Abstract
RNA-binding proteins (RPBs) are deeply involved in fundamental cellular processes in bacteria and are vital for their survival. Despite this, few studies have so far been dedicated to direct and global identification of bacterial RBPs. We have adapted the RNA interactome capture (RIC) technique, originally developed for eukaryotic systems, to globally identify RBPs in bacteria. RIC takes advantage of the base pairing potential of poly(A) tails to pull-down RNA–protein complexes. Overexpressing poly(A) polymerase I in Escherichia coli drastically increased transcriptome-wide RNA polyadenylation, enabling pull-down of crosslinked RNA–protein complexes using immobilized oligo(dT) as bait. With this approach, we identified 169 putative RBPs, roughly half of which are already annotated as RNA-binding. We experimentally verified the RNA-binding ability of a number of uncharacterized RBPs, including YhgF, which is exceptionally well conserved not only in bacteria, but also in archaea and eukaryotes. We identified YhgF RNA targets in vivo using CLIP-seq, verified specific binding in vitro, and reveal a putative role for YhgF in regulation of gene expression. Our findings present a simple and robust strategy for RBP identification in bacteria, provide a resource of new bacterial RBPs, and lay the foundation for further studies of the highly conserved RBP YhgF.
INTRODUCTION
RNA-binding proteins (RBPs) play key roles in many cellular processes in bacteria, which is illustrated by their intimate involvement in both transcription and translation. RBPs are also main players in post-transcriptional control, where they participate in RNA stabilization, regulation of translation, scaffolding, matchmaking, and termination (1). RBPs interact with their RNA targets through RNA-binding domains (RBDs), where well-known examples include the S1 domain, the KH domain, the cold shock domain, and the RNA recognition motif. However, it is worth noting that hundreds of RBPs lack known RBDs, suggesting that so far unknown modes of RNA–protein binding remain to be discovered and characterized (2).
Well-studied bacterial regulatory RBPs include Hfq, which facilitates base-pairing between small regulatory RNA (sRNA) and mRNA targets (3), and the RsmA/CsrA family of proteins that bind to mRNA 5’UTRs and regulate translation (4). Most of the currently well-studied RBPs, including Hfq (5) and CsrA (6), were identified due to their phenotypic effects rather than their ability to bind RNA. Their RNA-binding activity was subsequently revealed through studies aimed at understanding the mechanistic basis of the observed phenotypes (7,8). More recently, methods specifically dedicated to identifying RNA–protein complexes have proven successful for advancing the knowledge of new regulatory RBPs in various bacterial species, including ProQ in Salmonella enterica (9) and KhpB in Clostridioides difficile (10). These findings emphasize the value of approaches directly aimed at identifying RBPs, and highlights that far from all RBPs in bacteria have yet been discovered.
In eukaryotic systems, RBP discovery has been extensively addressed. For instance, the RNA Interactome Capture method (RIC) (11,12) globally identifies RBPs crosslinked to poly(A)-tailed eukaryotic RNAs using poly(dT) oligonucleotides as a bait. This strategy has identified several hundreds of proteins as putative RBPs in various eukaryotic species and cell types (reviewed in (13)). Surprisingly, these studies found that many of the identified RBPs had previously characterized cellular functions unrelated to RNA-binding. Among these were numerous metabolic enzymes, including all the enzymes in glycolysis and the citric acid cycle. This is in line with earlier findings that aconitases, both in eukaryotic and bacterial species, moonlight as RBPs by post-transcriptionally regulating expression of several genes, including their own expression (14–17). These findings led to the formulation of the REM-hypothesis, that suggests a comprehensive regulatory network based on interactions between RNA, enzymes, and metabolites (REM) (18). Despite the success of RIC, its application in bacteria has been hampered by the fact that, rather than protecting mRNA as in eukaryotes, poly(A) tails on bacterial RNA promote degradation and thus are both much shorter and less abundant than in eukaryotic cells. Instead, a hand-full of studies have applied alternative strategies to globally identify RBPs in bacteria. In TRAPP (19,20), silica beads were used to purify RBP-RNA complexes, in OOPS (21) and pTEX (20,22,23) variations of organic phase separation allowed isolation of RBPs from the remaining protein fraction, and in GRAD-seq and GradR, RNA–protein complexes were inferred from glycerol gradient sedimentation profiles (9,24).
In the present study, we have established RIC for direct identification of RBPs in Escherichia coli. Transient pulse-expression of poly(A) polymerase I (PAPI) was applied to broadly polyadenylate cellular transcripts, thereby circumventing the lack of extensive poly(A) tailing in bacteria. This allowed for oligo(dT)-based capture of crosslinked RNA–protein complexes, resulting in the identification of 169 putative RBPs. About half of these were already classified to bind RNA, and the RNA-binding activity of a dozen previously unknown RBPs was experimentally validated. Finally, the in vivo RNA ligands of the highly conserved but uncharacterized protein YhgF were determined, guiding the discovery of YhgF as a putative regulator of gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions
All strains used in this study are listed in Supplementary Table S1. The E. coli K-12 strain MG1655 was used for all experiments except for the PNK-assays where TOP10 (Invitrogen) and MC4100 (25) were used. All cultures were grown at 37°C with shaking at 180 rpm in M9 media supplemented with 0.4% glycerol and 0.1% casamino acids, unless otherwise specified. Antibiotics were added when appropriate at the following concentrations: ampicillin 100 μg/ml, chloramphenicol 30 μg/ml, kanamycin 50 μg/ml.
Strain construction
The yhgF deletion strain and the yhgF-3xFLAG strain were generated by λ Red recombineering. Plasmid pSim5-tet (26), containing the λ Red genes, was transformed into MG1655. Mutants were constructed in this strain as described (27). A kan-sacB cassette was inserted into the yhgF locus by selecting for kanamycin resistance followed by confirmation by PCR. The cassette was then replaced by counterselection on sucrose, with either a fragment designed to delete yhgF (except for the 3’-most 100 bp of the ORF in the chromosome, in order not to interfere with a putative sRNA expressed from the opposite strand, denoted STnc760 in Salmonella Typhimurium) or to add an N-terminal 3XFLAG tag. To reduce the risk of undesired genomic mutations, a two-step transduction was performed; first, the kan-sacB cassette and, next, the locus carrying the mutation, was moved to a wildtype strain by P1 transduction. All transductions were performed as described by Miller (28).
Plasmid construction
All plasmids used in the study are listed in Supplementary Table S2. pLA100 was constructed by linearizing plasmid pEH299 (29) by PCR using primers EHO-1054/-1055 followed by re-ligation. pTSS52 was constructed by replacing the NdeI and XbaI fragment in pLA100 with the yhgF ORF (primers EHO-1064/-1065). pEH499 was constructed by PCR amplification of the 2xStrepII-TEV-3xFLAG tandem affinity tag sequence from pEH299 (29) (primers JVO-11250/JVO-11251), followed by ligation into pBAD24 using the NcoI and HindIII sites. Plasmids expressing epitope-tagged putative RBPs for use in the PNK assay (pAK01-12) were constructed by first amplifying the relevant ORFs by PCR (primers listed in Supplementary Table S3). The PCR fragments were then inserted between the XbaI and NcoI sites in plasmid pEH499 by restriction enzyme cloning. The resulting plasmids encode the ORF of interest along with an additional codon inserted immediately downstream of the start codon and a 2xStrepII-TEV-3xFLAG tag at the C-terminal end. The pPAPI plasmid was constructed by PCR-mediated amplification of the Salmonella pcnB ORF (encoding PAPI), using oligos JVO-9893 and JOV-9894, followed by insertion between the NcoI and HindIII sites of the pBAD24 plasmid. Plasmid pTSS54 was constructed by inserting the yhgF ORF (primers EHO-1862/-1863) in plasmid pTYB11 (New England Biolabs) using the SapI and PstI sites, according the manufacturer's instructions.
Harvest of cultures for RNA interactome capture
E. coli MG1655 cells harboring plasmid pPAPI (for PAPI overexpression) was grown in M9 media until an OD600 of 0.35, where expression of PAPI was induced by addition of arabinose to a final concentration of 0.2% (w/v). After 30 min of induction, 250 OD600 units were harvested by rapid cooling in ice-water followed by centrifugation at 4600 g at 4°C for 30 min. The cell pellet was resuspended in 100 ml ice-cold PBS and divided in two halves, one of which was irradiated with UV-light (254 nm, 1 J/cm2) in a Stratalinker 1800 (Stratagene) device, while the other half was untreated. The samples were pelleted by centrifugation at 3500 g at 4°C for 15 min, flash frozen on dry ice and stored at -80°C until further processing.
