Abstract
The N terminus of the matrix (M) protein of vesicular stomatitis virus (VSV) and of other rhabdoviruses contains a highly conserved PPPY sequence (or PY motif) similar to the late (L) domains in the Gag proteins of some retroviruses. These L domains in retroviral Gag proteins are required for efficient release of virus particles. In this report, we show that mutations in the PPPY sequence of the VSV M protein reduce virus yield by blocking a late stage in virus budding. We also observed a delay in the ability of mutant viruses to cause inhibition of host gene expression compared to wild-type (WT) VSV. The effect of PY mutations on virus budding appears to be due to a block at a stage just prior to virion release, since electron microscopic examination of PPPA mutant-infected cells showed a large number of assembled virions at the plasma membrane trapped in the process of budding. Deletion of the glycoprotein (G) in addition to these mutations further reduced the virus yield to less than 1% of WT levels, and very few particles were assembled at the cell surface. This observation suggested that G protein aids in the initial stage of budding, presumably during the formation of the bud site. Overall, our results confirm that the PPPY sequence of the VSV M protein possesses L domain activity analogous to that of the retroviral Gag proteins.
The assembly and budding of enveloped negative-strand RNA viruses is a multistep process occurring at the plasma membrane of a host cell. Vesicular stomatitis virus (VSV), a prototype enveloped negative-strand RNA virus belonging to the family Rhabdoviridae, has long been utilized as a model for studying various steps in virus assembly and budding. Assembly is initiated when the matrix protein (M) binds to and condenses the nucleocapsid core into a tightly coiled helical structure in association with the inner leaflet of the plasma membrane. During budding, the condensed core becomes enveloped in a host-derived membrane highly enriched with the viral glycoprotein (G) and selectively depleted of most of the host proteins.
Recent studies have shown that while G protein contributes to high levels of virus budding, virus assembly and release can occur in the absence of G protein, but with reduced efficiency (28, 36, 37, 43). Thus, the condensed ribonucleocapsid core (RNP) complexed with M protein is sufficient to initiate and drive rhabdovirus budding. Also, in the absence of other viral proteins, VSV M protein can cause budding of lipid vesicles into the surrounding medium (20). In the case of rabies virus, deletion of M protein dramatically reduced virus yields, more than 10,000 fold. The ΔM particles that were produced were filamentous instead of having the characteristic bullet shape, confirming the earlier reports of the contribution of M protein to virus morphology (25, 30, 31).
Besides its role in virus assembly and budding, matrix protein also mediates most of the cytopathic effects (CPE) attributed to VSV infection (4, 6, 11, 38). Transient expression of M protein alone can cause cell rounding by disorganization of microtubules and intermediate filaments (24). It can also cause inhibition of host-directed transcription (4, 5, 32). Recently it was shown that VSV infection leads to the inactivation of transcription factor II D (TFIID), resulting in inhibition of RNA polymerase II-dependent transcription (48). A separate effect of M protein on host gene expression involves inhibition of Ran GTPase-mediated nuclear transport, although the exact mechanism of this effect is not known (18). M protein-induced CPE are genetically separable from the function of M protein in viral assembly (4), suggesting the existence of separate domains within M protein that carry out its multiple functions.
Previous studies have revealed that M protein is present at the inner surface of the plasma membrane of infected cells (3, 23, 27). Affinity labeling studies suggested that the N-terminal basic region of M protein was associated with the lipid bilayer (23). The N terminus of M protein also contains a PPPY sequence that is conserved among the matrix proteins of many rhabdoviruses (16, 17). This PPPY sequence closely resembles the PY motif in the late (L) domain of the Gag protein of Rous sarcoma virus (RSV), an avian retrovirus. The L domains of retroviruses are involved in the late stages of virus budding, specifically the fission event resulting in virus release (45–47). The L domains identified in the Gag proteins of other retroviruses have different amino acid sequences, but they can functionally substitute for the L domain of RSV and allow budding of chimeric particles (33, 35). The PPPY sequence in the RSV L domain matches the consensus proline-rich motif required for interaction with WW domains found in several regulatory and signal transduction proteins (9, 10, 26, 34). This observation has led to the suggestion that the PY motif in viral proteins may play a role in recruiting WW-containing cellular proteins for the purpose of assembly and budding of virus particles at the host membrane (12, 14).
Recently, it was suggested that the PY motif found in rhabdovirus M proteins might function as a late domain and contribute to virus budding (12). Alanine mutagenesis of the VSV PY motif drastically reduced the budding function of mutant proteins in an in vitro budding assay (12). Furthermore, it was demonstrated that the PY motifs of both VSV and rabies virus M proteins can interact with WW domains of certain cellular proteins in in vitro binding assays (17). Together, these studies suggested that the PY motif may be important in the budding of virus particles and that this process may involve cellular proteins.
In this study we have examined the contribution of the PY motif to VSV assembly and budding by recovering infectious virus containing point mutations within the PPPY sequence of M protein. Our results show that mutations in the PPPY motif reduced, but did not completely abolish, virus budding. Electron microscopy revealed that the mutants were blocked at a late stage of budding, apparently during the release of mature virions from the cell surface. Thus our studies confirm that the PPPY motif within VSV M protein is functionally similar to the L domains found in retroviral Gag proteins.
MATERIALS AND METHODS
Plasmids and oligonucleotide-directed mutagenesis.
