Abstract
Many viruses evolved mechanisms for capping the 5′-ends of their plus-strand RNAs as a means of hijacking the eukaryotic mRNA splicing/translation machinery. Although capping is critical for replication, the RNAs of these viruses have other essential functions including their requirement to be packaged as either genomes or pre-genomes into progeny viruses. Recent studies indicate that HIV-1 RNAs are segregated between splicing/translation and packaging functions by a mechanism that involves structural sequestration of the 5′-cap. Here we examined studies reported for other viruses and retrotransposons that require both selective packaging of their RNAs and 5′-RNA capping for host-mediated translation. Our findings suggest that viruses and retrotransposons have evolved multiple mechanisms to control 5′-cap accessibility, consistent with the hypothesis that removal or sequestration of the 5′ cap enables packageable RNAs to avoid capture by the cellular RNA processing and translation machinery.
Keywords: 5′-cap, cap sequestration, translation regulation, RNA packaging, RNA virus
Introduction
Eukaryotic messenger RNA (mRNA) transcribed by the DNA-dependent RNA polymerase II (Pol II) are co-transcriptionally capped at their 5′ ends by a 5′-triphosphate linked 7-methylguanosine residue (7mGpppN-, also known as the 5′-cap). Cap-synthesis is carried out by Pol II-associated capping enzymes and occurs early during transcription, likely after the incorporation of 10–20 nucleotides.[1] The 5′-cap is an essential post-transcriptional modification that plays critical roles in many RNA functionalities, including pre-mRNA splicing, 3′-polyadenylation, RNA nuclear export, resistance to 5′−3′ exonucleases, and translation initiation.[2] The functions of the 5′-cap are achieved by its interactions with various cap-binding proteins, mainly the cap-binding complex (CBC) in the nucleus and the eukaryotic translation initiation factor 4E (eIF4E) in the cytoplasm.[3] The cap-CBC interactions facilitate mRNA maturation and nuclear export, while eIF4E binding to the mRNA 5′-cap initiates translation. The predominant mechanism of eukaryotic translation initiation is cap-dependent: the pioneer round of translation is mediated by the cap-associated CBC protein, which is then replaced by eIF4E for the following steady-state translation.[2,4] In both cases, cap-associated CBC or eIF4E serves as the anchor site to recruit additional translation factors such as the eIF4G and RNA helicase eIF4A and eventually the 40S and 60S ribosomal subunits for successful translation initiation.[4,5] The 5′ cap also protects RNAs from degradation by cellular exoribonucleases.[6] Eukaryotic mRNAs are often methylated at the 2′-hydroxyl group (2′-O) of the first and second nucleotides following the 5′-cap (Figure 1a), and occasionally at the 2′-O positions of additional downstream nucleotides,[7] and RNAs lacking a 5′ cap structure or the 2′-O methylation(s) can be detected by host restriction factors and trigger immune responses as a means of responding to non-self RNAs.[8–10]
FIGURE 1. Viral RNA 5′-cap.
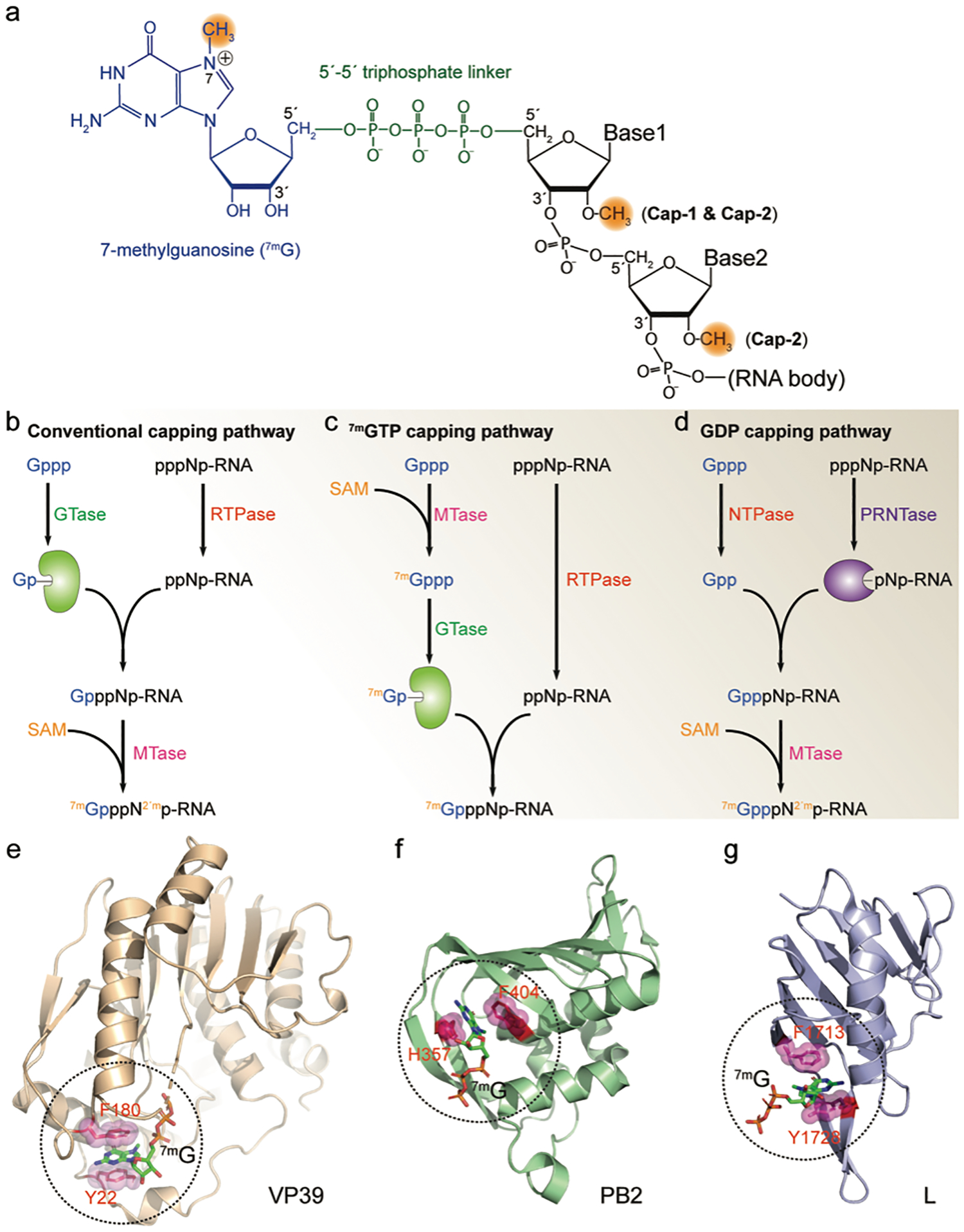
(a) Structure of the 5′-cap of viral mRNA. A 7-methylguanosine (blue) is linked to the RNA 5′-end through a 5′−5′ triphosphate linker (green). The methylations on the N7 and 2′-O are highlighted in orange. (b-d) Viral RNA capping mechanisms. All capping pathways start with GTP (Gppp) and the nascent RNA (pppNp-RNA). GTase, guanylyltransferase; RTPase, RNA triphosphatase; MTase, methyltransferase; SAM, S-Adenosyl methionine; NTPase, nucleoside triphosphatase; PRNTase, polyribonucleotidyl transferase; (e-g) Viral cap binding proteins and their cap-recognition mechanisms. In each protein, two residues with aromatic ring sandwich the 7-methylguanosine cap (7mG) through cation-π interactions. Additional hydrogen bonds further stabilize the protein/cap complex (not shown in the figure). VP39, vaccinia virus MTase (PDB ID 1AV6); PB2, influenza virus polymerase subunit (PDB ID 4C4B); L, Bunyavirus multifunctional polymerase (PDB ID 6QHG).
Viruses have evolved diverse mechanisms to co-opt the host translational machinery for viral protein production.[11,12] While some viruses use a viral protein covalently linked to the 5′-end of viral mRNA (viral protein genome-linked, VPg) and/or a cis-acting internal ribosomal entry site (IRES) RNA element (such as RNA viruses from the family of Picornaviridae),[13,14] the major way for a viral RNA to hijack the cellular translation machinery is to mimic the host cellular mRNAs by exploiting the cap-dependent translation initiation mechanism.[12] As discussed below, a diverse set of mechanisms have been utilized by different viruses to synthesize the cap structure at the 5′-end of their plus-strand RNAs. In addition to functioning as mRNA for the translation of structural and/or functional proteins, the viral plus-strand RNAs of all RNA viruses and some DNA viruses perform other essential functions during the replication. The plus-strand RNA of retroviruses (single-stranded positive-sense RNA virus) is selectively incorporated into assembling virions as the progeny genome (gRNA).[15] Plus-strand RNAs are also specifically packaged as pre-genomes in rotaviruses (double-stranded RNA virus)[16] and hepatitis B virus (HBV, double-stranded DNA virus).[17] In some RNA viruses, the plus-strand RNA is also critical for replicating the viral RNAs by serving as the template for the generation of the minus-strand RNA, such as in influenza viruses (single-stranded negative-sense RNA virus)[18] and flaviviruses (single-stranded positive-sense RNA virus).[19] Thus, the plus-strand RNAs of diverse viruses are essential for both translational and non-translational (replication and/or packaging of the viral genomes) functions.
