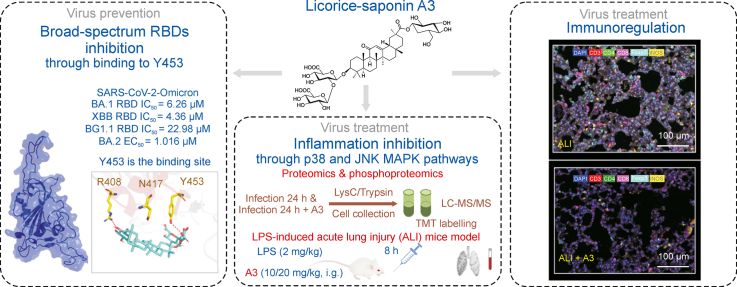
Abstract
Currently, human health due to corona virus disease 2019 (COVID-19) pandemic has been seriously threatened. The coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein plays a crucial role in virus transmission and several S-based therapeutic approaches have been approved for the treatment of COVID-19. However, the efficacy is compromised by the SARS-CoV-2 evolvement and mutation. Here we report the SARS-CoV-2 S protein receptor-binding domain (RBD) inhibitor licorice-saponin A3 (A3) could widely inhibit RBD of SARS-CoV-2 variants, including Beta, Delta, and Omicron BA.1, XBB and BQ1.1. Furthermore, A3 could potently inhibit SARS-CoV-2 Omicron virus in Vero E6 cells, with EC50 of 1.016 μM. The mechanism was related to binding with Y453 of RBD determined by hydrogen-deuterium exchange mass spectrometry (HDX-MS) analysis combined with quantum mechanics/molecular mechanics (QM/MM) simulations. Interestingly, phosphoproteomics analysis and multi fluorescent immunohistochemistry (mIHC) respectively indicated that A3 also inhibits host inflammation by directly modulating the JNK and p38 mitogen-activated protein kinase (MAPK) pathways and rebalancing the corresponding immune dysregulation. This work supports A3 as a promising broad-spectrum small molecule drug candidate for COVID-19.
Keywords: COVID-19, Inflammation, Licorice-saponin A3, Receptor-binding domain (RBD), SARS-CoV-2
Graphical abstract
Licorice-saponin A3 (A3) from the traditional Chinese herbal medicine licorice could widely inhibit RBDs of SARS-CoV-2 Beta, Delta and Omicron variants. Moreover, A3 can directly inhibit host inflammation by modulating the JNK and p38 MAPK pathways and rebalancing the corresponding immune dysregulation.
Highlights
-
•
Licorice-saponin A3 inhibits receptor-binding domains (RBDs) of SARS-CoV-2 variants BA.1/XBB/BQ1.1.
-
•
Y453 of Omicron BA.1 RBD is a critical binding site for the inhibitory activity of A3.
-
•
A3 exhibits anti-inflammatory activities through p38 and JNK MAPK pathways.
-
•
A3 could rebalance inflammation-induced immune dysregulation.
1. Introduction
Currently, 6.87 million deaths of corona virus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been reported till March 2023 [1,2]. While many vaccines have been developed for clinical use [3,4], new waves of infections due to SARS-CoV-2 variants have been challenged. These variants may ablate antibody binding and neutralization, resulting in diminished efficacy of antibodies derived from natural infections or vaccinations [5].
The spike (S) protein receptor-binding domain (RBD) of SARS-CoV-2 is a significant target to develop antiviral therapies [6,7]. Given the crucial role of RBD (aa 319–541) in recognizing the host cell receptor angiotensin converting enzyme 2 (ACE2), mutations may remarkably affect its binding to ACE2 [8], thus decreasing the protective ability of current vaccines and create a huge challenge in inhibiting diverse SARS-CoV-2 variants [9]. The first variant with an S mutation of D614G appeared in Germany and showed enhanced infectivity [10]. A number of SARS-CoV-2 variants, identified as variants of concern (VOCs), have been reported afterwards. First, Alpha (B.1.1.7) reported in September 2020 in Southeastern England spread quickly in 160 countries as of June 2021 [11]. In late 2020, Beta (1.351) and Gamma (P.1) both containing three mutations at RBD emerged respectively. Beta (1.351) has since spread to 113 countries with great concern about its immune escape [12,13]. After Delta (B.1.617.2) variant [14], a newly recognized variant Omicron (B.1.1.529), containing 26–32 aa mutations in the S protein, is the predominant variant globally with several sub-variants merging [15]. Compared to Delta, Omicron is more destructive due to the fast spread rate, immune escape potential, and high transmissibility.
Although antiviral therapies targeting S protein have been developed, various studies showed that neutralization efficiency of antibodies against SARS-CoV-2 VOCs was less effective [[16], [17], [18]]. Beta and Gamma variants with mutations K417 N/T, E484K, and N501Y at RBD decreased the neutralization capacity of antibodies activated by infection with vaccination, and showed resistance to neutralization by N-terminal domain-specific monoclonal antibodies [17,19]. The Delta variant with L452R mutation causes structural changes of RBD and promotes the interaction with ACE2, and has over 60% more transmissibility than Alpha. Recently, Omicron and its sub-variants carrying over 15 mutations in RBD are spreading rapidly in countries with high levels of population immunity, which demonstrates the limited antiviral potency of vaccination [20]. Thus, it will be of huge benefit to develop effective and broad-spectrum RBD inhibitors to suppress SARS-CoV-2 variants infection.
Thus far, several attractive therapies have been reported to block SARS-CoV-2 viral entry. For instance, the peptidomimetic compound N-0385, which abrogates the proteolytic activity of TMPRSS2, could inhibit the entry of SARS-CoV-2 variants into Calu-3 cells [21]. A novel fusion inhibitor of 5-Helix could target the S2 subunit HR2 domain of S protein to inhibit infection by pseudotyped SARS-CoV-2 and variants Delta and Omicron [22,23]. However, no potent broad-spectrum small molecule inhibitors for SARS-CoV-2 variants have been reported.
In addition to the direct coronavirus-induced injury to the respiratory system, more evidence reported the hyperinflammatory reaction and immune response triggered by SARS-CoV-2 infection. Several studies have shown that inflammation-related signaling pathways, as well as cytokines can be affected by SARS-CoV-2 and thus induce hyper inflammatory reactions [[24], [25], [26]]. The number and function of various subtypes of immune cells are severely affected in patients with COVID-19, which are associated with disease severity and can lead to severe complications [27]. These evidences suggest that regulation of host inflammatory levels and immune response may be effective strategies for COVID-19 treatment.
Herein, we report the natural product licorice-saponin A3 (A3) and glycyrrhetinic acid (GA) could widely inhibit RBDs of SARS-CoV-2 Beta, Delta, and Omicron variants. GA showed inhibitory activities against Omicron on Vero E6 cells with an inhibition rate of 26.5% at 1 μM, and A3 showed an EC50 of 1.016 μM. The antiviral mechanisms were elucidated by theoretical calculations and experiments. The anti-inflammatory activities of A3 were also evaluated.
2. Materials and methods
2.1. Cells and animals
Vero E6 cells (Vero C1008; CRL 1586, American Type Culture Collection, Manassas, VA, USA) were stored in high glucose Dulbecco's modified Eagle medium (DMEM) (GIBCO, Thermo Fisher, Waltham, MA, USA) with penicillin (100 IU/mL), 10% fetal bovine serum (FBS, GIBCO, Thermo Fisher) and streptomycin (100 μg/mL) at 37 °C in a 5% CO2 environment. SARS-CoV-2 RBDs including prototype, Beta, Delta and Omicron BA.1/XBB/BQ1.1 were provided by Prof. George Fu Gao (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China). ACE2 was from Novoprotein Technology Co., Ltd (Shanghai, China). SARS-CoV-2 Omicron BA.2 (CSTR:16,533.06IVCAS6.7600) was propagated in Vero E6 and titrated by standard plaque assay following the standard procedure. All experiments related to authentic SARS-CoV-2 were conducted at the Zhengdian Biosafety Level 3 (BSL3) facility of Wuhan Institute of Virology, China. GA (CAS No. 471-53-4, HPLC≥98%) and A3 (CAS No. 118325-22-7, HPLC≥98%) were obtained from a compound library of the authors’ laboratory [28]. PJ-34 (CAS No. 344458-19-1, HPLC≥98%) was purchased from Aladdin (Shanghai, China). The protocol for animal experiments was approved by the Animal Care and Use Committee of Peking University Health Science Center (Approval number: LA2021537).
