Abstract
This study aimed to investigate the role of long non-coding RNA maternally expressed gene 3 (lncRNA MEG3) in chemosensitivity of osteosarcoma (OS), and to reveal the possible underlying mechanisms. In this study, we found that the expression of lncRNA MEG3 was significantly lower in OS tissues and cell lines. Furthermore, lncRNA MEG3 overexpression enhanced chemosensitivity of OS by inhibiting cell proliferation, migration, autophagy, and promoting antitumor immunity. LncRNA MEG3 functioned as miR-21-5 sponge to regulate p53 expression in OS. Mechanically, lncRNA MEG3 promoted OS chemosensitivity by regulating antitumor immunity via miR-21-5p/p53 pathway and autophagy. Collectively, this study provided the evidence that lncRNA MEG3 might be a promising therapeutic target for OS chemoresistance.
Keywords: Antitumor immunity, Autophagy, Chemosensitivity, LncRNAMEG3, Osteosarcoma
Introduction
Osteosarcoma (OS) is one of the most common bone malignancies which affect mainly children and adolescents. In spite of great achievements in the treatment methods including operation and chemotherapy, overall survival of OS patients is still unsatisfying and its molecular mechanisms remain largely unknown.1 Cisplatin (DDP), as a common anti-tumor drug, has been widely suggested to be effective in solid tumors including OS. DDP has become the conventional agent for OS chemotherapy in daily clinical. Whereas, chemotherapy resistance is one of the major obstacles in OS treatment.2 The response rates of OS patients for DDP are less than 30% due to intrinsically resistance to chemotherapy.3 Furthermore, acquired drug resistance for OS patients for several months will further decrease therapy efficiency, leading to poor prognosis.4 Therefore, it is urgent to investigate the underlying mechanisms of chemotherapy resistance and find novel targets of OS treatment.
Long non-coding RNA (lncRNA) is one kind of novel endogenous RNAs longer than 200 nucleotides, without functional open reading frame. Several researches revealed that lncRNA plays a regulative role in gene expression, transcription and post-transcriptional processing.5,6 Moreover, the close relationships between disorder of lncRNA and cancers have attracted great attentions.7,8 LncRNA maternally expressed gene 3 (lncRNA MEG3) is a maternally expressed and imprinted gene with around 1.6 kb. Aberrant expression of lncRNA MEG3 in many cancers including OS have been investigated.9,10 Whereas, the further molecular mechanisms of lncRNA MEG3 in OS and its function in chemosensitivity are still unclear.
Immune cells in tumor microenvironment (TME) play a key role in tumor development, and immunotherapeutic strategies might be effective in the treatment of bone and soft tissue sarcoma such as OS.11,12 Immunotherapy such as cytotoxic T lymphocyte associated antigen 4 (CTLA4), programmed death-1 (PD-1) and programmed death ligand-1 (PD-L1) inhibitors were considered to be a promising therapy in various cancers.13, 14, 15, 16 Researchers have found that the tumor-infiltrating lymphocytes could influence the prognosis of cancer patients and efficacy of anti-tumor therapy.17,18 Accordingly, it is of great importance to find immune-related therapeutic targets in OS. Whereas, the expression levels of lncRNA as well as the immune infiltrates and prognostic values in OS have not been completely investigated.
In this study, we aimed to explore the effect of lncRNA MEG3 in chemosensitivity of OS and to reveal the underlying mechanisms. We investigated the expression of lncRNA MEG3 and correlation with prognosis of OS patients. Furthermore, the correlation of lncRNA MEG3 with tumor-infiltrating immune cells and marker genes of T cell exhaustion of bone and soft tissue sarcoma was conducted via the Gene Expression Profiling Interactive Analysis (GEPIA) database and the Tumor Immune Estimation Resource (TIMER) database. This study shed light on the vital role of lncRNA MEG3 in OS chemosensitivity as well as the underlying mechanisms between lncRNA MEG3 and tumor–immune interactions.
Materials and methods
Patient samples
This study was approved by the institutional review board and the medical ethics committee of Wuhan Union Hospital, Huazhong University of Science and Technology. We collected the medical data and samples of ten patients with osteosarcoma in our hospital from January 2018 to October 2020. Clinicopathological parameters of osteosarcoma patients were shown in Table S1.
Cell culture and treatment
Four human osteosarcoma cell lines (MNNG/HOS, U2OS, MG63, and SaOS-2) and normal human osteoblasts cell line (hFOB1.19) were obtained from the Cell Bank of China Academy of Sciences (Shanghai, China). All cells were maintained in a humidified incubator at 37 °C with a 5% CO2 atmosphere. DDP was purchased from MedChemExpress (MCE, China). The DDP group was exposed to DDP (5 μM) for 24 h.
Fluorescence in situ hybridization (FISH)
The lncRNA MEG3 FISH probe was provided by BOSTER (Wuhan, China), and the FISH assay was performed in accordance with the directions. Cells were fixed followed by prehybridization in PBS, and then hybridized at 37 °C for 30 min. Cells were also stained with 4′,6-diamidino-2-phenylindole (DAPI). The probe signals were detected using a fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Plasmid transfection of cell lines
The lncRNA MEG3 sequence was synthesized (based on the MEG3 sequence, GenBank™ NR_002766) and then subcloned into the pCDNA3.1 vector (GeneChem, Shanghai, China). Increased expression of lncRNA MEG3 was achieved by transfection of pCDNA-MEG3. Cells were harvested for following analyses 48 h after transfection.
Cell viability assay
Cell viability was assessed using a Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories Co. Ltd, Kumamoto, Japan). After treatments, the cell medium was replaced by 90 μl fresh α-MEM medium with 10 μl CCK-8 solution. After 1–4 h incubation at 37 °C, the absorbance was recorded at 450 nm.
