNA methylation and histone modifications by H1 leads to a dynamic chromatin landscape (16). Finally, to test the state of chromatin compaction of some of the selected differentially regulated genes in 3h1, we performed DNase I accessibility assays (42) (Figure 7B). We observed that WRKY29, HAC2 and WRKY38 were more accessible to DNase I digestion in 3h1, suggesting an open chromatin state for these genes (Figure 7C). However, the accessibility of HDA18 and WRKY62 was not significantly changed in 3h1, reflecting a more complex regulation of chromatin accessibility (Supplementary Figure S10A). Conversely, the overall accessibility of HAC2, WRKY29 and WRKY38 to some extent was restricted by flg22 treatment in 3h1 (Supplementary Figure S10B, C). Taken together, our data suggest that a multifaceted regulation of the flg22 or pathogen treatment did not significantly change the H3K4me3 and H3K27me3 levels by H1 histones via dynamic DNA methylation and H3K56 acetylation mainly accounts for the defective priming phenotype in 3h1 plants (Figure 7D).
Figure 7.
H1 controls H3K56ac levels in plants. (A) Western blot showing the H3K56ac levels in WT and 3h1 plants after water (mock) or flg22 treatment challenged with Pst DC3000 (6h). (B) Schematic representation showing the principle of the chromatin accessibility assay by DNase I digestion using PCR. Open chromatin is more frequently cut by limited DNase I digestion in small fragments giving reduced PCR signals while condensed chromatin which is less frequently cut gives strong PCR band signal. (C) DNase I accessibility PCR of HAC2, WRKY29 and WRKY38 in WT as compared to 3h1. DNase I treatment was performed for 5 min at 37°C. (D) Model showing the dynamic changes in epigenetic landscape in 3h1 after flg22 treatment as compared to WT Arabidopsis plants.
DISCUSSION
The role of linker histones is extensively studied in animal disease pathogenesis and progression (50). However, we lack an understanding of the role of H1 histones in plant immunity and disease. In this study, we aimed to understand the role of linker histone H1 in plant defense. The lack of developmental or pathogen phenotypes in single mutants of Arabidopsis H1.1 and H1.2 reflects the redundancy of the linker histones as already suggested in previous studies (17) (Figure 1). In contrast, animal studies showed that single H1 mutations lead to severe developmental defects or diseases (51). This can partly be explained by the cell type- and stage-specific isoforms present in animals, while three general isoforms of H1 exist in Arabidopsis (19). Accordingly, double h1.1h1.2 and triple mutant h1.1h1.2h.1.3 (3h1) plants show altered resistance to both bacterial and fungal pathogens. The developmental defects like stomatal spacing in 3h1 (17) do not seem to influence the bacterial load in plants, as reflected by almost equal bacterial counts at 3 hpi (Figure 1B). In 3h1, the enhanced disease resistance is associated with enhanced PTI induction as reflected by elevated ROS and MAPK activation (Figure 2A, B). The enhanced PTI marker gene expression and elevated defense hormone SA levels further support the resistance phenotype of 3h1 mutant plants. The higher levels of phytoalexins like camalexin and callose in 3h1 mutant plants in control conditions (as also suggested by transcriptome analysis Cluster 6,7 and 9, Figure 3B) could also explain the resistant phenotype of 3h1 (Figure 1). Our transcriptome analysis showed that 3h1 has differentially regulated transcriptome under normal conditions even in absence of any stress. This corroborates a previous study where almost 700 genes were found to be differentially regulated in 3h1 (17). The higher number of DEGs in our work is possibly due to using a different age of plants grown in pots and different growth conditions. Interestingly, after flg22 priming, WT plants show enhanced expression of defense-related genes like MAPKs and WRKYs compared to 3h1 (Cluster 1, Figure 4D) (47). The inability of flg22-treated 3h1 plants to mount a robust defense response after pathogen infection could lead to the observed non-priming phenotype (Figure 4A, B). This is also evident by the reduced camalexin and callose deposition in 3h1 after flg22 treatment (Supplementary Figure S3). The other major factors which correlate with the priming-deficient phenotype in 3h1 are the DNA methylation and histone acetylation changes in the mutant after flg22 treatment. An earlier study, using RNAi mediated silencing of H1, resulted in minor but significant changes in methylation of repetitive and single-copy DNA sequences in a stochastic manner (25). The higher methylation of TEs in all contexts in 3h1 mutant plants is in agreement with the methylation patterns observed for long TEs in the double mutant h1.1h1.2 (Figure 5) (23). A similar methylation profile as that of double h1.1h1.2 mutant was also observed around TEs under normal conditions for 3h1 (21). The presence of H1 histones might present a physical barrier to DNA methyltransferases at PCG, which are activated by flg22 priming. De novo DNA methylation by the RNA-directed DNA methylation (RdDM) pathway as well as cross-talk between maintenance DNA methylation pathways might explain the increase in PCG methylation in all contexts in 3h1 after flg22 treatment (52). However, the drop in CHG and CHH but not in CG methylation at the promoters of genes in 3h1 mutant plants is in accordance with the redistribution of RdDM activity from euchromatic to heterochromatic regions in H1 mutants (26) (Figure 5). The DNA methylation changes upon flg22 treatment in our study fit to the earlier reports of dynamic DNA methylation linked to differential gene expression in Arabidopsis after pathogen infection and PTI induction, although in our study flg22 treatment led to an overall increase of DNA methylation in WT, which was not described in previous reports (53,54). The effect of Pst DC3000 induced cell death could affect the overall DNA methylation levels in these studies; also the different time points, i.e. early time points of 3, 6 and 9 h for flg22 and late time point of 5d for SA treatment, could have different effects on the methylation landscape of the plants.