RNA interactome capture
This method is a modified version of the RIC protocol described by Castello et al. (11). Cell pellets were thawed and resuspended in 3 ml ice-cold lysis buffer (20 mM Tris–HCl (pH 7.5), 0.5% LiDS, 1 mM EDTA and 5 mM DTT). Each sample was divided into four 2 ml microcentrifuge tubes, each containing 1.3 g ice-cold 0.1 mm Zirconia beads (Biospec), followed by cell lysis using a FastPrep24 homogenizer for 20 s at 4.0 M/s. Cell lysates were centrifuged for 10 min at 16 000 g at 4°C and supernatants transferred to new microcentrifuge tubes. Centrifugation was repeated and supernatants from identical samples pooled in 50 ml Falcon tubes. Lysis buffer was added to a total volume of 20 ml per tube and a fraction from each sample was saved for later analysis.
Two milliliters of oligo d(T)25 magnetic beads (New England Biolabs), previously equilibrated by three washes with 3 x volume lysis buffer, was added to each tube, followed by incubation at 4°C with gentle rotation for 1 h. Subsequently, the beads were pelleted using magnets, and supernatants were removed. The beads were washed by resuspension in 20 ml ice-cold lysis buffer and incubated with gentle rotation for 5 min at 4°C. After removal of the supernatants, the beads were sequentially washed with wash buffers (20 mM Tris–HCl pH7.5, 1 mM EDTA and 5 mM DTT) containing decreasing amounts of salts and detergents: (i) 0.1% LiDS (w/vol) and 500 mM LiCl, (ii) 500 mM LiCl and (iii) 200 mM LiCl. Two washes were done with each buffer. RNA–protein complexes were eluted by moving the beads into microcentrifuge tubes, adding 270 μl of elution buffer (20 mM Tris–HCl pH7.5, 1 mM EDTA) and incubating at 55°C for 3 min. Beads were removed using magnets, and the eluates RNase-treated by addition of 6 μl RNase A/T1 mix (Thermo Scientific), 30 μl 10 x RNase buffer (100 mM Tris HCl pH 7.5, 1.5 M NaCl, 0.5% NP-40, 5 mM DTT) and incubation at 37°C for 90 min.
RNA-seq of polyadenylated total RNA
RNA-seq on total RNA from cultures overexpressing PAPI, before and after oligo(dT)-based purification were performed by Vertis Biotechnologie AG (Freising, Germany). Four biological replicates of each condition were depleted for ribosomal RNAs, subjected to CRISPR-Cas9-mediated removal of long poly(A) tails, and paired-end sequenced on an Illumina NextSeq 500 system using 2 × 75 bp read length. Adapter sequences were removed with SeqPurge (30). Read mapping were then performed with STAR (–alignIntronMax 1) (31) against E. coli MG1655 genome sequence (NCBI accession: NC_000913.3). Strand separated coverage files were generated with SAMtools (32), BEDTools genomecov (33) and bedgraphtobigwig script (www.encodeproject.org/software/bedgraphtobigwig/). Fractional read counts per gene were obtained with featureCounts (34) and were then rounded to the closest integer. Enrichment and depletion of transcripts after oligo(dT)-based purification were then analyzed with DESeq2 (35) (Supplementary Table S4).
Sample preparation for LC-MS
Lysates of the samples corresponding to 20 μg protein were loaded onto centrifugal 30 kD filter devices (Microcon-30 kDa; Merck, Darmstadt, Germany) for filter-aided sample preparation as described elsewhere (36). Briefly, the samples were washed with a buffer (pH 8.5) containing 100 mM Tris and 8 M urea. After reduction with 8 mM DTT and alkylation with 50 mM iodoacetamide, removal of excess iodoacetamide with 8 mM DTT was performed. After each incubation, samples were washed twice with the 100 mM Tris 8 M urea buffer. Before enzymatic digestion with trypsin (enzyme-to-protein ratio 1:50 (w/w) in a wet chamber at 37°C for 16 h, the samples were washed with 50 mM NH4HCO3 three times. The resulting peptides were washed from the filter with 50 mM NH4HCO3. Trifluoroacetic acid was added to the samples to a final concentration of 1% (v/v) and dried at 45°C. Finally, the samples were reconstituted in 3% acetonitrile and 0.1% formic acid (FA) in water to a final concentration of 150 ng/μl.
Liquid chromatography–mass spectrometry (LC–MS)
Tryptic peptides (loading amount 300 ng protein) were separated on a nanoAcquity UPLC system equipped with a C18 (5 μm, 180 μm × 20 mm) trap column followed by a HSS-T3 C18 (1.8 μm, 75 μm × 250 mm) analytical column (40°C) and further analyzed in positive ionization mode with the UDMSE approach (37,38) on a Synapt G2-Si HDMS mass spectrometer with electrospray ionization source (Waters Corporation, Manchester, UK). Mobile phase A contained 0.1% FA and 3% DMSO in water and mobile phase B 0.1% FA and 3% DMSO in acetonitrile. At a flow rate of 0.3 μl/min a gradient was run from 3–40% (v/v) over 120 min. Every 60 s a lock mass solution composed of 0.1 μM [Glu1]-fibrinopeptide B and 1 μM leu-enkephalin was introduced through the reference channel. Method quality control was performed with HeLa digest samples (Thermo Scientific) that were analyzed between the samples.
Data processing and label-free quantification of LC–MS data
Raw data was searched against a Uniprot database for E. coli K12 (2022_01 release) with inclusion of RNT1_ASPOR (Guanyl-specific ribonuclease T1) and RNAS1_BOVIN (Ribonuclease pancreatic) using ProteinLynx Global Server (PLGS) (version 3.0.3, Waters Corporation, Milford, MA, USA). The accepted false discovery rate was set to 0.01 obtained with parallel searching in a randomized database of the protein entries above. Minimum peptide matches per protein were 2 and minimum fragment ion matches per peptide and proteins 1 and 3, respectively. One missed cleavage per peptide was allowed. Trypsin was set as digest reagent, carbamidomethyl cysteine as fixed modification and methionine oxidation as variable modification.
Label-free quantification was done with TOP3 quantification using ISOQuant 1.8 including nonlinear retention time alignment, signal clustering based on accurate mass, retention and drift time, annotation of signal clusters using PLGS identifications, intensity normalization and protein isoform and homology filtering (37,38). The software settings are described in Supplementary Table S5. The resulting relative protein abundances were log2-transformed and further processed in R with the package limma (39) to estimate significant fold-changes in protein abundance.
RNA purification and northern blotting
RNA extraction and Northern blotting were performed as described (29) with the exceptions that (i) Church buffer (40) was used to block non-specific binding sites on the membrane, and (ii) membranes were washed three times in 1× SSC, 0.1% SDS.
Protein gels and western blotting
Gel staining and Western blot was used to detect proteins in the poly(A) pull-down experiment. After denaturation for 5 min at 95°C, samples were separated on Mini-PROTEAN TGX Stain-Free protein gels (BioRad). For total in-gel protein staining, QC Colloidal Coomassie Stain (BioRad) was used according to the manufacturer's protocol. For Western blot, proteins were transferred to PVDF membranes using the TransBlot TURBO transfer system (BioRad) according to the manufacturer's protocol. The membrane was blocked using TBS-T with 3% bovine serum albumin. FLAG-tagged Hfq was detected using an HRP-conjugated anti-FLAG antibody at 1:10 000 dilution (Sigma). Even gel loading was validated by probing the membranes with an HRP-conjugated anti-GroEL antibody (Sigma) at 1:50 000 dilution. Chemiluminescence was detected using Amersham ECL Prime reagents according to the supplier's protocol (Cytiva) on a ChemiDoc™ System (BioRad).