Colony formation assay
Cells (1 × 103 cells/well) were trypsinized into a single-cell suspension and seeded into 6-well plates. At the 10th day, cells were fixed with 4% paraformaldehyde for 15 min and then stained with 1% crystal violet. Clones containing more than 50 cells were counted using a grid.
Wound healing assay
Each well of a 6-well culture plate was seeded with cells to a final density of 1 × 105 cells/well and these cells were incubated for 24 h to permit cell adhesion and the formation of a confluent monolayer. Wounds were generated in the cell monolayer by scratching with a plastic pipette tip. Wound closure was monitored by collecting digitized images at 0 and 24 h after the scratch was performed.
Immunofluorescent detection of Beclin 1 expression
Fixed cells were treated with 1:200 diluted polyclonal rabbit anti-human IgG antibody against Beclin 1 (Proteintech, Wuhan, China) for 12 h in a humidifier at 4 °C. Following rinsing in the incubation buffer (twice for 5 min), the specimens were incubated with 1:100-diluted goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) conjugate (Proteintech, Wuhan, China) and propidium iodide (PI; Sigma–Aldrich) for 1 h at room temperature, and subsequently rinsed with PBS. Fluorescence was detected using a fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Western blot analysis
Cell protein lysates were separated in 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.22 μm PVDF membranes (Millipore, Massachusetts, USA). 5% skim milk containing 0.1% Tween-20 was utilized to block the PVDF membranes and then incubated with specific antibodies at 4 °C overnight. Horseradish peroxidase-linked secondary antibodies were added at a dilution ratio of 1:1000, and incubated at 37 °C for 1 h. The immunoreactive bands were visualized using ECL Kit (Thermo Fisher).
Xenograft tumor model
The study protocol was approved by the Committee on the Ethics of Animal Experiments of Huazhong University of Science and Technology. Nude mice (BALB/c, female, 4- to 5-week-old) were injected subcutaneously with 5 × 106 MNNG/HOS cells. All Mice were randomly divided into four groups and prepared for treatment with DDP or transfection of pCDNA-MEG3 or as controls when the subcutaneous tumor volume reached 50–60 mm3. For delivery of pCDNA-MEG3, 10 nmol pCDNA-MEG3 in 150 μL saline buffer was locally injected into the tumor mass every three days. Meanwhile, 2 mg/kg DDP was locally injected into the tumor at the same time. The animals were sacrificed, and tumors were harvested (measured and weighed) and fixed in 4% paraformaldehyde after three weeks.
Gene Expression Profiling Interactive Analysis (GEPIA)
The online database Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) was used to detect the prognosis value of lncRNA MEG3 in bone and soft tissue sarcoma. The tumor and normal tissue datasets were used for analysis. The correlation between the expressions of MEG3 and TP53, BECN1 and MAP1LC3B, and the expressions of TP53 and BECN1, MAP1LC3B in bone and soft tissue sarcoma were analyzed via GEPIA database.
TIMER database analysis
TIMER is a comprehensive database which investigates the immune infiltrates in various cancer types (https://cistrome.shinyapps.io/timer/). TIMER database is composed of 10,897 samples with 32 cancer types from The Cancer Genome Atlas (TCGA). In this study, we assessed the expression of MEG3 in various cancers and the correlation of MEG3, TP53, BECN1, and MAP1LC3B with the abundance of immune infiltrates, such as B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. SCNA module in the TIMER database provides the comparison of tumor infiltration levels among tumors with different somatic copy number alterations for a given gene. Moreover, correlations between the expression of MEG3, TP53, BECN1 and MAP1LC3B and gene markers of tumor-infiltrating immune cells such as exhausted T cells were explored via correlation modules. The gene expression level was displayed with log2 RSEM.
Statistical analysis
All experiments were repeated at least three times. The results of multiple experiments were presented as mean ± standard deviation (SD). Statistical analyses were performed using GraphPad 6.0 statistical software (GraphPad Software Inc., San Diego, CA). P values were calculated using one-way analysis of variance (ANOVA). The correlation of gene expression was assessed by Spearman's correlation and statistical significance. The gene expression level was displayed with log2 RSEM. A P-value of <0.05 was considered to indicate a statistically significant result.
Results
Expression of lncRNA MEG3 is dysregulated in OS
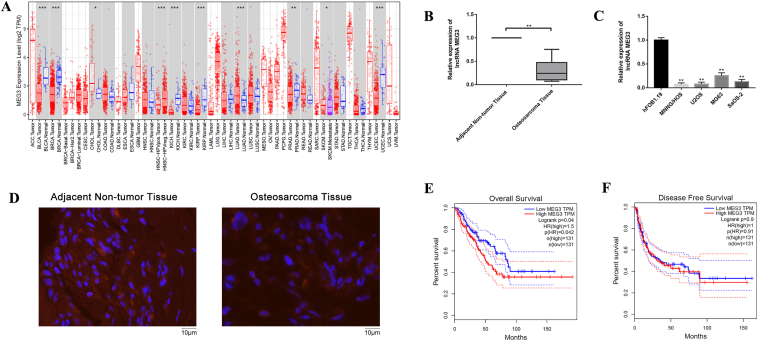
The expression levels of lncRNA MEG3 in multiple malignancies were shown in Figure 1A, by using the RNA-seq data in The Cancer Genome Atlas (TCGA). As shown in Figure 1B, lncRNA MEG3 expression was significantly decreased in OS tissues when compared with adjacent non-tumor tissues. We further detected the expression of lncRNA MEG3 in four OS cell lines (MNNG/HOS, U2OS, MG63, and SaOS-2) and human osteoblast cell line (hFOB1.19) via qRT-PCR. Results showed that the expressions of lncRNA MEG3 in four OS cell lines were significantly lower than that in hFOB1.19 cells (Fig. 1C). According to the results of FISH staining, the expression of lncRNA MEG3 was significantly decreased in OS tissues when compared with adjacent non-tumor tissues (Fig. 1D). We investigated whether lncRNA MEG3 was correlated with prognosis in bone and soft tissue sarcoma. In GEPIA database, the poor overall survival in bone and soft tissue sarcoma was shown to significantly correlate with higher lncRNA MEG3 level (HR = 1.50, P = 0.042, Fig. 1E), and no significant difference was observed with disease free survival (Fig. 1F). Therefore, the dysregulated expression of lncRNA MEG3 was liable to have great importance in osteosarcoma.