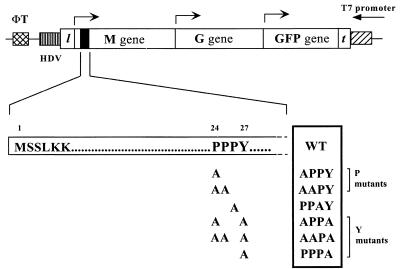
To generate mutations within the PPPY motif of M protein of VSV (Indiana serotype), we performed site-directed mutagenesis using an overlap PCR-based strategy. Six different sense strand, mutagenic oligonucleotides were synthesized containing the appropriate nucleotide changes for the indicated amino acid substitutions and also containing a silent mutation at nucleotide 2326 (G→A) which abolished the EarI site in the M gene. Each of the mutagenic primers was used in six separate PCRs together with a common antisense oligonucleotide downstream of the unique BglII site at nucleotide 2542 (M-2542 primer) to generate PCR products from the template pBS-MGF. The pBS-MGF plasmid carries a genomic sense RNA of VSV (Indiana serotype) containing the M, G, and green fluorescent protein (GFP) genes (previously referred to as pBS-GMF) (41). A schematic representation of the MGF minigenome is shown in Fig. 1. In a separate reaction, we used an antisense oligonucleotide that overlapped nucleotides 2320 to 2340 and a sense oligonucleotide complementary to the T7 terminator region (T7 term-1 primer), which is 180 bp downstream of a unique RsrII site (in the leader region), to amplify the 5′ end of the M gene. This PCR fragment overlapped the 5′ ends of the mutated PCR fragments. The PCR fragments were gel purified, each fragment carrying the mutation was used with the overlapping nonmutagenic fragment as templates, and the entire region was amplified using the M-2542 and T7 term-1 primers external to the PCR region. The resulting 500-bp PCR fragments were gel purified, digested with RsrII and BglII, and then used to replace the corresponding wild-type (WT) region in the pBS-MGF plasmid. Colonies carrying the correct plasmids were identified following digestion of miniprep DNA with EarI. The plasmids were then sequenced using the dideoxy sequencing method to ensure that only the specified mutations were introduced during the PCR amplification.
FIG. 1.
Schematic representation of PPXY mutations in the MGF minigenome. MGF stands for the M, G, and GFP minigenome. l and t denote the leader and trailer sequences, respectively. The hepatitis delta virus ribozyme sequence (HDV) and the T7 terminator sequence (ΦT) are represented by hatched boxes. Arrows indicate the direction of transcription for each gene. The solid box at the 5′ end of the M gene represents the N-terminal region of the matrix protein including the PPXY motif, which is enlarged below. The numbering shows the positions of amino acids with respect to the N terminus of M protein. Alanine substitutions were made for the proline and tyrosine residues either singly or in combinations. The name of each mutant is listed on the right. WT stands for the wild-type sequence of the PY motif in VSV M protein. The mutants were classified into two groups, P and Y mutants, based on the severity of their phenotypes.
Generation of full-length PPXY mutants.
Each of the six M genes carrying the specified mutations was subcloned into a plasmid encoding a modified VSV antigenome called ΔM-PLF. This is a full-length VSV cDNA in which the M coding region was replaced with a polylinker (PL) containing unique restriction sites (5′ AscI-XhoI-SmaI-EagI-AvrII 3′), while retaining the normal M gene transcriptional start and stop sequences. In addition, ΔM-PLF has a non-VSV reporter gene encoding GFP inserted between the G and L genes. The MGF mutants were digested with RcaI located in the 5′ untranslated region of the M gene and MluI located in the 5′ untranslated region of the G gene. Because RcaI is not a unique site, the remainder of the 5′ end of the M gene was obtained by digesting the WT pVSV FL-2 (+) (22) with XbaI located at the 3′ end of the P gene and RcaI. The fragments were then gel purified and used in a three-way ligation to replace the corresponding region of ΔM-PLF, which was digested with XbaI and MluI.
Recovery of PPXY mutants.
Recoveries of the minigenome mutants were performed as described previously (40) and monitored by detecting expression of GFP from the reporter gene. Full-length viruses were recovered from their cDNAs as described previously (22) with the following modifications. Briefly, BHK cells in 60-mm dishes were infected with vTF7-3 at a multiplicity of infection (MOI) of 10 for 1 h at 31°C in serum-free Dulbecco's modified Eagle's medium (DMEM) without penicillin and streptomycin. The cells were then cotransfected with plasmids encoding WT or mutant genomes together with 3, 5, and 1 μg of plasmids encoding the N, P, and L proteins, respectively, in the presence of 10 μg of 1-β-d-arabinofuranosylcytosine (araC)/ml. The supernatants were harvested 48 h posttransfection (p.i.), filtered through a 0.22-μm-pore-size filter (Millipore-GS) to remove vaccinia virus, and used to obtain plaque isolates from BHK cells. The mutations were confirmed by direct sequencing of reverse transcription-PCR (RT-PCR) amplification products generated using genomic RNA as the template.
Assembly and budding assay for PPXY mutants.
BHK-21 cells in 35-mm dishes were infected with either WT VSV or the mutant viruses at an MOI of 10 for 1 h. Following the adsorption period, the inoculum was removed, and the cells were washed three times with serum-free DMEM to remove any unadsorbed virus and then incubated with 2 ml of the same medium at 37°C. At 7 h p.i., the medium was removed and the cells were washed once with methionine-free DMEM. The cells were then labeled with 50 μCi of [35S]methionine in 1 ml of labeling medium (9 parts of Met-free DMEM plus 1 part of DMEM plus 5% fetal bovine serum [FBS]), using protein-labeling mix (Dupont, NEN) at 37°C for 8 h. At 15 h p.i., the supernatants were harvested and clarified by centrifuging at 1,260 × g for 5 min. The virions were then pelleted from the supernatant by ultracentrifugation through a 20% sucrose cushion (in 50 mM Tris–150 mM NaCl [pH 7.4]) at 45,000 rpm for 40 min in an AH-650 swinging bucket rotor (Sorvall). The pellets were resuspended in equal volumes of reducing sample buffer. One-tenth of each sample was resolved by electrophoresis on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel followed by autoradiography. The amounts of virus released from the WT and mutant infected cells were determined by quantitation of the amounts of N protein using the Storm 860 PhosphorImager and ImageQuant analytical software (both from Molecular Dynamics).
Growth kinetics of mutants.
BHK cells in 60-mm plates were infected with either WT VSV or the PPXY mutants at an MOI of 10 by adsorbing for 1 h at 37°C. The inoculum was removed, and following three washes with serum-free DMEM, the cells were incubated with 4 ml of DMEM plus 5% FBS at 37°C. At the designated time point, an aliquot of the supernatant was removed and the amount of infectious virus in the supernatant was determined by a standard plaque assay on BHK cells.
Host protein shutoff assay.
