Abstract
There are many complex eye diseases which are the leading causes of blindness, however, the pathogenesis of the complex eye diseases is not fully understood, especially the underlying molecular mechanisms of N6-methyladenosine (m6A) RNA methylation in the eye diseases have not been extensive clarified. Our review summarizes the latest advances in the studies of m6A modification in the pathogenesis of the complex eye diseases, including cornea disease, cataract, diabetic retinopathy, age-related macular degeneration, proliferative vitreoretinopathy, Graves’ disease, uveal melanoma, retinoblastoma, and traumatic optic neuropathy. We further discuss the possibility of developing m6A modification signatures as biomarkers for the diagnosis of the eye diseases, as well as potential therapeutic approaches.
Keywords: Degeneration, Eye diseases, Fibrosis, Inflammation, m6A RNA methylation, Tumor
Introduction
RNA methylation has become an important and rapidly developing in biomedical research.1, 2, 3 In the last five years, the study of RNA methylation, especially m6A methylation, has greatly promoted research on the expression of m6A content, the distribution of m6A modification factors in human tissues, and the significance of m6A modifications in cellular functions, and its potential as a biomarker. Physiologically, m6A RNA methylation plays a crucial role in regulating several biological processes, including gene expression, homeostasis maintenance, stem cell pluripotency and reprogramming, cell differentiation, proliferation, lipid and glucose metabolism, circadian rhythm, DNA damage repair, and stress responses.4, 5, 6 Furthermore, m6A modification also regulates pathological processes (including inflammation, angiogenesis, fibrosis, metabolic diseases, degeneration, tumorigenesis, etc.) and contributes to numerous human diseases.7, 8, 9, 10, 11, 12
There are numbers of factors which control the process of m6A methylation, including m6A writers (METTL3, METTL14, METTL16, WTAP, KIAA1429, RBM15, and ZFP217), m6A erasers (FTO and ALKBH5), and readers (YTHDF1,2,3, YTHDC1,2, eIF3, IGF2BP1,2,3, HNRNPA2B1, FMR1, and LRPPRC). METTL3 (methyltransferase-like 3) forms a complex with other m6A writers that catalyze m6A methylation. FTO (fat mass and obesity-associated protein) and ALKBH5 (AlkB homolog 5, RNA demethylase) remove the methylase. The reader proteins recognize methylated RNA. One of the important points is that the m6A modifications are dynamic and reversible.13 The basic principles of m6A modification as well as its role in normal cellular function,14, 15 and the role in the development of the complex human disease have been extensively reviewed in general.4,16, 17, 18, 19, 20, 21
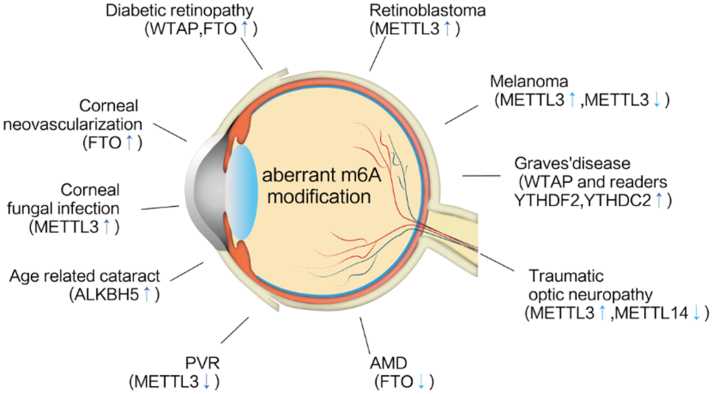
Although eye diseases and systemic diseases share many common characteristics, the eye has its own physiological characteristics, including tissue transparency, the transparency of the cornea, lens, and vitreous humor, the absence of blood vessels, and immune privilege that provides homeostatic mechanisms to limit immune responses in the eye. In addition to the cornea, the tolerance to the foreign antigen can be attributed to the blood–aqueous barrier, blood–retinal barrier (BRB), retinal pigmental epithelium (RPE), and anterior chamber-associated immune deviation.22,23 The two circulation systems in the retina have high metabolic rates and, therefore, the retina is at high risk for oxidative stress damage. Another unique feature of the eye is the special function of RPE cells that is essential for maintaining normal retinal function of transporting nutrition from the choroid to the retina and waste products to the choroidal circulation. Under pathological conditions, RPE cells may transdifferentiate into macrophage-like, immune-related and smooth muscle-like cells and overproduce cytokines and growth factors. Finally, the eye also has its own lymphatic system,24,25 suggesting that the eye has unique physiological and pathological characteristics. Therefore, it is crucial to uncover the role of m6A modifications, especially to gain insights into the pathogenesis of complex eye diseases (Fig. 1).
Figure 1.
The relevance of m6A modification to ocular diseases. The diagram depicts that m6A modification contributes to numerous ocular diseases including inflammatory, angiogenetic, age, fibrotic, traumatic, metabolic, and tumorigenic eye diseases.
The roles of m6A modification in the individual complex eye diseases
m6A modification and fungal keratitis
Corneal infections can be caused by bacterial, viral, or fungal invasion. Fungal keratitis is the most serious corneal infection associated with severe vision loss. Corneal fungal infections are complex and involve a variety of factors, including innate and adaptive immune responses and fungal virulence.26, 27, 28, 29 Fungal infection leads to corneal necrosis and remarkable loss of vision. Current understanding of fungal pathogenicity is limited, however, epigenetic factors may influence the pathogenesis of fungal infections.30,31 Another important aspect of the pathogenesis of fungal infections, especially in the cornea, is m6A modification. This is supported by the finding that the m6A transcript was altered after corneal fungal infection.32 The expression of total m6A and the methylase METTL3 was significantly increased in the cornea of Fusarium solani-infected mice, compared with the cornea in the absence of fungal infection.32 This suggests that the increased total m6A methylation is due to the up-regulated expression of METTL3, as METTL3 is a key m6A methylase. A comprehensive analysis of m6A changes in the cellular epitranscriptome revealed that of the 1137 m6A-modified mRNAs, 780 were hypermethylated, while 357 were hypomethylated under fungal infection conditions. The predominance of methylated mRNAs following fungal infection further supports the observation of aberrant m6A modifiers and a loss of balance between m6A methylation and demethylation. Importantly, in fungal-infected corneas, altered m6A modification was associated with the increased expression of the C–C family of chemotactic cytokines, particularly CCL8, CCL4, CCL2, and CCR1. The severe inflammatory response and corneal damage associated with fungal infections may be due to the overproduction of C–C chemokine, which can activate monocytes and T-cells, leading to a more severe inflammatory response in the cornea.33 Fungal infection also induces phosphorylation of PI3K/Akt in the corneal tissue of mice with fungal keratitis, which is associated with m6A modification.34 There are two conclusions to be drawn from the evidence presented here: m6A modification plays a role in the pathogenesis of fungal keratitis, and modulation of m6A methylation may be another approach to the treatment of this disease.
m6A modification and corneal neovascularization
Corneal neovascularization is characterized by the growth of blood vessels in the cornea. Persistent new blood vessels with scarring eventually threaten visual acuity due to the loss of corneal transparency.35 Corneal neovascularization can occur due to several reasons, including inflammation, ischemia, degeneration, and trauma. To test the hypothesis that corneal neovascularization is regulated by m6A methylation, an animal model is required. Suture placement, chemical burns, and transgenic mouse models are commonly used to induce corneal neovascularization.36,37 A mouse model of corneal neovascularization induced by suture was used to determine whether m6A modification plays a role in the regulation of corneal neovascularization.38 Strikingly, the expression of Fat mass and obesity-associated protein (FTO) in the corneas showing neovascularization was much higher than that in the control group, suggesting that FTO is dominantly expressed in the tissues of the corneal neovascularization model. Interestingly, total m6A levels were decreased in corneal neovascular lesions, and were associated with a significant upregulation of FTO in the cornea of the experimental mice. Furthermore, subconjunctival injection of short hairpin RNA (shRNA) targeting mouse FTO (FTO shRNA) in vivo showed that silencing FTO significantly inhibited the formation of new vessels in mouse corneas, confirming its role in neovascularization.38 Mechanistically, the inhibition of corneal neovascularization is due to the inhibition of endothelial cell proliferation, migration, and tube formation by knockdown of FTO as demonstrated in vitro. Contrary to this, FTO overexpression significantly promoted endothelial cell proliferation, migration, and tube formation. A study has found that focal adhesion kinase (FAK) can be activated by angiogenic growth factors TNF, VEGF, angiopoietin 1, and bFGF, which can trigger multiple downstream signaling pathways and participate in the pathological process of corneal neovascularization.39 Interestingly, by silencing YTHDF2, FTO knockdown-mediated reduction in vitro angiogenesis was partially reversed,38 suggesting that FTO-mediated corneal neovascularization may also be regulated by YTHDF2.
m6A modification and cataracts
Cataract, especially age-related cataract, is one of the leading causes of blindness worldwide. The formation of cataract is a complex pathological process.40,41 A variety of factors contribute to the formation of cataract, such as genetic defects, senescence, systemic or local inflammation, hormone abnormalities, metabolism abnormalities, drug abuse, oxidative stress, ER stress, and abnormal epigenetic factors.41, 42, 43 While surgical approaches do save sight, we still lack pharmacological strategies to treat or delay cataract formation because the exact mechanism of cataract formation is not known. Hence, it is critical to understand the pathogenesis of cataract. In addition to the roles of DNA methylation, histone modification, and non-coding RNA in cataract pathogenesis, RNA methylation, specifically m6A methylation, may contribute to understanding the molecular mechanism of cataract development.44, 45, 46 Several preliminary findings support the role of m6A modification in cataracts, including the abnormal expression of m6A writers (METTL3 and METTL14), m6A erasers (ALKBH5, FTO, and ALKBH5) and m6A readers (YTHDF1 and YTHDF2) in human lens epithelial cells, including age-related cataracts, diabetic cataracts, and cataracts associated with high myopia. It is reasonable to focus on lens epithelial cells to study cataract formation with RNA transcripts, as the integrity and metabolic activity of lens epithelial cells are critical for maintaining lens transparency, which depends on the normal transcriptome in lens epithelial cells. In addition, the expression of the m6A modification factors differs based on the type of cataract, and both ALKBH5 and METTL14 are significantly upregulated in human lens epithelial cells from patients with age-related cataracts. Furthermore, the increased expression of ALKBH5 induced by UV light was also confirmed in vitro in a human lens epithelial cell line.44 In terms of the relevance of m6A methylation to the pathogenesis of diabetic cataract, METTL3 was found to be highly expressed in human lens epithelial cells obtained from patients with diabetic cataract. Treatment of lens epithelial cells with high glucose induced upregulation of METTL3.46 Knockdown of METTL3 enhanced the proliferation of lens cells but inhibited lens epithelial cell apoptosis induced by high glucose.46 However, the expression of METTL3, ALKBH5, and FTO, but not METTL14, which was downregulated in the anterior capsule of the cataract lens in patients with high myopia, compared to simple nuclear cataracts.45 In addition, the m6A reader proteins YTHDF1 and YTHDF2 may also be involved in the pathogenesis of high myopia cataracts by affecting post-transcriptional gene expression.45 According to the studies, m6A modification plays a dynamic role in the pathogenesis of cataracts, and the expression of these factors varies depending on the differences in the inputs locally and systemically (the microenvironment at the time of cataract onset). It is worth noting that the target genes of m6A modification and cataract formation differ in different types of cataracts. Interestingly, in age-related cataracts, the involvement of circRNA m6A modification has been shown to be a major target for the pathogenesis of lens opacity.44 Compared with normal subjects, patients with age-related cataracts have reduced levels of m6A methylation of circRNAs in lens epithelial cells. In addition, lens cells with cataracts expressed downregulated m6A circRNAs compared with lens cells without cataracts. Most importantly, the upregulated ALKBH5 may contribute to the pathogenesis of age-related cataracts by modifying circRNAs and its downstream genes related to aging, DNA damage, DNA repair, oxidative stress, proteolysis, ubiquitination, apoptosis, and autophagy.44 This study demonstrated that cross-talk between m6A and circRNAs affects the pathogenesis of age-related cataracts.