Figure 1.
Expression of lncRNA MEG3 was dysregulated in OS. (A) The expression levels of lncRNA MEG3 in multiple malignancies were shown by using the RNA-seq data in The Cancer Genome Atlas (TCGA). (B) LncRNA MEG3 expression was significantly decreased in osteosarcoma tissues when compared with adjacent non-tumor tissues (∗∗P < 0.01). (C) The expression of lncRNA MEG3 in four OS cell lines (MNNG/HOS, U2OS, MG63, and SaOS-2) and human osteoblast cell line (hFOB1.19) via qRT-PCR (∗∗P < 0.01). (D) FISH staining was used to detect the expression of lncRNA MEG3 in adjacent non-tumor tissues and osteosarcoma tissues (E) In GEPIA database, the poor overall survival in bone and soft tissue sarcoma was shown to significantly correlate with higher lncRNA MEG3 level. (F) No significant difference was observed with disease free survival.
LncRNA MEG3 overexpression promotes chemosensitivity of OS cells
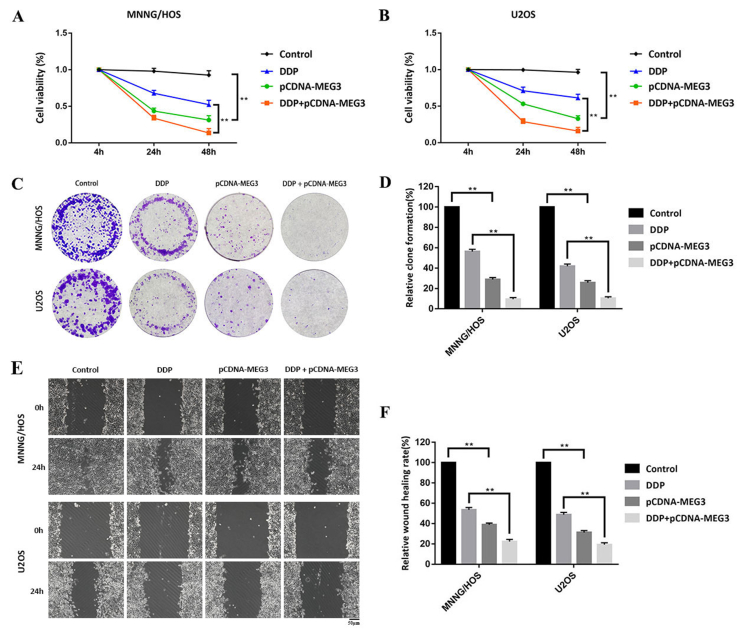
We transfected OS cells with pCDNA-MEG3 to overexpress lncRNA MEG3 in OS cells. In this way, we aimed to confirm the function of lncRNA MEG3 in human OS cells. CCK-8 assay showed that the cell viability of pCDNA-MEG3 group appeared to be significantly reduced compared with that of control group. Compared with DDP group, the cell viability of DDP + pCDNA-MEG3 group reduced significantly (Fig. 2A). The results were consistent in U2OS cells (Fig. 2B).
Figure 2.
LncRNA MEG3 overexpression promotes chemosensitivity of OS cells. We transfected OS cells with pCDNA-MEG3 to overexpress lncRNA MEG3 in OS cells. (A) CCK-8 assay showed that the cell viability of pCDNA-MEG3 group appeared to be significantly reduced compared with that of control group. Compared with DDP group, the cell viability of DDP + pCDNA-MEG3 group reduced significantly (∗∗P < 0.01). (B) The results were consistent in U2OS cells (∗∗P < 0.01). (C, D) In colony formation assay, lncRNA MEG3 overexpression prohibited proliferation ability of MNNG/HOS and U2OS cells when compared with control group. Moreover, compared with DDP group, the proliferation ability of DDP + pCDNA-MEG3 group reduced significantly (∗∗P < 0.01).(E, F) Comparison of the migration ability of MNNG/HOS and U2OS cells among all groups with scratch wound healing assay. The results revealed that pCDNA-MEG3 group displayed less migratory capacity when compared with that of control group. Compared with DDP group, the migration ability of DDP + pCDNA-MEG3 group reduced significantly (∗∗P < 0.01).
In colony formation assay, lncRNA MEG3 overexpression prohibited proliferation ability of MNNG/HOS and U2OS cells when compared with control group. Moreover, compared with DDP group, the proliferation ability of DDP + pCDNA-MEG3 group reduced significantly (Fig. 2C, D).
We compared the migration ability of MNNG/HOS and U2OS cells among all groups with scratch wound healing assay. The results revealed that pCDNA-MEG3 group displayed less migratory capacity when compared with that of control group. Compared with DDP group, the migration ability of DDP + pCDNA-MEG3 group reduced significantly (Fig. 2E, F).