PNK assay and CLIP-seq
The PNK assay was used to evaluate the RNA-binding potential of proteins identified in the poly(A) pull-down, and CLIP-seq was used to identify RNA-ligands of YhgF. For the PNK assay, the 3xFLAG-tagged proteins YajQ, YbcJ, YbeZ, YceD, YhgF, YibL, YicC, YifE, YihI, Bcp, GapA and Icd were expressed from plasmids, while ProQ, OmpA and YhgF, also 3xFLAG-tagged, were natively expressed from the chromosome. Bacterial cultures were grown to an OD600 of 0.5. Plasmid-expressed proteins were induced by addition of arabinose (0.2%) 30 min prior to harvest. Half of each culture (100 ml) was UV-light treated (800 mJ/cm2) to induce RNA–protein crosslinking, while the remaining half was placed on ice. Cells were pelleted by centrifugation at 3500 g, 4°C for 30 min. Each cell pellet was resuspended in NT-P buffer (50 mM NaH2PO4, 300 mM NaCl, 0.05% Tween, pH 8.0) with 2 U of Turbo DNase (Thermo Scientific), and divided into four 2 ml microcentrifuge tubes each containing 1.3 g ice-cold 0.1 mm Zirconia beads (Biospec), followed by lysis using a FastPrep24 homogenizer for 20 s at 4.0 M/s. Cell lysates were centrifuged for 10 min at 16 000 g and 4°C, and supernatants transferred to new microtubes. Centrifugation was repeated and identical samples were pooled in 15 ml tubes with addition of NP-T to a total volume of 5 ml. Then, 60 μl of anti-FLAG M2 magnetic beads (Sigma-Aldrich), prewashed three times in NP-T buffer, was added to each tube and the samples incubated at 4°C while gently rotating for 1 h. Beads were washed twice with 1 ml cold high-salt buffer (50 mM NaH2PO4, 1 M NaCl, 0.05% Tween, pH 8.0) and twice with cold NP-T buffer, using magnetic separation between washes. The beads were resuspended in 100 μl NP-T buffer containing 1 mM MgCl2 and 25 U benzonase nuclease (Thermo Fisher), and incubated for 10 min at 37°C, while shaking at 900 rpm. Subsequently, beads were washed once with high-salt buffer and twice with CIP-buffer (100 mM NaCl, 50 mM Tris–HCl pH 7.4, 10 mM MgCl2). The beads were then resuspended in 150 μl CIP buffer supplemented with 6 U calf intestinal alkaline phosphatase (Invitrogen) and incubated 30 min at 37°C, while shaking at 900 rpm. After this, the beads were washed once with high-salt buffer and twice with PNK buffer (50 mM Tris–HCl pH 7.4, 10 mM MgCl2, 0.1 mM spermidine), resuspended in 100 μl PNK buffer containing 10 U of T4 polynucleotide kinase (Thermo Fisher) and 10 μCi γ‐32P‐ATP, and incubated for 30 min at 37°C. After 5 min incubation in the presence of 20 μl ATP (10 mM), the beads were washed three times in NP-T buffer and then resuspended in 20 μl protein loading buffer (0.3 M Tris–HCl pH 6.8, 0.05% bromophenol blue, 10% glycerol, and 10% beta-mercaptoethanol). After 5 min denaturation at 95°C, the samples were separated on 14% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane. Radioactive signals were detected using autoradiography. For the PNK-assay, the membranes were subsequently used for Western blotting using anti-FLAG antibodies as described above. For CLIP-seq, the bands corresponding to the RNA–protein complexes were excised from the membrane and submerged in 400 μl PK buffer (50 mM Tris–HCl pH 7.9, 5 mM EDTA, 0.5% SDS) including 400 μg Proteinase K (Thermo Fisher) and 1 μl SuperaseIN. Samples were incubated for 1 h and 37°C, shaking at 1000 rpm, followed by a second one-hour incubation after addition of 100 μl PK buffer with 9 M urea. To isolate co-purified RNA, the samples were phenol-chloroform extracted and ethanol precipitated with 1 μl GlycoBlue (Thermo Fisher) per sample.
Library preparation and next generation sequencing
Libraries were prepared using the NEBNext multiplex small RNA library prep set for Illumina (New England Biolabs) according to the manufacturer's directions, except that 20 cycles of PCR was performed after reverse transcription. MinElute PCR purification kit (Qiagen) was used to purify and concentrate PCR samples. The amplified libraries were size-separated on 6% polyacrylamide gels, stained with SYBR gold (Thermo Fisher) and fragments of 140–250 bp excised from the gel. The DNA was purified using the elution buffer supplied with the kit and incubated overnight at 16°C while shaking at 1200 rpm. Gel fragments were removed by centrifugation through a Costar Spin-X column (0.45 μl cellulose membrane, Corning) and ethanol-precipitated using linear acrylamide (from kit) as co-precipitant. To amplify the DNA library, another nine rounds of PCR was performed followed by purification and concentration using the MinElute PCR purification kit. Next generation sequencing was performed at vertis Biotechnologie AG (Freising, Germany), using 75 bp paired-end sequencing on the Illumina NextSeq 500 system.
Analysis of CLIP-seq sequence data
Adapter trimming and merging of read pairs was performed with SeqPrep (https://github.com/jstjohn/SeqPrep). Pairs were merged if the resulting read was at least 12 nucleotides long with at least 12 bases overlapping. Prior to mapping, all rRNA operons +/- 50 bp except rrnH were masked from the E. coli K-12 substrain MG1655 genome sequence (NCBI accession: NC_000913.3). Read mapping was performed with bowtie 1.2.2 (41) allowing for 1 mismatch. Read coverage was analyzed by converting mapped reads to bigWig format with BEDTools genomecov (33) and bedGraphToBigWig (42).
Peak calling was performed with PEAKachu (43) run in adaptive mode using deseq normalization and adjusted P-value <0.05 as cutoff. Enriched transcripts were also identified by performing stranded read count per gene with featureCounts (34). Reads mapping sense to each gene were then used as input for analysis with DESeq2 (35). Hits with a positive log2(FC) and adjusted P-value <0.05 were considered to be enriched in the YhgF plus crosslink data.
Purification of YhgF
Tag-less recombinant YhgF was purified from E. coli BL21 (DE3) using the IMPACT system (New England Biolabs). Plasmid pTSS54 was transformed into BL21 (DE3), and the resulting strain (EHS-3275) was grown in LB supplemented with ampicillin at 37°C to an OD600 of 0.5. The culture was rapidly cooled on ice and kept overnight at 4°C. IPTG was added to a final concentration of 1 mM and the culture was incubated for 6 h at r.t. shaking at 220 rpm. Cells were pelleted by centrifugation, 3000 g for 30 min at 4°C, and the pellet was stored at -80°C until further processing.
After thawing, the cell pellet was resuspended in 50 ml lysis buffer (20 mM Tris–HCl pH 8.5, 500 mM NaCl, 0.1% Tween20), and 70 U of DNase I was added. Cells were lysed by French press at 15 kPSI, and the extract centrifuged at 4000 g for 30 min at 4°C. The cleared lysate was passed through a 0.2 μm filter, and lysis buffer was added to a total volume of 100 ml. The lysate was loaded onto a column containing 20 ml chitin resin, prewashed with 100 ml column buffer (20 mM Tris–HCl pH 8.5, 500 mM NaCl). The resin was washed with 100 ml column buffer by gravity flow. Subsequently, 30 ml cleavage buffer (20 mM Tris–HCl pH 8.5, 500 mM NaCl, 50 mM DTT) was added, the flow arrested, and the column incubated at 4°C overnight. Protein was eluted by addition of 30 ml cleavage buffer, collecting the flow-through. Pierce concentrator, PES, 30k MWCO (Thermo Scientific) columns were used for buffer exchange and concentration of the protein.
Electrophoretic mobility shift assay
Binding of YhgF to targets identified by CLIP-seq was examined using electrophoretic mobility shift assay (EMSA) with purified YhgF and in vitro transcribed RNA. DNA templates for in vitro transcription were made by PCR using primer sets EHO-1791 + EHO-1792, EHO-1802 + EHO-1803 and EHO-1793 + EHO-1794 as template for rmf, gapA and RyhB, respectively. All primers used in the study are listed in Supplementary Table S3. Transcription was performed o/n at 37°C using the MEGAscript T7 transcription kit (ThermoFisher). The resulting RNA was treated with TurboDNase (ThermoFischer) (0.1 unit/μl, 30 min at 37°C), gel-purified from denaturing polyacrylamide gels, dephosphorylated using calf intestinal alkaline phosphatase (30 min at 37°C), and 5′ radiolabeled using T4 polynucleotide kinase and [γ-32P] ATP. After each step the RNA was phenol/chloroform extracted and ethanol precipitated. The RNA was denatured for 2 min at 95°C before use. Binding reactions contained 100 mM KCl, 10 mM Tris–HCl pH 7.5, 1 mM DTT, 4% glycerol, 1 μM yeast tRNA, 1 nM labeled RNA and 0–1200 nM YhgF in a 10 μl reaction. Samples were incubated 5 min at 37°C and separated on 6% native polyacrylamide gels at 4°C, using 1x TBE as buffer. Radioactive signals were detected on a phosphorimager (Typhoon – Cytiva). For competition experiments, unlabeled competitor RNA was added to preformed complexes of rmf*-YhgF at concentrations of 0–800 nM.
RT-qPCR
For each sample, one μg of total RNA was DNase treated using 2U TURBO DNase (Thermo Fischer) in 20 μl for 20 min at 37°C, and purified with phenol; chloroform; isoamyl alcohol (ratio 25:24:1) and ethanol precipitation. cDNA was prepared using Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Fischer) with random hexamer primers according to the manufacturer's instructions. qPCR was performed using the Maxima SYBR Green/ROX qPCR kit (Thermo Fischer) for the rmf transcript (primers EHO-1681 + EHO-1682) and the reference transcript hcaT (44) (primers EHO-1698 + EHO-1699).
In vitro transcription assay
DNA templates for in vitro transcription were made by PCR for the genes rmf (primers EHO-1960 and EHO-1961) and cspE (primers EHO-1969 + EHO-1970). Each in vitro transcription reaction contained 15 nM DNA template, 200 μM ATP, GTP, CTP, 8 μm UTP, 0.3 pmol α-32P-UTP and 0,02 U/μl E. coli RNA Polymerase Holoenzyme (New England BioLabs) in 1× buffer (New England BioLabs). For each template, samples with and without the addition of purified YhgF to a concentration of 400 nM were included. The reactions were incubated at 37°C and 25 μl aliquots were inactivated at the indicated time points by addition of 175 μl H2O and 200 μl phenol; chloroform; isoamyl alcohol (ratio 25:24:1). After phenol extraction and ethanol precipitation, the RNA was size separated on a denaturing 6% PAA gel and radioactive signals were detected by autoradiography.