Linker H1 histones are normally associated with the compaction of chromatin and hinder the access of the transcription machinery to genes (12). This is also suggested by our data as we observe a higher number of differentially regulated genes in 3h1 under control conditions (Figure 5A). In the absence of H1, chromatin seems to be more accessible to enzymes for euchromatic histone modifications and antagonizes DNA methylation (23). For example, the histone acetyltransferase INCREASED DNA METHYLATION (IDM1) is important for preventing DNA hypermethylation, while the histone deacetylase HDA6 affects DNA methylation particularly in rDNA (55,56). We also observed that the histone acetyltransferase HAC2 and the deacetylase HDA18 were strongly upregulated in 3h1 mutant plants, which could promote chromatin decompaction, gene expression and influence DNA methylation (Figure 3D). This H1 dependent interplay between DNA methylation and histone modifiers prompted us to analyze the levels of histone marks in 3h1 before and after flg22 treatment. The higher levels of the H3K56ac activation mark in the absence of H1 (3h1) suggest the greater access of transcription factors to gene promoters (Figure 7A) (49). This makes sense as H3K56 is close to the exit-entry points of the nucleosomal DNA superhelix, which coincides with the position of linker histone H1. However, we observed minor changes in global H3K4me3 and H3K27me3 levels in 3h1 before and after flg22 treatment. In contrast, significant changes in H3K27me3 levels were observed in h1.1h1.2 double mutant especially in specially localized regions like telomeres (27). Lower levels of H3K4me3 and H3K27me3 were observed in H1 related mutants in previous studies taking into account the whole seedling or specialized tissues (17,42,57). These studies indicate that different tissues and developmental stages can exhibit variable dynamics of these histone marks. The different experimental setups can also reflect in the observed differences. As leaves of adult plants were used in this work, it will be interesting to further investigate the dynamics of these histone changes at different developmental stages (17,58). Paradoxically, flg22 treatment downregulates HAC2 and HDA18 expression in 3h1 mutant plants, possibly leading to reduced histone H3K56ac levels, which may allow DNA methylation to target promoters of defense-related genes and lead to the observed hypermethylation after bacterial infection. The intricate crosstalk between DNA methylation and histone acetylation in 3h1 plants before and after flg22 treatment can lead to structurally more dynamic chromatin accessibility to the transcription machinery. Taken together, we provide evidence for the role of H1 in governing dynamic changes in DNA methylation and histone acetylation during plant immunity.
DATA AVAILABILITY
RNA seq and methylation data generated for this study have been deposited in the NCBI under accession number PRJNA814075. Additional data related to this paper may be requested from the authors.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Kinga Rutowicz and Dr Célia Baroux from DPMB, University of Zurich, Switzerland for kindly sharing the single, double and triple mutants in H1 variants encoding genes, as described (17). We are very thankful to Prof. Steve Jacobson and Colette Picard at UCLA for providing the pipeline for DNA methylation analysis. We also thank Lea Faivre and Daniel Schubert at FU Berlin and Moussa Benhamed at IPS, Paris for fruitful insights. We thank Dr. Mubashir Ahmad from Ulm University for critically reading the manuscript. We also thank KAUST core lab for providing RNA seq and hormone quantification facilitates.
Author contributions: A.H.S. and H.H. conceptualized this study. A.H.S. performed most of the experiments. K.N., K.G. and M.G. analyzed RNA seq and Bisulfite seq data. NT performed some of the qPCR experiments. M.A.T. did phytohormone quantifications. H.A., M.A. and N.R. helped in performing initial patho-assays and data analysis. A.H.S., K.N., M.G. and H.H. wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Arsheed H Sheikh, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Kashif Nawaz, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Naheed Tabassum, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Marilia Almeida-Trapp, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Kiruthiga G Mariappan, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Hanna Alhoraibi, Department of Biochemistry, Faculty of Science, King Abdulaziz University, 21551, Jeddah, Saudi Arabia.