LncRNA MEG3 overexpression promotes antitumor immunity
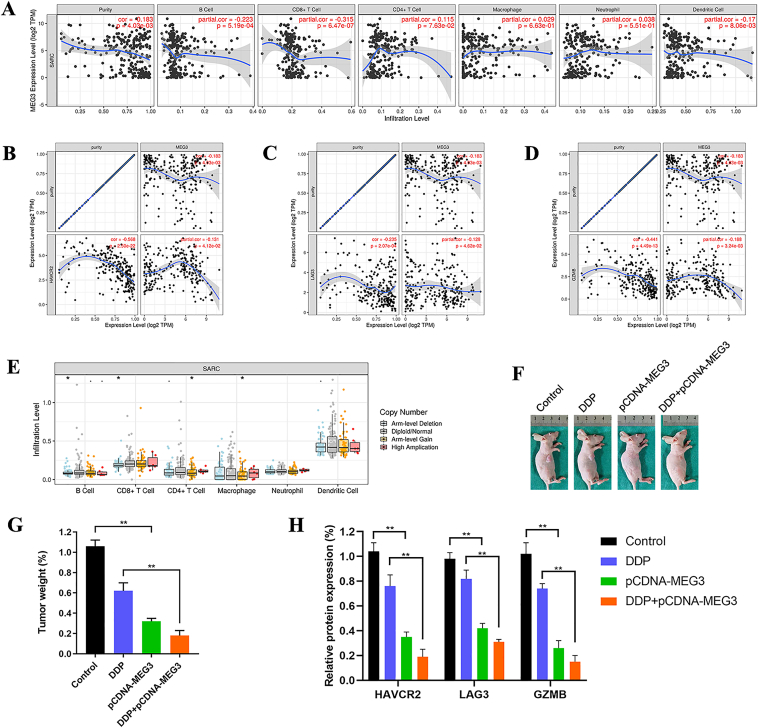
Tumor-infiltrating lymphocytes are independent predictors of sentinel lymph node status and survival in cancers. Therefore, we investigated whether lncRNA MEG3 was correlated with immune infiltration levels in bone and soft tissue sarcoma via the TIMER database. The results showed that lncRNA MEG3 had significant negative correlations with infiltrating levels of B cells (r = −0.223, P = 5.19e-04), CD8+ T cells (r = −0.315, P = 6.47e-07), and dendritic cells (r = −0.170, P = 8.06e-03) in bone and soft tissue sarcoma (Fig. 3A). SCNA module in the TIMER database provides the comparison of tumor infiltration levels among tumors with different somatic copy number alterations for a given gene. Figure 3E was presented to show the distributions of each immune subset at each copy number status of lncRNA MEG3 in bone and soft tissue sarcoma.
Figure 3.
LncRNA MEG3 overexpression promotes antitumor immunity. (A) LncRNA MEG3 had significant negative correlations with infiltrating levels of B cells (r = −0.223, P = 5.19e-04), CD8+ T cells (r = −0.315, P = 6.47e-07), and dendritic cells (r = −0.170, P = 8.06e-03) in bone and soft tissue sarcoma via the TIMER database. LncRNA MEG3 had a significant negative correlation with (B)TIM-3 (r = −0.131, P = 4.12e-02).(C)LAG3 (r = −0.128, P = 4.62e-02), and (D)GZMB (r = −0.188, P = 3.24e-03).(E) SCNA module in the TIMER database showed the distributions of each immune subset at each copy number status of lncRNA MEG3 in bone and soft tissue sarcoma (∗∗P < 0.01). (F) In xenograft tumor model, we randomly divided all mice into four groups including control group, DDP group, pCDNA-MEG3 group, and DDP + pCDNA-MEG3 group. (G) The weight of DDP + pCDNA-MEG3 group was significantly lower than that of DDP group (∗∗P < 0.01). (H) qRT-PCR showed that TIM-3, LAG3, and GZMB expression of pCDNA-MEG3 group significantly reduced compared with that of control group. Compared with DDP group, TIM-3, LAG3, and GZMB expression of DDP + pCDNA-MEG3 group reduced significantly (∗∗P < 0.01).
We also found significant correlations between lncRNA MEG3 and marker genes of T cell exhaustion, such as TIM-3 (HAVCR2), LAG3, and GZMB. TIM-3 is a crucial gene that regulates T cell exhaustion. LncRNA MEG3 had significantly negative correlations with TIM-3 (r = −0.131, P = 4.12e-02, Fig. 3B), LAG3 (r = −0.128, P = 4.62e-02, Fig. 3C), and GZMB (r = −0.188, P = 3.24e-03, Fig. 3D). It suggested that lncRNA MEG3 overexpression might reverse TIM-3 mediating T cell exhaustion.
In xenograft tumor model, we randomly divided all mice into four groups including control group, DDP group, pCDNA-MEG3 group, and DDP + pCDNA-MEG3 group (Fig. 3F). After harvesting and weighing the subcutaneous tumors, we found that the weight of DDP + pCDNA-MEG3 group was significantly lower than that of DDP group (Fig. 3G). qRT-PCR showed that TIM-3, LAG3, and GZMB expression of pCDNA-MEG3 group significantly reduced compared with that of control group. Compared with DDP group, TIM-3, LAG3, and GZMB expression of DDP + pCDNA-MEG3 group reduced significantly (Fig. 3H). The results were consistent with that of the TIMER database.