A major question is how processing/translation versus packaging functions of the plus-strand RNAs are regulated.[20] Influenza viruses produce two different types of plus-strand RNAs: a capped form that functions as mRNA and a noncapped form functions as the template for RNA replication.[18,21] Some retroviruses also have two distinct pools of full-length plus-strand RNA, one that is retained in the cell for translation (mRNA) and the other that is selected for packaging (genomic RNA, gRNA).[22,23] Recent studies in human immunodeficiency virus type-1 (HIV-1) revealed that the mRNA versus gRNA functions of the plus-strand RNA is governed by the accessibility of the 5′-cap, with cap-sequestration being an essential determinant for efficient packaging.[24,25] Here we summarize the role of the 5′-cap in regulating the function of viral plus-strand RNAs and emphasize its role in viral RNA packaging. We hypothesize that control of cap accessibility by de-capping or cap sequestration could be a general mechanism for expansion and control of plus-strand RNA function.
Viruses use diverse mechanisms to cap their plus-strand RNAs
Most viral mRNA 5′-caps are indistinguishable from that of the cellular mRNA: a 7-methylguanosine moiety appended to the viral RNA 5′-end through a triphosphate linker (7mGpppN-, cap-0) (Figure 1a). For a small number of viruses, the 5′-cap is hypermethylated with additional modification(s) at the N2 position of the cap residue[26–30] which is a hallmark of the cellular small nuclear, small nucleolar and telomerase RNAs.[31,32] The 2,2,7- trimethyl guanosine (TMG) cap of a subset of HIV-1 mRNAs was shown to initiate translation in a CBC-dependent manner rather than requiring eIF4E binding.[33,34] The extra methyl group(s) in the hypermethylated cap is added by a unique methyltransferase.[32,33]
Viruses have evolved multiple mechanisms for capping their RNAs.[11,35] The mRNAs of most nucleus-replicating DNA viruses (hepatitis B virus) and retroviruses (HIV-1) are synthesized by the host RNA polymerase II and capped by the normal cellular capping apparatus. For the capping of eukaryotic mRNAs (also called the conventional cap-synthesis pathway, Figure 1b), the RNA triphosphatase (RTPase) activity removes the g-phosphate from the terminus of the 5′-triphosphorylated mRNA, then the guanylyltransferase (GTase) activity catalyzes the transfer of a guanine monophosphate (GMP) to the 5′-diphosphorylated mRNA (GpppNN-), which is followed by the transfer of a methyl group from S-Adenosyl methionine (SAM) to the N7 position of the terminal guanine (7mGpppNpN-) by (guanine-7)-methyltransferase (N7MTase) and the nucleoside 2′-O position of the following nucleotides (7mGpppN2′mpN- or 7mGpppN2′mpN2′m) by (nucleoside 2′-O)-methyltransferase (2′OMTase). The collective catalytic reactions of RTase, GTase and MTases are referred to as the capping process, which are encoded either in separate polypeptides (for example in yeast) or as different domains of a single protein (such as the RTase-GTase in metazoans).[3,11]
Many viruses with a cytosolic replication cycle encode their own capping enzymes, some of which utilize the conventional capping mechanism of eukaryotic cells. The rotavirus VP3 multidomain protein contains all the RTPase, GTase, N7MTase and 2′OMTase activities[36] and the flavivirus NS3 protein has the RTPase activity while its RNA dependent RNA polymerase (RdRp) NS5 also performs the GTase and MTase reactions (Figure 1b).[37] Other viral capping enzymes have evolved unique “unconventional capping mechanisms” with cap synthesis pathways distinct from the eukaryotic capping mechanism. In alphaviruses, the N7MTase activity of the nsP1 replicase protein transfers a methyl group from SAM to the N7 position of GTP and then the GTPase activity of nsP1 appends the 7mGMP to the 5′-end of the diphosphorylated RNA generated by the RTPase activity of nsP2 (Figure 1c).[11] SARS-CoV-2 also employs an unconventional capping mechanism, in which its nsp12 protein transfers a guanine diphosphate moiety (GDP rather than a GMP) to the monophosphorylated viral RNA from the nsp9-p-RNA intermediate (also synthesized by nsp12), which is followed by the methylation by nsp14 (N7MTase) and nsp16 (2′OMTase) proteins.[38] Capping processes similar to SARS-CoV-2 have also been discovered in Rhabdoviridae-like viruses (Figure 1d).[11]
Some viruses do not encode their own capping enzymes but instead steal the 5′-cap from host mRNAs by a mechanism known as cap-snatching, which has been well-characterized in the single-stranded negative-sense RNA viruses from the families of Orthomyxoviridae and Bunyaviridae.[21,39] Cap-snatching is initiated by the binding of the cellular mRNA 5′-cap to the cap-binding site of a viral multi-functional protein, followed by the cleavage of the cellular mRNA at 1–20 nucleotides downstream of the 5′-cap. The resulting capped RNA fragment is then used to prime the transcription of the viral mRNA by its RdRp. Both the cap-binding and endonuclease activities are carried out by the viral polymerases in influenza virus (Orthomyxoviridae)[21] and bunyaviruses,[39] whereas the nucleocapsid (N) protein of Hantavirus is proposed to have the corresponding cap-binding site.[40] Cap-snatching enables efficient translation of viral mRNAs, while the de-capped host mRNAs are targeted for degradation.
Although the viral and cellular cap-binding proteins have no sequence homology and low overall structural similarity, most of them have similar cap-recognition mechanisms: two aromatic rings sandwich the cap moiety with cation-π stacking interactions, with binding further stabilized by specific intermolecular hydrogen bonds (Figure 1e–f). Although the Hantavirus N protein contains the potential cap binding site for cap-snatching,[40] no classical cap-binding pocket with aromatic residues has been identified in its three-dimensional structure,[41] suggesting a different cap-recognition mechanism. In addition, a distinct cap-binding mode has been reported for the cap-binding site of rotavirus VP1 protein (see below).[42] The binding of cellular proteins to the 5′-cap plays known roles in regulating viral translation, replication, and packaging, could conceivably also play an inhibitory role in the innate immune response.[43,44]
Cap sequestration is required for HIV-1 RNA packaging
HIV-1 is a retrovirus with single-stranded positive-sense RNA genome. Upon entering cells, the genomic RNA is copied into double-strand DNA by the viral reverse transcriptase and integrated into the host chromosome, conferring life-long infection.[45] The integrated proviral DNA is subsequently transcribed by the host RNA polymerase II and the RNAs co-transcriptionally capped using the cell’s messenger RNA-capping machinery as discussed above.[46] A fraction of the capped full-length HIV-1 RNA transcripts function as mRNAs that are translated into the viral Gag and Gag-Pol proteins, the protein building blocks of the virus. Gag-Pol contains peptide regions including the viral protease, reverse transcriptase and integrase, which are proteolytically liberated during viral maturation.[47] Full-length HIV-1 RNAs also serve as the genome for progeny viruses, and HIV-1 mRNA and gRNA functions were originally proposed to be conferred by a single RNA species.[48–50] However, early studies of murine leukemia virus[51] and more recent studies of HIV-1[52–54] suggest a mechanism in which the mRNAs and gRNAs exist in distinct, non-interconverting pools.
Like nearly all other retroviruses, HIV-1 specifically and efficiently packages two copies of its gRNA in the form of non-covalently linked dimer into the assembling virion, which is selected from a cellular milieu that contains a substantial excess of non-viral RNAs and viral spliced and unspliced mRNAs.[55–57] gRNA packaging by HIV-1 is mediated by specific interactions between the nucleocapsid (NC) domain of the viral Gag protein and the cis-acting RNA “packaging signal” located within a ~350 nt region at the 5′-end of the 9-kb genome (also known as the 5′-leader).[58–61] The 5′-leader is the most conserved region of the HIV-1 genome and contains multiple functionally important RNA elements. Studies indicate that the functions of the 5′-leader are modulated by its dimerization state, and that the monomeric and dimeric forms of the 5′-leader adopt distinct secondary and tertiary structures.[24,57,60,62–79] Over the past three decades, more than 20 secondary structures have been proposed for the 5ʹ-leader on the basis of mutagenesis experiments, free energy calculations, chemical probing, and NMR, and although there is consensus that transcriptional activation, primer binding, dimerization, and splicing activities are promoted by discrete hairpin structures (TAR, PBS, DIS and SD hairpins, respectively),[24,57,60,62–79] there has been little consensus regarding other structural features. This is likely due in part to the fact that the 5ʹ-leader adopts multiple structures both in vitro and in cells, which can confound analysis of chemical probing data.[79] Indeed, results of in-gel chemical probing of the dimeric 5′-leader[80] are more compatible with the NMR-derived structure than with models proposed on the basis of chemical probing and free energy predictions.[79] Additionally, it has recently been shown that a widely used method for probing nucleotide accessibility (selective 2′-hydroxyl acylation analyzed by primer extension, SHAPE)[81] does not robustly identify conformationally dynamic nucleotides.[82] SHAPE is potentially attractive because it enables structural probing of large RNAs not only in vitro, but also in living cells and viruses. However, the approach assumes that the probability of a chemical modification is reflected by the extent of termination or mutation that occurs at a modified site during reverse transcription (RT)-based readout, and it is now known that some modified bases are largely invisible to readout by RT termination or mutational mapping alone,[83] and that RT mutations and stops are poorly correlated, are subject to different biases, and represent largely discrete sets of information from chemical probing of RNA structure.[84] The use of multiple methods for detecting sites of nucleotide modification is likely to improve the reliability of RNA secondary structure predictions derived from chemical probing experiments.[84] The secondary structures outlined in Figure 2 are based on NMR methodologies[85] and the dimer structure appears consistent with a recent structural analysis by small angle X-ray scattering.[86]
FIGURE 2. Cap-sequestration promotes HIV-1 gRNA packaging.