BHK-21 cells in 35-mm dishes were infected with WT or mutant viruses at an MOI of 10 at 31°C. After 1 h, the inoculum was removed, and cells were washed twice with medium and incubated with 2 ml of DMEM plus 5% FBS at 37°C. At each time point, the medium was removed, and the cells were washed twice with Met-free DMEM and then incubated for an additional 10 min in the same medium. The cells were labeled for 15 min with 1 ml of methionine-free DMEM containing 50 μCi of [35S]methionine at 37°C. Following the pulse, the label was removed and cells were lysed with 1 ml of detergent solution (10 mM Tris [pH 7.4], 66 mM EDTA, 0.4% sodium deoxycholate, 1% Triton X-100, 0.05% sodium azide, and 200 U of aprotinin/ml). Cell extracts were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography. The degree of host shutoff was determined by quantitating the amount of 35S-labeled host proteins present in a small area of the gel mostly devoid of viral proteins by using a Storm 860 Phosphorimager (Molecular Dynamics). The shutoff of host protein synthesis was expressed as the percentage of the amount of total labeled host proteins in the uninfected sample (MOCK) detected at given time points in the assay.
Transmission electron microscopy.
BHK-21 cells were infected with either the WT or mutant viruses for 8 h at 37°C. Following infection, the cells were harvested and washed three times with phosphate-buffered saline (PBS). They were then fixed with 2% glutaraldehyde in PBS and postfixed with 1% osmium tetroxide (29). The fixed cells were dehydrated in a graded series of ethyl alcohol, stained with uranyl acetate, and embedded in Spurr resin. Ultrathin sections were cut on a Sorvall MT6000 ultramicrotome, and the sections were stained with Reynold's lead citrate before examination within a JEOL 1200 electron microscope.
Scanning electron microscopy.
BHK-21 cells were grown on plastic coverslips (Thermanox; Nunc) that were placed in six-well culture plates (Costar) and infected with WT and PPPA mutant viruses at an MOI of 10. At 8 h p.i., cells were rinsed with PBS, fixed with 5% glutaraldehyde in PBS, and postfixed with 2% osmium tetroxide in PBS. After dehydration in a graded series of ethanols, the cells were dried out with hexamethyldisilazane, coated with gold in a sputter-coater, and examined in the scanning mode of a JEOL 1200 EX II TEMscan electron microscope.
RESULTS
The PPXY motif is important but not essential for VSV budding.
A recent report by Harty et al. (17) suggested that the PPPY motif in VSV matrix (M) protein might be important for virus assembly and release, since mutations in this motif greatly reduced M-induced vesicle budding. To determine whether the PPXY motif was essential for the budding of virus particles, we first made use of a minigenome system (39). The minigenome used in this study was called MGF and consisted of genes encoding matrix protein (M) and glycoprotein (G) as well as a non-VSV reporter gene encoding GFP. Alanine substitutions were made either singly or in various combinations within the PPXY motif of the matrix protein (Fig. 1). If the PPXY motif was absolutely critical for virus budding, then the alanine substitutions would result in only single cells expressing M, G, and GFP, but no virus spread would occur. However, we found that none of the point mutations prevented the release and spread of infectious minivirus (data not shown).
Recovery and characterization of infectious full-length PPXY mutants.
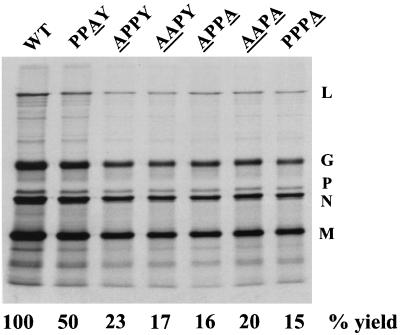
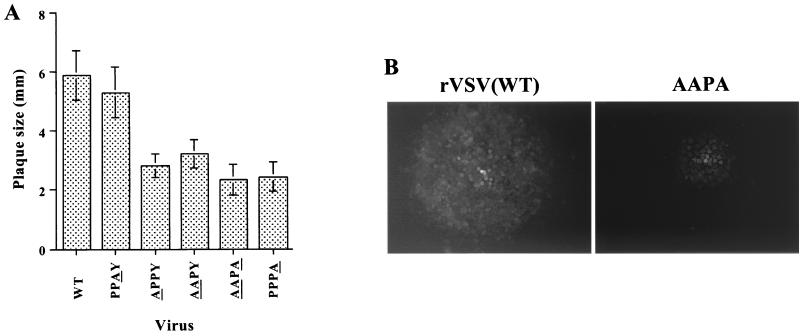
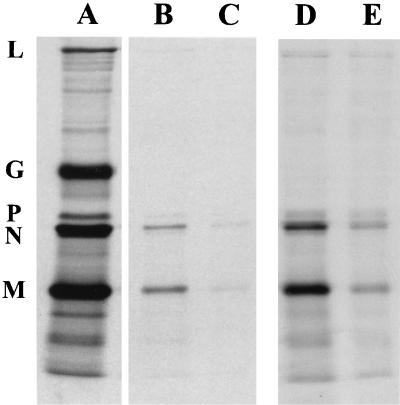
To obtain a more quantitative evaluation of the effect of mutations in the PPPY sequence, the mutations were subcloned into a modified full-length VSV cDNA (pVSV-GFP) that encodes GFP as a reporter gene. This construct will be referred to as WT in the subsequent experiments. The GFP gene was cloned between the G and L genes, making L the sixth gene. All of the PPXY mutants were recovered and could be passaged in cell culture, which supported our observations from the minigenome system. To determine the effects of the mutations on virus replication and budding, we analyzed the amount of virus released from cells infected with mutant viruses. Cells were infected with either the WT or the mutants, and the virus released in the culture media was analyzed by SDS-PAGE followed by autoradiography (Fig. 2). The mobilities of the five viral proteins were indistinguishable in the WT and mutant viruses, and all five VSV proteins were present in the mutant virions in similar proportions to that found in WT virus. However, the amount of virus released from the mutant infected cells, except for the PPAY mutant, was only 20% of that released from WT-infected cells. In comparison, cells infected with the PPAY mutant released virus at approximately 50% of the WT level. The reduced budding could not be relieved by growing the viruses at 31°C, indicating that the mutants were not temperature sensitive (data not shown). The plaque sizes of all mutants, except PPAY, were also smaller than that of the WT (Fig. 3A). Plaques formed by the AAPA mutant were about half the size of WT virus plaques (Fig. 3B) and never attained the size of WT plaques even after 48 h of incubation (data not shown). The plaque size of the PPAY mutant was about 80% that of the WT.