LncRNA MEG3 regulates p53 expression via sponging miR-21-5p in OS
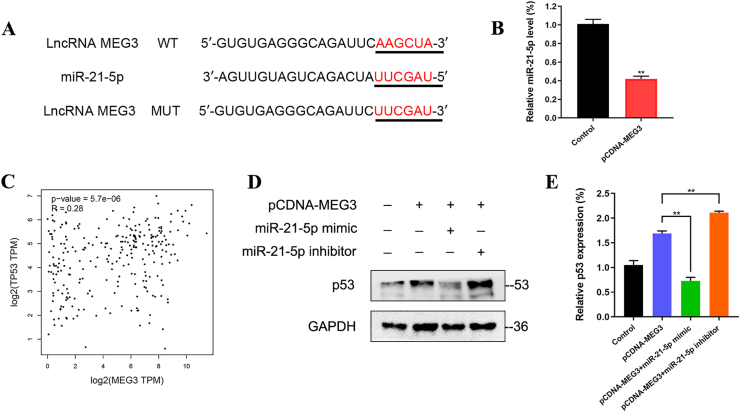
Previous studies have confirmed that lncRNA can function as miRNA sponges and subsequently regulate the function of the corresponding miRNA. To predict the potential target miRNAs of lncRNA MEG3, a bioinformatic analysis using the TargetScan database revealed that lncRNA MEG3 shares the miRNA response element with miR-21-5p (Fig. 4A). According to qRT-PCR, lncRNA MEG3 was significantly highly expressed, while the expression of miR-21-5p decreased correspondingly in OS cells of pCDNA-MEG3 group when compared with control group (Fig. 4B).
Figure 4.
LncRNA MEG3 regulates p53 expression via sponging miR-21-5p in OS. (A) A bioinformatic analysis using the TargetScan database revealed that lncRNA MEG3 shares the miRNA response element with miR-21-5p. (B) According to qRT-PCR, lncRNA MEG3 was significantly highly expressed, while the expression of miR-21-5p decreased correspondingly in OS cells of pCDNA-MEG3 group when compared with control group (∗∗P < 0.01). (C) GEPIA database showed that lncRNA MEG3 had a significant positive correlation with TP53 (r = 0.28, P = 5.7e-06). (D, E) Western blot indicated that pCDNA-MEG3+miR-21-5p mimic group was observed with a significantly decreased expression of p53 when compared with pCDNA-MEG3 group. Meanwhile, a significantly increased expression of p53 was observed in pCDNA-MEG3+miR-21-5p inhibitor group when compared with pCDNA-MEG3 group (∗∗P < 0.01).
GEPIA database showed that lncRNA MEG3 had a significantly positive correlation with TP53 (r = 0.28, P = 5.7e-06, Fig. 4C). We further investigated whether lncRNA MEG3 regulates p53 expression via sponging miR-21-5p. Western blot indicated that pCDNA-MEG3+miR-21-5p mimic group was observed with a significantly decreased expression of p53 when compared with pCDNA-MEG3 group. Meanwhile, a significantly increased expression of p53 was observed in pCDNA-MEG3+miR-21-5p inhibitor group when compared with pCDNA-MEG3 group (Fig. 4D, E).
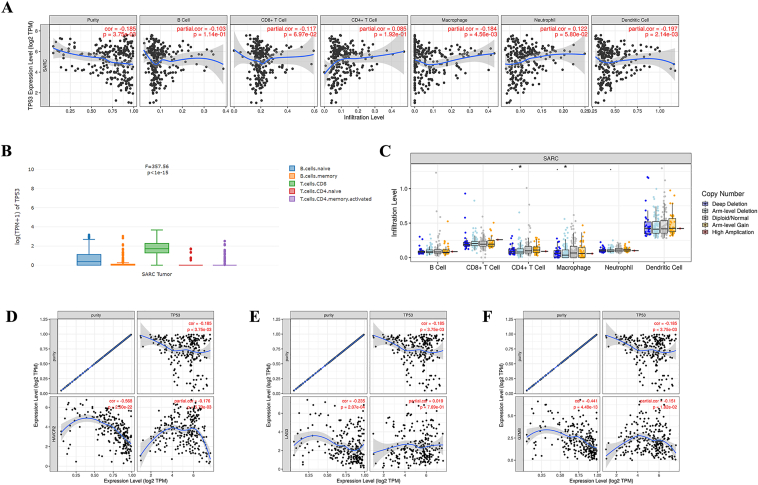
TP53 regulates tumor infiltration levels and T cell exhaustion
The results from TIMER database showed that TP53 had significant negative correlations with infiltrating levels of macrophage (r = −0.184, P = 4.56e-03), and dendritic cells (r = −0.197, P = 2.14e-03) in bone and soft tissue sarcoma (Fig. 5A). GEPIA2021 database indicated the TP53 gene expression in each cell type such as various types of B cells and T cells in bone and soft tissue sarcoma (Fig. 5B). Figure 5C was presented to show the distributions of each immune subset at each copy number status of TP53 in bone and soft tissue sarcoma.
Figure 5.
TP53 regulates tumor infiltration levels and T cell exhaustion. (A) The results from TIMER database showed that TP53 had significant negative correlations with infiltrating levels of macrophage (r = −0.184, P = 4.56e-03), and dendritic cells (r = −0.197, P = 2.14e-03) in bone and soft tissue sarcoma. (B) GEPIA2021 database indicated the TP53 gene expression in each cell type such as various types of B cells and T cells in bone and soft tissue sarcoma (C) SCNA module in the TIMER database showed the distributions of each immune subset at each copy number status of TP53 in bone and soft tissue sarcoma (∗∗P < 0.01). TP53 had significant negative correlations with (D)TIM-3 (r = −0.178, P = 5.79e-03), and (F)GZMB (r = −0.151, P = 1.82e-02) (E) No significant difference was observed between TP53 and LAG3.