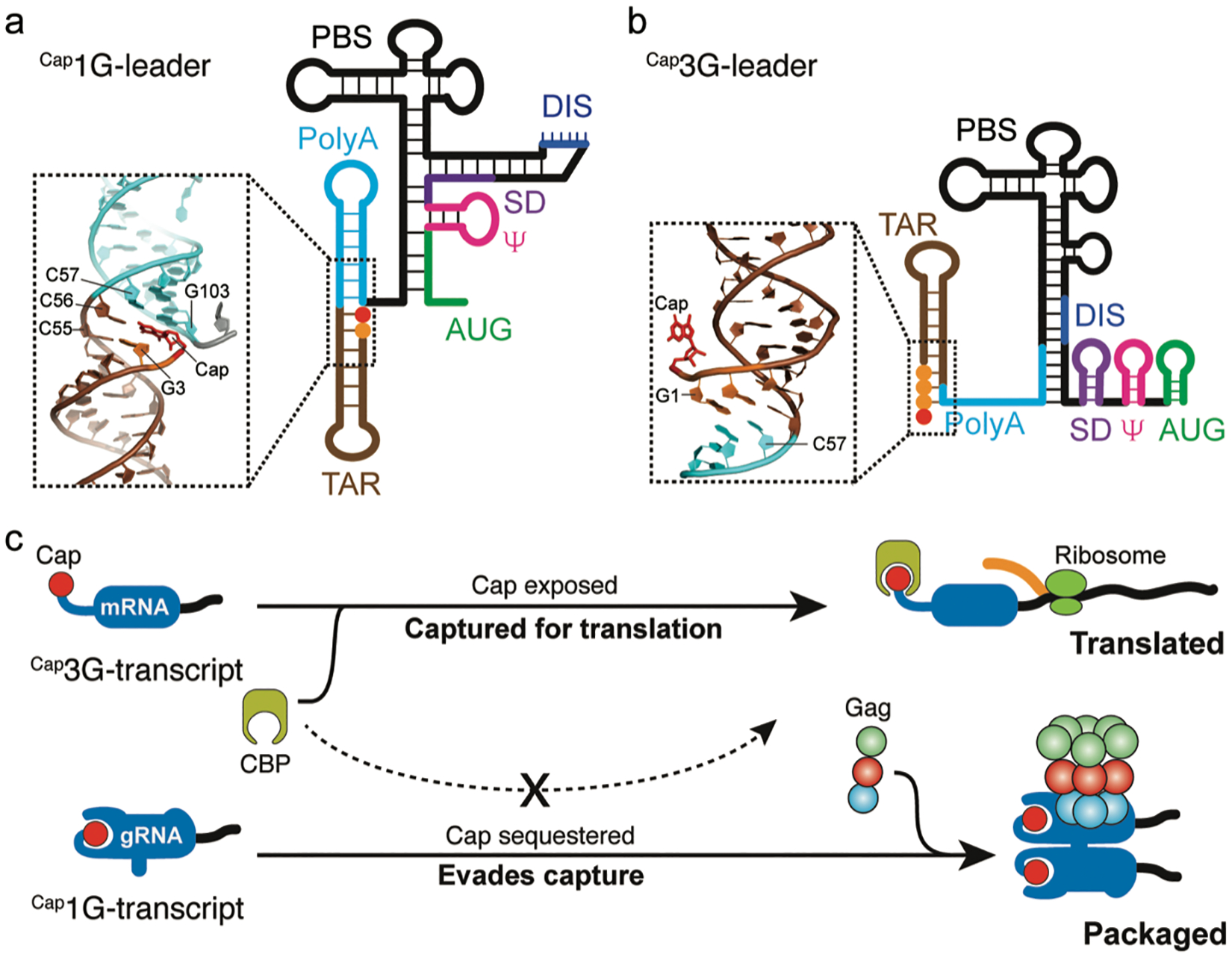
(a) Cartoon representation of the leader region of HIV-1 RNA transcript that starts with a single 5′-guanosine, derived by NMR (orange sphere, 1G-leader).[24,79] The 5′-cap (red sphere) is sequestered by the co-axial stacking of the TAR and PolyA helices (insert, PDB ID 6VVJ). The Cap1G-leader is dimerization competent with the dimerization initiation site (DIS) exposed and promotes packaging. (b) NMR-derived structural model of the leader region of HIV-1 RNA transcript with three 5′-guanosines (3G-leader).[24] The 5′-cap is exposed (insert, PDB ID 6VU1) and accessible to cellular cap-binding proteins. The Cap3G-leader adopts a monomeric conformation with the DIS region sequestered by intramolecular base-pairing. (c) Proposed working mechanism for HIV-1 selective gRNA packaging. The Cap3G-transcript exposes the 5′-cap which is recognized by the cellular cap-binding protein (CBP) for translation and thus retains in the cell and functions as mRNA. The Cap1G-transcript sequesters the 5′-cap to evade the capture by cellular translation machinery and is selected for packaging by viral Gag proteins. Adapted from [25], which is licensed under CC BY-NC-ND.
Although 5′-leader dimerization was initially thought to be regulated by a riboswitch-like mechanism of a single viral RNA transcript, recent studies indicate that the dimerization state of HIV-1 transcripts is controlled at the level of transcription by a heterogeneous transcriptional start site mechanism.[24,53,54] HIV-1 proviral DNA only contains one promoter, but heterogenous start site usage generates functionally distinct RNA pools that differ by the inclusion of one or two 5′-guanosines.[24,53,54] RNAs beginning with a single 5′-guanosine (1G) adopt the dimeric conformation that exposes high affinity NC binding sites and thus promotes its packaging as gRNA (Figure 2a). In contrast, HIV-1 RNAs that begin with two or three 5′-guanosines (2G or 3G, respectively; the dominant form is 3G) adopt a monomeric conformation with fewer NC binding sites and are retained in cells as mRNAs[24,54,78] (Figure 2b). Although both the 1G and 2G/3G forms of the RNA transcripts are capped co-transcriptionally, in vitro NMR experiments showed that the 5′-cap in the 1G form of the 5′-leader (Cap1G-L) is buried in a co-axial stacking structure of the TAR and PolyA hairpins and is thus inaccessible for the cap-binding eukaryotic translation initiation factors (Figure 2a). However, the 5′-cap in the 2G/3G form of the 5′-leader (Cap2G-L or Cap3G-L) is exposed and readily binds the cap-binding protein eIF4E (Figure 2b).[24] It was recently shown that a fraction of HIV-1 mRNAs are hypermethylated, which appears to enable translation initiation using the nuclear cap-binding protein rather than eIF4E.[33] It remains to be determined if cap hypermethylation is dependent on 5′-guanosine number.
The RNA determinants in the dimeric 5′-leader responsible for genome packaging have been extensively studied.[25,60,61,65,87–91] All of the high-affinity NC binding sites (ca. two dozen) are located in a ~150-nt region of the leader, which is the minimum region required for leader dimerization and is sufficient to promote packaging of heterologous RNA into HIV-1 virus-like particles.[60,61] This “core encapsidation signal” or ΨCES, which lacks that the TAR-PolyA helices and the PBS regions, adopts a tandem three-way junction structure.[61,79] Despite containing all the high-affinity NC binding sites, ΨCES is poorly packaged when assayed in competition with the wild-type 5′-leader. The additional determinants required for wild-type packaging were mapped to the TAR-PolyA tandem hairpins that sequester the 5′-cap.[24,25,92] Mutations in the full-length leader that expose the cap without affecting its dimerization or NC binding properties abrogate packaging, and addition of a cap-shedding ribozyme to ΨCES restores packaging efficiency to that of the native 5′-leader.[25] These findings collectively demonstrate that, in addition to NC-recognition, cap-sequestration is an essential determinant of HIV-1 RNA packaging. Since 5′-cap is critical for RNA splicing and translation, cap sequestration appears to function by preventing dominant negative cap-dependent capture by the cellular RNA processing and translation machinery (Figure 2c). Interestingly, cellular non-coding RNAs lack a 7-methylguanosine cap are also enriched in retroviruses,[93–95] consistent with the hypothesis that cap sequestration is required for RNA packaging. More recent studies confirmed that transcriptional start site heterogeneity and preferential packaging of specific 5′-Capped full-length RNA species are conserved properties among the primate lentiviruses.[96] Further studies are warranted to characterize the cap accessibility and its relationship to RNA fate determination in these viral RNAs. For retroviruses that do not exhibit transcriptional start site heterogeneity, we speculate that the originally proposed riboswitch-like mechanism that influences exposure of viral Gag binding sites[97] might also modulate cap accessibility (a hypothesis that has not yet been tested). A very recent study suggested that N6-methyladenosine deposition could play a role in regulating transcription versus packaging functions of individual HIV-1 RNA transcripts.[98] Although this mechanism is at odds with evidence that translation and packaging are mediated by different transcripts, the proposed role for epitranscriptomic regulation of HIV-1 RNA function is intriguing and deserves further exploration.
