Dear Editor,
The global coronavirus disease 2019 (COVID-19) pandemic, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has brought a profound impact on humanity. SARS-CoV-2 enters host cells through the interaction of its spike glycoprotein with angiotensin-converting enzyme 2 (ACE2) of the host and releases its viral RNA genome.1 The genome is further translated into two polyproteins, PP1a and PP1ab, that contain 16 non-structural proteins, in which the non-structural protein 5 (also called as 3C-Like protease, 3CLpro) plays a vital role in the life cycle of the virus.2 As a consequence, multiple peptide-like covalent 3CLpro inhibitors have been successively reported to show good 3CLpro inhibitory potencies and antiviral activities.3,4 However, the defects of peptidomimetic inhibitors about low membrane permeability and metabolic stability are obvious, in which the approved drug paxlovid (PF-07321332) from Pfizer must be co-administrated with ritonavir as a pharmacokinetic booster.3 Non-peptidic small molecule inhibitors may have more advantages in metabolic properties and are gradually attracting attention.5–7 Our research attempted to discover a novel non-peptidic and non-covalent small molecule 3CLpro inhibitor so as to give better drug-like properties and good antiviral activities.
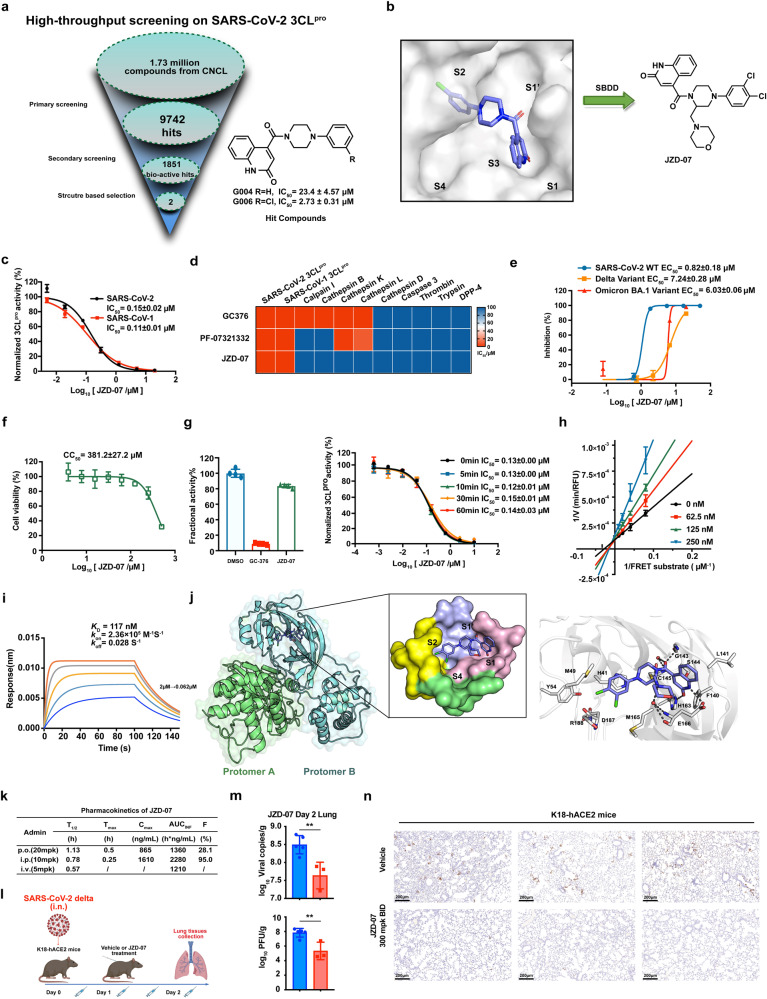
After screening more than 1,700,000 compounds from the Chinese National Compound Library,8 we identified 1851 hits targeting 3CLpro. Taken together with molecular docking and cell-based antiviral assay, we selected two structurally similar compounds, G006 and G004, for further optimization (Fig. 1a). The docking analyses for G006 demonstrated that the quinoline-2-one occupies the S1 site to form an important hydrogen bond with Glu166 and Gly143, the 3-chlorophenyl occupies the hydrophobic S2 site to form a π–π interaction with His41. (Fig. 1b and Supplementary Fig. 1). As we have seen, the active site was not fully occupied by G006, which may limit its inhibition potency against 3CLpro (Fig. 1b). Thus, we have attempted to introduce diversified side chains onto the core piperazine ring in the hope of filling these unoccupied cavities fully. Finally, JZD-07 outstripped its analogs, exhibiting excellent inhibitory activity against the 3CLpro of both SARS-CoV-2 and SARS-CoV-1, with IC50 of 0.15 μM and 0.11 μM, respectively (Fig. 1c).