We further found significant correlations between TP53 and marker genes of T cell exhaustion, such as TIM-3 (HAVCR2), LAG3, and GZMB. TP53 had significantly negative correlations with TIM-3 (r = −0.178, P = 5.79e-03, Fig. 5D), and GZMB (r = −0.151, P = 1.82e-02, Fig. 5F). No significant difference was observed between TP53 and LAG3 (Fig. 5E).
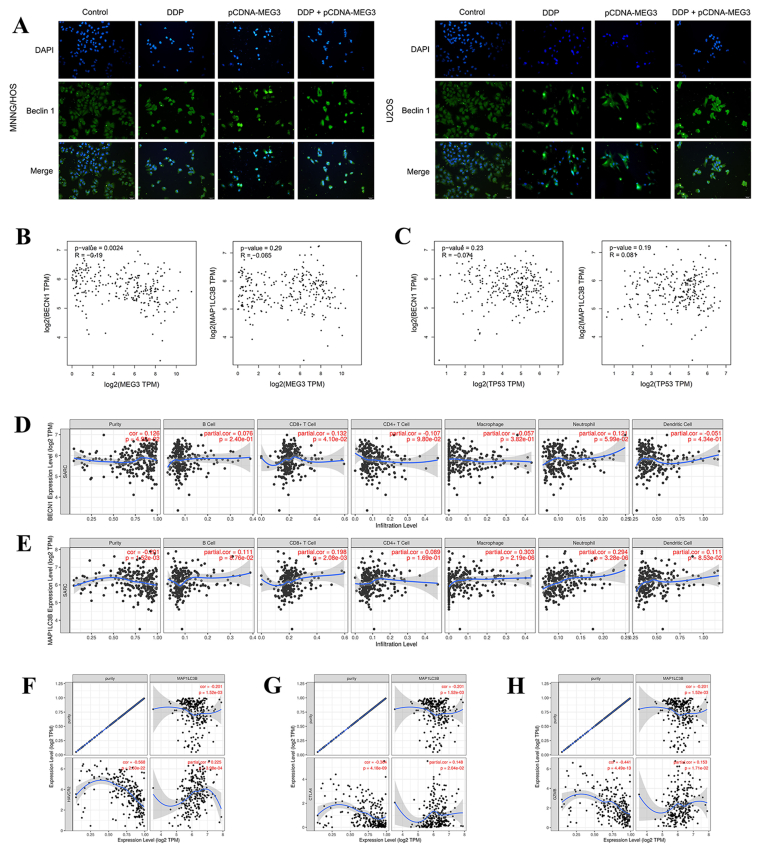
LncRNA MEG3 overexpression inhibits autophagy to regulate antitumor immunity
Beclin 1 and LC3B are markers of autophagy. By detecting the fluorescence level of Beclin 1, we found that Beclin 1 expression of DDP + pCDNA-MEG3 group reduced significantly when compared with DDP group (Fig. 6A). Furthermore, lncRNA MEG3 had a significantly negative correlation with Beclin 1 (BECN1) in bone and soft tissue sarcoma via GEPIA database (r = −0.19, P = 0.0024, Fig. 6B). No significant correlation was observed between TP53 and Beclin 1 or LC3B (MAP1LC3B) in bone and soft tissue sarcoma (Fig. 6C). TIMER database showed that both Beclin 1 (r = 0.132, P = 4.10e-02, Fig. 6D) and LC3B (r = 0.198, P = 2.08e-03, Fig. 6E) had significant positive correlations with infiltrating levels of CD8+ T cells in bone and soft tissue sarcoma. LC3B had a significantly positive correlation with TIM-3 (r = 0.225, P = 3.98e-04, Fig. 6F), CTLA4 (r = 0.148, P = 2.04e-02, Fig. 6G), and GZMB (r = 0.153, P = 1.71e-02, Fig. 6H). It suggested that LC3B might contribute to TIM-3 mediating T cell exhaustion.
Figure 6.
LncRNA MEG3 overexpression inhibits autophagy to regulate antitumor immunity. (A) By detecting the fluorescence level of Beclin 1, Beclin 1 expression of DDP + pCDNA-MEG3 group reduced significantly when compared with DDP group. (B) LncRNA MEG3 had a significantly negative correlation with Beclin 1 (BECN1) in bone and soft tissue sarcoma via GEPIA database (r = −0.19, P = 0.0024). (C) No significant correlation was observed between TP53 and Beclin 1 or LC3B (MAP1LC3B) in bone and soft tissue sarcoma. TIMER database showed that both (D) Beclin 1 (r = 0.132, P = 4.10e-02) and (E) LC3B (r = 0.198, P = 2.08e-03) had significant positive correlations with infiltrating levels of CD8+ T cells in bone and soft tissue sarcoma. LC3B had significant positive correlations with (F)TIM-3 (r = 0.225, P = 3.98e-04) (G)CTLA4 (r = 0.148, P = 2.04e-02), and (H)GZMB (r = 0.153, P = 1.71e-02).
Discussion
Operation and chemotherapy are two major approaches for OS clinical treatment.1 DDP is one of the four classical chemotherapy drugs (DDP, doxorubicin, methotrexate and ifosfamide) for OS, and its anti-tumor ability is mainly based on the combination with DNA to cause DNA damage.19 However, the response rates for DDP are less than 30% due to chemoresistance,3 which leads to decreased therapy efficiency and poor prognosis of OS patients.4 Different from the other three drugs, DDP is the only drug containing metal elements, which makes DDP free from cross-resistance with other drugs in some key pathways.20 Therefore, it is of great therapeutic value to separately investigate DDP sensitivity in OS. To sum up, chemoresistance is the major challenge in OS treatment. Due to lacking effective therapy strategy, chemoresistance and poor prognosis in OS, it is critical to explore the underlying mechanisms of chemoresistance and discover novel molecular targets for overcoming OS chemoresistance.
