SETD2 is the main transferase for the trimethylation of histone H3 at lysine 36 (H3K36me3) in mammals. SETD2 plays important role in repairing DNA double strand breaks and maintaining chromatin integrity.1 In renal carcinoma, SETD2 deficiency caused DNA replication fork instability and DNA damage.2 The absence of SETD2 was also reported to have strong tumor-promoting effects in lung adenocarcinomas (LUAD).3 However, the other roles of SETD2 remain poorly understood in LUAD. In this study, we utilized comprehensive omics-data analysis to determine that SETD2 was associated with DNA damage and immune-related signals in LUAD. SETD2 knockdown induced DNA damage and cGAS activation in LUAD cells, and reduced the number of cells at the G1 phase. Moreover, SETD2 deficiency was conducive to mutation burden, immune cell infiltration, and immunotherapy responses. High SETD2 expression was associated with high radiocurability. These findings suggest that SETD2 may be a promising biomarker of therapeutic responses for LUAD patients, and offer novel insights into immunotherapy and radiotherapy.
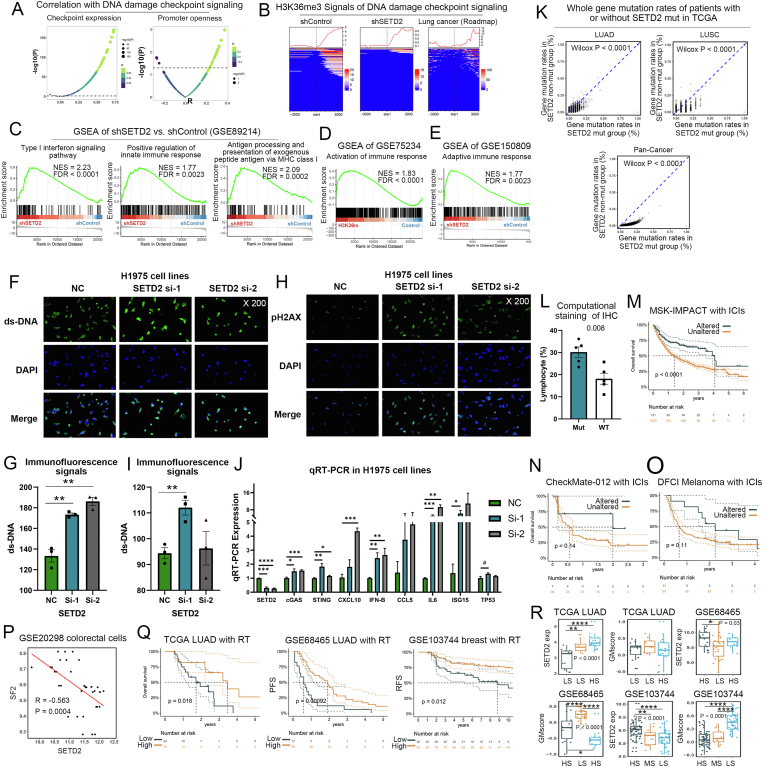
In the combined analysis of RNA sequencing (RNA-seq) from non-small cell lung cancer in The Cancer Genome Atlas (TCGA NSCLC), assay for transposase-accessible chromatin (ATAC) sequencing and H3K36me3 chromatin immunoprecipitation sequencing from GSE110318 and Roadmap confirmed that SETD2 was associated with expression and promoter opening of DNA damage checkpoints (Fig. 1A), and that their promoters were covered by H3K36me3 (Fig. 1B). The cGAS/STING pathway senses double-stranded DNA (ds-DNA) and activates the type I interferon signaling and innate immunity.4 We next analyzed the 3 RNA-seq datasets (GSE89214, GSE75234, and GSE150809) of shSETD2 or H3.3K36 mutation vs. controls. As expected, shSETD2 or H3.3K36 mutation promoted the type I interferon signaling and innate immunity (Fig. 1C–E).
Figure 1.