Interestingly, it was found that METTL3 can stabilize mRNA of the gene ICAM-1 by targeting the 3' UTR of ICAM-1. The ICAM1 gene encodes extracellular matrices such as COL6A3 (collagen alpha-3 (VI) chain), chitinase-3-like protein 1 (CHI3L1), and genes involved in ion transport are hypermethylated. Therefore, METTL3 may regulate the methylation status of ICAM-1 mRNA,46 suggesting that the ECM is regulated by m6A modification and contributes to the formation of diabetic cataracts.46
In the exploration of m6A target genes involved in the pathogenesis of high myopia-related cataracts, it was found that the main genes that up-regulated m6A mRNA markers were associated with ECM function. Among these genes, the CHI3L1-encoded protein YKL-40 plays a role in tissue remodeling and stress response to environmental changes, and is hypermethylated due to the downregulation of demethylase FTO and ALKBH5 and increased expression of METTL14.45 Taken together, these m6A modification factors may participate in the pathologic process associated with high myopia, such as reduced choroidal circulation, posterior scleral staphyloma, degeneration of retinal cells, and loss of visual acuity.45
The above findings suggest that the m6A modification may have an impact on cataracts formation. Nevertheless, a key question remains: what is the role of m6A methylation in the regulation of alpha-crystallin (A and B), a major structural protein critical for maintaining lens transparency, expression, integrity, and function? Is the cataract reversible by the regulation of m6A methylation?
m6A modification and diabetic retinopathy
Diabetic retinopathy (DR) is the most common and serious complication of diabetes.47 The major pathological feature of DR is the leakage of blood contents from micro vessels in the retina due to BRB rupture. Leakage can also occur through the RPE barrier in DR.48,49 The BRB breakage and new vessel growth may be regulated by epigenetic factors including aberrant DNA methylation of certain genes (POLG, AHRR, GIRP, GLRA1, and BCOR), histone modification (SUV39H2, EZH2, and H3K9), non-coding RNA, hyperglycemia-induced circular RNA.50, 51, 52, 53, 54 Another notable finding is that Kolwluru revealed the pathogenesis of DR, especially the metabolic memory associated with epigenetic modifications.50,51 Advanced DR may result in retinal fibrosis, detachment, and blindness. Anti-VEGF therapy has made significant progress in the treatment of DR. However, a large number of patients receiving anti-VEGF therapy did not experience significant improvement in vision because the pathogenesis of DR is not well understood.55 Recent advances in the study of m6A methylation have expanded the understanding of the pathogenesis of DR. It has been demonstrated that diabetes and its complications may be regulated by m6A methylation.46,56,57 DR is secondary to diabetes, so it would make sense to establish a link between them, as DR would not occur without diabetes. Since 2015, the reduction of m6A methylation has been recognized as a risk factor and biomarker for type 2 diabetes.56 The pioneering study by Shen et al showed that m6A methylation was globally altered in the blood of patients with type 2 diabetes (the most common form of diabetes) and experimental rats,56 a striking finding of this study was that the m6A methylation found in the peripheral blood of diabetic patients and diabetic rat models was significantly reduced compared to healthy individuals.56 In contrast, the expression of m6A demethylase FTO was significantly increased, suggesting that the reduction of m6A methylation may be due to the increased expression of FTO. A study published in 2019 supports this notion, in which the reduced m6A content in the blood is associated with the upregulation of FTO in patients with type 2 diabetes.57 In addition to blood, an important finding is that m6A methylases, especially METTL3 and METTL14, were much lower in the islets of patients with type 2 diabetes.58 Knockdown of METTL14 could induce hypomethylation, leading to the inhibition of islet cell duplication and impairment of the insulin/IGF1-Akt-PDX1 pathway in experimental diabetic mice, a phenomenon similar to that was seen in human type 2 diabetes. Interestingly, increased FTO expression was proportionate to the high glucose level in the serum of patients with type 2 diabetes. Mechanistically, it has been shown that high glucose levels stimulate FTO expression, whereas knockdown of FTO increases m6A methylation in hepatocytes. FTO may target the upregulation of the genes FOXO1, FASN, G6PC, and DGAT2, which regulate glucose and lipid metabolism and contribute to the pathogenesis of diabetes.57Modification of m6A in the islet β-cells plays a critical role in the pathogenesis of type 2 diabetes, therefore, the parameters of m6A modification may be used as biomarkers for the early diagnosis of type 2 diabetes and monitoring DR progression.
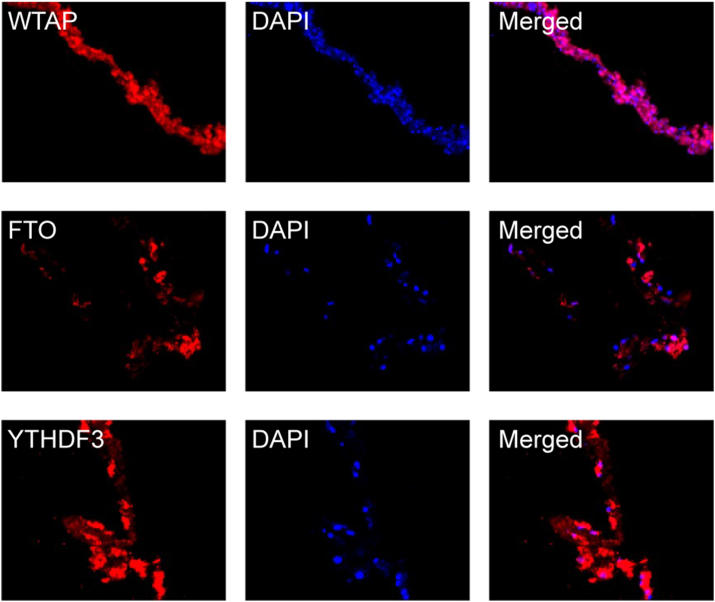
It is not surprising that DR is not a primary disease but a complication of diabetes. In fact, in our recent study, we found an abundant expression of WTAP, FTO, and YTHDF3 in human proliferative diabetic retinopathy (PDR) membranes (Fig. 2), which suggests that the involvement of m6A modification in DR pathogenesis. Importantly, we found that FTO, one of the factors expressed in human PDR membranes, may contribute to the damage of retinal microvessels under hyperglycemic conditions,57 as alterations in FTO are closely related to metabolic alterations.59,60 WTAP is involved in the deposition of m6A in mRNAs,61 YTHDF3 acts as an m6A reader, and both WTAP and YTHDF3 participate in transcriptional and post-transcriptional regulation of genes. WTAP and YTHDF3 play critical roles in RNA methylation and metabolism of m6A-modified mRNAs.62 However, whether the abnormal expression of WTAP and YTHFD3 is the cause or consequence of DR pathogenesis requires further investigation.
Figure 2.
The expression of WTAP, FTO, and YTHDF3 in surgically excised human PDR membranes by immunofluorescent staining. Red indicates positive staining for WTAP, FTO, and YTHDF3. The cell nuclei are stained blue (DAPI). The immunofluorescent reactivities of WTAP, FTO, and YTHDF3 are considerably higher. Original magnification 200 × . PDR membranes (n = 3) (Unpublished data).
Another important finding is the possible role of METTL3 and miR-25-3p in the pathogenesis of DR. It was found that miR-25-3p, a target gene of METTL3 and a miRNA that promotes cell survival and proliferation, was reduced in the blood of diabetic patients. High glucose downregulates m6A methylation (especially METTL3) and its target gene miR-25-3p, which may result in the upregulation of PTEN (associated with cell death), but decreases phosphorylated Akt (the cell survival signal), resulting in the dysfunction and death of RPE cells and contributing to the breakdown of the outer retinal barrier, which results in vascular leakage.63 This information suggests that reduced m6A methylation plays an important role in the pathogenesis of DR.
More research should be conducted to clarify the relationship between DR formation and m6A modification. Particularly, the mechanisms by which m6A modification modulates the loss of balance of vascular stimulators and inhibitors, affects retinal endothelial cell leakage, especially pericyte dysfunction and death, epigenetic-related metabolic memory, microaneurysms formation and key DR-inducing factors such as VEGF, PDGF receptors and PKC-δ.
m6A modification and age-related macular degeneration
Age-related macular degeneration (AMD) is the leading cause of vision loss in older adults. Although there are two types of AMD, geographic atrophy or macular neovascularization (MNV), the initial abnormality of the macula is similar.64,65 The primary tissue involved in this chronic degenerative disease is the retinal pigment epithelium–Bruch’s membrane–choriocapillaris complex. The pathogenesis of AMD is complex and regulated by many factors. It is well known that the risk factors of AMD include but are not limited to single nucleotide polymorphism CFH, ARMS2, IL-8, TIMP3, SLC16A8, RAD51B, VEGFA, and COL8A1;66, 67, 68 COL8A1-FILIP1L, IER3-DDR1, SLC16A8, TGF-β R1, RAD51B, ADAMTS9, and B3GALTL;69 additional 12 novel loci C4BPA-CD55, ZNF385B, ZBTB38, NFKB1, LINC00461, ADAM19, CPN1, ACSL5, CSK, RLBP1, CLUL1, and LBP;70 IL-17A;71 and RPE cells in aging, smoking, UV light damage, elevated serum lipid, and abnormal epigenetic factors.72, 73, 74 Polypoidal choroidal vasculopathy (PCV) is a subtype of MNV that has been recognized to be associated with some SNPs, such as SKIV2L. The rs5882 (GG) SNP in CETP and rs6982567 in GDF6 were not found in MNV patients.75, 76, 77 Genetic variations are responsible for the susceptibility to AMD, in no more than 50% of the population.77 The severity of AMD may be determined by genetics and the epigenome.78 All environmental risk factors may alter the epigenetic factors including m6A and therefore involve in the pathogenesis of AMD.74,79, 80, 81
Gneome-wide DNA methylation analysis of AMD patients84 and changes in VEGF methylation status68 suggests the relevance of DNA methylation to AMD may be related to abnormal methylation in anti-inflammatory gene, anti-oxidative gene, and anti-angiogenic factors such as clustering,82 glutathione S-transferase,79 IL-17RC80 and GPR15,83 and the aberrant methylation status of the genes SKI, GTF2H4, and TNXB.84 It has been suggested that smoking promotes AMD pathogenesis through DNA methylation. In the peripheral blood of patients with MNV, Shannath L Merbs’s team discovered significant differences in DNA methylation close to the ARMS-2 locus, as well as altered DNA methylation in the promoter region of protease serine 50 (PRSS50) through genome-wide DNA methylation analysis,81 These results expand our understanding that the alteration of epigenetics may be one of the risk factors for AMD.