Fig. 1.
Discovery of Novel Non-peptidic and Non-covalent Small Molecule SARS-CoV‑2 3CL Protease Inhibitors. a Flowchart of the High-throughput screening and hit validation process. b Docking of lead compounds with SARS-CoV-2 3CLpro and rationale design for the noncovalent SARS-CoV-2 3CLpro inhibitor. c Compound JZD-07 is a potent inhibitor of SARS-CoV-2 3CLpro as well as SARS-CoV-1 3CL.pro d Biochemical assay of several human proteases, compound JZD-07 showing great selectivity against human protease, especially Calpain I and Cathepsin family protein, the values plotted were the IC50 values from the FRET-based enzymatic assay shown as a heatmap. e Inhibition of original strain (blue), Delta variant (yellow), and Omicron BA.1. variant (red) replication by Compound JZD-07 in Vero E6 cells. Data are shown as mean ± SD, n = 3 biological replicates. f Cytotoxicity (CC50) to Vero E6 cells measured by CCK-8 assays. Data are shown as mean ± SD, n = 3 biological replicates. g Rapid dilution assay and time-dependent inhibition of SARS-CoV-2 3CLpro by Compound JZD-07. A 40 nM SARS-CoV-2 3CLpro was preincubated with Compound JZD-07 for various periods of time (0 min to 1 h) before the addition of 20 μM FRET substrate to initiate the enzymatic reaction. Data are shown as mean ± SD. h Lineweaver-Burk plot depicting the mode of inhibition for Compound JZD-07 on SARS-CoV-2 3CLpro activity using different inhibitor concentrations. i Compound JZD-07 to SARS-CoV-2 3CLpro measured by biolayer interferometry (BLI), BLI analysis showing the representative association and disassociation curves of Compound JZD-07 at series concentrations. j Binding mode of JZD-07 (PDB ID: 8GTV) with SARS-CoV-2 3CLpro revealed by co-crystal structure. The two protomers are shown in the green (protomer A) and cyan (protomer B) cartoon, respectively. JZD-07 is shown as slate sticks. The surface representation of JZD-07 interacting with S1ʹ-S4 subsites of SARS-CoV-2 3CLpro. The subsites are colored light blue (S1ʹ), light pink (S1), yellow (S2), and green (S4). Interactions of JZD-07 with the surrounding residues are also revealed by the crystal structures. Residues (gray), as well as JZD-07, are shown in the sticks. H-bonds are represented by black dashed lines. k Preliminary PK evaluation of compound JZD-07 in mice. Dosed orally at 20 mg/kg. Dosed intraperitoneally at 10 mg/kg. Dosed intravenously at 5 mg/kg. l In vivo efficacy of compound JZD-07 in K18-hACE2 mice infected with SARS-CoV-2 delta variant. Experiment flow diagram. m Viral RNA copies and viral titers in lung tissues of each group were determined on day 2 after the SARS-CoV-2 delta variant challenge (**P ≤ 0.01). n Immunohistochemistry staining targets viral nucleocapsid protein. (SARS-CoV-2 Nucleocapsid Protein (HL344) Rabbit mAb #26369, CST). Viral nucleocapsid proteins were stained in brown
To explore the selectivity of this 3CLpro inhibitor, we first examined its inhibitory activities against multiple closely related proteases, which indicated JZD-07 had almost no inhibition for human calpain I, cathepsin (B, D, K, and L), thrombin A, trypsin, caspase-3, DPP-4, and proteasome, while the control 3CLpro inhibitors GC-376 and PF-07321332 showed certain inhibitory potencies against different cathepsin proteins (Fig. 1d). We also evaluated the inhibitory potencies toward SARS-CoV-2 viral life cycle related targets, and JZD-07 had no inhibition against those enzymes with IC50 > 100 μM (Supplementary Table 1). These data demonstrate that JZD-07 is a highly specific 3CLpro inhibitor.