FIG. 2.
Assembly and budding profile of PPXY mutants. BHK-21 cells were infected with either the WT or mutant viruses at an MOI of 10. Cells were labeled at 7 h p.i. with [35S]methionine. At 15 h p.i., viruses were harvested from the supernatants by centrifugation and analyzed by SDS-PAGE followed by autoradiography. The positions of the five VSV proteins are indicated on the right. The mutated residue(s) in the PPPY sequence of each mutant is underlined. The percent yield of virus in this experiment is calculated based on the quantitation of N protein using ImageQuant software (Molecular Dynamics).
FIG. 3.
Comparison of the plaque sizes of WT and PPXY mutants in BHK-21 cells. (A) A standard plaque assay was performed for each virus on BHK-21 cells. At 13 h p.i., the sizes of 25 individual plaques were determined for each virus. The mean plaque size for each virus is shown. The residue(s) changed in each mutant is underlined. (B) Fluorescence micrograph of GFP-expressing cells comparing the plaque sizes of WT and the PPPA mutant (magnification, ×7.5).
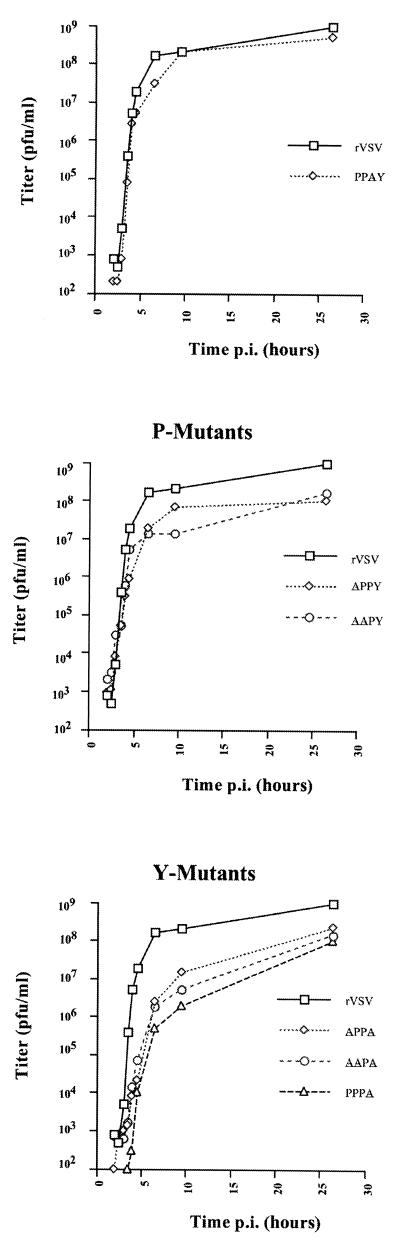
Growth curve of PPXY mutants.
We also performed one-step growth curves for each of the mutants as well as the WT virus (Fig. 4). The results show that there is a delay in virus release at early time points for all mutants except PPAY compared to the WT virus. The effect was more pronounced for the Y (APPA, AAPA, and PPPA) mutants compared to the P (APPY and AAPY) mutants. The delay was not relieved by performing the experiment at either a lower temperature (31°C) or a higher MOI (MOI, 50) (data not shown). The PPAY mutant, as expected, had growth kinetics similar to that of the WT virus, which correlates well with the observation that the third proline residue in this motif is not conserved among rhabdoviruses and therefore does not contribute significantly to the function of this motif.
FIG. 4.
Growth kinetics of PPXY mutants in BHK-21 cells. BHK-21 cells were infected with either the WT or the mutant viruses at an MOI of 10. At various times p.i., aliquots of supernatant were taken to determine the titer by a plaque assay on BHK cells. The kinetics for the P and Y mutants were determined simultaneously under similar conditions, but they were plotted individually to show the growth difference during early times of infection.
Morphogenesis of the PPPY mutants.