An important finding in the study of histone modification in AMD was the role of SIRT1 (a histone deacetylase converting enzyme) in the pathogenesis of AMD.85 The expression of SIRT1 was significantly reduced in the retina and RPE cells obtained from AMD patients compared with controls.86 We have shown that the activation of SIRT1 by resveratrol (RSV) suppresses HIF1α expression and VEGF production induced by cobalt chloride (CoCl2) in human RPE cells.87 Furthermore, macular neovascularization in a laser-induced mouse model is inhibited by the direct delivery of RSV to the vitreous mechanically,88 which may result from downregulation of VEGFR2 phosphorylation and inhibition of the HIF1α/VEGF/VEGFR2 signaling pathway and inhibition of VEGF production.87
In addition to SIRT1, we found that trichostatin A (TSA), an HDAC inhibitor for class I and II HDACs,89 inhibits experimental MNV induced by laser in mice by downregulating the key angiogenic factors HIF-1, VEGF, and VEGF receptor,73 as well as MNV-associated wound healing response and RPE epithelial-mesenchymal transdifferentiation in vitro. Inhibition of HDAC by TSA impaired angiogenesis and induced apoptosis in human vascular endothelial cells. The study's most significant finding was that anti-angiogenic neuroprotective PEDF is highly upregulated by the application of TSA.73
The role of miRNAs in the pathogenesis of AMD has also attracted the attention of researchers. Numerous miRNAs have been found in the vitreous of patients.90 RPE cell death has been reported to be associated with the downregulation of miR-23a.91 Increases of various miRNAs, such as miR-9, miR-125b, miR-146a, and miR-155 in the retina of AMD patients, may alter the regulation of inflammatory responses by CFH.92 Intravitreal injection of pre-miR-21 inhibited an experimental laser-induced murine model of CNV.93 A fundamental finding was the reduction of DICER1 expression in RPE cells, which led to the accumulation of Alu RNA, which resulted in RPE death and retinal changes similar to those seen in dry AMD in mice.94
m6A RNA methylation is another hot spot in epigenetic research, and it has been implicated in the pathogenesis of AMD.95 Amyloid-β1-40 induced RPE degeneration was found to be regulated by the m6A demethylase FTO.95 Amyloid-β1-40 is a major component of plaques in Alzheimer’s disease and is an important component of drusen, importantly, intravitreal injection of Amyloid-β1-40 induced RPE dysfunction and retinal degeneration.95, 96, 97, 98 Interestingly, increased FTO may protect against RPE dysfunction because RPE degeneration is intensified by the application of the FTO-specific inhibitor, meclofenamic acid (MA-1) to mice injected with amyloid-β1-40.95 Using the m6A-mRNA epi-transcriptomic microarray, it was demonstrated that PKA is the target gene of FTO. Further, inhibition of FTO exacerbates RPE cell degeneration through increased activation of the PKA/CREB signaling pathway.95 This implies that m6A modification may be involved in the pathogenesis of AMD. While this study provides some clues that aberrant m6A modification factors may contribute to AMD pathogenesis, it does not highlight the m6A writers, readers, and other m6A demethylases associated with AMD. Furthermore, these findings need to be validated in animal experiments and human specimens from patients with both dry and MNV.
AMD is a disease associated with aging. Thus, the dynamic changes in m6A modification must be tracked over a person’s lifetime to identify any changes before AMD develops. Moreover, the relevance of dynamic changes of m6A with regard to oxidative stress, inflammation, angiogenesis, senescence of RPE, and choroidal capillaries needs to be established to understand the pathogenesis of AMD and possible treatment options. AMD is a disease that involved a complex of RPE, Bruch’s membrane, and choroidal capillaries. Therefore, targeting only one of the members of the complex in the treatment of AMD may not be sufficient, and the pathologic effect on the Bruch’s membrane and choroidal vessels should be as important as RPE. Replacement of the Bruch’s membrane and choroidal capillaries may also need to be considered in addition to RPE transplantation.
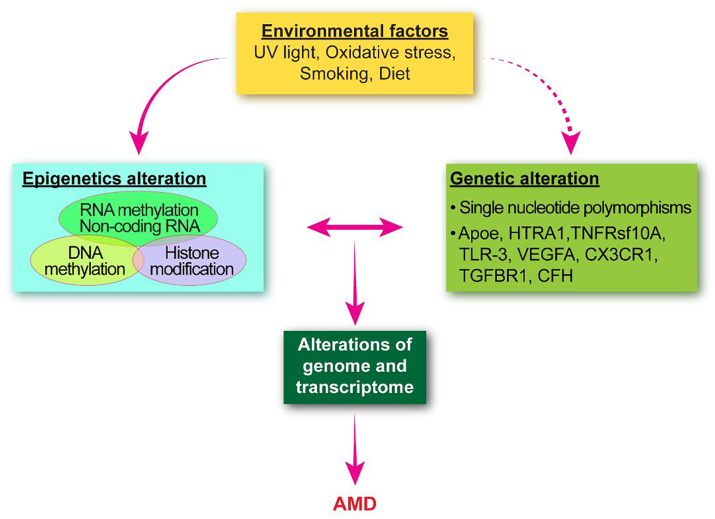
The interplay between environmental and epigenetic factors controls the activation or suppression of gene expression. These risk factors may accumulate throughout life and trigger changes in epigenetic factors, including DNA methylation, histone modification, non-coding RNA, and RNA methylation when a certain threshold is reached, which in turn contributes to the pathogenesis of AMD99 (Fig. 3).
Figure 3.
The contribution of epigenetic factors to the pathogenesis of AMD. The input of environmental factors may alter the epigenetic fingerprint, the Alterations in gene expression depend on the interaction of genetic (single nucleotide polymorphisms), epigenetic and environmental factors and thus the pathogenesis of AMD is regulated by multiple factors.
m6A modification and proliferative vitreoretinopathy
Proliferative vitreoretinopathy (PVR) is a long-term wound-healing response in the retina.100 Retinal pigment epithelial cells, glial cells (mainly Müller cells),101 and inflammatory cells (macrophages and lymphocytes) have been identified in PVR membranes. However, the crucial cell type involved in PVR formation is the RPE cell.100,102 RPE epithelial-mesenchymal transition (EMT) and associated proliferation, migration from the RPE monolayer, and overproduction of ECM lead to fibrotic membrane formation and tractional retinal detachment. The EMT process is dynamic and reversible. Therefore, the reverse process from mesenchymal to epithelial transition is called mesenchymal–epithelial transition (MET). Various factors trigger and promote PVR, among them was the hepatocyte growth factor (HGF), which participates in the disassembly of the monolayer of RPE, the induction of RPE behavior changes,103,104 and promotes the formation of the fibrotic membrane with TGF-β and CTGF.102,105,106 Furthermore, experiments showed that methyl-CpG binding protein 2 (MeCP2) and its phosphorylated form MeCP2-421 play an essential role in EMT in RPE cells and retinal fibrosis,107,108 the functional interaction between MeCP2 and TGF-β was revealed by chromatin immunoprecipitation assay to show that MeCP2 binds to the TGF-β gene promoter and contribute to the pathogenesis of PVR.108 However, in this section, we are focusing our discussion on the pathogenesis of PVR in m6A-related categories.
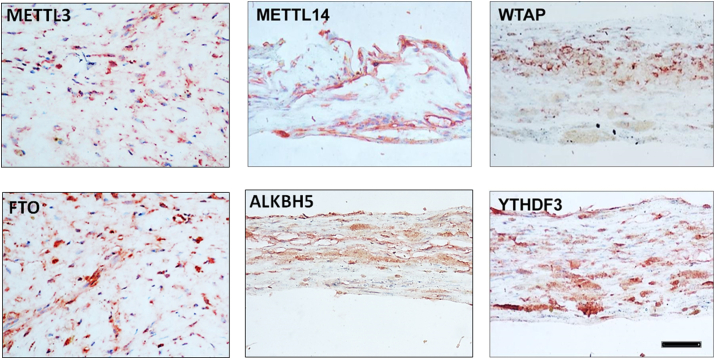
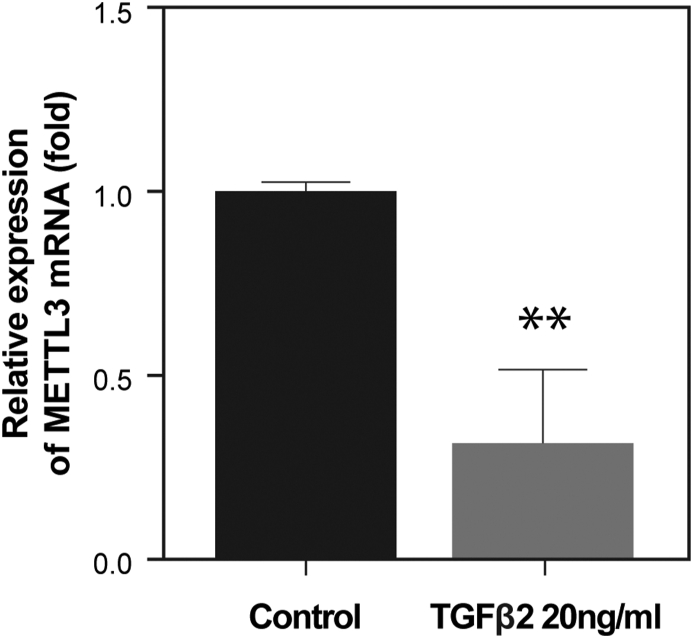
The pathogenesis of PVR may also be strictly regulated by m6A modifications. The development of PVR includes initiation, proliferative response, and fibrosis formation, each of which is influenced by several factors (inflammatory cytokines, growth factors, tissue remolding enzymes, and ECM). However, gene expression of these factors may be regulated by RNA methylation, especially m6A. To validate the hypothesis, we have to first answer whether there is an expression of m6A modification factors in human PVR membranes. Second, when discussing cellular and tissue EMT or fibrosis, we cannot avoid TGF-β in the induction of the pathological processes and the interaction of TGF-β with these factors. Here, we address the following points: We found evidence of the constitutive expressions of m6A writers (METTL14 and WTAP), erasers (FTO and ALKBH5), and reader protein (YTHDF3) in PVR membranes using immunohistochemical staining (Fig. 4). A recent study also supports the idea that m6A is involved in the development of PVR because the m6A modification factor (METTL3) in PVR membranes has been demonstrated.109 The expression of the key m6A writer METTL3 was considerably low in the PVR membranes compared with the normal RPE monolayer.109 In fact, METTL3 may function as an EMT and retinal fibrosis inhibitor, as METTL3 overexpression inhibited TGF-β-induced RPE cell proliferation and migration, whereas METTL3 knockdown enhanced RPE EMT.109 Importantly, the overexpression of METTL3 suppressed experimental PVR in rats.109 Mechanistically, decreased METTL3 expression may be due to the effect of TGF-β. It is known that human PVR membranes are enriched in TGF-β but reduced in METTL3.109, 110, 111 We further demonstrated that METTL3 was significantly inhibited in RPE cells after treatment with TGF-β (Fig. 5). In addition to the low expression of METTL3 as shown in previous publication109 and in Figure 4, abnormal FTO may also contribute to the pathogenesis of fibrosis, as increased FTO expression is demonstrated in renal fibrosis.112 In the condition of FTO deficiency, the response of α-SMA or CTGF to TGF-β stimulation is reduced.112 Interestingly, we found the immunoreactivities of FTO were relatively higher than METTL3 in PVR membranes (Fig. 4). These results suggest that a loss of balance between m6A methylase (METTL3) and demethylase (FTO) may contribute to the pathogenesis of PVR. Further studies are needed to reveal how METTL3 interacts with FTO and affects TGF-β signaling, including Smad 2/3 and others (such as snail and slug) in RPE, and how RPE EMT and retinal fibrosis are regulated by METTL3 and FTO in vitro and in vivo. Notably, the m6A reader proteins YTHDF may also be involved in the pathogenesis of PVR, we found that YTHDF3 was highly expressed in human PVR membranes (Fig. 4). The importance of YTHDF in cancer EMT was demonstrated by the induction of epithelial cell markers such as claudin 1 and ZO-1 by inhibition YTHDF family members, and silencing of YTHDF1 attenuated the EMT induced by TGF-β cells.113 Interestingly, YTHDF1 could activate the PI3K/AKT/mTOR signaling pathway, which is also one of the downstream signaling pathways of TGF-β and is closely related to the EMT process.114 Likewise, the importance of the m6A reader protein appears in the formation of EMT and fibrosis like PVR.