2.2. Surface plasmon resonance (SPR) assay
Biacore 8 K was performed for SPR study. RBDs were immobilized on the CM5 sensor chip, and the running buffer was PBS with 0.05% P20 and 5% DMSO (D8371, Solarbio, Beijing, China). RBD proteins were then diluted with sodium acetate solution (10 mM, pH 5.5) to a final concentration of 20 μg/mL. The final immobilized levels for RBDs were ∼10,000 RU (response units). In binding studies, compounds were used at calculated concentrations, and flowed with buffer (30 μL/min). Both the contact time and dissociation time were set to 60 s. Biacore evaluation software was used to analyze the data. The binding affinity of ligand-protein was evaluated with the equilibrium dissociation constants (KD). Kinetics analysis and steady state affinity fitting method were used.
2.3. Enzyme linked immunosorbent assay (ELISA)
The inhibition of drug candidates against SARS-CoV-2 RBDs was performed as reported in the literature [29]. Briefly, the S RBD proteins were diluted with coating liquid (NaHCO3, 2.93 mg/mL; Na2CO3, 1.59 mg/mL; pH 9.6) and incubated at 4 °C overnight. After washing with a wash solution (PBS with 0.05% Tween-20 (V/V)), the microplates were blocked with 2% bull serum albumin (BSA) in the wash solution at 37 °C for 40 min. After three times washing, the small molecule compounds were added at a final concentration of 0.625, 1.25, 2.5, 5, 10, 20, 40, and 80 μM (1 h before ACE2). Then, ACE2 was added and incubated for 30 min before being washed off. Enzyme-linked antibody SA-HRP (1:10,000) was subsequently conducted and tetramethyl benzidine was chosen as the chromogenic agent. Finally, the reaction was stopped with 1 M HCl. At OD450 nm, the enzyme activity was measured.
2.4. Pseudovirus-based neutralization assay
In pseudovirus-based neutralization assay [29], Beta, Delta and Omicron BA.1 pseudoviruses were provided by Prof. George Fu Gao (Institute of Microbiology, Chinese Academy of Sciences, China). Vero E6 cells were seeded at a density of 20,000 cells/well in 96-well plates. A3 and GA at different concentrations were respectively added with diluted VSV-SARS-CoV-2-EGFP virus (MOI = 0.5 PFU/mL). The mixture was cultivated at 37 °C for 1 h and then added into seeded Vero E6 cells. After 36 h of incubation, fluorescence images were measured by a CQ1 confocal image cytometer (Yokogawa, Tokyo, Japan) with a × 10 objective, and analyzed by a Harmony high-content analysis software (PerkinElmer, Akoya, MA, USA).
2.5. Antiviral assay
After being cultured with gradient concentrations of drugs overnight, Vero E6 cells in 48-well plates were infected by SARS-CoV-2 Omicron BA.2 (MOI = 0.01). After 24 h of infection, RNA was extracted from the cell supernatant using QIAamp viral RNA mini kit (52,906, Qiagen, Duesseldorf, Germany). The viral RNA was quantified by quantitative real time polymerase chain reaction (qRT-PCR) with a reported primer pair [29].
2.6. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) analysis
RBD proteins (1 mg/mL), and RBD proteins (1 mg/mL) with GA/A3 (1 mM) were respectively deuterium labeled with 100 mM phosphate buffer (D2O, pD 7.0, 20-fold dilution). After 0.083, 0.25, 1, 10, 30, 60, and 240 min, the quenching buffer (100 mM phosphate, 0.5 M tris(2-carboxyethyl)phosphine (TCEP), 4 M GdHCl, pH 2.0) was added to stop the labeling reaction. Samples were then injected and digested online on Waters ENZYMATE BEH pepsin columns (2.1 mm × 30 mm, 5 μm; Milford, MA, USA). The peptides were trapped and desalted on a VanGuard Pre-Column trap (ACQUITY UPLC BEH C18, 1.7 μm) for 3 min, eluted from the trap performing 15% acetonitrile at 100 μL/min, then separated by an ACQUITY UPLC BEH C18 column (1.0 mm × 100 mm, 1.7 μm, Waters). All mass spectra were acquired on a mass spectrometer (Waters Xevo G2). Peptides from an unlabeled protein were identified by applying ProteinLynx Global Server (PLGS) searches of a protein database including RBD sequences only. Peptides in relative deuterium levels were measured by subtracting the mass of the undeuterated control sample from that of the deuterium-labeled sample [30].
2.7. Proteomics and phosphoproteomics analysis
Vero E6 cells were incubated with or without 10 μM A3 for 8 h, and subsequently infected with SARS-CoV-2 for 24 h (MOI = 1). A high intensity ultrasonic processor (Scientz, Ningbo, China) was used to sonicate the samples in lysis buffer (8 mM urea, 1% protease inhibitor cocktail). The first digestion was performed overnight with a 1:50 trypsin-to-protein mass ratio. Tandem mass tags (TMT) reagents (Thermo Fisher Scientific) were used to label each channel of peptide, and each channel was incubated for 2 h at room temperature. We desalted the pooled samples using a Strata X C18 SPE column (Phenomenex, Torrance, CA, USA) and dried the samples by vacuum centrifugation. Solvent A (0.1% formic acid and 2% acetonitrile in water) was used to dissolve tryptic peptides, which were directly loaded onto a reversed-phase analytical column (25 cm length, 75 μm i. d.). Using solvent B (0.1% formic acid in 90% acetonitrile), peptides were separated from 5% to 25% over 60 min, 25%–35% in 22 min, up to 80% in 4 min, and then held at 80% for 4 min. The UPLC system (ThermoFisher Scientific) operated at a constant flow rate of 450 nL/min. Then the separated peptides were characterized by a Q Exactive™ HF-X mass spectrometer (ThermoFisher Scientific) with a nano-electrospray ion source [30].
2.8. Molecular docking and quantum mechanics/molecular mechanics (QM/MM) simulations
Omicron S protein (PDB ID: 7QTK) and SARS-CoV-2 S protein (PDB ID: 6M0J) were used as templates [31,32]. The structures of A3 and GA were optimized using density functional theory (DFT) at B3LYP/6-311G(d) [[33], [34], [35]]. Molecular docking of molecules and RBD was carried out by AutoDock Vina and AutoDocktools v1.5.6 software [[36], [37], [38]]. Furthermore, QM/MM simulations were applied by Gaussian16 to optimize the ligand and residues (400–422 and 453–470) from the docking model. The B3LYP/6-311G in DFT calculations was used to describe the QM atoms from A3 and Y453/Y453F, and the UFF force field was calculated to simulate the MM atoms [39].