Phylogenetic analysis
YhgF-like sequences were collected by blastp from RefSeq database using the E. coli YhgF sequence (HAZ8091882.1) as query. Matching sequences were required to cover >80% of the query, and sequence identity was above >32% for all recovered sequences. The selected sequences (Supplementary Table S6) were aligned by mafft-linsi v7.490 and trimmed by trimAI v1.4.rev15. The phylogeny was inferred by IQtree 2.2.0-beta using the LG + C20 + F + G mixture model with 1000 ultrafast bootstraps. The tree was visualized using Figtree v1.4.4.
Protein domain analysis
Conserved domain composition of putative RBPs was analyzed with Reverse Position Specific BLAST 2.12.0 + against the cdd_delta database using evalue < 0.01 as cut off. The result is presented in Supplementary Table S7.
RESULTS
RIC globally identifies RBPs in bacteria
The recent success of poly(A)- and crosslinking-dependent identification of RBPs in eukaryotic systems using the RNA interactome capture (RIC) method (12,45–47) inspired us to investigate the feasibility of this approach for identifying bacterial RBPs in a global and high-throughput manner. Since poly(A) tails in bacteria are scarce and signal RNA degradation rather than promoting stability—as in eukaryotes—we tested whether overexpression of PAPI would yield a sufficient degree of polyadenylation to allow capture of cellular RNAs and their associated RBPs using oligo(dT)-coated magnetic beads (Figure 1A). To this end, E. coli strain MG1655 harboring a PAPI overexpression vector was grown until mid-exponential phase in M9 minimal media, followed by induction of PAPI for thirty minutes. Northern blotting using an oligo(dT) probe showed a dramatic increase of polyadenylation in RNA harvested fifteen and thirty minutes after induction, compared to the pre-induction sample or the non-induced controls (Figure 1B). During this time, we observed little, if any, effect on bacterial growth rate (Supplementary Figure S1A). Thus, even though PAPI expression likely causes a certain level of stress, the applied conditions gave sufficient levels of polyadenylation without causing severe growth defects. To assess the grade of polyadenylation across the transcriptome, we performed RNA-seq on total RNA from cultures overexpressing PAPI, before and after oligo(dT)-based purification. Read counts for single genes were highly correlated between input and output samples (Figure 1C), indicating transcriptome-wide polyadenylation and purification on oligo(dT). Omitting PAPI overexpression resulted in strongly reduced recovery of RNA on oligo(dT) beads (Supplementary Figure S1B). Differential expression analysis based on four biological replicates revealed that only a small fraction of the transcriptome (71/4496 transcripts, Figure 1D, Supplementary Table S4) was significantly (fold change < 0.5, FDR-adjusted P-value < 0.01) depleted after oligo(dT) purification. The counter-selected transcripts included many tRNAs and type I toxin/antitoxin RNAs (Figure 1D). Possibly, PAPI failed to polyadenylate these transcripts due to limited access to the 3’ end, either due to extensive RNA structure (toxin/antitoxin transcripts) or the presence of an amino acid (tRNAs). We conclude that transcriptome-wide polyadenylation by PAPI enabled recovery of all cellular transcripts upon oligo(dT)-based purification, of which less than 2% were selectively depleted.
Figure 1.
RNA interactome capture in E. coli. (A) Schematic representation of the modified RIC protocol. E. coli cultures were grown in M9 minimal media supplemented with 0.4% glycerol, 0,1% casamino acids and 100 μg/ml ampicillin until an OD600 of 0.35 where expression of PAPI was induced from a plasmid harboring the pcnB gene by addition of arabinose to promote polyadenylation of RNA. After 30 min of induction, half of the culture was irradiated with UV-light (1 J/cm2, wavelength of 254 nm) and the other half was untreated, then cells from both fractions were lysed. Polyadenylated transcripts were captured along with crosslinked proteins, using oligo(dT)-covered beads. After elution, RNA was removed by addition of an RNase A/T1 mix and co-purifying proteins were identified using bottom-up proteomic mass spectrometry. (B) Northern blot with RNA harvested from cultures at different stages of PAPI induction along with empty vector control. The blot was probed using a radio-labeled oligo(dT) probe (EHO-1505) to visualize polyadenylated RNA. (C) RNA-seq analysis of global RNA composition before (lysate) and after (elution) purification with oligo(dT). Data points represent average read counts for single genes (n = 4496) based on four replicates for each condition. (D) Pie charts showing the RNA class distribution of all detected transcripts (left) and significantly (fold change < 0.5, FDR-adjusted p-value < 0.01) counter-selected RNA transcripts (right) according to DEseq analysis of total RNA before and after purification with oligo(dT). (E) Western blot detecting 3xFLAG-tagged Hfq using an anti-FLAG antibody in samples from the RIC experiment with and without UV-light crosslinking (XL). The membrane was stripped and re-probed using anti-GroEL. The image is an overlay with a white light caption of the same membrane, in order to visualize the ladder. (F) Coomassie-stained protein gel with samples harvested during the RIC experiment. The prominent band seen in the eluate samples derives from the added RNases.
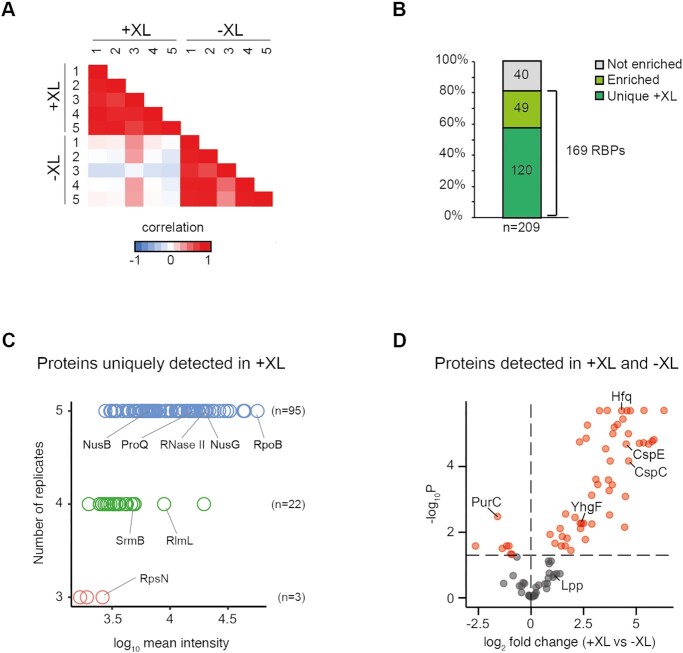
To test whether RBPs could be captured via polyadenylated RNA, cultures overexpressing PAPI were UV-irradiated to crosslink cellular RNA–protein complexes. For this, we used an E. coli strain expressing a C-terminally 3xFLAG tagged version of the well-studied RBP Hfq. After cell lysis, extracts were incubated with oligo(dT) beads to pull-down RNA and RNA–protein complexes, and the samples were analyzed by gel electrophoresis, Western blotting, and gel staining. Western blotting verified crosslinking-dependent co-purification of Hfq, indicating successful RBP co-purification (Figure 1C). Coomassie staining further revealed that the sample obtained from polyadenylated and crosslinked cultures yielded a number of different bands corresponding to proteins of different molecular masses (Figure 1D). This was not observed in control samples where either polyadenylation or crosslinking had been omitted. Next, we repeated the pull-down procedure with five independent cultures (biological replicates), each of which was divided into two halves, of which only one was crosslinked. After oligo(dT)-based purification and RNase treatment, the co-purified proteins were identified by bottom-up proteomic mass spectrometry. The protein content of crosslinked samples and non-crosslinked controls strongly correlated within each group, whereas no correlation could be observed between the groups (Figure 2A). In total, we identified 209 different proteins (Figure 2B and Supplementary Table S5), with the majority (120 proteins) exclusively found in the crosslinked samples. Of these, 79% (95 proteins) were found in all five replicates (Figure 2C). 49 out of the 89 remaining proteins were significantly enriched (FDR-adjusted P < 0.05) in the crosslinked compared to non-crosslinked samples (Figure 2D). Altogether, 169 proteins were uniquely found, or significantly enriched, in the crosslinked samples, which we therefore refer to as putative RBPs.
Figure 2.
RIC globally identifies RBPs in E. coli. (A) Pairwise correlation coefficients based on the intensity for each protein detected in each of the five crosslinked (+XL) and non-crosslinked (-XL) samples. (B) Percentage and absolute numbers of uniquely detected, enriched, or non-enriched proteins with respect to crosslinking among all detected proteins. (C) All proteins uniquely detected in crosslinked samples with respect to the number of replicates each protein was detected in (blue: all five replicates, green: four replicates, red: three replicates), and the intensity of each protein. (D) All proteins detected in both crosslinked and non-crosslinked samples plotted with respect to fold-change and multiple-testing adjusted statistical significance. Proteins with an adjusted p-value less than 0.05 are highlighted in red.