Retrotransposons package de-capped/uncapped RNAs into virus-like particles
Cap-dependent modulation of RNA packaging has also been reported in Long terminal repeat (LTR) retrotransposons, which are transposable DNA elements that replicate through RNA intermediates and are ubiquitously found in many eukaryotic genomes.[99] LTR retrotransposons are considered as the evolutionary progenitors of retroviruses. They replicate through a “copy-and-paste” mechanism in which their RNA transcripts are reverse transcribed back into cDNAs that are then inserted into new genomic loci.[100,101] The Ty1 element of Saccharomyces cerevisiae is among the best characterized LTR retrotransposons. Ty1 is 5.9 kilo-bases (kb) in length and contains two ~300 nucleotide terminal repeats with the sequences in between coding for the structural Gag protein and enzymes required for transposition (Figure 3a). The virus-like particle assembled from Gag proteins selectively packages the Ty1 RNA transcript as the template for reverse transcription. Ty1 RNA is transcribed by the host RNA polymerase II and is thus co-transcriptionally capped. Although most of the Ty1 RNAs in the cell are capped, the major form of RNA packaged into the virus like particle is uncapped, which is due to RNA branching and debranching events.[102] The 5′-cap of some Ty1 RNAs is removed by joining the 5′-end and a nucleotide close to the 3′-terminus through a 2′−5′ phosphodiester bond (Figure 3a). Breaking down of the 2′−5′ bond by the host RNA lariat debranching enzyme Dbr1 generates the linear RNA with a single phosphate group at the 5′-end, which is the RNA species enriched in the virus like particle.[102,103] While the branching process is required for 5′-cap removal, the debranching event has been suggested to play important roles in reverse transcription.[103,104] Thus, Ty1 RNA transcripts with 7-methylguanosine cap function as mRNA while the de-capped RNA transcripts containing a monophosphate 5′-end are likely selectively incorporated into the virus like particle as the template for reverse transcription.
FIGURE 3. Packaging of noncapped RNAs as the viral genomes.
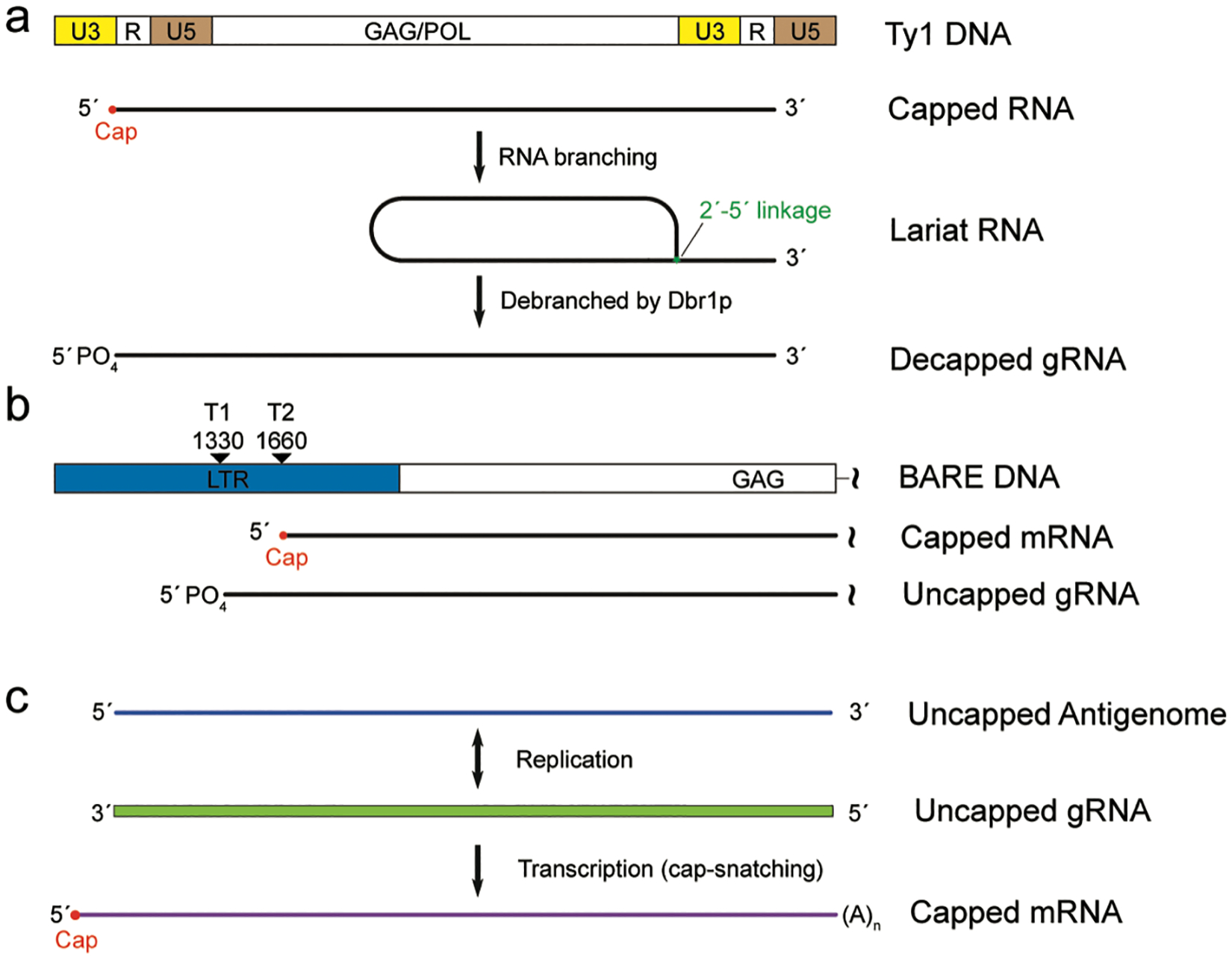
(a) Structural organization of the genomic DNA and RNA transcript of the Yeast Ty1 retrotransposon. R is the region repeated at the 5′ and 3′ end of the RNA transcript, U5 and U3 are the unique regions to the 5′- and 3′-end, respectively. Some of the Ty1 capped RNA transcript undergoes a branching process in which the RNA 5′-end is covalently linked to a 3′-proximal nucleotide through a 2′−5′ linkage. Debranching by the RNA lariat debranching enzyme Dbr1p generates RNA with 5′-monophospahte, which is accumulated in Ty1 VLPs (b) The retrotransposon BARE uses two distinct RNA pools for translation and packaging. The RNA transcript from promoter 2 (T2) is 5′-capped and 3′-polyadenylated to serve as the mRNA, while the RNA transcript from promoter 1 (T1) lacks 5′-cap and 3′-polyadenylation which is packaged into the VLP as gRNA. (c) Negative-strand RNA virus produces two distinct plus-strand RNAs, one is capped through cap-snatching and functions as mRNA and the other is not capped and serve as the template for transcription and replication. The negative-strand RNA genome is also not capped.
Selective packaging of uncapped RNA is also utilized by the BARE LTR retrotransposon, another Ty1/Copia superfamily member constituting over 10% of the barley genome. The BARE long terminal repeat region contains two sequential promoters separated by ~300 nt, which produce two groups of RNA transcripts with different lengths (Figure 3b).[105] RNAs generated from these two promoters share most of their sequences except that the longer transcripts from the first promoter (TATA-1) lack 5′-cap and 3′-polyadenylation modifications (Figure 3b). It was found that the uncapped TATA-1 transcripts are selectively packaged into virus-like particles while the capped TATA-2 transcripts are spliced and/or translated.[106] Thus, BARE LTR retrotransposon selectively packages its uncapped RNA transcripts, which are produced directly by transcription from a unique promoter. Generation and packaging of uncapped genomic RNAs were also discovered in RNA viruses including the single-stranded positive-sense RNA containing alphaviruses.[107,108] Significant amounts of uncapped genomic RNAs are produced early during infection, which has been shown to be critical for alphavirus pathogenicity.[108] It is thus conceivable that uncapped genomic RNAs increase the packaging efficiency by alphaviruses as well as other plus-strand RNA viruses.