Naganand Rayapuram, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Manuel Aranda, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
Martin Groth, Institute of Functional Epigenetics, Helmholtz Munich, 85764, Neuherberg, Germany.
Heribert Hirt, King Abdullah University of Science and Technology, KAUST, 23955 Thuwal, Saudi Arabia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
King Abdullah University of Science and Technology (KAUST) [BAS/1/1062-01-01 to H.H.]. Funding for open access charge: King Abdullah University of Science and Technology (KAUST) [BAS/1/1062-01-01 to H.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jones J.D.G., Dangl J.L.. The plant immune system. Nature. 2006; 444:323–329. [DOI] [PubMed] [Google Scholar]
- 2. Bigeard J., Colcombet J., Hirt H.. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant. 2015; 8:521–539. [DOI] [PubMed] [Google Scholar]
- 3. Berens M.L., Berry H.M., Mine A., Argueso C.T., Tsuda K.. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017; 55:401–425. [DOI] [PubMed] [Google Scholar]
- 4. Thomma B.P.H.J., Nürnberger T., Joosten M.H.A.J.. Of pamps and effectors: the blurred PTI-ETI dichotomy. Plant Cell. 2011; 23:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conrath U., Beckers G.J.M., Langenbach C.J.G., Jaskiewicz M.R.. Priming for enhanced defense. 2015; 53:97–119. [DOI] [PubMed] [Google Scholar]
- 6. Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T.. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007; 448:497–500. [DOI] [PubMed] [Google Scholar]
- 7. Gong B.Q., Guo J., Zhang N., Yao X., Wang H.B., Li J.F. Cross-microbial protection via priming a conserved immune Co-receptor through juxtamembrane phosphorylation in plants. Cell Host Microbe. 2019; 26:810–822. [DOI] [PubMed] [Google Scholar]
- 8. Birkenbihl R.P., Liu S., Somssich I.E.. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 2017; 38:1–9. [DOI] [PubMed] [Google Scholar]
- 9. Winkelmüller T.M., Entila F., Anver S., Piasecka A., Song B., Dahms E., Sakakibara H., Gan X., Kułak K., Sawikowska A.et al.. Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. Plant Cell. 2021; 33:1863–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kasinsky H.E., Lewis J.D., Dacks J.B., Ausló J.. Origin of H1 linker histones. FASEB J. 2001; 15:34–42. [DOI] [PubMed] [Google Scholar]
- 11. Zhou B.R., Feng H., Kato H., Dai L., Yang Y., Zhou Y., Bai Y.. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U. S. A. 2013; 110:19390–19395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fyodorov D.V., Zhou B.-R., Skoultchi A.I., Bai Y.. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018; 19:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hergeth S.P., Schneider R.. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015; 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I.. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005; 123:1199–1212. [DOI] [PubMed] [Google Scholar]
- 15. Rupp R.A.W., Becker P.B.. Gene regulation by histone H1: new links to DNA methylation. Cell. 2005; 123:1178–1179. [DOI] [PubMed] [Google Scholar]
- 16. Yang S.-M., Kim B.J., Toro L.N., Skoultchi A.I.. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. U. S. A. 2013; 110:1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rutowicz K., Lirski M., Mermaz B., Teano G., Schubert J., Mestiri I., Kroteń M.A., Fabrice T.N., Fritz S., Grob S.et al.. Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis. Genome Biol. 2019; 20:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen X., Yu L., Weir J.W., Gorovsky M.A.. Linker histories are not essential and affect chromatin condensation in vivo. Cell. 1995; 82:47–56. [DOI] [PubMed] [Google Scholar]
- 19. Kotliński M., Knizewski L., Muszewska A., Rutowicz K., Lirski M., Schmidt A., Baroux C., Ginalski K., Jerzmanowski A.. Phylogeny-based systematization of Arabidopsis proteins with histone H1 globular domain. Plant Physiol. 2017; 174:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. She W., Grimanelli D., Rutowicz K., Whitehead M.W.J., Puzio M., Kotliński M., Jerzmanowski A., Baroux C.. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development. 2013; 140:4008–4019. [DOI] [PubMed] [Google Scholar]
- 21. Rutowicz K., Puzio M., Halibart-Puzio J., Lirski M., Kotliński M., Kroteń M.A., Knizewski L., Lange B., Muszewska A., Śniegowska-Świerk K.et al.. A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis. Plant Physiol. 2015; 169:2080–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bourguet P., Picard C.L., Yelagandula R., Pélissier T., Lorković Z.J., Feng S., Pouch-Pélissier M.N., Schmücker A., Jacobsen S.E., Berger F.et al.. The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation. Nat. Commun. 2021; 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D.. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013; 153:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rea M., Zheng W., Chen M., Braud C., Bhangu D., Rognan T.N., Xiao W.. Histone H1 affects gene imprinting and DNA methylation in Arabidopsis. Plant J. 2012; 71:776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wierzbicki A.T., Jerzmanowski A.. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2005; 169:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choi J., Lyons D.B., Zilberman D.. Histone H1 prevents non-CG methylation-mediated small RNA biogenesis in Arabidopsis heterochromatin. Elife. 2021; 10:e72676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teano G., Concia L., Wolff L., Carron L., Biocanin I., Adamusová K., Fojtová M., Bourge M., Kramdi A., Colot V.et al.. Histone H1 protects telomeric repeats from H3K27me3 invasion in Arabidopsis. 2022; 21 January 2021, preprint: not peer reviewed 10.1101/2020.11.28.402172. [DOI] [PubMed]