2.9. Western blotting
Proteomics samples were determined using BCA Protein Content Assay Kit (AKPR017 Boxbio, Beijing, China). After boiling for 5 min, 20 μg of each sample was added to 10% sodium dodecyl sulfate (SDS)-polyacrylamide minigels (EpiZyme, Shanghai, China) and electrophoresed at 80 V (stacking gel) and 120 V (separating gel). A polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA) was then used to transfer proteins out of the gel, and was blocked for 1 h at room temperature in the blocking buffer (Beyotime, Shanghai, China). Primary and secondary antibodies were then incubated overnight at 4 °C on the membrane (nuclear factor-kappa-B (NF-κB) (AF5006, Affinity, Liyang, Jiangsu, China), 1:1000; PAK1/2/3 (2604 T, Cell Signaling, Boston, MA, USA), 1:1000; p38α (9218 T, Cell Signaling), 1:1000; c-Jun (BF0245, Affinity), 1:1000; JNK2 (DF2357, Affinity), 1:1000; STAT1 (AF6300, Affinity), 1:1000; MEK4 (4514 T, Cell Signaling), 1:1000). Signals were imaged by Bio-Rad ChemiDoc XRS. GAPDH was the loading control.
2.10. Lipopolysaccharide (LPS)-induced mice acute lung injury (ALI) model, pathological analysis, and pharmacokinetic study
Male BALB/c mice (20 g) were obtained from the Experimental Animal Center of Peking University Health Science Center (Beijing, China). Mice in the control, model, and A3 groups were treated with saline solution, LPS (2 mg/kg, i. n.), and LPS (2 mg/kg, i. n.) with A3 (10 and 20 mg/kg, i. g.), respectively. After 8 h, blood and lung tissue samples were collected. Small pieces of lung tissues of ALI model were fixed in 4% paraformaldehyde. After dehydration of the specimens in graded ethanol and embedding in paraplast, the sections were cut at 5-μm thickness. Then, hematoxylin and eosin (H&E) staining was performed to demonstrate the histological structures. Images were taken by WISLEAP (WS-10, Changzhou, Jiangsu, China).
For pharmacokinetic study, A3 (20 mg/kg) was given to BALB/c mice (20 g) by intragastric administration. Blood samples were collected at 1, 4, 8, 10, 12, 24, and 48 h, respectively, and centrifuged at 15,000 rpm (4 °C) for 30 min to obtain the plasma. Then 100 μL of plasma sample, 200 μL of methanol, and 100 μL internal standard solution (5 mg/mL astragaloside IV in methanol) were mixed. The mixture was vortexed and then centrifuged at 15,000 rpm for 30 min. The supernatant was separated, dried, and then dissolved in 100 μL of 50% methanol. The sample was centrifuged (15,000 rpm for 30 min), and then analyzed by Q-Exactive hybrid quadrupole Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA, USA) equipped with a heated electrospray ionization source (HESI). The conditions were as follows: negative ion mode; spray voltage, −3.5 kV; sheath gas flow rate, 45 arb; aux gas flow rate, 10 arb; capillary temperature, 350 °C; aux gas heater temperature, 4000 °C; and S-lens RF level, 60 V. Samples were separated on a Waters Acquity UPLC BEH C18 column (1.7 μm × 150 mm) (Waters). The mobile phase consisted of water containing 0.1% formic acid (V/V, A) and acetonitrile containing 0.1% formic acid (V/V, B). The gradient elution program was as follows: 0–5 min, 10%–30%; 5–8 min, 30%–50% B; 8–15 min, 50%–100% B; 15–18 min, 100%–10% B. The flow rate was 0.3 mL/min. The injection volume was 2 μL. A3 and astragaloside IV were determined by multiple reaction monitoring (MRM) in the negative ion mode, with MRM transitions of m/z 983.5 → 821.3 and 783.4 → 651.4, respectively.
2.11. Immunohistochemistry (IHC)
We applied the same lung tissues of ALI model at 5 μm for IHC. Following 10 min of incubation with 3% H2O2, the samples were incubated in citrate buffer (pH 8.0) for 3 min at high heat and 15 min at low heat for antigen retrieval. Sections were incubated with primary antibodies for 2 h after blocking with goat serum (tumour necrosis factor alpha (TNF-α) (Abcam, Cambridge, United Kingdom), 1:20; interleukin-6 (IL-6) (Abcam), 1:50; interleukin-1β (IL-1β) (Abcam), 1:250, interleukin-7 (IL-7) (Thermo Fisher), 1:100). Phosphate-buffered saline was used to wash the sections three times and anti-horseradish peroxidase (HRP) goat anti-rabbit IgG was used for incubation (PV-6001, ZSGB-BIO, Beijing, China). 3,3'-diaminobenzidine (DAB) was incubated for 2 min. The sections were then stained with hematoxylin after the reaction was stopped. Images were taken using WISLEAP (WS-10).
2.12. Multi fluorescent IHC (mIHC)
The 5-μm sections obtained from the lung tissues of ALI model were incubated in citrate buffer (pH 6.0, CD3 (Abcam), Foxp3 (Abcam) and iNOS (Affinity)) or ethylenediamine tetraacetic acid (EDTA) (pH 9.0, CD4 (Abcam) and CD8 (Abcam)) for 3 min high heat, and 15 min low heat (microwave oven). There were five sequential rounds of staining for each section, including a protein block with goat serum, a primary antibody (CD3, 1:200; CD4, 1:125; CD8, 1:75; iNOS, 1:75 and Foxp3, 1:375), and a secondary goat anti-rabbit IgG HRP stain. By using tyramide signal amplification (CD3, fluorescein isothiocyanate (FITC); CD4, Texas; CD8, CY5; iNOS, CY3 and Foxp3, Texas), each horseradish peroxidase-conjugated polymer covalently bound a different fluorophore. To remove bound antibodies before the next step in the sequence, additional antigen retrieval using heated citrate buffer (pH 6.0, CD3, Foxp3 and iNOS) or EDTA (pH 9.0, CD4 and CD8) was performed after this covalent reaction. The sections were counterstained with 4',6-diamidino-2-phenylindole (DAPI) (AlphaTSA, Beijing, China) and mounted with fluorescence mounting medium after completing all six sequential reactions. Vectra Polaris Imaging System version (PerkinElmer, Waltham, MA, USA) was used to image multiplex stained slides. Tissue sections stained with seven-plex fluorescence were scanned using the ×40 objective, with every 10 nm of the emission light spectrum across the range of each emission filter cube. Filter cubes used for multispectral imaging were DAPI (440–680 nm), FITC (520–680 nm), Cy3 (570–690 nm), Texas red (580–700 nm) and Cy5 (670–720 nm). A spectral library containing the emitting spectral peaks of all fluorophores was created using multispectral images obtained from single stained slides for each marker and associated fluorophore. This spectral library was then used to separate each multispectral image cube into its individual components (spectral unmixing) allowing for the color-based identification of all markers of interest in a single image using the inForm 2.6 image analysis software (Akoya, MA, USA). All spectrally unmixed and segmented images were subsequently subjected to a proprietary inForm active learning phenotyping algorithm, allowing for the phenotyping and subsequent quantifications of different cell phenotypes.
2.13. Limitations of the study
The complex crystal structure of SARS-CoV-2 RBD with A3 needs to be solved to further interpret the inhibition mechanisms. Moreover, SARS-CoV-2 infected animals treated with A3 should be investigated in future studies to confirm the anti-virus activity of A3 and further explain the anti-inflammatory capability of A3 in treating SARS-CoV-2. The anti-inflammation and immune regulation efficacy of A3 administered at different time points or with different doses in ALI and acute respiratory distress syndrome (ARDS) models are of vital importance in evaluating its druggability, which should also be studied in the future.
Moreover, it is of vital importance to construct different types of inflammation animal models to confirm the anti-inflammatory activity of A3. Analysis of more detailed subtypes of immune cells will also be helpful in explaining the immune regulation capability of A3, as well as interpreting the correlation between the immune regulation and inflammation inhibition.