Nowadays, a growing number of researches have showed the regulative roles of lncRNA in the development and malignancy of cancers.21,22 The dysregulated expression levels of lncRNA MEG3 had been observed in multiple malignancies in TCGA database. In this study, lncRNA MEG3 expression was confirmed to be significantly decreased in OS tissues and cell lines. Therefore, it could be inferred that lncRNA MEG3 might play a vital role in the biological processes of OS. For further studying, we transfected OS cells with pCDNA-MEG3 to increase the expression level of lncRNA MEG3 through human intervention.
Previous studies maintained that abnormal expression of lncRNA MEG3 inhibits proliferation, migration and invasion, and promotes apoptosis in human OS cells.9,10 LncRNA MEG3 is also involved in modulating chemoresistance in multiple types of human cancers. One study showed that lncRNA MEG3 promotes DDP sensitivity of non-small cell lung cancer (NSCLC) cells via miR-21-5p/SOX7 axis.23 Moreover, lncRNA MEG3 confers DDP-mediated apoptosis in glioma via inhibiting autophagy.24 LncRNA MEG3 also enhances oxaliplatin sensitivity in colorectal cancer25 and increases DDP sensitivity in bladder cancer.26 Therefore, several studies have investigated the relationship between lncRNA MEG3 and chemosensitivity, and lncRNA MEG3 might play a vital role in chemosensitivity of different cancers. In our study, we aimed to investigate whether overexpressing lncRNA MEG3 promotes chemosensitivity of OS. In order to confirm our speculation, we increased the expression level of lncRNA MEG3 during DDP treatment. The results showed that DDP + pCDNA-MEG3 group exhibited decreased cell viability, proliferation and migration ability when compared with DDP group. Mechanically, lncRNA MEG3 had significant negative correlations with infiltrating levels of CD8+ T cells, and dendritic cells in bone and soft tissue sarcoma. We also found significant negative correlations between lncRNA MEG3 and marker genes of T cell exhaustion. Therefore, the above results showed that lncRNA MEG3 overexpression might reverse T cell exhaustion and promote antitumor immunity. In vivo experiments further confirmed the regulative roles of lncRNA MEG3 in OS chemosensitivity and antitumor immunity.
In this study, lncRNA MEG3 might function as a sponge of miR-21-5p to regulate p53 expression. In OS, lncRNA MEG3 overexpression contributed to increased level of p53 during DDP treatment. TIMER database showed that TP53 had significant negative correlations with infiltrating levels and marker genes of T cell exhaustion in bone and soft tissue sarcoma. Accordingly, the above results indicated that lncRNA MEG3 might regulate antitumor immunity via miR-21-5p/p53 pathway in OS.
Autophagy exhibits a double-edged effect on cancer development.27, 28, 29 On the one hand, autophagy is prevented by oncoproteins including mutant p53.30 It helps inhibit the degradation of excessive protein in starved or stressed cancer cells. On the other hand, excessively activated autophagy might lead to autophagic programmed cell death or apoptosis.31,32 Autophagy has been reported to play the dual role in the regulation of OS chemoresistance by either promoting drug resistance or increasing drug sensitivity.33 Beclin 1 and LC3B are main markers of autophagy. By detecting the fluorescence level of Beclin 1, we found that Beclin 1 expression of DDP + pCDNA-MEG3 group reduced significantly when compared with DDP group. LncRNA MEG3 had a significantly negative correlation with Beclin 1 in bone and soft tissue sarcoma via GEPIA database, which indicated that autophagy is involved in the role of lncRNA MEG3 in OS chemosensitivity. According to TIMER database, LC3B might contribute to TIM-3 mediating T cell exhaustion in bone and soft tissue sarcoma. Therefore, lncRNA MEG3 overexpression might inhibit autophagy to regulate antitumor immunity in OS chemosensitivity. To sum up, lncRNA MEG3 overexpression might increase infiltrating levels of CD4+ T cells and reverse T cell exhaustion in OS. Mechanically, lncRNA MEG3 might regulate antitumor immunity via miR-21-5p/p53 pathway and autophagy, which promotes chemosensitivity of OS.
This study suggested that lncRNA MEG3 overexpression might promote chemosensitivity of OS by enhancing antitumor immunity via miR-21-5p/p53 pathway and autophagy. These findings provided the evidence that lncRNA MEG3 might be a promising therapeutic target for OS chemoresistance.
Author contributions
Xin Huang and Zhicai Zhang conceived and designed the project. Xin Huang and Weiyue Zhang carried out the experiments, performed statistical analyses and drafted the manuscript. Zhicai Zhang and Feifei Pu carried out the sample collection. Feifei Pu contributed to cells culture, reagent procurement and management.
Conflict of interests
Authors declare no conflict of interests.
Funding
This work is supported by The National Natural Science Foundation of China (No. 82072979).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.11.004.
Contributor Information
Xin Huang, Email: 2020xh0041@hust.edu.cn.
Feifei Pu, Email: pufeifeiemail@163.com.