Negative-Sense RNA viruses package uncapped RNAs
Uncapped genomic RNAs are also packaged into negative-sense RNA viruses (NSVs) such as influenza A virus (IAV) and Bunyaviruses. In these cases, the RNA genomes are antisense to their mRNAs and are thus not capped for translation.[18] After cells are infected, the NSV RdRp performs both replication and transcription using the same RNA template to generate two types of plus-strand RNAs with distinct functions. Replication by RdRp produces the plus-strand antigenome, which is the complementary copy of the negative-strand RNA genome.[18] The antigenome is further used as the template by RdRP to generate more negative-strand genomic RNAs that are packaged into the progeny viruses. Both the negative-strand gRNA and the plus-strand antigenome are uncapped. On the other hand, the plus-strand transcription products of the gRNA by RdRp are capped either by virally-encoded capping enzyme (non-segmented negative-strand RNA viruses) or a cap-snatching mechanism as mentioned above (segmented negative-strand RNA viruses)[21,39] and these capped plus-strand RNAs function as mRNA to be translated into viral proteins. Thus, the cap status of these plus-strand RNAs plays a critical role in modulating the RNA translation and replication/packaging function of the NSV plus-strand RNAs (Figure 3c).
Protein binding as a potential mechanism for cap sequestration and viral RNA packaging
The genome of rotaviruses is comprised of 11 segmented double-stranded RNA (dsRNA) fragments with their lengths ranging from 0.7 kb to 3.1 kb. During rotavirus assembly, each of the 11 genome fragments is selectively incorporated in its 5′-capped and non-polyadenylated plus-strand form.[16] In addition to functioning as the template for genome replication, each of the plus-strand RNAs generally contains a single open-reading frame (ORF) flanked by untranslated regions, and these RNAs functions as the mRNA for one of the structural proteins (VPs) or non-structural proteins (NSPs) of the virus. Rotavirus plus-strand RNAs likely contain sequence and structure elements that promote their packaging. Computational studies and RNase mapping experiments suggest that 5′−3′ base-pairing of the plus-strand RNA termini creates a panhandle structure with a highly conserved single-stranded 3′-terminus (“UGUGACC” for group A rotavirus, Figure 4a). This 3′-consensus sequence (3′-CS) is directly recognized by the viral RdRp (VP1), which is believed to facilitate the packaging of plus-strand RNAs into the viral particle.[109,110] The core shell of the viral particle is composed of viral VP2 protein and engagement of VP2 activates the enzymatic activity of VP1 to initiate the synthesis of the minus-strand RNA and the production of the dsRNA genome within the virial core particle.[16] VP1 is associated with the viral capping enzyme (VP3), which adds the 7-methyl guanosine cap to the plus-strand RNA in the next round of replication.[36] Importantly, a cap-binding site has been identified on the surface of the VP1 polymerase, and the VP1/5′-cap interaction in the panhandle structure of the plus-strand RNAs has been proposed to further stabilize the VP1/3′-CS interactions for RNA packaging.[16,42] While the 5′-cap is associated with the viral VP1 polymerase when the plus-strand RNAs function as the replication templates for packaging, the 5′-cap is bound by eIF4E for translation initiation when the plus-strand RNAs function as mRNA, in which the 3′-CS is occupied by the viral NSP3 protein instead of VP1 (Figure 4a).[111,112] NSP3 is thought to be a surrogate of the polyA binding protein, which promotes translation by enhancing 5′−3′ communication of the mRNAs through its binding to both the 3′-polyA signal and the 5′-cap associated eukaryotic initiation factors eIF4G.[113,114] Since packaging and translation are conflicting functions of the rotavirus plus-strand RNAs and the 5′-cap is critical for translation initiation, it seems plausible that sequestration of the 5′-cap by the viral VP1 polymerase may suppress the translation activity of the plus-strand RNAs, similar to the RNA structure-dependent cap sequestration observed for HIV-1.[25]
FIGURE 4. Potential protein-mediated cap-sequestration may contribute to gRNA packaging in other viruses.
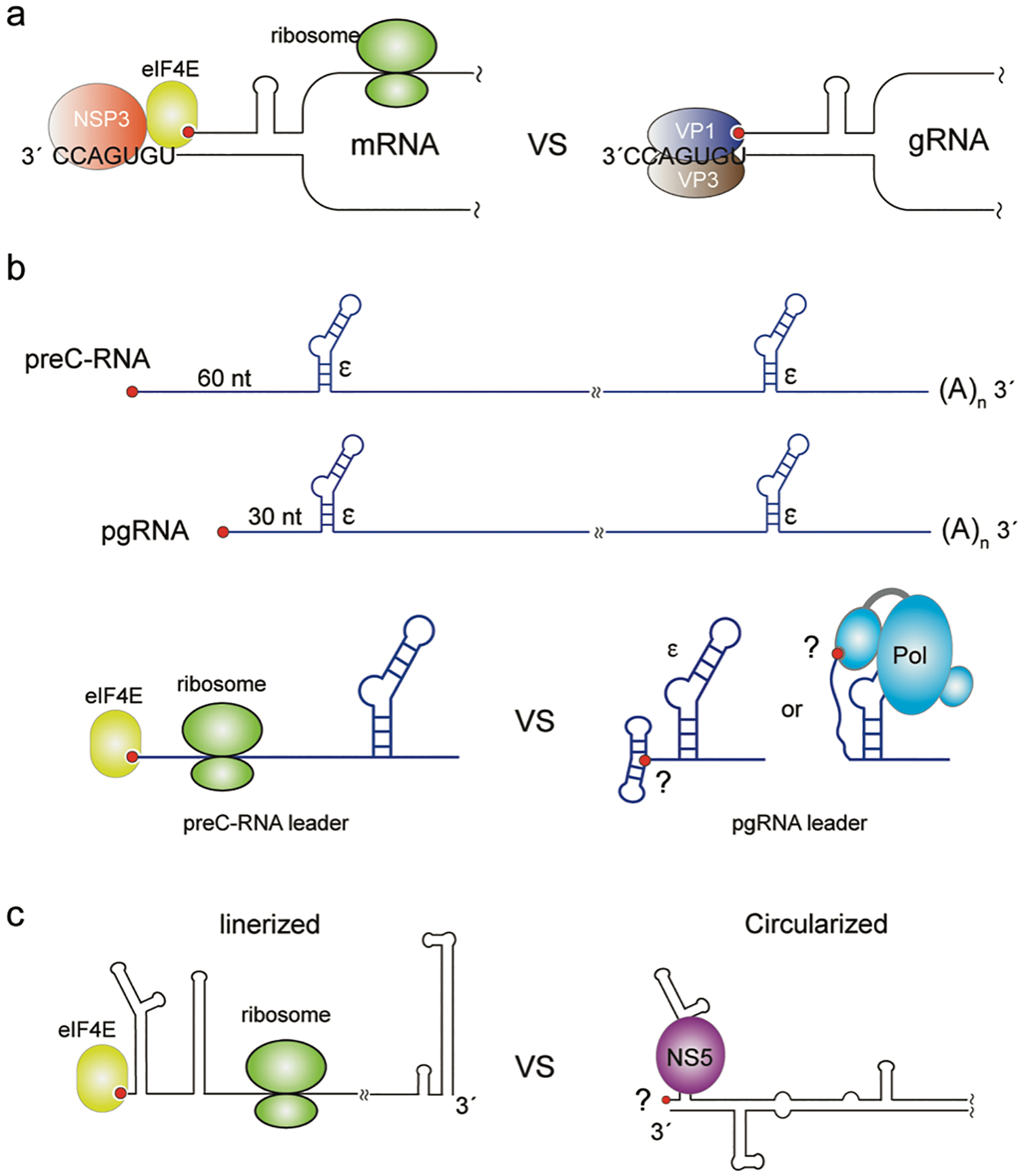
(a) The Rotavirus plus-strand RNA forms a panhandle structure due to base-pairing of the 5′- and 3′-ends. When functioning as mRNA, the 5′-cap and 3′-consensus sequence (UGUGACC) are bound by eIF4E and NSP3, respectively, which promotes efficient translation. When functioning as gRNA, the 5′-cap and 3′-CS sequence are occupied by the VP1-VP3 polymerase/capping enzyme complex. (b) Diagram of HBV preC-RNA and pgRNA transcripts. Both RNAs are ~3.5 kb and the preC-RNA has an additional ~30 nt at the 5′-end. The ε-elements required for packaging is labeled. The bottom cartoon schematic shows a proposed mechanism for the distinct functions of preC-RNA and pgRNA. The 5′-cap of preC-RNA is accessible for eIF4E to initiate translation. The 5′-cap of pgRNA is proposed to be sequestered by 5′-RNA elements or by the viral polymerase. (c) The flavivirus plus-strand RNA adopts two distinct conformations: the linear conformation promotes translation, and the circularized conformation suppresses translation and promotes RNA replication/packaging. The binding of viral polymerase NS5 to a 5′-cap proximal site in the circularized conformation is required for translation suppression and RNA replication/packaging.