3. Results and discussion
3.1. Inhibitory activities of A3 and GA against RBDs of SARS-CoV-2
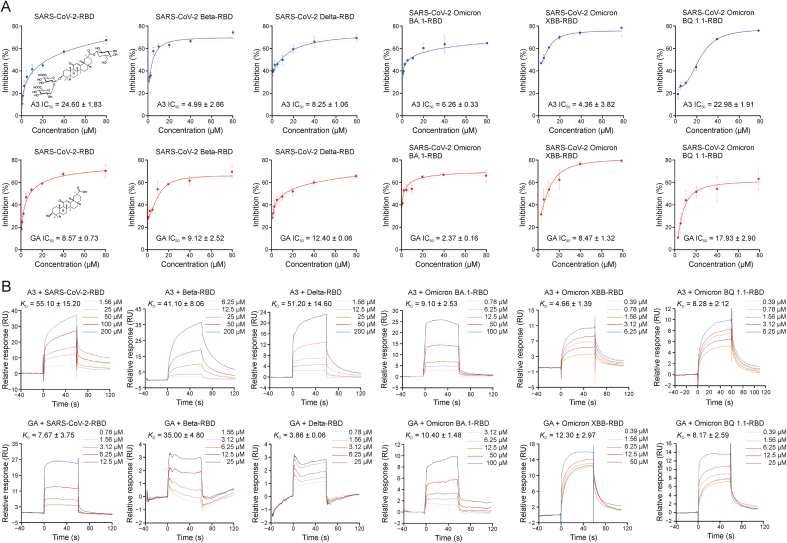
Potent inhibitory activities of A3 and GA against the prototype SARS-CoV-2 RBD have been reported [29]. Herein, we showed that A3 and GA could inhibit RBDs of SARS-CoV-2 variants Beta, Delta, and Omicron BA.1/XBB/BQ1.1 evaluated by measuring the binding capacity between A3/GA and RBDs of the three SARS-CoV-2 variants by ELISA, respectively. Both A3 and GA exhibited strong inhibitory activities against the RBDs of Beta (A3, IC50 = 4.99 ± 2.86 μM; GA, IC50 = 9.12 ± 2.52 μM), Delta (A3, IC50 = 8.25 ± 1.06 μM; GA, IC50 = 12.4 ± 0.06 μM), Omicron BA.1 (A3, IC50 = 6.26 ± 0.33 μM; GA, IC50 = 2.37 ± 0.16 μM), Omicron XBB (A3, IC50 = 4.36 ± 3.82 μM; GA, IC50 = 8.47 ± 1.32 μM), and Omicron BQ1.1 (A3, IC50 = 22.98 ± 1.91 μM; GA, IC50 = 17.93 ± 2.9 μM) (Fig. 1A). Subsequently, the binding of A3 and GA with RBDs of Beta, Delta and Omicron variants were determined by SPR, respectively. Not surprisingly, both GA and A3 showed binding capabilities with RBDs of SARS-CoV-2 and the five variants. Particularly, A3 presented increased binding affinity to Omicron variants, with KD values of 9.1, 4.66, and 8.28 μM, respectively (Fig. 1B).
Fig. 1.
Enzyme linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR) assays of licorice-saponin A3 (A3) and glycyrrhetinic acid (GA) binding with S receptor-binding domains (RBDs) of different severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants. (A) Inhibitory activities of A3 (upper) or GA (lower) against the S RBD of SARS-CoV-2 and its variants Beta, Delta, and Omicron BA.1/XBB/BQ1.1 by ELISA. RBD proteins of SARS-CoV-2 (0.2 μg/mL), Beta (0.3 μg/mL), Delta (0.2 μg/mL) and Omicron BA.1/XBB/BQ1.1 (0.5 μg/mL) were respectively coated onto 96-well microplates. After incubation with A3 or GA for 1 h, ACE2 at the same concentration as RBD protein was added and incubated with the S protein for 30 min. (B) SPR analysis of A3 (upper) or GA (lower) binding to RBDs of SARS-CoV-2, or SARS-CoV-2 variants (Beta, Delta, and Omicron BA.1/XBB/BQ1.1).
3.2. Weak inhibitory activities of A3 and GA against SARS-CoV RBD
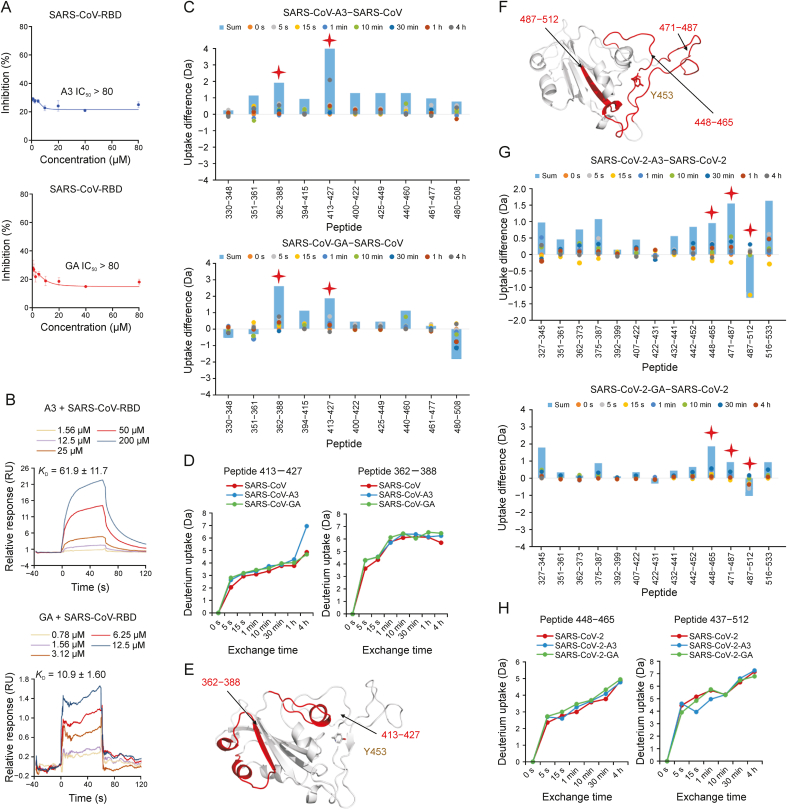
SARS-CoV is another human coronavirus, and possesses a highly similar gene sequence to SARS-CoV-2 [40]. Most proteins of these two viruses share high homology (95%–100%) [41]. However, there is only 60% similarity between the RBD sequences of SARS-CoV-2 and SARS-CoV (Fig. S1). Considering the abundant licorice compound glycyrrhizic acid (GA-gg) had been reported as an active compound to inhibit replication of SARS-CoV in 2003 [42], we tested the inhibitory activities of A3, GA, and GA-gg, against SARS-CoV RBD. Consistently, GA-gg could inhibit SARS-CoV RBD with IC50 of 62.95 ± 7.02 μM (Fig. S2). However, both A3 and GA showed weak inhibitory activities against SARS-CoV RBD with IC50 > 80 μM (Fig. 2A), though SPR indicated A3 and GA could target SARS-CoV RBD with KD values of 61.9 and 10.9 μM, respectively (Fig. 2B).
Fig. 2.
Binding mechanisms of licorice-saponin A3 (A3) targeting S receptor-binding domains (RBDs) of severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2. (A) Inhibitory activities of A3 (upper) and glycyrrhetinic acid (GA) (lower) against the S RBD of SARS-CoV, performed by enzyme linked immunosorbent assay (ELISA). RBD protein of SARS-CoV (0.3 μg/mL) was coated onto 96-well microplates. After incubation with test drugs (1 h), angiotensin converting enzyme 2 (ACE2) at the same concentration as RBD protein was added and incubated with the S protein for 30 min. (B) Surface plasmon resonance (SPR) analysis of A3 (upper) and GA (lower) binding to SARS-CoV RBD. (C) Deuterium uptake differences of SARS-CoV RBD with A3 (upper) and GA (lower), respectively. Sum is total uptakes from 0 s to 4 h. (D) Deuterium uptake plots of two peptides 413–427 and 362–388 of SARS-CoV RBD with A3 and GA. (E) Location of peptides 413–427 and 362–388 in SARS-CoV S RBD. (F) Location of peptides 448–465, 471–487, and 487–512 in SARS-CoV-2 S RBD. (G) Deuterium uptake differences of SARS-CoV-2 RBD with A3 (upper) and GA (lower), respectively. Sum is total uptakes from 0 s to 4 h. (H) Deuterium uptake plots of two peptides 448–465 and 487–512 of SARS-CoV-2 RBD with A3 and GA.