Zhicai Zhang, Email: zhicaizhang@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Luetke A., Meyers P.A., Lewis I., et al. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40(4):523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Hattinger C.M., Patrizio M.P., Fantoni L., et al. Drug resistance in osteosarcoma: emerging biomarkers, therapeutic targets and treatment strategies. Cancers. 2021;13(12):e2878. doi: 10.3390/cancers13122878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim M., Jung J.Y., Choi S., et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13(1):149–168. doi: 10.1080/15548627.2016.1239676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilienthal I., Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int J Mol Sci. 2020;21(18):e6885. doi: 10.3390/ijms21186885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.He R.Z., Luo D.X., Mo Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6(1):6–15. doi: 10.1016/j.gendis.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:e38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahadi A. Functional roles of lncRNAs in the pathogenesis and progression of cancer. Genes Dis. 2021;8(4):424–437. doi: 10.1016/j.gendis.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Wang J., Li J.W., et al. LncRNA MEG3 inhibits proliferation and promotes apoptosis of osteosarcoma cells through regulating Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(2):581–590. doi: 10.26355/eurrev_202001_20034. [DOI] [PubMed] [Google Scholar]
- 10.Shen B., Zhou N., Hu T., et al. LncRNA MEG3 negatively modified osteosarcoma development through regulation of miR-361-5p and FoxM1. J Cell Physiol. 2019;234(8):13464–13480. doi: 10.1002/jcp.28026. [DOI] [PubMed] [Google Scholar]
- 11.Huang X., Hu H., Zhang W., et al. Prognostic value of prognostic nutritional index and systemic immune-inflammation index in patients with osteosarcoma. J Cell Physiol. 2019;234(10):18408–18414. doi: 10.1002/jcp.28476. [DOI] [PubMed] [Google Scholar]
- 12.Huang X., Zhang W., Zhang Z., et al. Prognostic value of programmed cell death 1 ligand-1 (PD-L1) or PD-1 expression in patients with osteosarcoma: a meta-analysis. J Cancer. 2018;9(14):2525–2531. doi: 10.7150/jca.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbee M.S., Ogunniyi A., Horvat T.Z., et al. Current status and future directions of the immune checkpoint inhibitors ipilimumab, pembrolizumab, and nivolumab in oncology. Ann Pharmacother. 2015;49(8):907–937. doi: 10.1177/1060028015586218. [DOI] [PubMed] [Google Scholar]
- 14.Ackermann C.J., Adderley H., Ortega-Franco A., et al. First-line immune checkpoint inhibition for advanced non-small-cell lung cancer: state of the art and future directions. Drugs. 2020;80(17):1783–1797. doi: 10.1007/s40265-020-01409-6. [DOI] [PubMed] [Google Scholar]
- 15.Shen H., Yang E.S., Conry M., et al. Predictive biomarkers for immune checkpoint blockade and opportunities for combination therapies. Genes Dis. 2019;6(3):232–246. doi: 10.1016/j.gendis.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang X., Liu G., Li Y., et al. Immune checkpoint: the novel target for antitumor therapy. Genes Dis. 2021;8(1):25–37. doi: 10.1016/j.gendis.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waniczek D., Lorenc Z., Śnietura M., et al. Tumor-associated macrophages and regulatory T cells infiltration and the clinical outcome in colorectal cancer. Arch Immunol Ther Exp. 2017;65(5):445–454. doi: 10.1007/s00005-017-0463-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H., Liu H., Shen Z., et al. Tumor-infiltrating neutrophils is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric cancer. Ann Surg. 2018;267(2):311–318. doi: 10.1097/SLA.0000000000002058. [DOI] [PubMed] [Google Scholar]
- 19.Isakoff M.S., Bielack S.S., Meltzer P., et al. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan C., Kang J., Hwang J.S., et al. Cisplatin-mediated activation of glucocorticoid receptor induces platinum resistance via MAST1. Nat Commun. 2021;12(1):e4960. doi: 10.1038/s41467-021-24845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Q., Lin Z., Xu J., et al. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting β-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018;9(3):e253. doi: 10.1038/s41419-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin L., Cai Q., Wang S., et al. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018;9(10):e1017. doi: 10.1038/s41419-018-1064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P., Chen D., Ma H., et al. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. OncoTargets Ther. 2017;10:5137–5149. doi: 10.2147/OTT.S146423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma B., Gao Z., Lou J., et al. Long non-coding RNA MEG3 contributes to cisplatin-induced apoptosis via inhibition of autophagy in human glioma cells. Mol Med Rep. 2017;16(3):2946–2952. doi: 10.3892/mmr.2017.6897. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Li H., Zhang L., et al. Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis. Biomed Pharmacother. 2018;106:1607–1615. doi: 10.1016/j.biopha.2018.07.131. [DOI] [PubMed] [Google Scholar]
- 26.Feng S.Q., Zhang X.Y., Fan H.T., et al. Up-regulation of LncRNA MEG3 inhibits cell migration and invasion and enhances cisplatin chemosensitivity in bladder cancer cells. Neoplasma. 2018;65(6):925–932. doi: 10.4149/neo_2018_180125N55. [DOI] [PubMed] [Google Scholar]
- 27.Mowers E.E., Sharifi M.N., Macleod K.F. Autophagy in cancer metastasis. Oncogene. 2017;36(12):1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wirawan E., Vanden Berghe T., Lippens S., et al. Autophagy: for better or for worse. Cell Res. 2012;22(1):43–61. doi: 10.1038/cr.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mrakovcic M., Fröhlich L.F. p53-mediated molecular control of autophagy in tumor cells. Biomolecules. 2018;8(2):e14. doi: 10.3390/biom8020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol. 2016;16(11):661–675. doi: 10.1038/nri.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y.X., Yu H.Y., Lv J.Y., et al. Targeting autophagy is a promising therapeutic strategy to overcome chemoresistance and reduce metastasis in osteosarcoma. Int J Oncol. 2019;55(6):1213–1222. doi: 10.3892/ijo.2019.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.