Viral polymerase-mediated suppression of translation to promote plus-strand RNA packaging has been reported for hepatitis B virus (HBV), the prototypic member of the Hepadnaviriae. HBV is a small enveloped virus that contains a partially double-stranded DNA genome (also called the relaxed circular DNA or rcDNA), which is repaired into a covalently closed circular DNA genome (cccDNA) in the nucleus of infected cells.[17] Transcription by the host RNA Pol-II generates 5′-capped and 3′-polyadenylated viral RNAs, including the pregenomic RNA (pgRNA) and other viral mRNAs. Concomitant with viral capsid assembly, one copy of the pgRNA is selectively incorporated and then reverse transcribed into the rcDNA genome by the virally encoded polymerase (HBV-Pol or P protein). Selective encapsidation of pgRNA is mediated by specific interactions between the HBV-Pol and a cis-acting encapsidation signal (called the ε-element) located at the 5′-end of the 5′-capped pgRNA (Figure 4b).[115–117] Because of the terminal redundancy of pgRNA and the co-terminal nature of all viral transcripts, the ε-element is also present at the 3′-ends of all the sub-genomic viral mRNAs, which however does not direct selective RNA packaging.[117] More intriguingly, the packaging deficient mRNA of pre-core protein (preC-RNA) only differs from pgRNA by an additional ~30-nt at its 5′-end, which is also full-length and contains all the sequences of pgRNA (Figure 4b). In vivo packaging experiments with RNA constructs containing the ε-element inserted at different locations revealed that the ε-element and its appropriate distance (~30 nt in length) to the 5′-cap are both critical for wild-type packaging efficiency.[118] It was also shown that the binding of the HBV-Pol to the ε-element suppresses the translation of pgRNA, which is also dependent on the proximity between the 5′-cap and the ε-element in a similar manner.[119,120] Therefore, translation suppression of pgRNA by HBV-Pol and its selective encapsidation appear to be mechanistically coupled. We propose that the HBV pgRNA translation suppression and its selective packaging may be due to the inaccessibility of its 5′-cap and envisage a mechanism in which the distance between the 5′-cap the ε-element is critical for the potential cap sequestration in the HBV-Pol/pgRNA complex (Figure 4). One possibility is that the leader region of the pgRNA could facilitate RNA structure-based cap sequestration. A second possibility is that HBV-Pol may possess a cap-binding site similar to the above-mentioned rotavirus polymerase VP1 (protein-mediated cap-sequestration). It is also possible that the binding of HBV-Pol may interfere with the subsequent assembly process of the translation machinery rather than the 5′-cap/eIF4E recognition.
Translation suppression by the binding of viral polymerase to the 5′ proximal site of the plus-strand RNA has also been observed for flaviviruses. The genome of flaviviruses is a ~11 kb single-stranded positive-sense RNA encoding a single large polypeptide (C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) that is co- and post-translationally cleaved into the mature viral proteins.[19] In addition to the function as mRNA, the viral gRNA serves as the template for RNA synthesis and is also selectively packaged into progeny viral particles.[121] The gRNA of flaviviruses contains a 5′-cap and a conserved 3′-stem loop structure (3′-SL) instead of a polyadenylation signal.[19,122] The highly structured 5′-UTR (~100 nt) and 3′-UTR (~400–700 nt) play important roles in flavivirus RNA translation and replication, such as the recognition by the viral RdRp NS5. It was shown that flavivirus RNA packaging requires the presence of a replication-competent NS5 polymerase.[123] It was also found that the flavivirus gRNA adopts two distinct conformations to cope with its multi-functions: a linear conformation that promotes translation, and a circularized conformation that suppresses translation and promotes RNA replication/packaging (Figure 4c).[124–126] gRNA circularization is mediated by long-range interactions between RNA elements in the terminal UTRs and the binding of NS5 is necessary for the translation suppression in the circularized gRNA (Figure 4c).[127] The requirement of NS5 for flavivirus RNA packaging and NS5-mediated translation suppression is reminiscent of behaviors described above for other viruses and suggests the possibility of an RNA structure or NS5 dependent 5′-cap sequestration mechanism.
Summary
The recent finding that RNA packaging by HIV-1 is strongly attenuated by cap exposure supports the hypothesis that cap dependent capture by the host RNA processing and translation machinery is a dominant negative determinant of packaging.[25] Examination of the RNA packaging properties of other viruses that require 5′ RNA capping for replication suggests a common overall mechanism for control of cap-dependent functions (trafficking/processing/translation) versus packaging fates. In each case examined, packaging appears to be promoted by processes that have the potential to hide or remove the 5′ cap. Thus, HIV-1 uses structural changes associated with heterogeneous transcriptional start site usage to modulate cap exposure; de-capped or noncapped RNAs are selectively packaged into retrovirus-like retrotransposons; and the binding of viral protein factors such as the polymerase in rotavirus, HBV and flaviviruses, which is critical for viral RNA packaging, may be attributable to a direct cap sequestration function. We hypothesize that 5′-cap sequestration might be a general strategy used by viruses to inhibit capture by the cellular RNA translation machinery and thereby enable packaging of plus-strand RNAs as genomes or pre-genomes of progeny viruses.
ACKNOWLEDGEMENTS
This work was supported by NIH grant AI150498.
ABBREVIATIONS
- Pol II
RNA polymerase II
- eIF4E
eukaryotic translation initiation factor 4E
- CBC
cap-binding complex
- gRNA
genomic RNA
- pgRNA
pre-genomic RNA
- preC-RNA
pre-core protein mRNA
- dsRNA
double-stranded RNA
- HIV-1
human immunodeficiency virus type-1
- HBV
hepatitis B virus
- NSV
negative-sense RNA virus
- IAV
influenza A virus
- RTPase
RNA triphosphatase
- GTase
guanylyltransferase
- MTase
methyltransferase
- NTPase
nucleoside-triphosphatase
- PRNTase
polyribonucleotidyl transferase
- RdRp
RNA-dependent RNA polymerase
- SAM
S Adenosyl methionine
- NC
nucleocapsid
- LTR
long terminal repeat
- ORF
open-reading frame
- NSPs
non-structural proteins
- 3′-CS
3′-consensus sequence
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERNCES
- 1.Kim HJ, Jeong SH, Heo JH, Jeong SJ, Kim ST, Youn HD, Han JW, Lee HW, & Cho EJ (2004). mRNA capping enzyme activity is coupled to an early transcription elongation. Mol Cell Biol, 24, 6184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramanathan A, Robb GB, & Chan SH (2016). mRNA capping: biological functions and applications. Nucleic Acids Res, 44, 7511–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topisirovic I, Svitkin YV, Sonenberg N, & Shatkin AJ (2011). Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA, 2, 277–98. [DOI] [PubMed] [Google Scholar]
- 4.Maquat LE, Tarn WY, & Isken O (2010). The pioneer round of translation: features and functions. Cell, 142, 368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson RJ, Hellen CU, & Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews Molecular cell biology, 11, 113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houseley J, & Tollervey D (2009). The many pathways of RNA degradation. Cell, 136, 763–76. [DOI] [PubMed] [Google Scholar]
- 7.Perry KL, Watkins KP, & Agabian N (1987). Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc Natl Acad Sci U S A, 84, 8190–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung DW, & Amarasinghe GK (2016). When your cap matters: structural insights into self vs non-self recognition of 5’ RNA by immunomodulatory host proteins. Curr Opin Struct Biol, 36, 133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M Jr., Shi PY, & Diamond MS (2010). 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature, 468, 452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markiewicz L, Drazkowska K, & Sikorski PJ (2021). Tricks and threats of RNA viruses - towards understanding the fate of viral RNA. RNA Biol, 18, 669–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decroly E, Ferron F, Lescar J, & Canard B (2011). Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol, 10, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaafar ZA, & Kieft JS (2019). Viral RNA structure-based strategies to manipulate translation. Nat Rev Microbiol, 17, 110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodfellow I (2011). The genome-linked protein VPg of vertebrate viruses - a multifaceted protein. Curr Opin Virol, 1, 355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, & Roberts L (2005). Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO reports, 6, 968–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuzembayeva M, Dilley K, Sardo L, & Hu W-S (2014). Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology, 454–455, 362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald SM, & Patton JT (2011). Assortment and packaging of the segmented rotavirus genome. Trends Microbiol, 19, 136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukuda S, & Watashi K (2020). Hepatitis B virus biology and life cycle. Antiviral Res, 182, 104925. [DOI] [PubMed] [Google Scholar]