- 28. Rayapuram N., Jarad M., Alhoraibi H.M., Bigeard J., Abulfaraj A.A., Völz R., Mariappan K.G., Almeida-Trapp M., Schlöffel M., Lastrucci E.et al.. Chromatin phosphoproteomics unravels a function for AT-hook motif nuclear localized protein AHL13 in PAMP-triggered immunity. Proc. Natl. Acad. Sci. U. S. A. 2021; 118:e2004670118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranf S., Eschen-Lippold L., Pecher P., Lee J., Scheel D.. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 2011; 68:100–113. [DOI] [PubMed] [Google Scholar]
- 30. Trapp M.A., De Souza G.D., Rodrigues-Filho E., Boland W., Mithöfer A.. Validated method for phytohormone quantification in plants. Front. Plant Sci. 2014; 5:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y., Li X., Fan B., Zhu C., Chen Z.. Regulation and function of defense-related callose deposition in plants. Int. J. Mol. Sci. 2021; 22:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z.. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic. Acids. Res. 2017; 45:W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E.. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008; 452:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic. Acids. Res. 2014; 42:W187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramirez-Prado J.S., Latrasse D., Benhamed M.. Histone modification ChIP-seq on Arabidopsis thaliana plantlets. Bio Protoc. 2021; 11:e4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shu H., Gruissem W., Hennig L.. Measuring arabidopsis chromatin accessibility using dnase I-polymerase chain reaction and dnase I-chip assays. Plant Physiol. 2013; 162:1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Torres M.A., Dangl J.L., Jones J.D.G.. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U. S. A. 2002; 99:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim K.C., Lai Z., Fan B., Chen Z.. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008; 20:2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piasecka A., Jedrzejczak-Rey N., Bednarek P.. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 2015; 206:948–964. [DOI] [PubMed] [Google Scholar]
- 46. Mauch-Mani B., Baccelli I., Luna E., Flors V.. Defense priming: an adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017; 68:485–512. [DOI] [PubMed] [Google Scholar]
- 47. Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J.. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002; 415:977–983. [DOI] [PubMed] [Google Scholar]
- 48. Willcockson M.A., Healton S.E., Weiss C.N., Bartholdy B.A., Botbol Y., Mishra L.N., Sidhwani D.S., Wilson T.J., Pinto H.B., Maron M.I.et al.. H1 histones control the epigenetic landscape by local chromatin compaction. Nature. 2020; 589:293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernier M., Luo Y., Nwokelo K.C., Goodwin M., Dreher S.J., Zhang P., Parthun M.R., Fondufe-Mittendorf Y., Ottesen J.J., Poirier M.G.. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat. Commun. 2015; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ye X., Feng C., Gao T., Mu G., Zhu W., Yang Y.. Linker histone in diseases. Int J Biol Sci. 2017; 13:1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Behrends M., Engmann O.. Linker histone H1.5 is an underestimated factor in differentiation and carcinogenesis. Environ. Epigenetics. 2020; 6:dvaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Du J., Johnson L.M., Jacobsen S.E., Patel D.J.. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015; 16:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu A., Lepère G., Jay F., Wang J., Bapaume L., Wang Y., Abraham A.L., Penterman J., Fischer R.L., Voinnet O.et al.. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. U. S. A. 2013; 110:2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R.. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U. S. A. 2012; 109:E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qian W., Miki D., Zhang H., Liu Y., Zhang X., Tang K., Kan Y., La H., Li X., Li S.et al.. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012; 336:1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I.et al.. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004; 16:1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. She W., Baroux C.. Chromatin dynamics in pollen mother cells underpin a common scenario at the somatic-to-reproductive fate transition of both the male and female lineages in Arabidopsis. Front. Plant Sci. 2015; 6:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Geeven G., Zhu Y., Kim B.J., Bartholdy B.A., Yang S.M., Macfarlan T.S., Gifford W.D., Pfaff S.L., Verstegen M.J.A.M., Pinto H.et al.. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol. 2015; 16:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA seq and methylation data generated for this study have been deposited in the NCBI under accession number PRJNA814075. Additional data related to this paper may be requested from the authors.