Figure 4.
Expression of m6A writers (METTL3, METTL14, and WTAP), erasers (FTO and ALKBH5), and readers (YTHDF3) in human PVR membranes (from surgical excision). Red chromogen staining indicates the immunoreactivity of m6A modification factors, and blue indicates the nuclear contrast staining. magnification 200 × . PVR membranes (n = 3) (Unpublished data).
Figure 5.
Effects of TGF-β on the mRNA expression of m6A methylases METTL3 in RPE cells (ARPE-19). ARPE-19 cells were treated with TGF-β2 (20 ng/ml, MedChemexpress, New Jersey, USA) for 24 h, and total RNA was isolated to analyze the expression of the genes by real-time PCR. The expression of METTL3 was significantly inhibited by adding TGF-β compared with control (t-test, ∗∗P < 0.01) (Unpublished data).
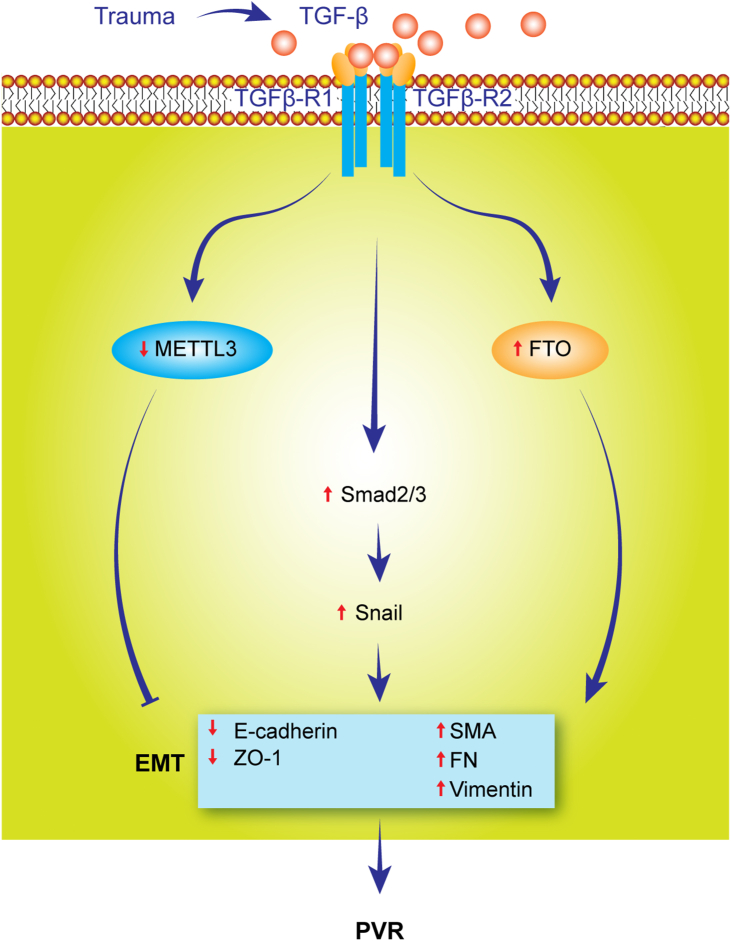
Reports mentioned above highlight the critical role of m6A in regulating the pathogenesis of PVR. Since m6A modification is involved in numerous pathological processes, it is likely that the expression of PVR regulators, in addition to TGF-β, such as HGF, MeCP2, and SIRT1, etc., is under the regulation of m6A. Therefore, our view is that targeting METTL3, FTO, or YTHDF3 may interfere with the pathological processes in PVR, such as EMT and fibrosis (Fig. 6).
Figure 6.
The prospective role of m6A modification in the pathogenesis of PVR. Retinal trauma induces the synthesis and secretion of TGF-β. TGF-β promotes the pathogenesis of PVR through the following pathways:1. activating snail through the Smad 2/3 pathways; 2. increasing FTO expression; 3. downregulating the expression of METTL3 (the negative regulator of EMT and fibrosis), which results in the repression of epithelial marker genes (E-cadherin and ZO-1) expression but enhances the mesenchymal gene (αSMA, FN, vimentin) expression and mesenchymal phenotype.
m6A modification and graves’ disease
Graves’ ophthalmopathy (GO) is a disorder of autoimmune origin. However, the pathogenic mechanisms of GO have not been fully elucidated.115,116 Several autoimmune and inflammatory factors contribute to the pathogenesis of GO, including retroocular fibroblasts, adipocytes, and extraocular muscles (EOMs).117, 118, 119, 120, 121, 122 When researching on the components contributing to the orbit immune response, we might need to pay more attention to EOMs since they represent one of the major effector cells in GO and are crucial to inflammatory responses and dysfunction of ocular motion.123 Importantly, autoantibodies against EOM antigens have been demonstrated.124 Even though cumulative evidence suggests that epigenetic factors, including altered genome-wide DNA methylation, histone modification in PBMCs, and orbit fibroblasts from patients with GO, could be involved in the development of GO,122,125,126 so far it is still unclear what the role of epigenetics is in the pathogenesis of GO, particularly in connection with m6A methylation. In a previous study, we demonstrated that m6A modification was involved in the pathogenesis of GO using surgically excised EOMs.123 Consequently the m6A modification factors were altered, which displayed a significantly increased expression of m6A globally in the specimens of EOMs obtained from patients with GO as compared with controls without GO. There was an upregulation of the m6A writer WTAP and readers YTHDF2, YTHDC2, and ELF3, suggesting that m6A modification may be a critical regulator of GO pathogenesis.123
Other important evidence is that the expression of abnormal m6A modification factors is associated with local hyperinflammatory responses in EOM. Gene ontology analysis demonstrated that the top 10 upregulated mRNAs were functionally involved in immune and inflammatory responses. We identified 12 of the 19 pathways involved in immune and inflammatory responses by KEGG analysis of biological pathways.123 Amongst the m6A modification factors, we demonstrated that WTAP and YTHDF1/2 might play more important roles in GO than others.123 We found that the expression of WTAP and levels of YTHDF1/2 are high in the EOMs of patients with GO and these proteins activate many signaling pathways, including the mTOR, Wnt signaling, and NF-κB signaling pathways.114,123,127, 128, 129 These were associated with inflammatory response, suggesting that WTAP and YTHDF1/2 could play a role in GO pathogenesis. It was also noted that ECM gene expression in EOMs of GO patients may also be regulated by m6A. In our study, the increase in m6A methylation is related to the downregulation of ECM (collagen 1A1, collagen 1A2, and collagen 2A1) in EOMs of GO patients.
m6A modification in melanoma and retinoblastoma
The most common malignant intraocular tumors in adults and children are uveal melanoma (UM) and retinoblastoma (RB), respectively. The metastasis rate of UM is as high as 50%, and the genetic mutation is different from that of cutaneous melanomas.130,131 In people with UM, nine genetic mutations have been identified: GNAQ, GNA11, BAP1, SF3B1, EIF1AX, CYSLTR2, SRSF2, MAPKAPK5, and PLCB4.132,133 Abnormal epigenetic factors,134,135 such as DNA methylation, are generally considered to be the major epigenetic factors mediating the development of UM. RASSF1A and p16INK4a are the key genes for hypermethylation and, thus lose their inhibitory effect on UM development.134 Among the non-coding RNAs, six miRNAs (let-7b, miR-199a, miR-199a, miR-143, miR-193b, and miR-652) were found to be the major miRNAs expressed in UM.136 Some are oncogenes, and some are tumor suppressor genes.137 Histone modifications were also associated with melanoma development, along with decreased levels of H4K5ac and H4K8ac, and increased methylation of H3K9me2 and H3K36me3 in UM cells.138 Despite the progress in understanding the pathogenesis of UM, the differences in metastasis, response to treatment, and prognosis of UM remain unclear, and the possibility of developing a clinical therapeutic approach for UM relies heavily on understanding its development and progression. Therefore, m6A methylation may provide additional insights into the pathogenesis and therapeutic implications of UM.
The expression of m6A modification factors is variable in patients with UM. There was a remarkable increase in the methylation of m6A in specimens obtained from patients with UM and in the UM-derived cell lines (M17, M21, M23, SP6.5, and MP38) compared with normal uveal melanocytes.139 This was associated with upregulation of METTL3 and decreased expression of ALKBH5. However, significantly enhanced expression of the demethylase ALKBH5 (both mRNA and protein), but not the m6A methylase, was observed in human UM cell lines (MuM-2B and C918).140 Interestingly, a significant decrease in m6A was found in UM tissue and UM cell lines OCM1, OCM1a, and OM431,141 which was accompanied by the upregulation of ALKBH5 and downregulation of METTL3. The m6A fingerprints vary widely, and the different results obtained by different research groups may be due to the following reasons: Specimens were from different stages of UM. The profile of m6A may differ between early and advanced UM because RNA modification is dynamic and individual-specific. Additionally, different areas (tumor central or border) were obtained for the m6A analysis, and thus the results may also differ. Another possibility is that the detection system for m6A expression may be different. Therefore, these conclusions need to be validated using the same criteria in the future.