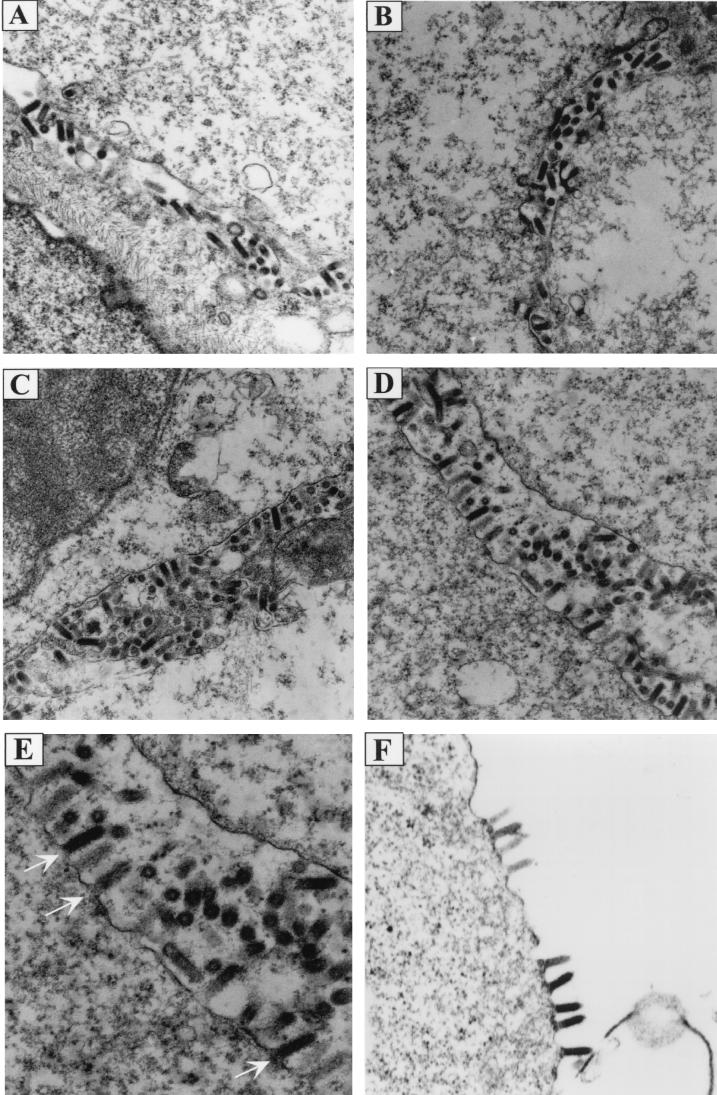
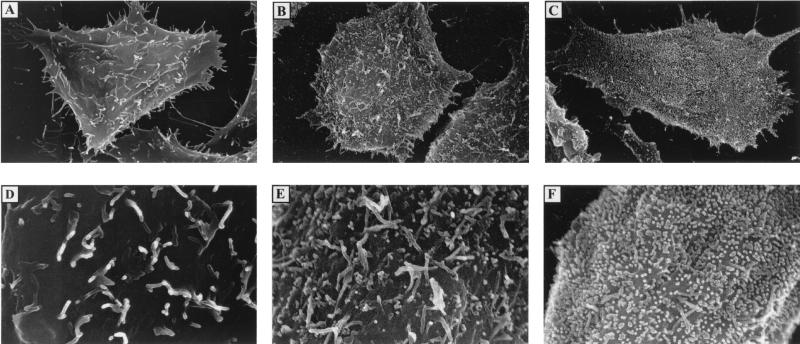
The above results indicated that the ability of the PPPY mutants to produce virus was affected. There are several possibilities which could explain the phenotype of these mutants. The mutations could inhibit (i) the association of M with RNPs, (ii) the association of RNP-M complexes with the plasma membrane, or (iii) virus release from the cell surface. To distinguish between these possibilities, thin-section electron-microscopic analysis was performed on BHK cells infected with either the WT or the mutant viruses. Thin sections of cells infected with WT virus or ΔG virus (virus lacking G), each of which has an intact PPPY sequence, had predominantly mature extracellular virions present (Fig. 5A and B). However, electron micrographs of cells infected with the AAPY or the AAPA mutant showed a larger proportion of virions assembled at the cell surface, apparently in the process of budding (Fig. 5C and D). Quantification of the budding virions revealed a threefold increase in the number of AAPY and AAPA mutant virions that were not yet released from the plasma membrane (e.g., budding) compared to the WT or the ΔG virions (Table 1). Therefore, these mutants were able to condense RNP cores and associate with the plasma membrane similarly to WT virus, but it appeared that virus release was inhibited and consequently virions accumulated on the cell surface. This is very clearly seen in the scanning electron micrographs shown in Fig. 6. Uninfected BHK cells have microvilli projecting from an otherwise smooth cell surface (Fig. 6A and D). Cells infected with WT virus showed only a few virions budding from the cell (Fig. 6B and E). On the other hand, the entire surface of the cell infected with the PPPA mutant was covered with virions presumably trapped in the process of budding (Fig. 6C and F). Thus, the PPPY sequence appears to be involved in the late budding step of virus morphogenesis, most likely the pinching off of mature virions from the cell surface which is analogous to the L domain activity in the retroviral Gag proteins.
FIG. 5.
Electron micrographs showing morphologies of PPXY mutants. BHK-21 cells infected with WT or mutant viruses were fixed at 8 h p.i., and thin sections of cells were examined under the electron microscope to determine the stage(s) at which morphogenesis of PPXY mutants was blocked. (A) WT; (B) ΔG-GFP; (C) AAPY; (D and E) PPPA; (F) ΔG-AAPA. Magnifications: ×15,000 for panels A through D; ×18,000 for panel F. Panel E) is an enlargement of panel D. Arrows in panel E point to the budding virions attached to the plasma membrane of the infected cell.
TABLE 1.
Quantification of virions budding from the plasma membrane
| Virus | No. of budding virions/3 μm of membranea |
|---|---|
| WT-VSV | 9.1 ± 4.6 |
| ΔG-VSV | 9.2 ± 2.6 |
| AAPY | 22.5 ± 7.8 |
| PPPA | 22.5 ± 5.7 |
The number of virions budding from a 3-μm linear section of the cell surface was counted. Each value is the average number of virions counted from five to eight individual micrographs, and standard deviations are given. For WT-VSV, n = 8; for ΔG-VSV, n = 5; for AAPY, n = 7; and for PPPA, n = 6.
FIG. 6.
Scanning electron micrographs of WT virus and the PPPA mutant. BHK-21 cells on plastic coverslips were infected with either WT virus or the PPPA mutant at an MOI of 10. At 8 h p.i., cells were fixed and processed as described in Materials and Methods. (A and D) Uninfected cells; (B and E) WT infected cells; (C and F) PPPA mutant-infected cells. Magnifications: ×4,000 for panels A through C; ×12,000 for panels D through F.
Effects of the PPXY mutations in the absence of glycoprotein.
Because the mutations in the PPPY sequence did not completely abolish virus budding, the residual budding activity could be attributed to another region(s) in M or to spontaneous release of virus accumulated at the cell surface over time. Alternatively, G protein may affect virus release either by acting in concert with the PPXY motif of M protein or by acting alone. To assess the role of G protein in the budding process, we recovered an AAPA mutant that lacked G protein (ΔG-AAPA) and performed a similar analysis of virus assembly and release. If G contributes to virus release, then in ΔG-AAPA infected cells, all virions would be trapped at the cell surface in the process of budding and very few, if any, would be released into the medium. As shown in Fig. 7, ΔG-AAPA was severely affected in its ability to produce virus; less than 1% of virus was released compared to WT-VSV, even though the mutant infected cells synthesized WT levels of viral proteins (data not shown). However, thin-section electron microscopy indicated that in the absence of G protein, fewer particles were assembled at the surface, and of those that did, most were not released (Fig. 5F). Thus, G protein does not account for the low levels of virions released from the PPXY mutants; instead, it appears that G protein is involved in an early stage of virus budding prior to the step of virion release.