To elucidate the binding mechanisms between A3/GA and SARS-CoV RBD, we employed HDX-MS to determine protein conformation changes. Deuterium uptakes of A3 and GA with SARS-CoV RBD increased for majority of the peptides from 330 to 503 (Fig. 2C), demonstrating that uptake of the ligand-protein structures was slower than the individual SARS-CoV protein. It is noteworthy that deuterium uptakes of peptides 362–388 and 413–427 increased remarkably, indicating that binding residues of A3 and GA with SARS-CoV RBD may locate in these two peptides (Figs. 2C and D). Y453 had been considered as a crucial residue for inhibitory activity of A3 and GA against SARS-CoV-2 RBD (Fig. 2E) [29]. As Y453 is not located within these two peptides, this may be the reason why A3 and GA could bind to SARS-CoV RBD but showed weak inhibitory activities.
3.3. A3 inhibits SARS-CoV-2 Omicron variants through binding site Y453 of RBD
We had reported A3 and GA could potently inhibit SARS-CoV-2 in Vero E6 cells with EC50 of 75 nM and 3.17 μM, respectively. One inhibitory mechanism was hydrogen bonding with Y453 of RBD [29]. To confirm the significance of Y453, we conducted HDX-MS analysis. The receptor-binding motif (RBM) of S protein to bind with ACE2 consists of residues 437 to 508. According to our experimental results, the uptake of peptides 448–512 which mainly located in the RBM showed obvious changes upon A3 or GA treatment (Figs. 2F and G). These changes may be caused by hydrogen binding of A3 or GA with Y453 (Fig. 2H) [29].
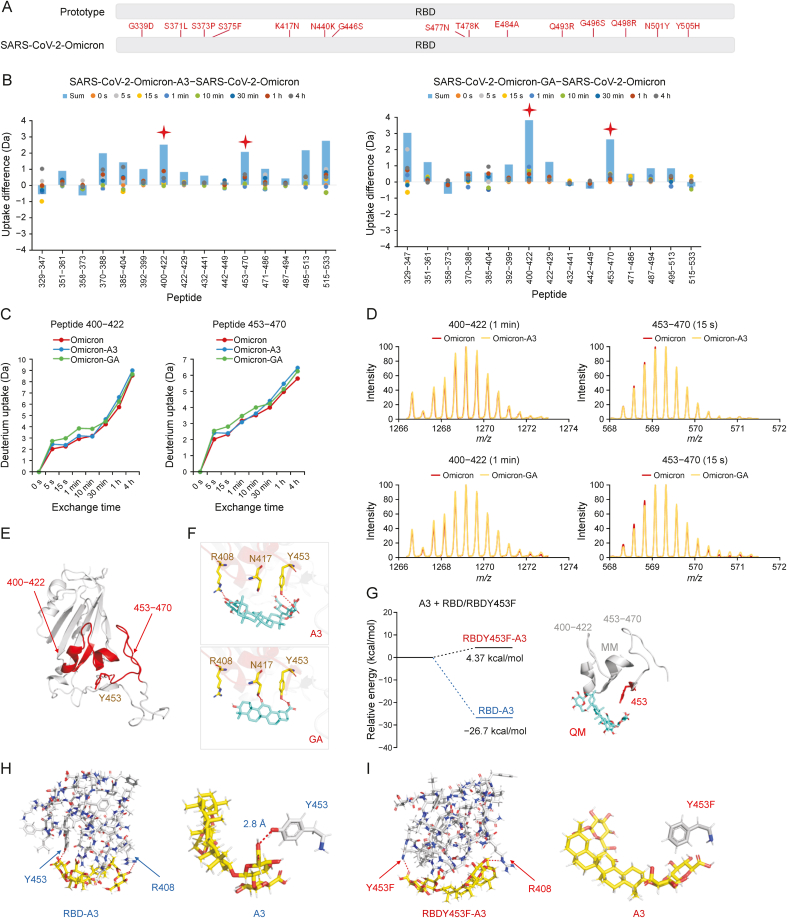
The SARS-CoV-2 Omicron BA.1 carries 15 mutations (Fig. 3A), but not including Y453. Both A3 and GA showed noticeable inhibitory activities against the RBD of Omicron (Fig. 1A). HDX-MS analysis indicated the uptake of peptide 453–470 with A3 and GA was remarkably increased (Figs. 3B−D). Subsequently, we used the structure of Omicron RBD (PDB ID: 7QTK) to simulate binding with A3. Two hydrogen bonds were respectively formed between A3 and Y453/R408, and between GA and Y453/N417 (Figs. 3E and F). A3 showed higher inhibitions against SARS-CoV-2 Beta, Delta, and Omicron BA.1 pseudoviruses than GA (Fig. S3). Particularly, A3 showed an EC50 of 1.016 μM against SARS-CoV-2 Omicron in Vero E6 cells (Fig. S4).
Fig. 3.
Binding mechanisms of licorice-saponin A3 (A3) and glycyrrhetinic acid (GA) targeting the S receptor-binding domain (RBD) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.1. (A) 15 mutations of Omicron S RBD compared to wild type SARS-CoV-2. (B) Deuterium uptake differences between Omicron RBD, and Omicron RBD with A3 or GA. Sum is total uptakes from 0 s to 4 h. (C) Deuterium uptake plots of peptides 400–422 and 453–470 of Omicron RBD, Omicron RBD with A3, and Omicron RBD with GA. (D) Mass spectra of peptides 400–422 and 453–470 of Omicron RBD and Omicron RBD with A3/GA. (E) Location of peptides 400–422 and 453–470 in Omicron RBD. (F) Molecular docking of A3 and GA with Omicron RBD, respectively. Hydrogen bonds (red dashes) are shown. (G) Relative energies of A3 with RBD (peptides 400–422 and 453–470), and A3 with RBDY453F (peptides 400–422 and 453–470) computed by quantum mechanics/molecular mechanics (QM/MM). (H,I) Binding modes of A3 with active residues of RBD (H) and RBDY453F (I), respectively. Hydrogen bonds (red dashes) are shown.
To confirm the significance of Y453 in the binding between A3 and Omicron RBD, we constructed the Y453F mutant. QM/MM calculations demonstrate the binding energy of A3 with peptide 453–470 of Omicron RBD was −26.7 kcal/mol, which was much lower than that with peptide 453–470 of the Omicron Y453F mutant (4.37 kcal/mol) (Fig. 3G), indicating a stable confirmation with Omicron. Moreover, the phenolic hydroxyl group of Y453 could form a hydrogen bond with –OH of C (6’)−OOH of A3 (2.8 Å), while no hydrogen bond could form for the Y453F mutant (Figs. 3H and I). These results confirmed that Y453 was a crucial site in the binding between A3 and Omicron RBD.