Characteristics of identified RBPs
Of the 169 putative RBPs, 82 are classified as ‘RNA-binding’ according to Gene Ontology (GO) terminology. These include proteins in the small and large ribosomal subunits, RNA polymerase subunits, ribonucleases, RNA modifying enzymes, tRNA synthetases, transcription termination factors, cold-shock proteins and sRNA-binding RBPs such as Hfq and ProQ (Figure 3A and B, Supplementary Table S5). Searching for conserved protein domains in the 169 putative RBPs revealed that all domains identified in five proteins or more are RNA-related (Supplementary Table S7), further indicating that RIC strongly enriches proteins that interact with RNA. Among the proteins identified in the pull-down with known functions not related to RNA-binding, many have enzymatic activity. For instance, we find that enzymes involved in glycolysis and the TCA-cycle are significantly overrepresented (Figure 3A and Supplementary Figure S2), and in total 87 of the 169 proteins are categorized as having catalytic activity. These findings are in line with observations from eukaryotic organisms as well as other studies from prokaryotes, where many metabolic proteins were identified as putative RBPs (48,49). We compared our findings to two other RBP identification studies in E. coli, which employed the TRAPP (19) or OOPS (21) methods, respectively. The overlap of identified proteins between the methods is very high, with 116 proteins identified in all three studies (Figure 3C). However, there is a large difference in numbers of identified proteins, with TRAPP identifying more than three times the number of putative RBPs identified in each of the other methods. Interestingly, although our approach identifies fewer proteins than TRAPP, the majority of proteins identified by our RIC protocol are among the most significant proteins identified by TRAPP (Supplementary Figure S3) and the majority (123 out of 169) of proteins are shared with OOPS (Figure 3C).
Figure 3.

Functional classification of putative RBPs identified by RIC. (A) Selected groups of statistically enriched Gene Ontology categories in the pull-down, including number of proteins from each category found in the pull-down compared to the total number of proteins in that category. Data, including statistics (Fisher's exact test with FDR correction), was collected at the Geneontology server (http://geneontology.org) using the enrichment analysis tool. FDR: false discovery rate. (B) Venn diagram showing the overlap between putative RBPs identified by RIC and selected Gene Ontology categories listed in (A). (C) Venn diagram illustrating the overlap between the RIC dataset with datasets from two other RBP identification studies in E. coli; TRAPP (using the dataset acquired using UV-light of 800 mJ /cm2) and OOPS. See main text for references.
In summary, we have established a method that allows for global identification of bacterial RBPs using polyadenylation, UV-crosslinking and oligo(dT) purification. Using this method in E. coli we identified 169 RNA-interacting proteins, of which roughly half were already annotated as RNA-binding.
Validation of RNA-binding activity of putative RBPs
The strong enrichment for known RNA-binding proteins indicates that our method is working as intended. To further probe the performance of the approach, we evaluated the RNA-binding ability of a subset of proteins not previously known to interact with RNA (Figure 4A). Of the 86 proteins not classified as RNA-binding, 11 were uncharacterized, with a protein name starting with ‘Y’. Predictions revealed that some of these may contain RNA-binding domains (see Figure 4B).
Figure 4.
RNA-binding activity of putative RBPs identified by RIC. (A) PNK assay to test RNA-binding activity of fourteen different proteins identified by RIC, plus the positive control ProQ. Bacterial cultures were grown and harvested at similar conditions as in the RIC experiment. UV-treated: XL+; non-treated: XL-. After radioactive labeling of co-purifying RNA, the samples were size-separated on polyacrylamide gels, transferred to nitrocellulose membranes, and subjected to autoradiography. Subsequently, the membranes were used for Western blotting with an anti-FLAG antibody, as seen in the lower panel. Uncropped versions of the membrane images are presented in Supplementary Figure S4. (B) List of the uncharacterized proteins tested in (A), including size and predicted domains according to Pfam. Domains shown in green are predicted to have RNA-binding activity (DUF1732 (68), S4 (69), DUF520 (70) and S1 (71)).
We tested the RNA-binding activity of ten uncharacterized proteins (YajQ, YbcJ, YbeZ, YceD, YhgF, YibL, YicC, YifE, YajC and YihI), three enzymes (Bcp, GapA and Icd), and outer membrane protein A (OmpA), as well as the well-known RBP ProQ as a positive control, using the PNK assay (50). In these assays, all proteins were 3xFLAG-tagged. OmpA, YhgF, and ProQ were expressed from their native loci on the chromosome, while the remaining proteins were expressed from plasmids. After culturing, UV-crosslinking, and immunoprecipitation, during which copurified RNAs were trimmed and radioactively labeled, the samples were separated on protein gels, transferred to membranes, and analyzed by autoradiography. Since the RNA is radioactively labeled, only proteins carrying a crosslinked RNA moiety will emit a signal, as illustrated by the control protein ProQ (Figure 4A). To control for protein expression and loading, the membranes were subsequently subjected to Western blotting with an anti-FLAG antibody, showing successful immunoprecipitation of each protein (Figure 4A). Using the same growth conditions as in the initial RIC experiment, we obtained strong radioactive signals in crosslinked samples for 11 of the 14 putative RBPs, strongly indicating that these proteins possess RNA-binding activity in vivo. Three of the tested proteins, OmpA, YajC and YicC, did not yield a strong radioactive signal. However, longer exposure times revealed weaker crosslinking-dependent signals for YajC and YicC (Supplementary Figure S4). Taken together, we tested the RNA-binding capability of 14 proteins previously not known to interact with RNA, and verified RNA-binding for 11, possibly 13, of them, indicating that our modified RIC method identifies RBPs with high stringency.
Analysis of YhgF–RNA interactions in vivo
Out of the ten uncharacterized proteins analyzed by the PNK assay (Figure 4), YhgF (also known as Tex in some bacterial species), caught our attention. It is required for toxin regulation in Bordetella pertussis (51) and Clostridium perfringens (52), and plays a role in virulence in Pseudomonas aeruginosa (53), Streptococcus pneumoniae (54) and Burkholderia pseudomallei (55). However, the molecular function of YhgF-like proteins is still unknown. YhgF contains several predicted domains, including an S1 domain which is present in many well-studied RBPs. Although the YhgF homolog in Pseudomonas, for which two crystal structures are available, has been shown to possess RNA binding activity in vitro (53), in vivo targets of members of this protein family have not been identified in any organism. To this end, we performed CLIP-seq using an E. coli strain carrying a chromosomally 3xFLAG tagged yhgF allele (Figure 5). Since RNA-binding by YhgF increased upon entry into stationary phase as compared to exponential growth in the PNK-assay (see Supplementary Figure S4), we harvested cells from three biological replicates at an OD600 of ∼1.7 (Figure 5A). In CLIP-seq, RNA fragments bound by the assayed protein are protected from digestion prior to purification and RNA-seq library preparation, thereby revealing high-affinity RNA-binding sites. To identify such sites, we applied the peak-calling algorithm PEAKachu. Manual inspection of the identified peaks and the mapped CLIP-seq reads revealed broad distribution over the entire length of transcripts (see Figure 5D for examples), in contrast to the sharp single peaks obtained RBPs such as Hfq, ProQ, and CsrA (29,56). We therefore also analyzed the data using regular differential expression analysis with DEseq2. In total, 52 different RNAs were significantly (FDR-adjusted P-value < 0.05) enriched in the crosslinked samples (Supplementary Table S8), including both mRNAs, sRNAs (e.g. RdlA, RyhB, and CsrC), and a few tRNAs (Figure 5B, C). Read coverage plots from some of the most highly enriched RNAs are shown in Figure 5D. The genes encoding YhgF-bound RNAs do not seem to be functionally related, since GO terminology analysis of the 52 enriched targets did not yield any significant enrichment. Meta-gene analysis of mRNA peaks revealed YhgF binding along the whole transcript body, with a slight enrichment of binding towards 3’ ends (Figure 5E).
Figure 5.
Identification of YhgF RNA-ligands in vivo using CLIP-seq. (A) Autoradiography image of the membrane from which the YhgF co-purifying RNA was excised. Equal parts of EHS-2291 (YhgF-3xFLAG) cultures were either crosslinked by applying UV-light (XL+) or not treated (XL−). YhgF was immunoprecipitated and co-purifying RNA was partly digested and radiolabeled with 32P. The samples were separated on polyacrylamide gels and transferred to nitrocellulose membranes. The RNA was prepared from the membrane pieces highlighted by red boxes. Co-purifying RNA was identified by deep sequencing. The experiment was performed in independent biological triplicates. (B) Plot showing all annotated genes detected in the CLIP-experiment by relative abundance (fold change) between crosslinked and non-crosslinked samples versus the average expression among all samples (baseMean). Genes significantly enriched (adjusted p-value < 0.05) in crosslinked samples are shown in red. The top enriched genes are indicated by names. (C) Pie chart illustrating the significantly enriched transcripts presented in (B) according to RNA class. (D) Visualization of the reads mapping to the transcripts for rmf, lpp, RyhB and RdlA. Black: non-crosslinked (-XL) samples, red: crosslinked (+XL) samples. The extent of the ORFs is indicated with grey boxes. The extent of transcription units is indicated by arrows (transcription start site) and rings (termination site). (E) Meta-gene analysis of relative CLIP-seq peak distribution along all detected ORFs.