Based on the inhibitory potency against 3CLpro, we conducted an antivirus assay of JZD-07 in Vero E6 cells. JZD-07 showed good inhibitory activity against the SARS-CoV-2 original virus with EC50 of 0.82 μM and also showed dose-dependent inhibition on the replication of delta and omicron BA.1. variant with EC50 values of 7.24 μM and 6.03 μM, respectively (Fig. 1e and Supplementary Fig. 2). Besides, JZD-07 exhibited good antivirus activities against HCoV-OC43 and HCoV-229E (Supplementary Fig. 2). Meanwhile, this compound almost has no effect on cell viability with the CC50 in Vero E6 cell over 300 μM (Fig. 1f). These results indicate that JZD-07 has good antiviral activity in vitro and almost has no cytotoxicity.
To elucidate the mechanism of action of this non-peptidic small molecule inhibitor, the enzyme kinetics and drug binding-kinetics profiles were studied in detail. Firstly, the inhibition reversibility was investigated by rapid dilution assay. JZD-07 (10 μM) or an irreversible 3CLpro inhibitor (GC-376, 2 μM) were incubated with SARS-CoV-2 3CLpro (2 μM) for 20 min, the activities of 3CLpro incubated with JZD-07 recovered more than 70% after 100-fold dilution. Meanwhile, the IC50 values did not change with preincubation time (Fig. 1g), which indicates that the inhibition against 3CLpro is reversible. Furthermore, we determined the kinetic parameters using different concentrations of JZD-07. After global fits of the respective enzyme inhibition curves, JZD-07 is shown to be a competitive inhibitor with a Ki value of 0.117 μM (Fig. 1h), comparable to the reported 3CLpro inhibitor CCF981 and more potent than ML-188 (Supplementary Fig. 3). Recently, the importance of drug-binding kinetics profiles for drug design have been increasingly recognized. We utilized bio-layer interferometry (BLI) assay to measure the binding kinetics and affinity parameters for our compound (Fig. 1i and Supplementary Fig. 4). An overall kon value of JZD-07 is 2.36 × 105 M−1 S,−1 the dissociation behavior is very slow with koff = 0.028 S−1, and the KD value of JZD-07 is 117 nM. Taken together, the results of enzymatic inhibition and BLI data indicate that JZD-07 is a high affinity, slow-dissociation, and reversible competitive 3CLpro inhibitor.
To elucidate the precise binding modes of the inhibitor with protease, we successfully determined the crystal structure of SARS-CoV-2 3CLpro in complex with JZD-07 at a resolution of 1.80 Å (Supplementary Table 2). The binding modes showed JZD-07 non-covalently bounded into the substrate-binding site and occupied S1’, S1, and S2 subsites (Fig. 1j). The 2-quinolinone group fitted into the S1 subsite by forming hydrogen bonds with the side chains of His163 and Glu166. The carbonyl oxygen atom between the piperazine ring and quinoline ring fixed the conformation of the flexible oxyanion loop by making hydrogen bonds with the main chain of Gly143 as well as a water molecule which simultaneously contacted with the main chain of Cys145. The O-dichlorobenzene ring is deeply inserted into the S2 subsite by making extensive hydrophobic interactions with multiple residues His41/Met49/Tyr54/Met165/Asp187/Arg188. Notably, it also formed π–π stacking interactions with the side chain of catalytic His41. The morpholinyl group of JZD-07 oriented to the S1 subsite, producing water-mediated hydrogen bonds and hydrophobic interactions with the residue Glu166. These results together provided the molecular details JZD-07 recognized by SARS-CoV-2 3CLpro, which is consistent with its excellent 3CLpro inhibitory activities.
Considering our compound is a novel non-covalent and non-peptidic 3CLpro inhibitor, we further explored the potential advantages of PK profiles. Oral administration of 20 mg/kg of JZD-07 could give favorable PK parameters with an oral bioavailability of 28.1%. Intraperitoneal administration of 10 mg/kg of JZD-07 also showed an excellent PK profile with an AUC of 2280 h*ng/mL and a Cmax of 1610 ng/mL (Fig. 1k). Besides, we have launched an acute toxicity experiment, and the results indicate that JZD-07 has a good safety profile (Supplementary Fig. 6). These favorable PK and safety characteristics indicated JZD-07 might be suitable for the treatment of SARS-CoV-2 in vivo infection model.