- 18.Ferhadian D, Contrant M, Printz-Schweigert A, Smyth RP, Paillart JC, & Marquet R (2018). Structural and Functional Motifs in Influenza Virus RNAs. Frontiers in microbiology, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrows NJ, Campos RK, Liao KC, Prasanth KR, Soto-Acosta R, Yeh SC, Schott-Lerner G, Pompon J, Sessions OM, Bradrick SS, & Garcia-Blanco MA (2018). Biochemistry and Molecular Biology of Flaviviruses. Chemical reviews, 118, 4448–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boersma S, Rabouw HH, Bruurs LJM, Pavlovic T, van Vliet ALW, Beumer J, Clevers H, van Kuppeveld FJM, & Tanenbaum ME (2020). Translation and Replication Dynamics of Single RNA Viruses. Cell, 183, 1930–45 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Te Velthuis AJW, Grimes JM, & Fodor E (2021). Structural insights into RNA polymerases of negative-sense RNA viruses. Nat Rev Microbiol, 19, 303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messer LI, Levin JG, & Chattopadhyay SK (1981). Metabolism of viral RNA in murine leukemia virus-infected cells: evidence for differential stability of viral message and virion precursor RNA. J Virol, 40, 683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mougel M, Akkawi C, Chamontin C, Feuillard J, Pessel-Vivares L, Socol M, & Laine S (2020). NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA. RNA Biol, 17, 528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JD, Kharytonchyk S, Chaudry I, Iyer AS, Carter H, Becker G, Desai Y, Glang L, Choi SH, Singh K, Lopresti MW, Orellana M, Rodriguez T, Oboh U, Hijji J, Ghinger FG, Stewart K, Francis D, Edwards B, Chen P, Case DA, Telesnitsky A, & Summers MF (2020). Structural basis for transcriptional start site control of HIV-1 RNA fate. Science, 368, 413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding P, Kharytonchyk S, Kuo N, Cannistraci E, Flores H, Chaudhary R, Sarkar M, Dong X, Telesnitsky A, & Summers MF (2021). 5’-Cap sequestration is an essential determinant of HIV-1 genome packaging. Proc Natl Acad Sci U S A, 118, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.HsuChen CC, & Dubin DT (1976). Di-and trimethylated congeners of 7-methylguanine in Sindbis virus mRNA. Nature, 264, 190–1. [DOI] [PubMed] [Google Scholar]
- 27.van Duijn LP, Kasperaitis M, Ameling C, & Voorma HO (1986). Additional methylation at the N(2)-position of the cap of 26S Semliki Forest virus late mRNA and initiation of translation. Virus research, 5, 61–6. [DOI] [PubMed] [Google Scholar]
- 28.Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, & Shi PY (2008). West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol, 82, 4295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benarroch D, Qiu ZR, Schwer B, & Shuman S (2009). Characterization of a mimivirus RNA cap guanine-N2 methyltransferase. RNA, 15, 666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yedavalli VS, & Jeang KT (2010). Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci U S A, 107, 14787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamond AL (1990). The trimethyl-guanosine cap is a nuclear targeting signal for snRNPs. Trends Biochem Sci, 15, 451–2. [DOI] [PubMed] [Google Scholar]
- 32.Buemi V, Schillaci O, Santorsola M, Bonazza D, Broccia PV, Zappone A, Bottin C, Dell’Omo G, Kengne S, Cacchione S, Raffa GD, Piazza S, di Fagagna FD, Benetti R, Cortale M, Zanconati F, Del Sal G, & Schoeftner S (2022). TGS1 mediates 2,2,7-trimethyl guanosine capping of the human telomerase RNA to direct telomerase dependent telomere maintenance. Nature communications, 13, 2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh G, Seufzer B, Song Z, Zucko D, Heng X, & Boris-Lawrie K (2022). HIV-1 hypermethylated guanosine cap licenses specialized translation unaffected by mTOR. Proc Natl Acad Sci U S A, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boris-Lawrie K, Singh G, Osmer PS, Zucko D, Staller S, & Heng X (2022). Anomalous HIV-1 RNA, How Cap-Methylation Segregates Viral Transcripts by Form and Function. Viruses, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh A, & Lima CD (2010). Enzymology of RNA cap synthesis. Wiley Interdiscip Rev RNA, 1, 152–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar D, Yu X, Crawford SE, Moreno R, Jakana J, Sankaran B, Anish R, Kaundal S, Hu L, Estes MK, Wang Z, & Prasad BVV (2020). 2.7 A cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity. Sci Adv, 6, eaay6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeedi BJ, & Geiss BJ (2013). Regulation of flavivirus RNA synthesis and capping. Wiley Interdiscip Rev RNA, 4, 723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park GJ, Osinski A, Hernandez G, Eitson JL, Majumdar A, Tonelli M, HenzlerWildman K, Pawlowski K, Chen Z, Li Y, Schoggins JW, & Tagliabracci VS (2022). The mechanism of RNA capping by SARS-CoV-2. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olschewski S, Cusack S, & Rosenthal M (2020). The Cap-Snatching Mechanism of Bunyaviruses. Trends Microbiol, 28, 293–303. [DOI] [PubMed] [Google Scholar]
- 40.Mir MA, Sheema S, Haseeb A, & Haque A (2010). Hantavirus nucleocapsid protein has distinct m7G cap- and RNA-binding sites. J Biol Chem, 285, 11357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olal D, & Daumke O (2016). Structure of the Hantavirus Nucleoprotein Provides Insights into the Mechanism of RNA Encapsidation. Cell Rep, 14, 2092–9. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, & Harrison SC (2008). Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure, 16, 1678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikkert M (2020). Innate Immune Evasion by Human Respiratory RNA Viruses. J Innate Immun, 12, 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyde JL, & Diamond MS (2015). Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology, 479–480, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coffin JM, Hughes SH, & Varmus HE (1997). Retroviruses Plainview, N.Y., Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- 46.Wilusz J (2013). Putting an ‘End’ to HIV mRNAs: capping and polyadenylation as potential therapeutic targets. AIDS Res Ther, 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freed EO (2015). HIV-1 assembly, release and maturation. Nat Rev Microbiol, 13, 484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorman N, & Lever A (2000). Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J Virol, 74, 11413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poon DT, Chertova EN, & Ott DE (2002). Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology, 293, 368–78. [DOI] [PubMed] [Google Scholar]
- 50.Liang C, Hu J, Russell RS, & Wainberg MA (2002). Translation of Pr55(gag) augments packaging of human immunodeficiency virus type 1 RNA in a cis-acting manner. AIDS Res Hum Retroviruses, 18, 1117–26. [DOI] [PubMed] [Google Scholar]
- 51.Levin JG, & Rosenak MJ (1976). Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci U S A, 73, 1154–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Liu Y, Wu B, Nikolaitchik OA, Mohan PR, Chen J, Pathak VK, & Hu WS (2020). Visualizing the translation and packaging of HIV-1 full-length RNA. Proc Natl Acad Sci U S A, 117, 6145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masuda T, Sato Y, Huang YL, Koi S, Takahata T, Hasegawa A, Kawai G, & Kannagi M (2015). Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep, 5, 17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kharytonchyk S, Monti S, Smaldino PJ, Van V, Bolden NC, Brown JD, Russo E, Swanson C, Shuey A, Telesnitsky A, & Summers MF (2016). Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci U S A, 113, 13378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berkowitz R, Fisher J, & Goff SP (1996). RNA packaging. Curr Top Microbiol Immun, 214, 177–218. [DOI] [PubMed] [Google Scholar]
- 56.Johnson SF, & Telesnitsky A (2010). Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Souza V, & Summers MF (2005). How retroviruses select their genomes. Nature Reviews Microbiology, 3, 643–55. [DOI] [PubMed] [Google Scholar]
- 58.Mailler E, Bernacchi S, Marquet R, Paillart JC, Vivet-Boudou V, & Smyth RP (2016). The Life-Cycle of the HIV-1 Gag-RNA Complex. Viruses, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olson ED, & Musier-Forsyth K (2019). Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly. Semin Cell Dev Biol, 86, 129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, & Summers MF (2012). Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol, 417, 224–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding P, Kharytonchyk S, Waller A, Mbaekwe U, Basappa S, Kuo N, Frank HM, Quasney C, Kidane A, Swanson C, Van V, Sarkar M, Cannistraci E, Chaudhary R, Flores H, Telesnitsky A, & Summers MF (2020). Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proceedings of the National Academy of Sciences, 117, 17737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrison GP, & Lever AML (1992). The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. Journal of Virology, 66, 4144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baudin F, Marquet R, Isel C, Darlix J-L, Ehresmann B, & Ehresmann C (1993). Functional sites in the 5’ region of human immunodeficiency virus type 1 RNA form defined structural domains. Journal of Molecular Biology, 229, 382–97. [DOI] [PubMed] [Google Scholar]
- 64.Clever J, Sassetti C, & Parslow TG (1995). RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of Human Immunodeficiency Virus Type 1. J Virol, 69, 2101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McBride MS, & Panganiban AT (1996). The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol, 70, 2963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miele G, Mouland A, Harrison GP, Cohen E, & Lever AM (1996). The human immunodeficiency virus type 1 5’ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. Journal of virology, 70, 944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clever JL, Miranda J,D, & Parslow TG (2002). RNA structure and packaging signals in the 5’ leader region of the human immunodeficiency virus type 1 genome. J Virol, 76, 12381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abbink TEM, & Berkhout B (2003). A novel long distance base-pairing interaction in Human Immunodeficiency Virus Type 1 RNA occludes the Gag start codon. J Biol Chem, 278, 11601–11. [DOI] [PubMed] [Google Scholar]
- 69.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, & Kjems J (2004). RNA interactions in the 5’ region of the HIV-1 genome. J Mol Biol, 336, 369–79. [DOI] [PubMed] [Google Scholar]
- 70.Paillart JC, Dettenhofer M, Yu X-F, Ehresmann C, Ehresmann B, & Marquet R (2004). First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem, 279, 48397–403. [DOI] [PubMed] [Google Scholar]
- 71.Paillart J-C, Shehu-Xhilaga M, Marquet R, & Mak J (2004). Dimerization of retroviral RNA genomes: An inseparable pair. Nature Reviews Microbiology, 2, 461–72. [DOI] [PubMed] [Google Scholar]