To verify that the CLIP-seq data faithfully reported on preferential YhgF binding, we performed electromobility shift assays (EMSA) using purified YhgF and in vitro transcribed RNA. We chose two of the most highly enriched transcripts in the CLIP-seq dataset: the rmf mRNA, which encodes a ribosome hibernation factor, and the sRNA RyhB. The 5’UTR of gapA mRNA, for which no YhgF binding site (peak) was detected by CLIP-seq (Supplementary Table S8), was used as negative control. The EMSAs revealed binding between YhgF and all tested RNAs, however, the affinity for rmf and RyhB was much higher than for the gapA 5’UTR (Figure 6A, Supplementary Figure S5), with an estimated KD for the YhgF-rmf interaction between 150 and 300 nM. The difference in affinity was further demonstrated by challenging the rmf-YhgF complex with unlabeled rmf or gapA 5’UTR. Unlabeled rmf efficiently competed off the radioactively labeled rmf from YhgF, while the non-target gapA 5’UTR failed to do so, consistent with rmf being a specific YhgF ligand (Figure 6B). We noted that experiments with rmf resulted in a slowly migrating complex that appears to be stuck in the well during electrophoresis. We interpreted this as unspecific clumping of protein and thus disregarded it when estimating the KD of the interaction. We also noted that the binding between rmf and YhgF appears to be highly cooperative, as the rmf RNA migrates as from almost completely unbound to almost completely bound over a single doubling of YhgF concentration (150–300 nM).
Figure 6.

YhgF binds to rmf mRNA and regulates Rmf expression. (A) Migration of in vitro transcribed and radioactively labeled rmf mRNA or gapA 5’UTR (negative control) in a non-denaturing gel after incubation with increasing concentrations of purified YhgF (0, 75, 150, 300, 600 and 1200 nM). (B) Competition assay between rmf mRNA and gapA 5’UTR. A preformed complex of labeled rmf and YhgF (200 nM), was challenged with increasing concentrations of unlabeled competitor RNA (50, 100, 200, 400 and 800 nM), The gel includes control lanes with labeled rmf only and the preformed complex of YhgF and labeled rmf without competitor. (C) In vivo levels of rmf mRNA in WT, yhgF deletion, and yhgF overexpression strains. Relative mRNA levels were determined by RT-qPCR. Bars represent average values based on six biological replicates for each strain. Error bars represent standard deviation. Statistical significance was determined using a two-tailed t test.
Since the biological function of YhgF is unknown, we asked if YhgF affects expression of its RNA ligands. To this end, we monitored rmf mRNA steady-state levels by qPCR upon deletion or overexpression of yhgF. While yhgF deletion did not affect rmf mRNA beyond wild-type levels, overexpression of YhgF lead to significantly (P < 0.05) increased levels (Figure 6C). In summary, our results indicate that YhgF is an RBP that interacts with mRNAs and sRNAs in E. coli, and may affect the levels of its RNA targets in vivo.
YhgF is a highly conserved protein
YhgF is annotated to contain five functional domains (Figure 7A). Interestingly, the same domain arrangement is present in the two human proteins SRBD1 and SPT6, which both share a high degree of sequence conservation with YhgF (Supplementary Figure S6). Moreover, alignment of AlphaFold-generated structures indicates that YhgF and SRBD1 adopt highly similar folds (Figure 7B), suggesting that YhgF-like proteins have been conserved over great evolutionary distances. To investigate this, we searched in all bacterial phyla and in the major groups of archaeal and eukaryotic organisms for YhgF homologs with an amino acid sequence identity of at least 32% covering more than 80% of the YhgF sequence (Supplementary Table S6). Strikingly, we identified homologous proteins in 26 of the 30 probed bacterial phyla, in all major archaeal groups (Euryarchaeota, Proteoarchaeota and DPANN), and in most eukaryotic supergroups (Figure 7C). This remarkably high degree of conservation of YhgF-like proteins likely indicates an ancient and important function.
Figure 7.
The YhgF protein family is conserved in all domains of life. (A) Domain organization of E. coli YhgF and the human proteins SRBD1 and SPT6 according to Pfam annotations. C- and N-terminal extensions of SPT6 are not shown. (B) Structural alignment (RMSD: 3.7 over 544 residues) of YhgF (residues 1–722) and human SRBD1 (residues 211–995). The alignment was generated using Pymol (cealign command) with AlphaFold structures downloaded from Uniprot. (C) Phylogenetic tree built from protein sequences homologous to YhgF identified by BLAST searches within major evolutionary groups of bacterial, archaeal, and eukaryotic organisms.
DISCUSSION
In the present study we have established a protocol for RBP identification in bacteria based on the RIC method (Figure 1). Through purification of in vivo polyadenylated and UV-crosslinked total RNA, we identified 169 RNA-interacting proteins in E. coli, half of which were previously classified as RBPs (Figures 2 and 3). We verified cellular RNA-binding activity for a dozen previously unknown RBPs, the majority of which lack canonical RNA-binding domains (Figure 4). Finally, we analyzed in vivo and in vitro RNA-binding of the uncharacterized protein YhgF (Figures 5 and 6), a highly conserved protein present in all domains of life (Figure 7), and observed that overexpression of YhgF lead to increased levels of one of its identified mRNA ligands (Figure 6).
The global RBP identification studies conducted in bacteria so far have resulted in very different numbers of detected putative RBPs. The PTex and OOPS methods, both relying on organic phase separation, identified 172 and 364 putative RBPs in Salmonella and E. coli, respectively (21,22). The TRAPP method, which relies on purification of RNA–protein complexes using silica beads, detected between 322 and 1106 putative E. coli RPBs depending on the dose of UV-light applied to the cells (19). These discrepancies may not only reflect intrinsic differences in the approaches, but also effects of the chosen growth conditions. For instance, proteins that moonlight as RBPs show condition-dependent RNA-binding activity (16,57,58), and the total number of expressed RBPs will vary between condition. For these reasons, it is difficult to determine the fraction of total proteins that can act as RBPs.
In this study, we detect ∼4% of the proteome as putative RBPs. Based on the above-mentioned studies in both bacteria and eukaryotes, where approximately 5–20% of proteins are estimated to be RBPs, it is possible that our results underestimate the total number of RBPs in E. coli. In line with this, it is clear that calculating significant enrichment in UV-treated samples versus non-treated samples resulted in removal of some known RBPs. For instance, the RNA modification proteins RluC, RlmN, RlmA, TruA, and Tgt, the aminoacyl-tRNA synthetase HisRS, and the RNase P protein subunit RnpA, were not significantly enriched in the crosslinked samples, due to similar abundances in crosslinked and non-crosslinked samples (see Supplementary Table S5). Possibly, the interactions of these proteins with their RNA-targets are so strong that they persist during the pull-down even without crosslinking. Although the degree of false positives produced by the different methods is difficult to estimate, we argue that that our dataset likely contains a small fraction of falsely identified RBPs, based on the following: (i) half of all identified proteins are previously verified RBPs (Figure 2), (ii) 60% of all proteins were uniquely detected in crosslinked samples (Figure 2), (iii) the in vivo RNA-binding activity of eleven of fourteen tested proteins were verified in the PNK assays (Figure 4), while two of the three ‘non-binders’ actually showed weak RNA-binding activity (Supplementary Figure S4), tentatively suggesting >90% of the tested proteins to be true RBPs. A possible source of false positives in RBP identification approaches is that UV-light not only crosslinks RNA–protein interactions, but also, to some extent, DNA-protein complexes (59). In this respect, RIC should be superior to other methods, since it relies on capturing extended single stranded poly(A) tails, considerably limiting the risk of enriching for DNA–protein complexes. Another advantage with RIC is that it relies on a simple and well-understood principle; base-pairing between poly(A) RNA sequences and poly(dT) oligonucleotides. Hence, pull-down of RNA–protein complexes does not rely on the nature of the protein or RNA, as long as the 3’ end of the RNA can be polyadenylated. In contrast, the chemistry underlying RBP-enrichment using organic phase separation is not completely understood. For instance, it is unclear how each individual protein's chemical properties affect its behavior in these assays. Similarly, silica-based purification may result in false positives due to specific interaction with DNA (60), as well as selective binding of disordered protein regions (61).
On the other hand, RBP identification methods relying on organic separation (21,22), binding to silica beads (19), or RNase-sensitivity in gradient sedimentation (24), do not require any genetic manipulation, while our RIC protocol for bacteria relies on overexpression of PAPI. It will therefore be applicable in any bacterial species in which ectopic overexpression can be achieved, but obviously less useful in genetically intractable species. However, implementing polyadenylation in cell lysates using commercially available PAPI could circumvent this limitation. Another limitation with RIC is the potential impact on global gene expression caused by PAPI overexpression. Although we did not observe a significant effect on bacterial growth within the assayed time after PAPI induction (Supplementary Figure S1A), PAPI-dependent changes in the levels of specific transcripts cannot be ruled out. The advantages and limitations of different methods notwithstanding, the development of RBP identification methods relying on distinct purification principles strengthens the evidence for proteins identified by all methods being bona fide RBPs.