Regardless of whether m6A modification factors are increased or decreased, manipulating the expression of m6A modification affects the proliferation, migration, invasion, and colony formation behaviors of UM cells. Knockdown of METTL3 negatively regulated tumor cell activity, and overexpression of METTL3 promoted tumor cell invasiveness,139 suggesting that increased m6A methylation is associated with tumorigenesis in UM. After injection of UM cells with ALKBH5 knockdown, UM tumor growth (volume, weight) and tumor cell duplication were inhibited in an experimental animal. Furthermore, loss of ALKBH5 facilitated the cell cycle arrest and promoted UM cell apoptosis, showing increased cleaved caspase-3.140 There is also evidence that m6A modification factors regulate UM tumorigenesis. METTL3 knockdown promotes normal melanocyte proliferation, colony formation, and reduction of apoptosis, which share some of the characteristics of UM cells.141 Knockdown of ALKBH5 inhibits colony formation in UM cells, whereas silencing of ALKBH5 increases m6A methylation and reduces tumor cell colony formation.141 These results suggest that UM tumorigenesis is mediated by m6A modification.
Predicting UM progression and prognosis is always challenging, and the characteristics of UM histology, cytogenetic, and gene expression profiles may help improve the prognosis of UM.142,143 Because m6A has been recognized as a novel parameter for UM progression and prognosis, its modification factors can serve as indicators. Reduced total m6A levels were often associated with an advanced stage of ocular melanoma and a poor prognosis.141 Interestingly, in another report, high METTL3, and enhanced c-Met expression could be used as a predictor of recurrence and UM aggressiveness.139 Another study suggested that increased ALKBH5 expression may be an indicator of poor prognosis in UM.140 The Cancer Genome Atlas dataset and Gene Expression Omnibus analysis validated the idea that m6A regulators may be prognostic indicators for UM.144
Targeting m6A modification factors and their downstream genes may provide a new therapeutic target for the treatment of UM. Using a chemical m6A inhibitor (cycloleucine) or directly silencing METTL3 could inhibit UM cell proliferation, migration, colony formation, and invasion through suppression of the oncogene c-met expression and Akt activation.139 ALKBH5 may provide another target for inhibiting tumor behavior of UM cells in vitro, as ALKBH5 silencing in UM was remarkably inhibited in an animal model by the regulation of FOXM1 and the EMT pathway.140 A significant finding is that m6A methylation increases the expression of the tumor suppressor gene HINT2, which is also the target of the m6A reader YTH family. These studies suggest that the use of m6A regulators may be valuable for the treatment of human melanoma, especially those resistant to anti-PD1 therapy.131
Similar to UM, RB may lead to vision loss and even death, while exploring the role of m6A modification in the pathogenesis of RB, it was demonstrated that m6A modification factor METTL3 does contribute to the development of RB.145 This notion is supported by evidence that METTL3 mRNA and protein expression is higher in Y79 and WERI-Rb-1 cell lines compared with ARPE-19 cells, and importantly, high expression of METTL3 was detected in specimens from RB patients. Functionally, RB cell replication and invasion are tightly regulated by METTL3; notably, METTL3 overexpression promotes RB development while knockdown METTL3 inhibits the tumor growth in a mouse model, and the result suggested that m6A methylation plays a key role in the pathogenesis of RB.145
m6A modification and traumatic optic neuropathy
Traumatic optic neuropathy (TON) is often observed after traumatic optic nerve injury or secondary to traumatic brain injury (TBI).146 TON is characterized by an inflammatory response, ischemia, and subsequent neurodegeneration, leading to severe vision loss. Many questions and challenges remain in the study of TON, such as the pathogenesis, the rescue of dying optic ganglia cells, and optic nerve regeneration. Considering the complex nature of TON, dysregulation of m6A RNA methylation may be involved in traumatic optic nerve injury147 and optic nerve regeneration.148,149 Although the following evidence supports the role of m6A in the pathogenesis of TON, there are some differences in the expression of the parameters of m6A between primary TON and TBI, indicating that the distribution of m6A modification is variable and dynamic in different regions of neural tissue. Study results showed that the RNA methylation and expression levels of 175 mRNAs in the cerebral cortex of rats were significantly altered after TBI,150 and that METTL14 and FTO were downregulated. Furthermore, a genome-wide comparison of m6A-tagged transcripts in the hippocampus of TBI mice detected a total of 922 m6A modified sites,151 most of which (552) were downregulated. The mRNA expression of m6A and METTL3 significantly decreased. Unlike TBI, Qu et al found that m6A methylases (METTL3 and WTAP), as well as demethylases (FTO and ALKBH5), were upregulated in the retina of experimental rat TON. Similarly, the large part (2,810) m6A peaks were upregulated, while 689 m6A peaks were downregulated, as shown by MeRIP-seq.152 Gene ontology analysis revealed that altered m6A peaks were highly correlated components of the cellular development and differentiation machinery of the nervous system.152 Interestingly, most m6A-tagged transcripts are linked to inflammatory signaling pathways, such as TNF, MAPK, and NF-κB pathways.152
The MAPK signaling pathway is the main pathway of TBI, and the Sarm1/MAPK pathway can disrupt the energy balance of axons, leading to the depletion of adenosine triphosphate (ATP) depletion before axon injury and promoting the progression of axonal injury.153 NF-κB is a key transcription factor that regulates inflammation in central nervous system injury and regulates the expression of pro-inflammatory and pro-apoptotic genes.154 The results suggest that m6A RNA methylation may be crucial to the pathogenesis of TON, that regulating m6A modification may limit injury-induced inflammatory responses and cell death in the optic nerve, and that the MAPK and NF-κB signaling pathways can be manipulated to achieve neuroprotective effects.152, 153, 154, 155, 156
Concluding remarks
This review mentions the following concepts: mRNA is not only a message carrier but also a transcript regulator, and epigenetic research is taking place in the epitranscriptome era; m6A modification affects translation, which, in turn, affects ocular cell function and characteristics; Abnormalities in m6A modifications may contribute to the pathogenesis of complex eye diseases (Table 1). The interplay between RNA methylation and other epigenetic factors may play a role in the pathogenesis of eye diseases; m6A modification can be used as a new marker for the early detection, diagnosis, and treatment of eye diseases; The future research direction of m6A modification in the eye diseases.
Table 1.
Summary of the role m6A modifications in complex eye diseases.
| Eye Diseases | Altered m6A Factors | Expression | Roles in Eye Diseases |
|---|---|---|---|
| Fungal Keratitis | Total m6A, METTL3 | Up | Promote the inflammation |
| Corneal Neovascularization | FTO | Up | Promote the neovascularization |
| Age-Related Cataract | ALKBH5, METTL14 | Up | Contribute to the cataract formation |
| Diabetic Cataract | METTL3 | Up | Contribute to the cataract formation |
| Cataract with High Myopia | METTL3, FTO, ALKBH5 | Down | Contribute to the pathogenesis of the cataract |
| METTL14 | Up | ||
| Diabetic Retinopathy | WTAP, FTO, and YTHDF3 | Up | Contribute to the pathogenesis of DR |
| METTL3 | Down | ||
| AMD | FTO | Up | Protect RPE from degeneration |
| PVR | METTL3 | Down | Contribute to the pathogenies of PVR |
| FTO, YTHDF3 | Up | ||
| Graves' Ophthalmopathy | WTAP, YTHDF2, YTHDC2 | Up | Contribute to the pathogenies of GO |
| Uveal Melanoma (UM) | Total m6A, METTL3 | Up | Promote UM genesis |
| Total m6A, METTL3 | Down | Contribute to UM genesis | |
| Retinoblastoma | METTL3 | Up | Promote the RB genesis |
| Traumatic Optic Neuropathy | METTL3, WTAP, FTO | Up | Contribute to TON |
Although the progression of the study of the relevance of m6A modification to the pathogenesis of complex eye diseases is very encouraging, the comprehensive and extensive understanding of how the m6A modification contributes to complex eye diseases remains to be explored. The following are the main challenges we face:
-
A.
m6A modification is under the control of methyltransferase, demethylase, and m6A recognition protein. Therefore, the phenotypic changes of an ocular cell in complex eye diseases may represent the effects of various m6A modification combinations, however, additional studies are required to obtain such information.
-
B.
Whether the m6A methyltransferase and demethylase play a role like Yin and Yang physiologically in maintaining normal eye function and whether the loss of the balance of Yin and Yang contribute to complex eye diseases, need to be determined.
-
C.
Since aberrant expression of cytokines and growth factors is associated with many ocular diseases, how do cytokines and growth factors regulate m6A expression in ocular cells and ocular pathological conditions and vice versa?
-
D.
What are the interplays between m6A and genetic factors, and m6A with other epigenetic factors (DNA methylation, histone modification and non-coding RNA) in the pathogenesis of eye diseases?
-
E.
Since oxidative stress damage in ocular cells may be closely related to mitochondrial and ER stress, cross-talk between organelles may be related to many eye diseases.157, 158, 159, 160, 161 Therefore, what is the role of m6A modification in mitochondrial, endoplasmic reticulum, and Golgi stress in eye diseases?
-
F.
Could m6A modification factors be employed as a biomarker for the early detection of eye disease and the development of a new therapeutic approach?
-
G.
A variety of signaling pathways are involved in m6A-mediated modification. Is there any cross-talk among the signaling pathways mediated by m6A modification related to eye diseases? Targeting of m6A regulators and related pathways has been suggested as a novel therapeutic approach for the treatment of eye diseases. However, which m6A modification factor is specific to particular ocular diseases?
Author contributions
Xiaohua Li, Conceptualization; Writing- original draft, Data analysis, Visualization, Supervision.
Bingyun Ma: Conceptualization, Writing- original draft, Data analysis, Visualization.
Wenfang Zhang: Review & editing, Data analysis.
Zongming Song: Review & editing, Data analysis.
Xiaodan Zhang, Xue Li, Mengyu Liao, Xueru Zhao, Mei Du, Jinguo Yu: Data producing, Review & editing, Visualization.
Shikun He: Conceptualization, Writing-original draft, Data analysis, Visualization, Supervision.
Hua Yan: Conceptualization, Review & editing, Data analysis, Visualization, Supervision.
Conflict of interests
The authors declare no conflict of interests.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81873681, 2018; 81770952, 2017) and the Basic Research Project of Henan Eye Hospital, China (No. 20JCQN005, 2020).
Acknowledgements
We thank Min Yuan and Ruijie Yin (Henan Eye Hospital) for their technique assistance.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.008.
Contributor Information
Xiaohua Li, Email: xhl_6116@163.com.