3.4. A3 regulates the mitogen-activated protein kinase (MAPK) signaling pathway of SARS-CoV-2 infected host cells
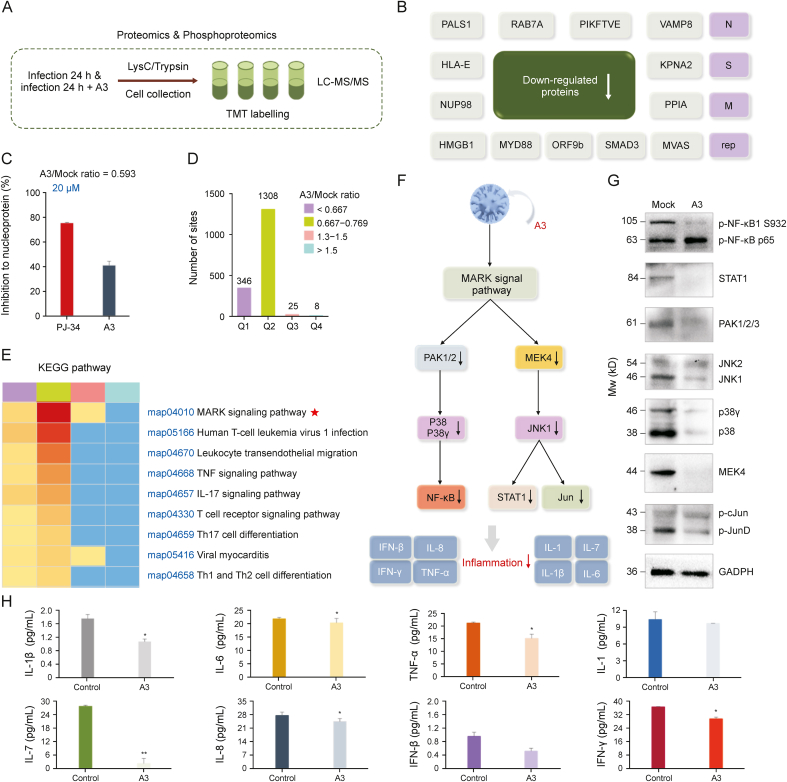
To explore the effects of A3 on SARS-CoV-2 infected host cells, we conducted proteomics analysis of Vero E6 cells infected by SARS-CoV-2 with the treatment of A3 (10 μM, pretreated for 8 h and infected for 24 h) (Fig. 4A). The expression levels of 1 protein in virus and 20 proteins in host cells were significantly altered after A3 treatment (Fig. S5A). We further conducted phosphoproteomic analysis, according to the significant cellular regulatory mechanism of protein phosphorylation [43]. The results showed 17 down-regulated phosphoproteins in virus, together with 1070 down-regulated and 31 up-regulated phosphoproteins in host cells (fold change >1.3) (Fig. S5B). Among the 17 down-regulated proteins, aside from the S protein, we identified two other structural proteins including the membrane protein (M) and the nucleoprotein (N), which are both targets for SARS-CoV-2 viral inhibition (Fig. 4B) [7]. Moreover, one popular functional protein of replicase polyprotein 1 ab (rep) was determined [44]. Subsequently, enrichment analysis of biological processes performed by gene ontology annotations (P value < 0.05) was conducted, and several biological activities of the proteins were listed (Figs. S6 and S7). In order to evaluate whether A3 could inhibit the down-regulated proteins, we measured the inhibitory activity of A3 against nucleoprotein whose quantity was decreased with an A3/mock ratio of 0.593 by ELISA. As a result, A3 could inhibit nucleoprotein by 41.1% at 20 μM (Fig. 4C). These data demonstrated that A3 could not only down-regulate the protein expression, but also inhibit their activities.
Fig. 4.
Phosphoproteomics profiling of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected cells treated with licorice-saponin A3 (A3). (A) Experimental scheme. Vero E6 cells were infected with SARS-CoV-2 (MOI = 1) in the presence or absence of 10 μM A3 for 24 h. Proteins were prepared using lysis buffer (8 M urea, 1% protease inhibitor cocktail) and trypsin. (B) Down-regulated viral proteins after A3 treatment (fold change >1.3), popularly reported proteins presented in pink color. (C) Inhibitory activity of A3 against nucleoprotein at 20 μM determined by enzyme linked immunosorbent assay (ELISA) kit (Abclonal, https://abclonal.com.cn/), and the positive control was PJ-34 [50]. (D) Four categories Q1–Q4 were divided by A3/mock ratio (<0.667, 0.667–0.769, 1.3–1.5 and > 1.5), and the numbers of their phosphoproteomic sites were presented. Mock group: Vero E6 cells infected by SARS-CoV-2 for 24 h; A3 group: Vero E6 cells infected by SARS-CoV-2 in the presence of A3 (10 μM) for 24 h, n = 3. (E) Enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for categories Q1–Q4. (F) Mitogen-activated protein kinase (MAPK) signaling pathway analysis to explain the anti-inflammatory mechanism of A3. (G) Expression of proteins participated in inflammation-related MAPK signaling pathway in SARS-CoV-2 infected Vero E6 cells before and after the treatment of A3. Mock group: Vero E6 cells infected by SARS-CoV-2 for 24 h; A3 group: Vero E6 cells infected by SARS-CoV-2 in the presence of A3 (10 μM) for 24 h, n = 3. (H) Interleukin (IL)-1, tumor necrosis factor (TNF)-α, IL-6, IL-1β, IL-8, IL-7, function of interferons (IFN)-β and IFN-γ of proteomic samples were measured by monkey ELISA kit (MEIMIAN, www.mmbio.cn). Vero E6 cells respectively treated with SARS-CoV-2 in the absence (control), and presence of A3 for 24 h. ∗P < 0.05, ∗∗P < 0.01, compared with the control group, n = 3. LC-MS/MS: liquid chromatography coupled to tandem mass spectrometry; TMT: tandem mass tags.
Next, phosphoproteins in the host cells regulated by A3 in phosphoproteomic analysis were studied. GO annotations indicated that 128 proteins were related to immune system (Fig. S8), and Kyoto Encyclopedia of Genes and Genomes functional enrichment analysis presented that A3 remarkably regulated inflammation through MAPK signaling pathway in four categories Q1–Q4 divided by A3/mock ratio (Figs. 4D and E).
Among the proteins related to anti-inflammation (Table S1), PAK, p38/p38γ, NF-κB, MEK4, JNK, STAT1 and Jun respectively participated in the c-Jun amino-terminal kinases 1/2/3 and p38 (α, β, γ, and δ) MAPK signaling pathways were obviously down-regulated. Thus, we deduced the anti-inflammatory activity of A3 was related to the JNK and p38 MAPK pathways (Fig. 4F). Consistently, Western blotting analysis confirmed that PAK1/2, p38 (α, β) and NF-κB in the p38 MAPK pathway, and MEK4, JNK, STAT1 and Jun in JNK1/2/3 were remarkably regulated (Figs. 4G and S9–S16). Because the expression of IL-6, TNF-α, and IL-1β could be suppressed by inhibiting NF-κB and JNK activation [26], the protein samples of Vero E6 cells treated with SARS-CoV-2 (control), and SARS-CoV-2 combined with A3 were respectively measured by ELISA [30]. As a result, cytokines including IL-1β, IL-6, TNF-α, IL-1, IL-7, IL-8, IFN-β, and IFN-γ were all decreased significantly upon A3 treatment (Fig. 4H). The above results indicated that A3 possessed anti-inflammatory activities of SARS-CoV-2 infected host cells through regulating key pro-inflammation proteins involved in the MAPK signaling pathway.