In the end, we investigated the antiviral potency for JZD-07 in K18-hACE2 mice challenged with the SARS-CoV-2 delta variant. After 2 h of challenge, the mice were injected intraperitoneally with JZD-07 or vehicle (day 0). Then mice were administrated twice at 8 h intervals on day 1. On day 2, the mice were euthanized 4 h after receiving one compound or vehicle treatment (Fig. 1l). Lung tissues were collected for the detection of viral RNA and viral titer, histopathological analysis and immunostaining of lungs were also evaluated. Compared with the vehicle group, the viral copy loads of the lung in the JZD-07 (300 mg/kg bid) treated group were significantly reduced by nearly 1.0 log on day 2. More importantly, the viral titers of the lung of the JZD-07 treated group showed 2.0 log reduction (Fig. 1m). Immunohistochemistry staining targeting viral nucleocapsid protein demonstrated that JZD-07 suppressed viral titer in mice lungs at day 2 (Fig. 1n). All these results indicate JZD-07 has a good therapeutic potential against SARS-CoV-2 delta variant in vivo.
Therefore, we here reported our discovery of a novel non-peptidic and non-covalent small molecule SARS-CoV-2 3CLpro inhibitor. Based on the hit compounds from high-throughput screening studies and a structure-based drug design strategy, compound JZD-07 stands out and shows an excellent inhibitory potency against 3CLpro. It also exhibits good anti-SARS-CoV-2 activity in Vero E6 cells without cytotoxicity. Mechanistic studies and X-ray crystal structure analysis have shown that JZD-07 is a highly selective, slow-dissociation, non-covalent, and reversible competitive 3CLpro inhibitor. JZD-07 also displays favorable pharmacokinetic profiles in mice with good plasma exposure and bioavailability. More importantly, JZD-07 exhibits a good therapeutic potential against the SARS-CoV-2 delta variant in vivo assay. Overall, these new findings offer a good drug candidate for the clinical treatment of COVID-19.
Supplementary information
Acknowledgements
We thank the staff from the BL19U1 beamline of the National Facility for Protein Science in Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility for assistance during data collection. This work was supported by the National Natural Science Foundation of China (82151219 and 82130105), the Strategic Priority Research Program of the Chinese Academy of Sciences (SIMM010109 and SIMM010111), the Qiusuo Outstanding Youth Project of Lingang Laboratory (LG-QS-202205-02).
Author contributions
Z.D.J. performed research on the design and synthesis of compounds. B.F. performed research on the inhibition of protease assay and the mechanism of action studies. Y.M.Z. and L.K.Z. evaluated the antiviral activities at the cellular level and in vivo. T.Q.N. and H.X.S. explored the crystal structure of compounds. H.L. and J.L. gave important guidance for this study. Y. Zang and Y. Zhou designed the study and drafted the paper. All authors read and approved the contents of the paper.
Data availability
All data supporting the findings of this study are available within the article or from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics declarations
All the K18-hACE2 mice were cared for following the recommendations of the National Institutes of Health Guidelines for the Care and Use of Experimental Animals. Viral infections were performed in a biosafety level 3 (BSL-3) facility. (Ethics number: WIVA25202202).
Footnotes
These authors contributed equally: Zhidong Jiang, Bo Feng, Yumin Zhang, Tianqing Nie
Change history
10/13/2023
A Correction to this paper has been published: 10.1038/s41392-023-01665-4
Contributor Information
Haixia Su, Email: suhaixia1@simm.ac.cn.
Leike Zhang, Email: zhangleike@wh.iov.cn.
Yi Zang, Email: yzang@lglab.ac.cn.
Yu Zhou, Email: zhouyu@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-023-01482-9.
References
- 1.Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin Z, et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 3.Owen DR, et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 4.Dai W, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unoh Y, et al. Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem. 2022;65:6499–6512. doi: 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao S, et al. Discovery and crystallographic studies of nonpeptidic piperazine derivatives as covalent SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2022;65:16902–16917. doi: 10.1021/acs.jmedchem.2c01716. [DOI] [PubMed] [Google Scholar]
- 7.Gao S, et al. Discovery and crystallographic studies of trisubstituted piperazine derivatives as non-covalent SARS-CoV-2 main protease inhibitors with high target specificity and low toxicity. J. Med. Chem. 2022;65:13343–13364. doi: 10.1021/acs.jmedchem.2c01146. [DOI] [PubMed] [Google Scholar]
- 8.Zang Y, et al. High-throughput screening of SARS-CoV-2 main and papain-like protease inhibitors. Protein Cell. 2023;14:17–27. doi: 10.1093/procel/pwac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article or from the corresponding author upon reasonable request.