Shikun He, Email: shikunhe@usc.edu.
Hua Yan, Email: zyyyanhua@tmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Saletore Y., Meyer K., Korlach J., et al. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13(10):175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer K.D., Saletore Y., Zumbo P., et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C., Hu Y., Zhou B., et al. The role of m6A modification in physiology and disease. Cell Death Dis. 2020;11(11):960. doi: 10.1038/s41419-020-03143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertero A., Brown S., Madrigal P., et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature. 2018;555(7695):256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jian D., Wang Y., Jian L., et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics. 2020;10(20):8939–8956. doi: 10.7150/thno.45178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L., Li Y., He Y., et al. Knockdown of m6A reader IGF2BP3 inhibited hypoxia-induced cell migration and angiogenesis by regulating hypoxia inducible factor-1α in stomach cancer. Front Oncol. 2021;11:711207. doi: 10.3389/fonc.2021.711207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Z., Huang N., Liu L., et al. Dynamic analysis of m6A methylation spectroscopy during progression and reversal of hepatic fibrosis. Epigenomics. 2020;12(19):1707–1723. doi: 10.2217/epi-2019-0365. [DOI] [PubMed] [Google Scholar]
- 10.Kumari N., Karmakar A., Ahamad Khan M.M., et al. The potential role of m6A RNA methylation in diabetic retinopathy. Exp Eye Res. 2021;208:108616. doi: 10.1016/j.exer.2021.108616. [DOI] [PubMed] [Google Scholar]
- 11.Yen Y.P., Chen J.A. The m6A epitranscriptome on neural development and degeneration. J Biomed Sci. 2021;28(1):40. doi: 10.1186/s12929-021-00734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han H., Fan G., Song S., et al. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood. 2021;137(12):1603–1614. doi: 10.1182/blood.2019003764. [DOI] [PubMed] [Google Scholar]
- 13.Cao G., Li H.B., Yin Z., et al. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6(4):160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Li K., Cai J., et al. Landscape and regulation of m6A and m6Am methylome across human and mouse tissues. Mol Cell. 2020;77(2):426–440. doi: 10.1016/j.molcel.2019.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Qin Y., Li L., Luo E., et al. Role of m6A RNA methylation in cardiovascular disease (Review) Int J Mol Med. 2020;46(6):1958–1972. doi: 10.3892/ijmm.2020.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F., Cheng W., Zhao F., et al. Association of N6-methyladenosine with viruses and virally induced diseases. Front Biosci (Landmark Ed). 2020;25(6):1184–1201. doi: 10.2741/4852. [DOI] [PubMed] [Google Scholar]
- 18.He L., Li H., Wu A., et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han M., Liu Z., Xu Y., et al. Abnormality of m6A mRNA methylation is involved in Alzheimer's disease. Front Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang X., Liu B., Nie Z., et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubey P.K., Patil M., Singh S., et al. Increased m6A-RNA methylation and FTO suppression is associated with myocardial inflammation and dysfunction during endotoxemia in mice. Mol Cell Biochem. 2022;477(1):129–141. doi: 10.1007/s11010-021-04267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor A.W. Ocular immune privilege. Eye (Lond). 2009;23(10):1885–1889. doi: 10.1038/eye.2008.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forrester J.V., Dick A.D., McMenamin P.G., et al. 4th ed. Saunders Elsevier; Edinburgh: 2015. The Eye: Basic Science in Practice. [Google Scholar]
- 24.Yücel Y.H., Johnston M.G., Ly T., et al. Identification of lymphatics in the ciliary body of the human eye: a novel “uveolymphatic” outflow pathway. Exp Eye Res. 2009;89(5):810–819. doi: 10.1016/j.exer.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Kim M., Johnston M.G., Gupta N., et al. A model to measure lymphatic drainage from the eye. Exp Eye Res. 2011;93(5):586–591. doi: 10.1016/j.exer.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 26.van Burik J.A., Magee P.T. Aspects of fungal pathogenesis in humans. Annu Rev Microbiol. 2001;55:743–772. doi: 10.1146/annurev.micro.55.1.743. [DOI] [PubMed] [Google Scholar]
- 27.Vemuganti G.K., Garg P., Gopinathan U., et al. Evaluation of agent and host factors in progression of mycotic keratitis: a histologic and microbiologic study of 167 corneal buttons. Ophthalmology. 2002;109(8):1538–1546. doi: 10.1016/s0161-6420(02)01088-6. [DOI] [PubMed] [Google Scholar]
- 28.Ansari Z., Miller D., Galor A. Current thoughts in fungal keratitis: diagnosis and treatment. Curr Fungal Infect Rep. 2013;7(3):209–218. doi: 10.1007/s12281-013-0150-110.1007/s12281-013-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahmoudi S., Masoomi A., Ahmadikia K., et al. Fungal keratitis: an overview of clinical and laboratory aspects. Mycoses. 2018;61(12):916–930. doi: 10.1111/myc.12822. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery M.L., Fuller K.K. Experimental models for fungal keratitis: an overview of principles and protocols. Cells. 2020;9(7):1713. doi: 10.3390/cells9071713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nai Y.S., Huang Y.C., Yen M.R., et al. Diversity of fungal DNA methyltransferases and their association with DNA methylation patterns. Front Microbiol. 2021;11:616922. doi: 10.3389/fmicb.2020.616922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J., Lin Y. Fusarium infection alters the m6A-modified transcript landscape in the cornea. Exp Eye Res. 2020;200:108216. doi: 10.1016/j.exer.2020.108216. [DOI] [PubMed] [Google Scholar]
- 33.Tajbakhsh A., Fazeli M., Rezaee M., et al. Prevalence of CCR5delta32 in northeastern Iran. BMC Med Genet. 2019;20(1):184. doi: 10.1186/s12881-019-0913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonifazi P., D'Angelo C., Zagarella S., et al. Intranasally delivered siRNA targeting PI3K/Akt/mTOR inflammatory pathways protects from aspergillosis. Mucosal Immunol. 2010;3(2):193–205. doi: 10.1038/mi.2009.130. [DOI] [PubMed] [Google Scholar]
- 35.Sharif Z., Sharif W. Corneal neovascularization: updates on pathophysiology, investigations & management. Rom J Ophthalmol. 2019;63(1):15–22. [PMC free article] [PubMed] [Google Scholar]
- 36.Jia C., Zhu W., Ren S., et al. Comparison of genome-wide gene expression in suture- and alkali burn-induced murine corneal neovascularization. Mol Vis. 2011;17:2386–2399. [PMC free article] [PubMed] [Google Scholar]
- 37.Kather J.N., Kroll J. Transgenic mouse models of corneal neovascularization: new perspectives for angiogenesis research. Invest Ophthalmol Vis Sci. 2014;55(11):7637–7651. doi: 10.1167/iovs.14-15430. [DOI] [PubMed] [Google Scholar]
- 38.Shan K., Zhou R.M., Xiang J., et al. FTO regulates ocular angiogenesis via m6A-YTHDF2-dependent mechanism. Exp Eye Res. 2020;197:108107. doi: 10.1016/j.exer.2020.108107. [DOI] [PubMed] [Google Scholar]
- 39.Kornberg L.J., Shaw L.C., Spoerri P.E., et al. Focal adhesion kinase overexpression induces enhanced pathological retinal angiogenesis. Invest Ophthalmol Vis Sci. 2004;45(12):4463–4469. doi: 10.1167/iovs.03-1201. [DOI] [PubMed] [Google Scholar]
- 40.Lam D., Rao S.K., Ratra V., et al. Cataract. Nat Rev Dis Primers. 2015;1:15014. doi: 10.1038/nrdp.2015.14. [DOI] [PubMed] [Google Scholar]
- 41.Periyasamy P., Shinohara T. Age-related cataracts: role of unfolded protein response, Ca 2+ mobilization, epigenetic DNA modifications, and loss of Nrf2/Keap1 dependent cytoprotection. Prog Retin Eye Res. 2017;60:1–19. doi: 10.1016/j.preteyeres.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilliard A., Mendonca P., Russell T.D., et al. The protective effects of flavonoids in cataract formation through the activation of Nrf2 and the inhibition of MMP-9. Nutrients. 2020;12(12):3651. doi: 10.3390/nu12123651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Su D., Sun Z., et al. MDM2 phosphorylation mediates H2O2-induced lens epithelial cells apoptosis and age-related cataract. Biochem Biophys Res Commun. 2020;528(1):112–119. doi: 10.1016/j.bbrc.2020.05.060. [DOI] [PubMed] [Google Scholar]
- 44.Li P., Yu H., Zhang G., et al. Identification and characterization of N6-methyladenosine CircRNAs and methyltransferases in the lens epithelium cells from age-related cataract. Invest Ophthalmol Vis Sci. 2020;61(10):13. doi: 10.1167/iovs.61.10.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen K., Zhang Y., Li Y., et al. Comprehensive analysis of transcriptome-wide m6A methylome in the anterior capsule of the lens of high myopia patients. Epigenetics. 2021;16(9):955–968. doi: 10.1080/15592294.2020.1834917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Liu J., Zhao S., et al. N6-methyladenosine METTL3 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract. Mol Ther Nucleic Acids. 2020;20:111–116. doi: 10.1016/j.omtn.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duh E.J., Sun J.K., Stitt A.W. Diabetic retinopathy: current understanding, mechanisms, and treatment strategies. JCI Insight. 2017;2(14):e93751. doi: 10.1172/jci.insight.93751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu H.Z., Le Y.Z. Significance of outer blood-retina barrier breakdown in diabetes and ischemia. Invest Ophthalmol Vis Sci. 2011;52(5):2160–2164. doi: 10.1167/iovs.10-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H.Z., Song Z., Fu S., et al. RPE barrier breakdown in diabetic retinopathy: seeing is believing. J Ocul Biol Dis Infor. 2011;4(1–2):83–92. doi: 10.1007/s12177-011-9068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Q., Kowluru R.A. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110(6):1306–1313. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong Q., Kowluru R.A. Epigenetic modification of Sod2 in the development of diabetic retinopathy and in the metabolic memory: role of histone methylation. Invest Ophthalmol Vis Sci. 2013;54(1):244–250. doi: 10.1167/iovs.12-10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tewari S., Zhong Q., Santos J.M., et al. Mitochondria DNA replication and DNA methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012;53(8):4881. doi: 10.1167/iovs.12-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agardh E., Lundstig A., Perfilyev A., et al. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015;13:182. doi: 10.1186/s12916-015-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X., Zhao L., Hambly B., et al. Diabetic retinopathy: reversibility of epigenetic modifications and new therapeutic targets. Cell Biosci. 2017;7:42. doi: 10.1186/s13578-017-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W., Lo A.C.Y. Diabetic retinopathy: pathophysiology and treatments. Int J Mol Sci. 2018;19(6):1816. doi: 10.3390/ijms19061816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen F., Huang W., Huang J.T., et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab. 2015;100(1):E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y., Shen F., Huang W., et al. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J Clin Endocrinol Metab. 2019;104(3):665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 58.De Jesus D.F., Zhang Z., Kahraman S., et al. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat Metab. 