3.5. Host-directed anti-inflammatory activity of A3 in vivo
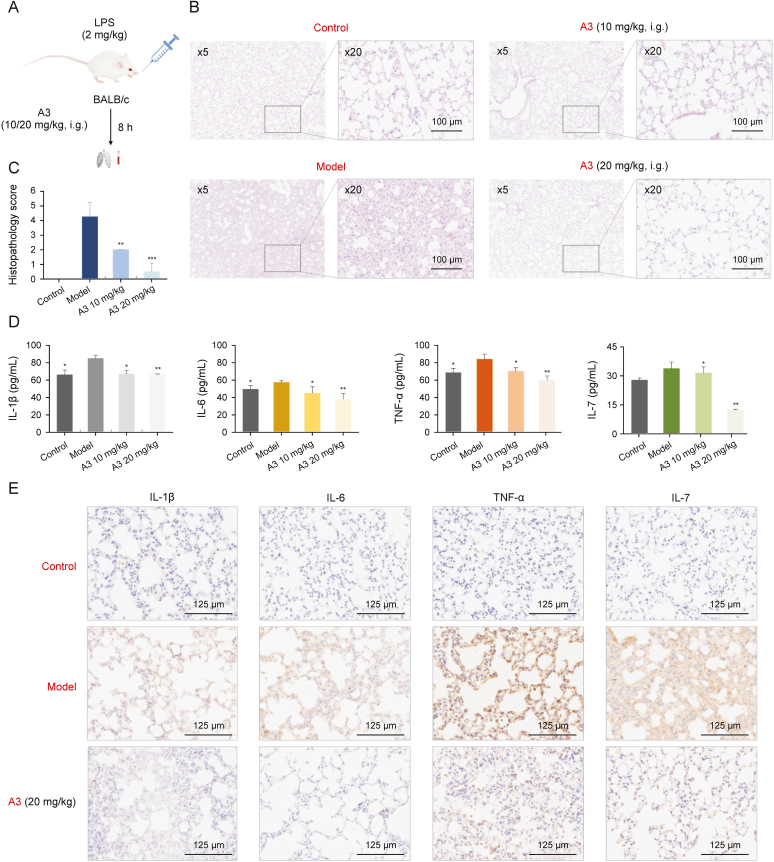
To further evaluate the anti-inflammatory potential and to explore the anti-inflammatory mechanisms of A3 in vivo, we constructed an ALI mice model as demonstrated in Fig. 5A [45]. After treatment with 2 mg/kg LPS by nasal intubation drip (i.t.), the two groups of mice were respectively treated with A3 at a dose of 10 mg/kg (low dose) or 20 mg/kg (high dose) by intragastric administration (i.g.). Mice treated with saline without LPS administration were used as the control. The mice were sacrificed 8 h after LPS treatment, and histopathology changes of the lung tissues were examined by H&E staining (Figs. 5B and S17). After analyzing and scoring the histopathology injuries of the excised lung tissue sections (Fig. 5C), we found the injury levels in lung section of the ALI mice treated with A3 were obviously alleviated. Moreover, the high-dose group showed more potent anti-inflammatory effects than the low-dose group.
Fig. 5.
Anti-inflammatory effects of licorice-saponin A3 (A3) in lipopolysaccharide (LPS)-induced mice acute lung injury (ALI) mice model. (A) Experimental scheme to construct LPS-induced ALI mice model. (B) Representative histopathology of lung sections of ALI mice with or without A3 administration. Control group: healthy mice treated with saline solution; Model group: LPS-induced ALI mice without A3 administration; A3 groups: LPS-induced ALI mice with A3 administration in a dose of 10 or 20 mg/kg, i. g., respectively, n = 4. The scale bars represent 500 μm for × 5 magnification images and 100 μm for × 20 magnification images. (C) Pathology evaluation for the therapeutic effects of A3 in LPS-induced ALI as shown in Fig. 5B, n = 4, ∗∗P < 0.01, ∗∗∗P < 0.001. (D) Tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-1β and IL-7 of blood samples were measured by mouse enzyme linked immunosorbent assay (ELISA) kit (MEIMIAN, www.mmbio.cn). ∗P < 0.05, ∗∗P < 0.01 compared with the control group, n = 4. (E) Representative immunohistochemistry (IHC) of lung sections of LPS-induced ALI mice with or without A3 administration for TNF-α, IL-6, IL-1β and IL-7 analysis.
Subsequently, the levels of proinflammatory cytokines IL-1β, IL-6, TNF-α and IL-7 of blood samples from ALI mice treated with A3 were determined (Fig. 5D). IL-7 was decreased significantly upon A3 treatment. Moreover, high-dose A3 (20 mg/kg) was more effective than the low-dose (10 mg/kg). Then, IHC staining of ALI mice lung section was conducted (Fig. 5E), IL-1β, IL-6, TNF-α and IL-7 were remarkably down-regulated after A3 treatment. The results provided solid evidence that A3 could remarkably inhibit inflammation by directly down-regulating the pro-inflammatory cytokines in host cells.
3.6. Host-directed immunoregulatory activity of A3
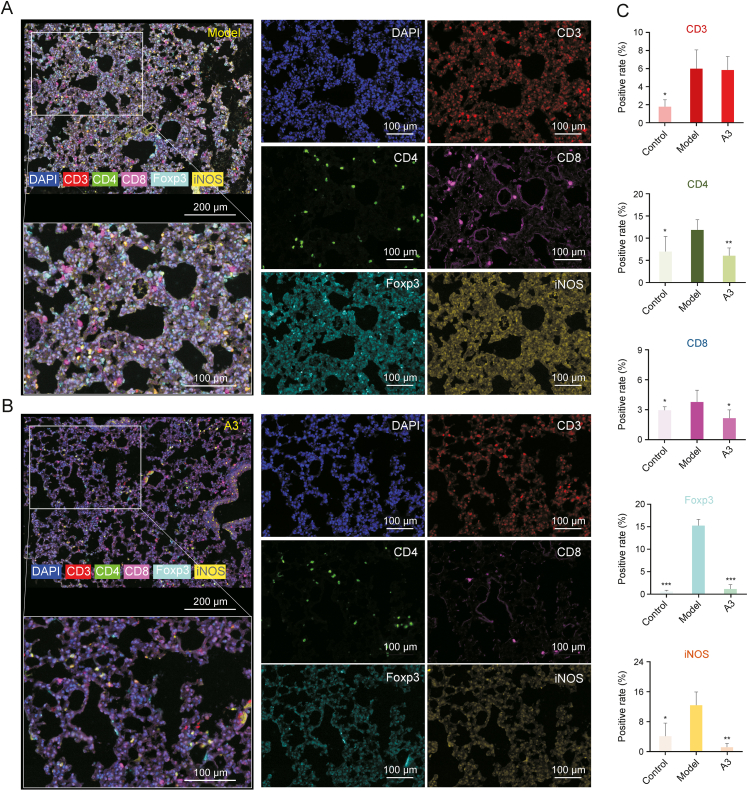
As p38 and JNK play critical roles in the innate and adaptive immune systems [46,47], and immune-modulating approaches are commonly used in inflammation treatment, we studied the infiltrates of different subtypes of immune cells in the lung tissues of ALI mice. T-cell subpopulations including CD3+ total T cells, CD4+ T helper cells (Th cells), CD8+ cytotoxic T lymphocytes (CTL), and Foxp3+ regulatory T cells (Tregs), as well as iNOS + M1 macrophages of the lung tissue samples from treated or untreated ALI mice as well as control mice were analyzed by multiplex fluorescent immunohistochemistry (mIHC) staining (Fig. 6). Multispectral imaging followed by spectral unmixing of five biomarkers in lung tissue samples of the three groups could provide the spatial distribution information and positive rates of each immune cell subtype (Figs. 6A, 6B and S18). The results showed the CD4+ Th cells, CD8+ CTL and iNOS + M1 macrophages, as well as the immune-suppressive Foxp3+ Tregs in lungs of ALI mice were generally improved compared with that in the control group without LPS, and were remarkably decreased after A3 treatment (n = 3) (Fig. 6C). This finding suggested LPS-induced inflammation caused a broad immune dysregulation and that A3 could rebalance the immune system.
Fig. 6.