SETD2 deficiency induced DNA damage, promoted the cGAS/STING signaling, enhanced genomic mutations and immune cell infiltration, and was related to immunotherapy and radiotherapy. (A) Correlation between SETD2 expressions and ATAC signals of DNA damage checkpoints. (B) H3K36me3 signals of DNA damage checkpoints. (C) GSEA of shSETD2 vs. control in GSE89214. (D) GSEA of control vs. H3.3K36 mutations in GSE75234. (E) GSEA of shSETD2 vs. control in GSE150809. (F) Representative immunofluorescence (200 ×) indicated increased ds-DNA in siSETD2 H1975 cells. (G) The quantitative results of ds-DNA immunofluorescence. (H) Representative immunofluorescence (200 ×) indicated increased pH2AX in siSETD2 H1975 cells. (I) The quantitative results of pH2AX immunofluorescence. (J) qRT-PCR of cGAS/STING pathway components in H1975 cells with or without siSETD2. (K) The frequent genomic mutations in patients with or without SETD2 mutations in TCGA cohorts. (L) Proportion of lymphocyte infiltration of patients with or without SETD2 mutations in the TCGA LUAD cohort. (M–O) Kaplan–Meier analysis of SETD2 mutation vs. WT group in the ICI cohorts. (P) Correlation between SETD2 expression and survival fraction at 2 Gy in GSE20298. (Q) Kaplan–Meier analysis of SETD2 expression in the 3 cohorts with radiotherapy. (R) SETD2 expression and GMscore of patient clusters in the 3 cohorts with radiotherapy.
H1975 cells had higher levels of SETD2 than the other NSCLC cells (Fig. S1). Immunofluorescence indicated that SETD2 knockdown significantly increased ds-DNA and pH2AX in H1975 cells, suggesting more DNA damage (Fig. 1F–I). As demonstrated in Figure 1J, siSETD2 had high efficiency, and SETD2 deficiency induced the cGAS/STING pathway. Moreover, SETD2 was associated with cell cycle checkpoints in TCGA NSCLC cohort (Fig. S2A, B). SETD2 had the lowest expression in the G2/M cluster from single cell RNA-seq data GSE131907 (Fig. S2C, D). Less siSETD2 cells were arrested at the G1 phase compared with the control group (Fig. S2E, F).
The negative regulation of DNA damage by SETD2 may be potentially beneficial to genomic stability. We next explored the effects of SETD2 mutations on genomic mutation burden, which was a sign of genomic instability. In TCGA pan-cancer datasets, 513 SETD2 mutation sites were found, of which 188 sites with annotations were possibly oncogenic (Fig. S3), suggesting that SETD2 mutations might change its tumor suppressor function. Moreover, we found that driver mutations, but not variant of uncertain significance, were significantly associated with lower expression levels of SETD2 in TCGA cohorts (Fig. S4). The whole gene mutation rates were increased in patients with SETD2 mutations in the TCGA LUAD, LUSC, and pan-cancer cohorts (Fig. 1K). In MSK-IMPACT cohort (pan-cancer, n = 10,945; LUAD, n = 1357; LUSC, n = 170), patients with SETD2 non-synonymous mutations had high genomic mutations (Fig. S5A). Similar results were found in the other 3 independent data sets (Broad, LUAD, n = 183; MSS Mixed Solid Tumors, pan-cancer, n = 249; Cancer Cell Line Encyclopedia, pan-cancer, n = 1739). In colon cancer of the CPTAC-2 cohort, SETD2-mutated patients had higher percentages of microsatellite instability (Fig. S5B).
Next, we fairly checked the relations between each cut-off point of SETD2 expressions and genome altered fractions. In Cancer Cell Line Encyclopedia pan-cancer data (n = 1739) and NSCLC data (n = 135, Fig. S6), low expression of SETD2 was associated with increased genome altered fraction in the majority of cut-off points and all significant points. Similar results were also found in TCGA NSCLC (Fig. S7).
The cGAS/STING pathway activation and unstable genome may favor immune infiltration. A total of 100 pathological slides were obtained from 10 randomly selected patients with or without SETD2 mutations in TCGA LUAD cohort. We selected 10 images of 910 × 910 pixels in the region of tumor cells for each patient. The representative images were shown in Figure S8. Patients with SETD2 mutations had higher proportions of lymphocyte infiltration (Fig. 1L) and fewer tumor cells, but they were not related to macrophages and stromal cells (Fig. S9).