2019;1(8):765–774. doi: 10.1038/s42255-019-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Church C., Moir L., McMurray F., et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42(12):1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C., Xiang Q., Liu W., et al. Co-expression network revealed roles of RNA m6A methylation in human β-cell of type 2 diabetes mellitus. Front Cell Dev Biol. 2021;9:651142. doi: 10.3389/fcell.2021.651142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horiuchi K., Umetani M., Minami T., et al. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci U S A. 2006;103(46):17278–17283. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi H., Wang X., Lu Z., et al. YTHDF3 facilitates translation and decay of N 6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zha X., Xi X., Fan X., et al. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. Aging (Albany NY) 2020;12(9):8137–8150. doi: 10.18632/aging.103130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyers K.J., Liu Z., Millen A.E., et al. Joint associations of diet, lifestyle, and genes with age-related macular degeneration. Ophthalmology. 2015;122(11):2286–2294. doi: 10.1016/j.ophtha.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yonekawa Y., Miller J.W., Kim I.K. Age-related macular degeneration: advances in management and diagnosis. J Clin Med. 2015;4(2):343–359. doi: 10.3390/jcm4020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen W., Stambolian D., Edwards A.O., et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107(16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lambert N.G., ElShelmani H., Singh M.K., et al. Risk factors and biomarkers of age-related macular degeneration. Prog Retin Eye Res. 2016;54:64–102. doi: 10.1016/j.preteyeres.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cascella R., Strafella C., Caputo V., et al. Towards the application of precision medicine in Age-Related Macular Degeneration. Prog Retin Eye Res. 2018;63:132–146. doi: 10.1016/j.preteyeres.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Fritsche L.G., Chen W., Schu M., et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han X., Gharahkhani P., Mitchell P., et al. Genome-wide meta-analysis identifies novel loci associated with age-related macular degeneration. J Hum Genet. 2020;65(8):657–665. doi: 10.1038/s10038-020-0750-x. [DOI] [PubMed] [Google Scholar]
- 71.Zhang S., Liu Y., Lu S., et al. Genetic variants of interleukin 17A are functionally associated with increased risk of age-related macular degeneration. Inflammation. 2015;38(2):658–663. doi: 10.1007/s10753-014-9973-3. [DOI] [PubMed] [Google Scholar]
- 72.van Lookeren Campagne M., LeCouter J., Yaspan B.L., et al. Mechanisms of age-related macular degeneration and therapeutic opportunities. J Pathol. 2014;232(2):151–164. doi: 10.1002/path.4266. [DOI] [PubMed] [Google Scholar]
- 73.Chan N., He S., Spee C.K., et al. Attenuation of choroidal neovascularization by histone deacetylase inhibitor. PLoS One. 2015;10(3):e0120587. doi: 10.1371/journal.pone.0120587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gemenetzi M., Lotery A.J. Epigenetics in age-related macular degeneration: new discoveries and future perspectives. Cell Mol Life Sci. 2020;77(5):807–818. doi: 10.1007/s00018-019-03421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu K., Chen L.J., Tam P.O., et al. Associations of the C2-CFB-RDBP-SKIV2L locus with age-related macular degeneration and polypoidal choroidal vasculopathy. Ophthalmology. 2013;120(4):837–843. doi: 10.1016/j.ophtha.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 76.Zhang X., Li M., Wen F., et al. Different impact of high-density lipoprotein-related genetic variants on polypoidal choroidal vasculopathy and neovascular age-related macular degeneration in a Chinese Han population. Exp Eye Res. 2013;108:16–22. doi: 10.1016/j.exer.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 77.Ji Y., Zhang X., Wu K., et al. Association of rs6982567 near GDF6 with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Han Chinese cohort. BMC Ophthalmol. 2014;14:140. doi: 10.1186/1471-2415-14-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seddon J.M., Reynolds R., Shah H.R., et al. Smoking, dietary betaine, methionine, and vitamin D in monozygotic twins with discordant macular degeneration: epigenetic implications. Ophthalmology. 2011;118(7):1386–1394. doi: 10.1016/j.ophtha.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hunter A., Spechler P.A., Cwanger A., et al. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012;53(4):2089–2105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei L., Liu B., Tuo J., et al. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012;2(5):1151–1158. doi: 10.1016/j.celrep.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oliver V.F., Jaffe A.E., Song J., et al. Differential DNA methylation identified in the blood and retina of AMD patients. Epigenetics. 2015;10(8):698–707. doi: 10.1080/15592294.2015.1060388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suuronen T., Nuutinen T., Ryhänen T., et al. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357(2):397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- 83.Kõks G., Uudelepp M.L., Limbach M., et al. Smoking-induced expression of the GPR15 gene indicates its potential role in chronic inflammatory pathologies. Am J Pathol. 2015;185(11):2898–2906. doi: 10.1016/j.ajpath.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 84.Porter L.F., Saptarshi N., Fang Y., et al. Whole-genome methylation profiling of the retinal pigment epithelium of individuals with age-related macular degeneration reveals differential methylation of the SKI, GTF2H4, and TNXB genes. Clin Epigenet. 2019;11(1):6. doi: 10.1186/s13148-019-0608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balaiya S., Abu-Amero K.K., Kondkar A.A., et al. Sirtuins expression and their role in retinal diseases. Oxid Med Cell Longev. 2017;2017:3187594. doi: 10.1155/2017/3187594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng C.H., Cherng J.Y., Chiou G.Y., et al. Delivery of Oct4 and SirT1 with cationic polyurethanes-short branch PEI to aged retinal pigment epithelium. Biomaterials. 2011;32(34):9077–9088. doi: 10.1016/j.biomaterials.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 87.Zhang H., He S., Spee C., et al. SIRT1 mediated inhibition of VEGF/VEGFR2 signaling by Resveratrol and its relevance to choroidal neovascularization. Cytokine. 2015;76(2):549–552. doi: 10.1016/j.cyto.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H.M., Li X.H., Chen M., et al. Intravitreal injection of resveratrol inhibits laser-induced murine choroidal neovascularization. Int J Ophthalmol. 2020;13(6):886–892. doi: 10.18240/ijo.2020.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsing C.H., Hung S.K., Chen Y.C., et al. Histone deacetylase inhibitor trichostatin A ameliorated endotoxin-induced neuroinflammation and cognitive dysfunction. Mediat Inflamm. 2015;2015:163140. doi: 10.1155/2015/163140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ménard C., Rezende F.A., Miloudi K., et al. MicroRNA signatures in vitreous humour and plasma of patients with exudative AMD. Oncotarget. 2016;7(15):19171–19184. doi: 10.18632/oncotarget.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin H., Qian J., Castillo A.C., et al. Effect of miR-23 on oxidant-induced injury in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2011;52(9):6308–6314. doi: 10.1167/iovs.10-6632. [DOI] [PubMed] [Google Scholar]
- 92.Askou A.L., Alsing S., Holmgaard A., et al. Dissecting microRNA dysregulation in age-related macular degeneration: new targets for eye gene therapy. Acta Ophthalmol. 2018;96(1):9–23. doi: 10.1111/aos.13407. [DOI] [PubMed] [Google Scholar]
- 93.Sabatel C., Malvaux L., Bovy N., et al. MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One. 2011;6(2):e16979. doi: 10.1371/journal.pone.0016979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaneko H., Dridi S., Tarallo V., et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu Y., Chen J., Sun J., et al. 2020. FTO Alleviates Aβ1-40 Induced Retinal Pigment Epithelium Degeneration via PKA/CREB Signaling Pathway. [Google Scholar]
- 96.Ding J.D., Johnson L.V., Herrmann R., et al. Anti-amyloid therapy protects against retinal pigmented epithelium damage and vision loss in a model of age-related macular degeneration. Proc Natl Acad Sci U S A. 2011;108(28):E279–E287. doi: 10.1073/pnas.1100901108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ratnayaka J.A., Serpell L.C., Lotery A.J. Dementia of the eye: the role of amyloid beta in retinal degeneration. Eye (Lond). 2015;29(8):1013–1026. doi: 10.1038/eye.2015.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lynn S.A., Keeling E., Munday R., et al. The complexities underlying age-related macular degeneration: could amyloid beta play an important role? Neural Regen Res. 2017;12(4):538–548. doi: 10.4103/1673-5374.205083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X., He S., Zhao M. An updated review of the epigenetic mechanism underlying the pathogenesis of age-related macular degeneration. Aging Dis. 2020;11(5):1219–1234. doi: 10.14336/AD.2019.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mudhar H.S. A brief review of the histopathology of proliferative vitreoretinopathy (PVR) Eye (Lond). 2020;34(2):246–250. doi: 10.1038/s41433-019-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bringmann A., Pannicke T., Grosche J., et al. Müller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25(4):397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 102.Agrawal R.N., He S., et al. In vivo models of proliferative vitreoretinopathy. Nat Protoc. 2007;2(1):67–77. doi: 10.1038/nprot.2007.4. [DOI] [PubMed] [Google Scholar]
- 103.Jin M., Barron E., He S., et al. Regulation of RPE intercellular junction integrity and function by hepatocyte growth factor. Invest Ophthalmol Vis Sci. 2002;43(8):2782–2790. [PubMed] [Google Scholar]
- 104.Jin M., Chen Y., He S., et al. Hepatocyte growth factor and its role in the pathogenesis of retinal detachment. Invest Ophthalmol Vis Sci. 2004;45(1):323–329. doi: 10.1167/iovs.03-0355. [DOI] [PubMed] [Google Scholar]
- 105.Hinton D.R., He S., Jin M.L., et al. Novel growth factors involved in the pathogenesis of proliferative vitreoretinopathy. Eye (Lond). 2002;16(4):422–428. doi: 10.1038/sj.eye.6700190. [DOI] [PubMed] [Google Scholar]
- 106.He S., Yaung J., Kim Y.H., et al. Endoplasmic reticulum stress induced by oxidative stress in retinal pigment epithelial cells. Graefes Arch Clin Exp Ophthalmol. 2008;246(5):677–683. doi: 10.1007/s00417-008-0770-2. [DOI] [PubMed] [Google Scholar]
- 107.He S., Barron E., Ishikawa K., et al. Inhibition of DNA methylation and methyl-CpG-binding protein 2 suppresses RPE transdifferentiation: relevance to proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2015;56(9):5579–5589. doi: 10.1167/iovs.14-16258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li X., Li X., He S., et al. MeCP2-421-mediated RPE epithelial-mesenchymal transition and its relevance to the pathogenesis of proliferative vitreoretinopathy. J Cell Mol Med. 2020;24(16):9420–9427. doi: 10.1111/jcmm.15602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma X., Long C., Wang F., et al. METTL3 attenuates proliferative vitreoretinopathy and epithelial-mesenchymal transition of retinal pigment epithelial cells via wnt/β-catenin pathway. J Cell Mol Med. 2021;25(9):4220–4234. doi: 10.1111/jcmm.16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bochaton-Piallat M.L., Kapetanios A.D., Donati G., et al. TGF-beta1, TGF-beta receptor II and ED-A fibronectin expression in myofibroblast of vitreoretinopathy. Invest Ophthalmol Vis Sci. 2000;41(8):2336–2342. [PubMed] [Google Scholar]