FIG. 7.
Assembly phenotype of AAPA mutant in the absence of glycoprotein. Virions released from cells infected with either ΔG-GFP or ΔG-PPPA were compared to those released from WT-infected cells. Virions were prepared and analyzed on SDS-polyacrylamide gels as described for Fig. 2. (A) WT; (B) ΔG-GFP; (C) ΔG-PPPA; (D and E) longer exposures of panels B and C.
Inhibition of host protein synthesis by PPXY mutants.
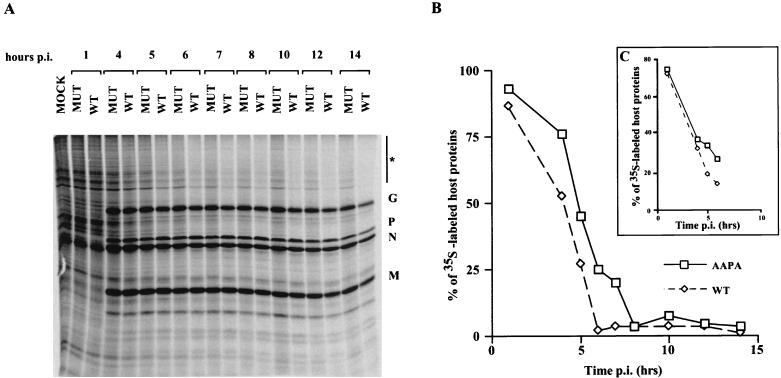
In addition to its role in the assembly and budding of virus, matrix protein has been shown to cause cell rounding and inhibition of host gene expression. However, relatively little is known about the regions of M protein involved in these CPE. Therefore, we wanted to determine whether the mutations in the PPXY motif would have any effect on viral cytopathogenesis. Examination of cells by phase-contrast microscopy indicated that the extent as well as the kinetics of cell rounding in BHK cells was unaltered during infection by mutants (data not shown). We also examined whether there was any difference in the ability of the mutants to inhibit host cell gene expression. BHK cells were infected with either the WT virus or mutant viruses and then pulse-labeled with [35S]methionine at several times p.i. The crude cell lysates were then analyzed by SDS-PAGE as described previously (21). We found that the kinetics of host shutoff was delayed at early times p.i. compared to that for WT virus (Fig. 8). WT virus caused rapid inhibition of host gene expression, with >90% inhibition seen by 6 h p.i. In contrast, this level of inhibition was not seen until approximately 8 h p.i. with the mutants, and the delayed host shutoff effect was not relieved by using a higher MOI (MOI, 50) (Fig. 8, inset panel C).
FIG. 8.
Kinetics of host protein synthesis shutoff by the AAPA mutant. Cells were infected with either WT virus or the AAPA mutant (MUT) virus at an MOI of 10. At the times indicated, the cells were pulse-labeled for 15 min with [35S]methionine and lysed in detergent buffer. A portion of the cell lysate was analyzed by SDS-PAGE followed by autoradiography. Labeled uninfected cells (MOCK) served as a control. (B) The amount of 35S-labeled host proteins in each lane was determined by quantitating a representative area on the gel (indicated by the asterisk in panel A) with a STORM Phosphorimager and ImageQuant software (Molecular Dynamics). The amount of host proteins in the uninfected cells (control) was considered to be 100%. The extent of host shutoff was expressed as a percentage of the amount of host proteins in the control. The extent of host shutoff caused by the AAPA mutant at an MOI of 50 is shown in the inset (C).
DISCUSSION
Recently it was shown that the N-terminal 74 amino acids of VSV M protein containing the PPPY sequence could functionally replace the L domain in the Gag protein of RSV (12). Deletion or point mutations in this motif drastically reduced the budding activity of the M-Gag chimeras in a transient expression system. Based on these results, it was suggested that the PPPY motif might be the putative L domain of VSV M protein, functioning in a similar fashion to the L domains of retroviral Gag proteins. In this report we have analyzed the effects of mutations in the PPPY sequence found in the N terminus of VSV M protein on virus assembly and budding. All PPPY mutants synthesized WT levels of all viral proteins in the infected cells (Fig. 8). They also appeared to condense RNP cores normally, since the virions had the characteristic bullet-shaped morphology, similar to that of WT particles. The virions assembled at the plasma membrane, indicating that there was no defect in the association of condensed RNPs with the plasma membrane. However, the final stage of budding, i.e., the release of particles from the cell surface, was inefficient in these mutants. Transmission electron microscopy of mutant-infected cells showed a large number of virions at the cell surface apparently trapped in the process of budding compared to the WT virus. This is more dramatically seen in the scanning electron micrographs, wherein the entire surface of the mutant-infected cell is covered with such trapped virion particles. Based on these micrographs, it appears that VSV budding is not restricted to a specific region on the cell surface, at least under the experimental conditions used here.
Of all the mutants generated, the PPAY mutant gave the least severe phenotype; this mutant showed growth kinetics similar to that of the WT and gave virus yields about 50% of WT levels. This was not surprising, since the third proline residue is the least conserved in this motif among all the rhabdoviruses compared to date (17). While low levels of virions were obtained for all the other mutants (approximately 15 to 20% of WT levels), we found a more pronounced lag during early times of virus infection for mutants with tyrosine substitutions, either singly or in conjunction with the first or second proline (Y mutants). These results suggest that the tyrosine residue is particularly important for the function of this motif. A similar delay in the kinetics of budding was observed for the RSV AD2 mutants lacking the PPPYV sequence (45). A more severe phenotype was observed for the PY mutants of Mason-Pfizer monkey virus (M-PMV), wherein no virus particles were detected in the culture medium of cells infected with a tyrosine mutant (47). The differences in the severity of the phenotypes of the Y mutants in VSV and M-PMV indicate that the mechanism of budding for rhabdoviruses, though similar in many ways to those of retroviruses, has its own unique features.