Clinically, Kaplan–Meier analysis indicated that the upregulated SETD2 was associated with longer survival in TCGA LUAD, GSE50081 (NSCLC, n = 172), and Raponi (LUSC, n = 130) cohorts (Fig. S10). Using multivariate Cox regression to correct covariates (pathological stage, age, and gender) in TCGA LUAD, SETD2 expression (P = 0.16), age (P = 0.13) and pathological stage were related to the prognosis (Table S1). High immune infiltration and mutation burden may be beneficial to immune checkpoint inhibitor (ICI) responses. As expected, the SETD2 mutation group showed the favorable prognosis in the ICI cohorts (MSK-IMPACT with anti-PD-1/PD-L1/CTLA-4, pan-cancer, n = 1661; CheckMate-012 with anti-PD1 plus anti-CTLA-4, NSCLC, n = 75; DFCI GSE4573 with anti-CTLA-4, melanoma, n = 110, Fig. 1M−O). Patients with low SETD2 expression had the favorable prognosis in the DFCI cohort with immunotherapy (Fig. S11). High expression of SETD2 attenuated the immunotherapy responses in all the significant cut-off points of SETD2 in the GSE35640 cohort (melanoma with MAGE-A3, n = 65, Fig. S12).
Since DNA damage is related to radiation, we speculated that SETD2 was associated with radiotherapy responses. SETD2 expression showed a strong negative correlation with the survival fraction at 2 Gy in colorectal cancer cell data GSE20298 (P = 0.0004, Fig. 1P). The high levels of SETD2 were correlated with superior prognosis in the 3 cohorts with radiotherapy (Fig. 1Q). In terms of multivariate Cox regression, SETD2 expression and pathological stages were the predictors of radiocurability (Table S2).
Previous studies suggested that an unstable genome contributed to radiotherapy resistance.5 To expand the use scenario of genomic mutation, we developed a transcriptome-based genomic mutation score (GMscore). The design process was outlined in Figure S13A. Using only TCGA LUAD for training, our GMscore significantly discriminated between the high- and low-frequent mutation groups in TCGA LUAD, LUSC, BRCA, and SKCM cohorts (Fig. S13B). In the 3 cohorts with radiotherapy, patients were divided into 3 groups (Fig. S13C). The high survival group was associated with the highest SETD2 expression and lowest GMscore in comparison with the middle and low survival groups (Fig. 1R; Fig. S13D).
Generally, our work demonstrated that defective SETD2 induced DNA damage, activated the cGAS/STING pathway, and reduced G1 phase cells in LUAD. SETD2 downregulation favored immune infiltration and immunotherapy responses. Moreover, deficient SETD2 was positively correlated with genomic mutations, which was associated with unfavorable radiotherapy prognosis.
Conflict of interests
The authors declare that they have no competing interests.
Funding
This work was supported by National Natural Science Foundation, China (No. 81972852, 81800429); Key Research & Development Project of Hubei Province, China (No. 2020BCA069); Nature Science Foundation of Hubei Province, China (No. 2020CFB612); Young and Middle-Aged Medical Backbone Talents of Wuhan, China (No. WHQG201902); Application Foundation Frontier Project of Wuhan, China (No. 2020020601012221); Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University, China (No. ZNJC201922, ZNJC202007).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.02.016.
Contributor Information
Yan Gong, Email: yan.gong@whu.edu.cn.
Conghua Xie, Email: chxie_65@whu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Carvalho S., Vítor A.C., Sridhara S.C., et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife. 2014;3:e02482. doi: 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanu N., Grönroos E., Martinez P., et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34(46):5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter D.M., Venancio O.S., Buza E.L., et al. Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 2017;77(7):1719–1729. doi: 10.1158/0008-5472.CAN-16-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang B.S., Han W., Kim I.A. Tumor mutation burden, immune checkpoint crosstalk and radiosensitivity in single-cell RNA sequencing data of breast cancer. Radiother Oncol. 2020;142:202–209. doi: 10.1016/j.radonc.2019.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.