- 111.Kita T., Hata Y., Arita R., et al. Role of TGF-beta in proliferative vitreoretinal diseases and ROCK as a therapeutic target. Proc Natl Acad Sci U S A. 2008;105(45):17504–17509. doi: 10.1073/pnas.0804054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang C.Y., Shie S.S., Tsai M.L., et al. FTO modulates fibrogenic responses in obstructive nephropathy. Sci Rep. 2016;6:18874. doi: 10.1038/srep18874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin X., Chai G., Wu Y., et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Luo X., Cao M., Gao F., et al. YTHDF1 promotes hepatocellular carcinoma progression via activating PI3K/AKT/mTOR signaling pathway and inducing epithelial-mesenchymal transition. Exp Hematol Oncol. 2021;10(1):35. doi: 10.1186/s40164-021-00227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bahn R.S. Graves' ophthalmopathy. N Engl J Med. 2010;362(8):726–738. doi: 10.1056/NEJMra0905750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bartalena L., Piantanida E., Gallo D., et al. Epidemiology, natural history, risk factors, and prevention of Graves' orbitopathy. Front Endocrinol (Lausanne) 2020;11:615993. doi: 10.3389/fendo.2020.615993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bahn R.S. Current insights into the pathogenesis of Graves' ophthalmopathy. Horm Metab Res. 2015;47(10):773–778. doi: 10.1055/s-0035-1555762. [DOI] [PubMed] [Google Scholar]
- 118.Dik W.A., Virakul S., van Steensel L. Current perspectives on the role of orbital fibroblasts in the pathogenesis of Graves' ophthalmopathy. Exp Eye Res. 2016;142:83–91. doi: 10.1016/j.exer.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 119.Lee J.S., Chae M.K., Kikkawa D.O., et al. Glycogen synthase kinase-3β mediates proinflammatory cytokine secretion and adipogenesis in orbital fibroblasts from patients with Graves' orbitopathy. Invest Ophthalmol Vis Sci. 2020;61(8):51. doi: 10.1167/iovs.61.8.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wei Y., Li N., Zhao L., et al. MicroRNAs and autoimmune-mediated eye diseases. Front Cell Dev Biol. 2020;8:818. doi: 10.3389/fcell.2020.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei Y.H., Liao S.L., Wang S.H., et al. Simvastatin and ROCK inhibitor Y-27632 inhibit myofibroblast differentiation of Graves' ophthalmopathy-derived orbital fibroblasts via RhoA-mediated ERK and p38 signaling pathways. Front Endocrinol (Lausanne) 2021;11:607968. doi: 10.3389/fendo.2020.607968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Virakul S., Somparn P., Pisitkun T., et al. Integrative analysis of proteomics and DNA methylation in orbital fibroblasts from Graves' ophthalmopathy. Front Endocrinol (Lausanne) 2021;11:619989. doi: 10.3389/fendo.2020.619989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu L., Li S., He S., et al. The critical role of m6A methylation in the pathogenesis of Graves' ophthalmopathy. Eye Vis (Lond). 2020;7(1):55. doi: 10.1186/s40662-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kodama K., Sikorska H., Bandy-Dafoe P., et al. Demonstration of a circulating autoantibody against a soluble eye-muscle antigen in Graves' ophthalmopathy. Lancet. 1982;2(8312):1353–1356. doi: 10.1016/s0140-6736(82)91267-3. [DOI] [PubMed] [Google Scholar]
- 125.Cai T.T., Muhali F.S., Song R.H., et al. Genome-wide DNA methylation analysis in Graves' disease. Genomics. 2015;105(4):204–210. doi: 10.1016/j.ygeno.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 126.Yan N., Zhou J.Z., Zhang J.A., et al. Histone hypoacetylation and increased histone deacetylases in peripheral blood mononuclear cells from patients with Graves' disease. Mol Cell Endocrinol. 2015;414:143–147. doi: 10.1016/j.mce.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 127.Yu R., Li Q., Feng Z., et al. m6A reader YTHDF2 regulates LPS-induced inflammatory response. Int J Mol Sci. 2019;20(6):1323. doi: 10.3390/ijms20061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Q., Wang C., Dong W., et al. WTAP facilitates progression of endometrial cancer via CAV-1/NF-κB axis. Cell Biol Int. 2021;45(6):1269–1277. doi: 10.1002/cbin.11570. [DOI] [PubMed] [Google Scholar]
- 129.Luo J., Xu T., Sun K. N6-methyladenosine RNA modification in inflammation: roles, mechanisms, and applications. Front Cell Dev Biol. 2021;9:670711. doi: 10.3389/fcell.2021.670711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jager M.J., Shields C.L., Cebulla C.M., et al. Uveal melanoma. Nat Rev Dis Prim. 2020;6(1):24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 132.Robertson A.G., Shih J., Yau C., et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32(2):204–220. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bakhoum M.F., Esmaeli B. Molecular characteristics of uveal melanoma: insights from the cancer genome atlas (TCGA) project. Cancers (Basel) 2019;11(8):1061. doi: 10.3390/cancers11081061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li Y., Jia R., Ge S. Role of epigenetics in uveal melanoma. Int J Biol Sci. 2017;13(4):426–433. doi: 10.7150/ijbs.18331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chokhachi Baradaran P., Kozovska Z., Furdova A., et al. Targeting epigenetic modifications in uveal melanoma. Int J Mol Sci. 2020;21(15):5314. doi: 10.3390/ijms21155314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Worley L.A., Long M.D., Onken M.D., et al. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18(3):184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 137.Bande M., Fernandez-Diaz D., Fernandez-Marta B., et al. The role of non-coding RNAs in uveal melanoma. Cancers. 2020;12(10):2944. doi: 10.3390/cancers12102944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Azevedo H., Pessoa G.C., de Luna Vitorino F.N., et al. Gene co-expression and histone modification signatures are associated with melanoma progression, epithelial-to-mesenchymal transition, and metastasis. Clin Epigenet. 2020;12(1):127. doi: 10.1186/s13148-020-00910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Luo G., Xu W., Zhao Y., et al. RNA m6A methylation regulates uveal melanoma cell proliferation, migration, and invasion by targeting c-Met. J Cell Physiol. 2020;235(10):7107–7119. doi: 10.1002/jcp.29608. [DOI] [PubMed] [Google Scholar]
- 140.Hao L., Yin J., Yang H., et al. ALKBH5-mediated m6A demethylation of FOXM1 mRNA promotes progression of uveal melanoma. Aging (Albany NY) 2021;13(3):4045–4062. doi: 10.18632/aging.202371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jia R., Chai P., Wang S., et al. m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18(1):161. doi: 10.1186/s12943-019-1088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaliki S., Shields C.L., Shields J.A. Uveal melanoma: estimating prognosis. Indian J Ophthalmol. 2015;63(2):93–102. doi: 10.4103/0301-4738.154367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shields C.L., Say E.A.T., Hasanreisoglu M., et al. Cytogenetic abnormalities in uveal melanoma based on tumor features and size in 1059 patients: the 2016 W. Richard green lecture. Ophthalmology. 2017;124(5):609–618. doi: 10.1016/j.ophtha.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 144.Tang J., Wan Q., Lu J. The prognostic values of m6A RNA methylation regulators in uveal melanoma. BMC Cancer. 2020;20(1):674. doi: 10.1186/s12885-020-07159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang H., Zhang P., Long C., et al. m6A methyltransferase METTL3 promotes retinoblastoma progression via PI3K/AKT/mTOR pathway. J Cell Mol Med. 2020;24(21):12368–12378. doi: 10.1111/jcmm.15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guy W.M., Soparkar C.N., Alford E.L., et al. Traumatic optic neuropathy and second optic nerve injuries. JAMA Ophthalmol. 2014;132(5):567–571. doi: 10.1001/jamaophthalmol.2014.82. [DOI] [PubMed] [Google Scholar]
- 147.Du K., Zhang L., Lee T., et al. m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56(3):1596–1606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 148.Boles N.C., Temple S. Epimetronomics: m6A marks the tempo of corticogenesis. Neuron. 2017;96(4):718–720. doi: 10.1016/j.neuron.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 149.Zhou H., Wang B., Sun H., et al. Epigenetic regulations in neural stem cells and neurological diseases. Stem Cell Int. 2018;2018:6087143. doi: 10.1155/2018/6087143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yu J., Zhang Y., Ma H., et al. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol Brain. 2020;13(1):11. doi: 10.1186/s13041-020-0554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Y., Mao J., Wang X., et al. Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics. 2019;11(7):805–819. doi: 10.2217/epi-2019-0002. [DOI] [PubMed] [Google Scholar]
- 152.Qu X., Zhu K., Li Z., et al. The alteration of M6A-tagged transcript profiles in the retina of rats after traumatic optic neuropathy. Front Genet. 2021;12:628841. doi: 10.3389/fgene.2021.628841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang J., Wu Z., Renier N., et al. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell. 2015;160(1–2):161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Howell J.A., Bidwell G.L., 3rd Targeting the NF-κB pathway for therapy of ischemic stroke. Ther Deliv. 2020;11(2):113–123. doi: 10.4155/tde-2019-0075. [DOI] [PubMed] [Google Scholar]
- 155.Weng Y.L., Wang X., An R., et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97(2):313–325. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang C., Wang Y., Peng Y., et al. METTL3 regulates inflammatory pain by modulating m6A-dependent pri-miR-365-3p processing. Faseb J. 2020;34(1):122–132. doi: 10.1096/fj.201901555R. [DOI] [PubMed] [Google Scholar]
- 157.Ouyang Y.B., Xu L.J., Emery J.F., et al. Overexpressing GRP78 influences Ca2+ handling and function of mitochondria in astrocytes after ischemia-like stress. Mitochondrion. 2011;11(2):279–286. doi: 10.1016/j.mito.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dou G., Sreekumar P.G., Spee C., et al. Deficiency of αB crystallin augments ER stress-induced apoptosis by enhancing mitochondrial dysfunction. Free Radic Biol Med. 2012;53(5):1111–1122. doi: 10.1016/j.freeradbiomed.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Osborne N.N., Núñez-Álvarez C., Del Olmo-Aguado S. The effect of visual blue light on mitochondrial function associated with retinal ganglions cells. Exp Eye Res. 2014;128:8–14. doi: 10.1016/j.exer.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 160.Senft D., Ronai Z.A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015;40(3):141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Osborne N.N., Núñez-Álvarez C., Joglar B., et al. Glaucoma: focus on mitochondria in relation to pathogenesis and neuroprotection. Eur J Pharmacol. 2016;787:127–133. doi: 10.1016/j.ejphar.2016.04.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.