We used the PNK assay to verify the RNA-binding potential of a number of proteins previously unknown to bind RNA. 14 different proteins were tested, of which all except YicC, YajC and OmpA gave strong radioactive signals indicative of RNA-binding (Figure 4). The apparent lack of RNA-binding for these proteins could be due to the addition of the 3xFLAG tag that render the proteins unable to interact with RNA. The weak crosslinking-dependent signals obtained for YicC and YajC suggest that the tagged proteins may possess some residual RNA-binding activity. Interestingly, a recent study showed that YicC regulates stability of the sRNA RyhB, presumably through a direct interaction (62).
All of the three well-characterized enzymes tested (Bcp, Icd, and GapA) show positive signals for RNA-binding (Figure 4). This finding is congruent with similar studies in eukaryotic organisms, where many core house-keeping enzymes moonlight as RBPs (13,48,49). Our results suggest that this alternative function probably has an ancient origin and is conserved between very distant groups in the tree of life. Regarding the uncharacterized proteins identified as putative RBPs in RIC, RNA-binding activity was supported for eight of the ten tested proteins (Figure 4). This strongly suggests that even in the extensively studied bacterium E. coli, novel RBPs are still there to be discovered.
Among the uncharacterized putative RBPs, YhgF stood out due to having five predicted domains, including the well-characterized S1 domain, and a remarkable high degree of conservation across all domains of life (Figure 7). Using CLIP-seq we identified its cellular RNA-targets (Figure 5), and we could verify binding to the rmf mRNA and the sRNA RyhB in vitro (Figure 6 and Supplementary Figure S5). Overexpression of YhgF lead to increased rmf mRNA levels, suggesting that YhgF might affect the transcription or the stability of one of its RNA targets. The human YhgF homolog SPT6 interacts with RNA polymerase (RNAP) II during transcription elongation and is critical for transcription processivity (63). Providing purified YhgF during in vitro transcription did not result in an apparent effect on rmf mRNA synthesis (Supplementary Figure S7), however, additional factors may be needed to reconstitute such an effect. Interestingly, WebFlags analysis (64) revealed that the yhgF gene is flanked by greB, encoding a transcription elongation factor that alleviates RNAP stalling (reviewed in (65)), in many enterobacterial species (Supplementary Figure S8). In addition, thermal proteome profiling suggests a physical association between YhgF and GreB (66). The mechanistic details of YhgF’s role in gene expression and regulation will be an important subject for future studies.
Taken together, we have adapted the RNA interactome capture technique to bacteria and identified a large number of RNA-interacting proteins in E. coli. The method is simple to perform and produces a low fraction of false-positives. This study, in conjunction with other recently published similar studies, suggests that bacteria encode many more RBPs than previously assumed. Together, these findings lay the foundation for substantially expanding the roles of RNA-interacting proteins in bacteria.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (67) partner repository with the dataset identifier PXD032939 and 10.6019/PXD032939.
The RNA-seq data of polyadenylated and oligo(dT)-purified total RNA, and the YhgF CLIP-seq data, are available at the NCBI Gene Expression Omnibus with the accession numbers GSE217569 and GSE198953, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Liis Andresen for construction of plasmid pLA100 and for valuable input regarding the RIC method, and Gerhart Wagner for carefully reading and commenting on the manuscript.
Author contributions: T.S.S. and E.H. wrote the manuscript, conceived and designed the research. T.S.S., A.D.K., F.A.S., E.T.J. and E.H. designed the experiments. T.S.S., A.D.K. and F.A.S. performed the experiments. T.S.S., A.D.K., F.A.S., J.K., J.J., E.T.J. and E.H. analyzed the data. E.H., S.K., E.T.J. and P.E.A. contributed with funding acquisition, methodology, resources and supervision. All authors commented on the manuscript and approved the submitted version.
Contributor Information
Thomas Søndergaard Stenum, Microbiology and Immunology, Department of Cell and Molecular Biology, Biomedical Centre, Uppsala University, Box 596, 75124 Uppsala, Sweden.
Ankith D Kumar, Microbiology and Immunology, Department of Cell and Molecular Biology, Biomedical Centre, Uppsala University, Box 596, 75124 Uppsala, Sweden.
Friederike A Sandbaumhüter, Medical Mass Spectrometry, Department of Pharmaceutical Biosciences, Biomedical Centre, Uppsala University, Box 591, 75124 Uppsala, Sweden.
Jonas Kjellin, Microbiology and Immunology, Department of Cell and Molecular Biology, Biomedical Centre, Uppsala University, Box 596, 75124 Uppsala, Sweden.
Jon Jerlström-Hultqvist, Microbiology and Immunology, Department of Cell and Molecular Biology, Biomedical Centre, Uppsala University, Box 596, 75124 Uppsala, Sweden.
Per E Andrén, Medical Mass Spectrometry, Department of Pharmaceutical Biosciences, Biomedical Centre, Uppsala University, Box 591, 75124 Uppsala, Sweden; Science for Life Laboratory, Spatial Mass Spectrometry, Biomedical Centre, Uppsala University, Box 591, 75124 Uppsala, Sweden.
Sanna Koskiniemi, Microbiology and Immunology, Department of Cell and Molecular Biology, Biomedical Centre, Uppsala University, Box 596, 75124 Uppsala, Sweden.
Erik T Jansson, Medical Mass Spectrometry, Department of Pharmaceutical Biosciences, Biomedical Centre, Uppsala University, Box 591, 75124 Uppsala, Sweden.
Erik Holmqvist, Microbiology and Immunology, Department of Cell and Molecular Biology, Biomedical Centre, Uppsala University, Box 596, 75124 Uppsala, Sweden.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Foundation for Strategic Research [ICA16-0021 to E.H., ICA16-0010 to E.T.J., RIF14-0078 to P.E.A.]; Swedish Research Council [2016-03656, 2021-04657 to E.H., 2018-03988 to E.T.J., 2018-03320 to P.E.A.]; Science for Life Laboratory (to P.E.A.). Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Holmqvist E., Vogel J.. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018; 16:601–615. [DOI] [PubMed] [Google Scholar]
- 2. Corley M., Burns M.C., Yeo G.W.. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell. 2020; 78:9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Santiago-Frangos A., Woodson S.A.. Hfq chaperone brings speed dating to bacterial sRNA. WIREs RNA. 2018; 9:e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Romeo T., Babitzke P.. Global regulation by CsrA and its RNA antagonists. Microbiol. Spectrum. 2018; 6: 10.1128/microbiolspec.rwr-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Franze de Fernandez M.T., Eoyang L., August J.T.. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968; 219:588–590. [DOI] [PubMed] [Google Scholar]
- 6. Romeo T., Gong M., Liu M.Y., Brun-Zinkernagel A.M.. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1993; 175:4744–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jay G., Kaempfer R.. Host interference with viral gene expression: mode of action of bacterial factor i. J. Mol. Biol. 1974; 82:193–212. [DOI] [PubMed] [Google Scholar]
- 8. Liu M.Y., Romeo T.. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 1997; 179:4639–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smirnov A., Förstner K.U., Holmqvist E., Otto A., Günster R., Becher D., Reinhardt R., Vogel J.. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lamm-Schmidt V., Fuchs M., Sulzer J., Gerovac M., Hör J., Dersch P., Vogel J., Faber F.. Grad-seq identifies KhpB as a global RNA-binding protein in Clostridioides difficile that regulates toxin production. Microlife. 2021; 2: 10.1093/femsml/uqab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castello A., Horos R., Strein C., Fischer B., Eichelbaum K., Steinmetz L.M., Krijgsveld J., Hentze M.W.. System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc. 2013; 8:491–500. [DOI] [PubMed] [Google Scholar]
- 12. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M.et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 13. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 14. Tang Y., Guest J.R.. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology. 1999; 145:3069–3079. [DOI] [PubMed] [Google Scholar]
- 15. Tang Y., Quail M.A., Artymiuk P.J., Guest J.R., Green J.. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology. 2002; 148:1027–1037. [DOI] [PubMed] [Google Scholar]
- 16. Hentze M.W., Argos P.. Homology between IRE-BP, a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991; 19:1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hentze M.W., Kühn L.C.. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. 1996; 93:8175–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hentze M.W., Preiss T.. The REM phase of gene regulation. Trends Biochem. Sci. 2010; 35:423–426. [DOI] [PubMed] [Google Scholar]