Although disrupting the motif caused most of the virus particles to be trapped at the cell surface, it did not completely block the release of virus (20% of virus was still released from the cell). This leaky phenotype has been observed for some late domain mutants in RSV (except Y mutants) and M-PMV. The leakiness may be due to a spontaneous release of some particles from the cell surface. Alternatively, the amount of budding observed could also be attributed to redundant signals in M protein, as suggested earlier (17).
The ΔG-AAPA mutant, which lacks the glycoprotein in addition to having a mutated PPPY sequence, showed a drastically reduced virus yield, releasing less than 1% of WT levels. The electron micrographs of ΔG-AAPA mutant-infected cells showed very few particles at the plasma membrane; most of these seemed to be trapped in the process of pinching off from the cell surface (Fig. 5F). On the other hand, a ΔG-VSV virus that has an intact PPXY motif was rapidly released into the extracellular milieu, similarly to the WT virus (Fig. 5A and B). Thus it appears that in the absence of G protein, few virus particles are able to assemble at the cell surface, and when the absence of G protein is combined with the AAPA mutations, those particles that do assemble are not released. Based on these observations, we propose that G protein does not influence virion release per se but instead is involved at an earlier step in budding. The recent observation that a very short domain in the membrane proximal “stem” region of the ectodomain of VSV-G protein can confer efficient virus assembly and budding suggests that only a portion of G is sufficient for this event (36). Therefore, it will be interesting to determine if the stem region can rescue the ΔG-AAPA mutant and restore its phenotype to that of the AAPA mutant.
The polyproline sequence containing the core consensus of XPPXY was first identified as the main ligand for the WW domain, a protein interaction module found in a variety of cytoskeletal and regulatory proteins (2, 7, 8, 10, 19, 42). Subsequently, the identification of a conserved PPXY motif in several retroviruses (45, 47), and recently in the matrix proteins of rhabdoviruses (17), has led to a working hypothesis that the PPXY motif, or its functional analogues in the L domain of viruses, serves to bind cellular proteins containing WW domains, thereby recruiting them to the site of virus budding (12, 14). It was reported previously that amino acids 17 to 33 in VSV M protein, which includes the PY motif, mediate interaction with WW domains of specific cellular proteins, and point mutations in this motif significantly decreased this binding as measured in an in vitro far-Western blotting assay (17). Since the PPPY mutants have a leaky phenotype, it appears that there might be an alternative way of recruiting these cellular proteins to the budding site. Another possibility is that the mutations in the PPPY sequence did not completely abrogate binding to WW domains, but affected the affinity of binding. This possibility is supported by the data obtained from the solution structure of the WW domain of Yap65 complexed with a synthetic polyproline peptide containing the core motif PPXY (26). The mutants did not have an altered two-dimensional structure, but the binding affinities changed depending on the residue altered, with the tyrosine substitution having the most severe binding defect. Reduced binding affinity could therefore lead to recruitment of fewer molecules of host proteins, thus accounting for the low level of virus release in these mutants.
Our experiments also revealed that the disruption of the PPPY sequence affected the kinetics of host shutoff in the infected cells. The tyrosine mutants showed a delay in the inhibition of host gene expression, which was not relieved by infection at a higher multiplicity (Fig. 8C). This defect could reflect a general impairment of M protein function because of incorrect conformation caused by replacement of an aromatic residue (tyrosine) with a nonaromatic hydrophobic residue (alanine). However, cell rounding was unaffected, suggesting that it may not be the conformational change which is affecting the host shutoff ability. An alternative explanation could be that a specific interaction(s) between the M protein and the host factors resulting in the host shutoff might have been affected. The ability of M protein to inhibit RNA polymerase (RNAP) II-dependent transcription has been well recognized (1, 4, 13, 18, 32). It was also shown that M protein inhibits nuclear import of transcription factors in VSV-infected cells (18). This effect is separate from the inhibition of host transcription by inactivation of TFIID, since the levels of TFIID in the nuclear extracts of infected cells remain unchanged (48). The inhibition of TFIID activity is proposed to be an indirect effect of M protein, since nuclear extracts from VSV-infected cells as well as purified M protein failed to cause a reproducible inhibition of TFIID activity (48). It has been demonstrated that the function of M protein in viral assembly is genetically separable from its role in the inhibition of host-directed expression (5). This suggests that there might be different domains within M protein which mediate these different functions. Although the activity of viral assembly has been largely associated with N-terminal sequences of the protein, the domains involved in viral cytopathogenesis have not yet been identified. The effect of PY mutations on the inhibition of host gene expression suggests that this motif might be involved in this activity of the protein. Sequence analysis shows that the RNAP II C-terminal domain (CTD) contains several PY motifs with a consensus “XSPXY”. It has been shown that the CTD of mammalian RNAP II binds efficiently to the YAP WW domains as well as to mNEDD4-WW2, the mammalian homologue of the ubiquitin ligase Rsp5 in a far-Western assay (15). Interestingly, the WW domains from YAP and Nedd4 also interacted strongly and specifically with the PY motifs of both VSV and rabies virus M proteins (17). This coincidental observation suggests that VSV M protein may indirectly associate with the CTD of RNAP II via one or more of these WW-containing proteins. Alternatively, the PPPY sequence in M protein may disrupt the interaction between the RNAP II CTD and YAP or Yap-like proteins, thereby interfering in one or more signaling pathways. The mutations in the PY motif may therefore affect such an action of M protein. However, the effect was not very drastic, suggesting again that there might be more than one region of M protein involved in its cytopathic role. The implication of such a potential association between M protein and RNAP II for the inhibition of host gene expression needs to be addressed in order to enhance our understanding of how M protein contributes to the different CPE during VSV infection.
ACKNOWLEDGMENTS
We greatly appreciate the technical assistance of Carolyn Matthews (UT Memphis) and Donna Davis (St. Jude Children's Research Hospital). We thank Tim Higgins for assistance in figure preparation. We also thank Clint Robison, Eswaraka Jeetendra, and Indrani Halder for helpful comments after reading the manuscript. Oligonucleotides were synthesized by the Molecular Resource Center (MRC) at the University of Tennessee at Memphis.
This work was supported by NIH grant GM-53726 to M.A.W.