- 72.Russell RS, Liang C, & Wainberg MA (2004). Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Greatorex J (2004). The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology, 1, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abbink TEM, & Berkhout B (2008). RNA structure modulates splicing efficiency at the human Immunodeficiency Virus Type 1 major splice donor. J Virol, 82, 3090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jablonski JA, Buratti E, Stuani C, & Caputi M (2008). The secondary structure of the human immunodeficiency virus type 1 transcript modulates viral splicing and infectivity. J Virol, 82, 8038–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, & Weeks KM (2008). High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol, 6, 883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr., Swanstrom R, Burch CL, & Weeks KM (2009). Architecture and secondary structure of an entire HIV-1 RNA genome. Nature, 460, 711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu K, Heng X, Garyu L, Monti S, Garcia E, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Sachin Divakaruni S, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, & Summers MF (2011). NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science, 344, 242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, & Summers MF (2015). Structure of the HIV-1 RNA packaging signal. Science, 348, 917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kenyon JC, Prestwood LJ, Le Grice SF, & Lever AM (2013). In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res, 41, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkinson KA, Merino EJ, & Weeks KM (2006). Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Prot, 1, 1610–6. [DOI] [PubMed] [Google Scholar]
- 82.Merino EJ, Wilkinson KA, Coughlan JL, & Weeks KM (2005). RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc, 127, 4223–31. [DOI] [PubMed] [Google Scholar]
- 83.Sexton AN, Wang PY, Rutenberg-Schoenberg M, & Simon MD (2017). Interpreting Reverse Transcriptase Termination and Mutation Events for Greater Insight into the Chemical Probing of RNA. Biochemistry, 56, 4713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busan S, Weidmann CA, Sengupta A, & Weeks KM (2019). Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies. Biochemistry, 58, 2655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wüthrich K (1986). NMR of Proteins and Nucleic Acids New York, John Wiley & Sons. [Google Scholar]
- 86.Cantara WA, Pathirage C, Hatterschide J, Olson ED, & Musier-Forsyth K (2022). Phosphomimetic S207D Lysyl–tRNA Synthetase Binds HIV-1 5′UTR in an Open Conformation and Increases RNA Dynamics. Viruses, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lever AML, Göttlinger HG, Haseltine WA, & Sodroski JG (1989). Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. Journal of Virology, 63, 4085–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aldovini A, & Young RA (1990). Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. Journal of Virology, 64, 1920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clavel F, & Orenstein JM (1990). A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. Journal of Virology, 64, 5230–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clever JL, & Parslow TG (1997). Mutant Human Immunodeficiency Virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol, 71, 3407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berkhout B (1996). Structure and function of the human immunodeficiency virus leader RNA. Prog Nucl Acid Res and Mol Biol: Academic Press, Inc. p 1–34. [DOI] [PubMed] [Google Scholar]
- 92.Jones CP, Cantara WA, Olson ED, & Musier-Forsyth K (2014). Small-angle X-ray scattering-derived structure of the HIV-1 5’ UTR reveals 3D tRNA mimicry. Proc Natl Acad Sci U S A, 111, 3395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Onafuwa-Nuga AA, King SR, & Telesnitsky A (2005). Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J Virol, 79, 13528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eckwahl MJ, Arnion H, Kharytonchyk S, Zang T, Bieniasz PD, Telesnitsky A, & Wolin SL (2016). Analysis of the human immunodeficiency virus-1 RNA packageome. RNA, 22, 1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Telesnitsky A, & Wolin SL (2016). The Host RNAs in Retroviral Particles. Viruses, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rawson JMO, Nikolaitchik OA, Shakya S, Keele BF, Pathak VK, & Hu WS (2022). Transcription Start Site Heterogeneity and Preferential Packaging of Specific Full-Length RNA Species Are Conserved Features of Primate Lentiviruses. Microbiol Spectr, e0105322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.D’Souza V, & Summers MF (2004). Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature, 431, 586–90. [DOI] [PubMed] [Google Scholar]
- 98.Pereira-Montecinos C, Toro-Ascuy D, Ananias-Saez C, Gaete-Argel A, Rojas-Fuentes C, Riquelme-Barrios S, Rojas-Araya B, Garcia-de-Gracia F, Aguilera-Cortes P, Chnaiderman J, Acevedo ML, Valiente-Echeverria F, & Soto-Rifo R (2022). Epitranscriptomic regulation of HIV-1 full-length RNA packaging. Nucleic Acids Res, 50, 2302–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Havecker ER, Gao X, & Voytas DF (2004). The diversity of LTR retrotransposons. Genome Biol, 5, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Curcio MJ, Lutz S, & Lesage P (2015). The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol Spectr, 3, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sandmeyer S, Patterson K, & Bilanchone V (2015). Ty3, a Position-specific Retrotransposon in Budding Yeast. Microbiol Spectr, 3, MDNA3–0057-2014. [DOI] [PubMed] [Google Scholar]
- 102.Cheng Z, & Menees TM (2004). RNA branching and debranching in the yeast retrovirus-like element Ty1. Science, 303, 240–3. [DOI] [PubMed] [Google Scholar]
- 103.Menees TM (2020). RNA Lariat Debranching Enzyme as a Retroviral and Long-Terminal-Repeat Retrotransposon Host Factor. Annu Rev Virol, 7, 189–202. [DOI] [PubMed] [Google Scholar]
- 104.Karst SM, Rutz ML, & Menees TM (2000). The yeast retrotransposons Ty1 and Ty3 require the RNA Lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem Biophys Res Commun, 268, 112–7. [DOI] [PubMed] [Google Scholar]
- 105.Chang W, & Schulman AH (2008). BARE retrotransposons produce multiple groups of rarely polyadenylated transcripts from two differentially regulated promoters. Plant J, 56, 40–50. [DOI] [PubMed] [Google Scholar]
- 106.Chang W, Jaaskelainen M, Li SP, & Schulman AH (2013). BARE retrotransposons are translated and replicated via distinct RNA pools. PLoS One, 8, e72270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sokoloski KJ, Haist KC, Morrison TE, Mukhopadhyay S, & Hardy RW (2015). Noncapped Alphavirus Genomic RNAs and Their Role during Infection. J Virol, 89, 6080–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.LaPointe AT, Landers VD, Westcott CE, & Sokoloski KJ (2020). Production of Noncapped Genomic RNAs Is Critical to Sindbis Virus Disease and Pathogenicity. mBio, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guglielmi KM, McDonald SM, & Patton JT (2010). Mechanism of intraparticle synthesis of the rotavirus double-stranded RNA genome. J Biol Chem, 285, 18123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Patton JT (1996). Rotavirus VP1 alone specifically binds to the 3’ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J Virol, 70, 7940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vende P, Piron M, Castagne N, & Poncet D (2000). Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3’ end. J Virol, 74, 7064–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deo RC, Groft CM, Rajashankar KR, & Burley SK (2002). Recognition of the rotavirus mRNA 3’ consensus by an asymmetric NSP3 homodimer. Cell, 108, 71–81. [DOI] [PubMed] [Google Scholar]
- 113.Groft CM, & Burley SK (2002). Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell, 9, 1273–83. [DOI] [PubMed] [Google Scholar]
- 114.Vicens Q, Kieft JS, & Rissland OS (2018). Revisiting the Closed-Loop Model and the Nature of mRNA 5’−3’ Communication. Mol Cell, 72, 805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Junker-Niepmann M, Bartenschlager R, & Schaller H (1990). A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J, 9, 3389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bartenschlager R, Junker-Niepmann M, & Schaller H (1990). The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol, 64, 5324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pollack JR, & Ganem D (1993). An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol, 67, 3254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jeong JK, Yoon GS, & Ryu WS (2000). Evidence that the 5’-end cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J Virol, 74, 5502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ryu DK, Kim S, & Ryu WS (2008). Hepatitis B virus polymerase suppresses translation of pregenomic RNA via a mechanism involving its interaction with 5’ stem-loop structure. Virology, 373, 112–23. [DOI] [PubMed] [Google Scholar]
- 120.Ryu DK, Ahn BY, & Ryu WS (2010). Proximity between the cap and 5’ epsilon stem-loop structure is critical for the suppression of pgRNA translation by the hepatitis B viral polymerase. Virology, 406, 56–64. [DOI] [PubMed] [Google Scholar]
- 121.Mazeaud C, Freppel W, & Chatel-Chaix L (2018). The Multiples Fates of the Flavivirus RNA Genome During Pathogenesis. Front Genet, 9, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ng WC, Soto-Acosta R, Bradrick SS, Garcia-Blanco MA, & Ooi EE (2017). The 5’ and 3’ Untranslated Regions of the Flaviviral Genome. Viruses, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khromykh AA, Varnavski AN, Sedlak PL, & Westaway EG (2001). Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J Virol, 75, 4633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, Zhao H, Yu JY, & Qin CF (2016). Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sanford TJ, Mears HV, Fajardo T, Locker N, & Sweeney TR (2019). Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res, 47, 9789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu ZY, & Qin CF (2020). Structure and function of cis-acting RNA elements of flavivirus. Rev Med Virol, 30, e2092. [DOI] [PubMed] [Google Scholar]
- 127.Fajardo T, Sanford TJ, Mears HV, Jasper A, Storrie S, Mansur DS, & Sweeney TR (2020). The flavivirus polymerase NS5 regulates translation of viral genomic RNA. Nucleic Acids Res, 48, 5081–93. [DOI] [PMC free article] [PubMed] [Google Scholar]