- 19. Shchepachev V., Bresson S., Spanos C., Petfalski E., Fischer L., Rappsilber J., Tollervey D.. Defining the RNA interactome by total RNA -associated protein purification. Mol. Syst. Biol. 2019; 15:e8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu L.-C., Arede P., Li W., Urdaneta E.C., Ivanova I., McKellar S.W., Wills J.C., Fröhlich T., von Kriegsheim A., Beckmann B.M.et al.. The RNA-bound proteome of MRSA reveals post-transcriptional roles for helix-turn-helix DNA-binding and Rossmann-fold proteins. Nat. Commun. 2022; 13:2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Queiroz R.M.L., Smith T., Villanueva E., Marti-Solano M., Monti M., Pizzinga M., Mirea D.-M., Ramakrishna M., Harvey R.F., Dezi V.et al.. Comprehensive identification of RNA–protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 2019; 37:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Urdaneta E.C., Vieira-Vieira C.H., Hick T., Wessels H.-H., Figini D., Moschall R., Medenbach J., Ohler U., Granneman S., Selbach M.et al.. Purification of cross-linked RNA–protein complexes by phenol-toluol extraction. Nat. Commun. 2019; 10:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith T., Villanueva E., Queiroz R.M.L., Dawson C.S., Elzek M., Urdaneta E.C., Willis A.E., Beckmann B.M., Krijgsveld J., Lilley K.S.. Organic phase separation opens up new opportunities to interrogate the RNA-binding proteome. Curr. Opin. Chem. Biol. 2020; 54:70–75. [DOI] [PubMed] [Google Scholar]
- 24. Gerovac M., El Mouali Y., Kuper J., Kisker C., Barquist L., Vogel J.. Global discovery of bacterial RNA-binding proteins by RNase-sensitive gradient profiles reports a new FinO domain protein. RNA. 2020; 26:1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Casadaban M.J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 1976; 104:541–555. [DOI] [PubMed] [Google Scholar]
- 26. Koskiniemi S., Pränting M., Gullberg E., Näsvall J., Andersson D.I.. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol. Microbiol. 2011; 80:1464–1478. [DOI] [PubMed] [Google Scholar]
- 27. Sawitzke J.A., Thomason L.C., Costantino N., Bubunenko M., Datta S., Court D.L.. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007; 421:171–199. [DOI] [PubMed] [Google Scholar]
- 28. Miller J.H. Experiments in Molecular Genetics. 1972; Cold Spring Harbor, NY: ColdSpring Harbor Laboratory Press. [Google Scholar]
- 29. Holmqvist E., Li L., Bischler T., Barquist L., Vogel J.. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol. Cell. 2018; 70:971–982. [DOI] [PubMed] [Google Scholar]
- 30. Sturm M., Schroeder C., Bauer P.. SeqPurge: highly-sensitive adapter trimming for paired-end NGS data. BMC Bioinf. 2016; 17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 35. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sandbaumhüter F.A., Nezhyva M., Eriksson O., Engberg A., Kreuger J., Andrén P.E., Jansson E.T.. Well-plate μFASP for proteomic analysis of single pancreatic islets. J. Proteome Res. 2022; 21:1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Distler U., Kuharev J., Navarro P., Tenzer S.. Label-free quantification in ion mobility–enhanced data-independent acquisition proteomics. Nat. Protoc. 2016; 11:795–812. [DOI] [PubMed] [Google Scholar]
- 38. Distler U., Kuharev J., Navarro P., Levin Y., Schild H., Tenzer S.. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods. 2014; 11:167–170. [DOI] [PubMed] [Google Scholar]
- 39. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Church G.M., Gilbert W.. Genomic sequencing. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010; 26:2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bischler T., Förstner K.U., Maticzka D., Wright P.R.. PEAKachu: a peak calling tool for CLIP/RIP-seqdata (V0.2.0). 2021; Zenodo 10.5281/zenodo.4669966. [DOI]
- 44. Zhou K., Zhou L., Lim Q.’E., Zou R., Stephanopoulos G., Too H.-P.. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011; 12:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baltz A.G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M.et al.. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 46. Mitchell S.F., Jain S., She M., Parker R.. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013; 20:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.-M., Foehr S., Curk T., Krijgsveld J., Hentze M.W.. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016; 63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Castello A., Hentze M.W., Preiss T.. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 2015; 26:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.-M., Schwarzl T., Curk T., Foehr S., Huber W.et al.. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015; 6:10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tawk C., Sharan M., Eulalio A., Vogel J.. A systematic analysis of the RNA-targeting potential of secreted bacterial effector proteins. Sci. Rep. 2017; 7:9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuchs T.M., Deppisch H., Scarlato V., Gross R.. A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J. Bacteriol. 1996; 178:4445–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abe K., Obana N., Nakamura K.. Effects of depletion of RNA-binding protein Tex on the expression of toxin genes in Clostridium perfringens. Biosci. Biotechnol. Biochem. 2010; 74:1564–1571. [DOI] [PubMed] [Google Scholar]
- 53. Johnson S.J., Close D., Robinson H., Vallet-Gely I., Dove S.L., Hill C.P.. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J. Mol. Biol. 2008; 377:1460–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He X., Thornton J., Carmicle-Davis S., McDaniel L.S.. Tex, a putative transcriptional accessory factor, is involved in pathogen fitness in Streptococcus pneumoniae. Microb. Pathog. 2006; 41:199–206. [DOI] [PubMed] [Google Scholar]
- 55. Moule M.G., Spink N., Willcocks S., Lim J., Guerra-Assunção J.A., Cia F., Champion O.L., Senior N.J., Atkins H.S., Clark T.et al.. Characterization of new virulence factors involved in the intracellular growth and survival of Burkholderia pseudomallei. Infect. Immun. 2016; 84:701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J.. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016; 35:991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nagy E., Rigby W.F.C.. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD+-binding region (Rossmann Fold). J. Biol. Chem. 1995; 270:2755–2763. [DOI] [PubMed] [Google Scholar]
- 58. Chu E., Koeller D.M., Casey J.L., Drake J.C., Chabner B.A., Elwood P.C., Zinn S., Allegra C.J.. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:8977–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stützer A., Welp L.M., Raabe M., Sachsenberg T., Kappert C., Wulf A., Lau A.M., David S.-S., Chernev A., Kramer K.et al.. Analysis of protein-DNA interactions in chromatin by UV induced cross-linking and mass spectrometry. Nat. Commun. 2020; 11:5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J.. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990; 28:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klein G., Mathé C., Biola-Clier M., Devineau S., Drouineau E., Hatem E., Marichal L., Alonso B., Gaillard J.-C., Lagniel G.et al.. RNA-binding proteins are a major target of silica nanoparticles in cell extracts. Nanotoxicology. 2016; 10:1555–1564. [DOI] [PubMed] [Google Scholar]
- 62. Chen J., To L., de Mets F., Luo X., Majdalani N., Tai C.-H., Gottesman S.. A fluorescence-based genetic screen reveals diverse mechanisms silencing small RNA signaling in E. coli. Proc. Natl. Acad. Sci. 2021; 118:e2106964118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Narain A., Bhandare P., Adhikari B., Backes S., Eilers M., Dölken L., Schlosser A., Erhard F., Baluapuri A., Wolf E.. Targeted protein degradation reveals a direct role of SPT6 in RNAPII elongation and termination. Mol. Cell. 2021; 81:3110–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saha C.K., Sanches Pires R., Brolin H., Delannoy M., Atkinson G.C.. FlaGs and webFlaGs: discovering novel biology through the analysis of gene neighbourhood conservation. Bioinformatics. 2021; 37:1312–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deighan P., Hochschild A.. Conformational toggle triggers a modulator of RNA polymerase activity. Trends Biochem. Sci. 2006; 31:424–426. [DOI] [PubMed] [Google Scholar]
- 66. Mateus A., Hevler J., Bobonis J., Kurzawa N., Shah M., Mitosch K., Goemans C.V., Helm D., Stein F., Typas A.et al.. The functional proteome landscape of Escherichia coli. Nature. 2020; 588:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M.et al.. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022; 50:D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rigden D.J. Ab initio modeling led annotation suggests nucleic acid binding function for many DUFs. OMICS. 2011; 15:431–438. [DOI] [PubMed] [Google Scholar]
- 69. Volpon L., Lievre C., Osborne M.J., Gandhi S., Iannuzzi P., Larocque R., Cygler M., Gehring K., Ekiel I.. The solution structure of YbcJ from Escherichia coli reveals a recently discovered αL motif involved in RNA binding. J. Bacteriol. 2003; 185:4204–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Teplyakov A., Obmolova G., Bir N., Reddy P., Howard A.J., Gilliland G.L.. Crystal structure of the YajQ protein from Haemophilus influenzae reveals a tandem of RNP-like domains. J. Struct. Funct. Genomics. 2003; 4:1–9. [DOI] [PubMed] [Google Scholar]
- 71. Bycroft M., Hubbard T.J., Proctor M., Freund S.M., Murzin A.G.. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid–binding fold. Cell. 1997; 88:235–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (67) partner repository with the dataset identifier PXD032939 and 10.6019/PXD032939.
The RNA-seq data of polyadenylated and oligo(dT)-purified total RNA, and the YhgF CLIP-seq data, are available at the NCBI Gene Expression Omnibus with the accession numbers GSE217569 and GSE198953, respectively.