REFERENCES
- 1.Ahmed M, Lyles D S. Identification of a consensus mutation in M protein of vesicular stomatitis virus from persistently infected cells that affects inhibition of host-directed gene expression. Virology. 1997;237:378–388. doi: 10.1006/viro.1997.8808. [DOI] [PubMed] [Google Scholar]
- 2.Andre B, Springael J Y. WWP, a new amino acid motif present in single or multiple copies in various proteins including dystrophin and the SH3-binding Yes-associated protein YAP65. Biochem Biophys Res Commun. 1994;205:1201–1205. doi: 10.1006/bbrc.1994.2793. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann J E, Fusco P J. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J Cell Biol. 1988;107:1707–1715. doi: 10.1083/jcb.107.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black B L, Lyles D S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black B L, Rhodes R B, McKenzie M, Lyles D S. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J Virol. 1993;67:4814–4821. doi: 10.1128/jvi.67.8.4814-4821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blondel D, Harmison G G, Schubert M. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J Virol. 1990;64:1716–1725. doi: 10.1128/jvi.64.4.1716-1725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 8.Chan D C, Bedford M T, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H I, Einbond A, Kwak S J, Linn H, Koepf E, Peterson S, Kelly J W, Sudol M. Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 10.Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulon P, Deutsch V, Lafay F, Martinet Edelist C, Wyers F, Herman R C, Flamand A. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J Gen Virol. 1990;71:991–996. doi: 10.1099/0022-1317-71-4-991. [DOI] [PubMed] [Google Scholar]
- 12.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferran M C, Lucas Lenard J M. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 15.Gavva N R, Gavva R, Ermekova K, Sudol M, Shen C J. Interaction of WW domains with hematopoietic transcription factor p45/NF-E2 and RNA polymerase II. J Biol Chem. 1997;272:24105–24108. doi: 10.1074/jbc.272.39.24105. [DOI] [PubMed] [Google Scholar]
- 16.Gill D S, Banerjee A K. Complete nucleotide sequence of the matrix protein mRNA of vesicular stomatitis virus (New Jersey serotype) Virology. 1986;150:308–312. doi: 10.1016/0042-6822(86)90293-x. [DOI] [PubMed] [Google Scholar]
- 17.Harty R N, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Her L S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Bucher P. The rsp5-domain is shared by proteins of diverse functions. FEBS Lett. 1995;358:153–157. doi: 10.1016/0014-5793(94)01415-w. [DOI] [PubMed] [Google Scholar]
- 20.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretzschmar E, Peluso R, Schnell M J, Whitt M A, Rose J K. Normal replication of vesicular stomatitis virus without C proteins. Virology. 1996;216:309–316. doi: 10.1006/viro.1996.0066. [DOI] [PubMed] [Google Scholar]
- 22.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenard J, Vanderoef R. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J Virol. 1990;64:3486–3491. doi: 10.1128/jvi.64.7.3486-3491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyles D S, McKenzie M O. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology. 1997;229:77–89. doi: 10.1006/viro.1996.8415. [DOI] [PubMed] [Google Scholar]
- 25.Lyles D S, McKenzie M O, Kaptur P E, Grant K W, Jerome W G. Complementation of M gene mutants of vesicular stomatitis virus by plasmid-derived M protein converts spherical extracellular particles into native bullet shapes. Virology. 1996;217:76–87. doi: 10.1006/viro.1996.0095. [DOI] [PubMed] [Google Scholar]
- 26.Macias M J, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 27.McCreedy B J, Jr, Lyles D S. Distribution of M protein and nucleocapsid protein of vesicular stomatitis virus in infected cell plasma membranes. Virus Res. 1989;14:189–205. doi: 10.1016/0168-1702(89)90001-4. [DOI] [PubMed] [Google Scholar]
- 28.Mebatsion T, Konig M, Conzelmann K K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 29.Murti K G, Brown P S, Kumagai M, Campana D. Molecular interactions between human B-cell progenitors and the bone marrow microenvironment. Exp Cell Res. 1996;226:47–58. doi: 10.1006/excr.1996.0201. [DOI] [PubMed] [Google Scholar]
- 30.Newcomb W W, Brown J C. Role of the vesicular stomatitis virus matrix protein in maintaining the viral nucleocapsid in the condensed form found in native virions. J Virol. 1981;39:295–299. doi: 10.1128/jvi.39.1.295-299.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb W W, Tobin G J, McGowan J J, Brown J C. In vitro reassembly of vesicular stomatitis virus skeletons. J Virol. 1982;41:1055–1062. doi: 10.1128/jvi.41.3.1055-1062.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paik S Y, Banerjea A C, Harmison G G, Chen C J, Schubert M. Inducible and conditional inhibition of human immunodeficiency virus proviral expression by vesicular stomatitis virus matrix protein. J Virol. 1995;69:3529–3537. doi: 10.1128/jvi.69.6.3529-3537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parent L J, Wilson C B, Resh M D, Wills J W. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J Virol. 1996;70:1016–1026. doi: 10.1128/jvi.70.2.1016-1026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 35.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robison C S, Whitt M A. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J Virol. 2000;74:2239–2246. doi: 10.1128/jvi.74.5.2239-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 38.Simon K O, Whitaker-Dowling P A, Youngner J S, Widnell C C. Sequential disassembly of the cytoskeleton in BHK21 cells infected with vesicular stomatitis virus. Virology. 1990;177:289–297. doi: 10.1016/0042-6822(90)90482-7. [DOI] [PubMed] [Google Scholar]
- 39.Stillman E A, Rose J K, Whitt M A. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J Virol. 1995;69:2946–2953. doi: 10.1128/jvi.69.5.2946-2953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stillman E A, Whitt M A. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73:7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 43.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitt M A, Buonocore L, Rose J K, Ciccarone V, Chytil A, Gebeyehu G. TransfectACE reagent: transient transfection frequencies greater than 90% Focus. 1991;13:8–12. [Google Scholar]
- 45.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan H, Yoza B K, Lyles D S. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology. 1998;251:383–392. doi: 10.1006/viro.1998.9413. [DOI] [PubMed] [Google Scholar]