Multi-spectral imaging and quantification analysis of the immune phenotype in lung tissues of lipopolysaccharide (LPS)-induced mice acute lung injury (ALI) mice treated by licorice-saponin A3 (A3). (A, B) Representative images displaying the lung tissues after multispectral imaging (raw image) and after spectral unmixing (composite image). Mice in the model (A) and A3 groups (B) were respectively administered LPS (2 mg/kg, i. t.), and LPS (2 mg/kg, i. t.) coupled with A3 (20 mg/kg, i. g.). The markers included CD3 (membrane, fluorescein isothiocyanate (FITC), red), CD4 (membrane, Texas, green), CD8 (membrane, CY5, blue), iNOS (membrane, CY3, cyan) and Foxp3 (nuclear, Texas, orange). The scale bars represent 100 μm for the single channel images. (C) Positive rate of the five markers in the control, model, and A3 groups. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared with the control group, n = 3.
4. Discussion
In this work, we report the natural product A3 from the traditional herbal medicine licorice (Gan-Cao) could inhibit RBDs of SARS-CoV-2 variants Alpha, Delta, and Omicron BA.1/XBB/BQ1.1. Particularly, A3 could potently inhibit SARS-CoV-2 Omicron in Vero E6 cells with an EC50 of 1.016 μM (Fig. S4). While several peptide or protein therapies could inhibit the entry of SARS-CoV-2 variants [21,22,48], A3 is the first broad-spectrum small molecule inhibitor of RBDs.
Considering that Y453 of the S protein RBD had been reported as a key amino acid in the hydrogen bonding between A3 and SARS-CoV-2 RBD [29], we explored the significance of Y453 in the inhibition of SARS-CoV-2 Omicron variant by A3. HDX-MS analysis and molecular docking revealed that A3 inhibited the Omicron RBD also through binding to Y453. Moreover, QM/MM calculations indicated the binding energy of A3 with peptide 453–470 of Omicron RBD (−26.7 kcal/mol) was much lower than that with Omicron Y453F mutant (4.37 kcal/mol) (Fig. 3G), suggesting a stable binding between A3 and Omicron RBD mediated by Y453.
For severe COVID-19 patients, overproduction of early response proinflammatory cytokines IL-1β, IL-6, and TNF-α can result in cytokine storm with symptoms like fever, cough, and diarrhea [26]. In the SARS-CoV-2-induced hyperinflammatory reaction, several reports have shown that the IL-1–IL-6 axis is the most biologically relevant signal [49]. Since protein phosphorylation is an important cellular regulatory mechanism, including inflammation [42], phosphoproteomic analysis of Vero E6 cells treated with A3 and then infected by SARS-CoV-2 was conducted to explore the anti-inflammatory mechanisms of A3 for COVID-19. Surprisingly, inflammation related proteins in JNK and p38 MAPK signaling pathways, including PAK, p38/p38γ, NF-κB, MEK4, JNK, STAT1 and Jun (Table S1), were all down-regulated by A3. Moreover, A3 could significantly and dose-dependently decrease the levels of IL-1β, IL-6 and TNF-α in both blood samples and lung tissues of mice in the ALI mice model. These data collectively suggested that A3 could directly inhibit inflammation by regulating key proinflammation proteins involved in the JNK and p38 MAPK signaling pathways.
Dysregulation of host immune responses is also a feature prominently in COVID-19 and immunomodulatory is another COVID-19 treatment strategy besides antiviral agents, mainly used for inhibiting the SARS-CoV-2-induced hyperinflammatory reaction [48]. Inspired by this, we explored the potential host-directed therapeutic capabilities of A3 by studying the immune phenotype in lung tissues of LPS-induced ALI mice treated with A3. Spatial visualization and quantification of several subtypes of immune cells infiltrating in lungs of ALI mice, including the immune activable CD4+ Th cells, CD8+ CTL and iNOS + M1 macrophages, as well as the immune-suppressive Foxp3+ Tregs. Results suggested that the four subtypes of immune cells were generally decreased after A3 treatment (n = 3) (Fig. 6C), indicating that A3 could restore the immune dysregulation of host induced by inflammation, further demonstrating the anti-inflammatory activities of A3 [48].
Furthermore, we monitored the pharmacokinetics of A3 in mice after 20 mg/kg administration (i.g.) (Fig. S19), and found that A3 was metabolically stable with a half-life of 7.13 h. Majority of the drugs were present in the circulation in its parent form. Furthermore, A3 showed little toxicity in both the cell line and the mice model, suggesting a fine biocompatibility.
5. Conclusion
In this work, we report that the natural product A3 could widely inhibit RBDs of SARS-CoV-2 variants, including Beta, Delta, and Omicron BA.1/XBB/BQ1.1. Furthermore, we find that A3 could directly suppress host inflammation by modulating the JNK and p38 MAPK pathways, thus restoring the corresponding immune dysregulation. Given its good safety and druggability, A3 could be a promising broad-spectrum small molecule drug candidate for the prevention and treatment of COVID-19.
CRediT author statement
Yang Yi: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - Original draft preparation, Reviewing and Editing, Visualization, Funding acquisition; Wenzhe Li: Methodology, Validation, Formal analysis, Data curation, Writing - Reviewing and Editing, Funding acquisition; Kefang Liu: Conceptualization, Methodology, Validation, Formal analysis, Data curation; Heng Xue: Validation, Formal analysis, Data curation; Rong Yu: Validation; Meng Zhang: Validation; Yang-Oujie Bao: Validation; Xinyuan Lai: Validation; Jingjing Fan: Validation; Yuxi Huang: Validation; Jing Wang: Validation; Xiaomeng Shi: Validation; Junhua Li: Validation; Hongping Wei: Formal analysis, Data curation; Kuanhui Xiang: Formal analysis, Data curation; Linjie Li: Validation; Rong Zhang: Validation; Xin Zhao: Formal analysis, Data curation; Xue Qiao: Formal analysis, Funding acquisition; Hang Yang: Formal analysis, Data curation, Writing - Reviewing and Editing, Supervision, Funding acquisition. Min Ye: Conceptualization, Methodology, Formal analysis, Writing - Reviewing and Editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant Nos.: 81891010/81891011, 81725023, 82003614, 82173950, 31770192, 32070187, 32161133003 and 82003681) and China Postdoctoral Science Foundation (Grant No: 2022T150029). We thank Professor George Fu Gao from CAS Institute of Microbiology for helpful and constructive suggestions. We also thank Tao Du, Lun Wang, Jin Xiong, and the entire running team from Zhengdian Biosafety Level 3 Laboratory of Wuhan Institute of Virology for technical support. We thank the Wroclaw Center for Networking and Super Computing for providing generous computer time, and Jingjie PTM BioLab Co., Ltd. (Hangzhou, China) for proteomics mass spectrometry analysis.
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.05.011.
Contributor Information
Hang Yang, Email: yangh@wh.iov.cn.
Min Ye, Email: yemin@bjmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2022. https://covid19.who.int (WHO Coronavirus (COVID-19) Dashboard, World Health Organization). [Google Scholar]
- 3.Xu K., Dai L., Gao G.F. Humoral and cellular immunity and the safety of COVID-19 vaccines: A summary of data published by 21 May 2021. Int. Immunol. 2021;33:529–540. doi: 10.1093/intimm/dxab061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu K., Gao P., Liu S., et al. Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2. Cell. 2022;185:2265–2278. doi: 10.1016/j.cell.2022.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plante J.A., Mitchell B.M., Plante K.S., et al. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y., Wang F., Shen C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;12:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naqvi A.A.T., Fatima K., Mohammad T., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P., Nair M., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 10.Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021;81:27–28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tegally H., Wilkinson E., Giovanetti M., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 13.Sabino E.C., Buss L.F., Carvalho M.P.S., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Xiao T., Cai Y., et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mannar D., Saville J.W., Zhu X., et al. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier D.A., Marco A., Ferreira I.A.T.M., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2022;608:1514. doi: 10.1038/s41586-022-05103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Beltran W.F., Lam E.C., Denis K.S., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang M., Wu L., Zheng A., et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3. Immunity. 2022;55:1501–1504. doi: 10.1016/j.immuni.2022.06.005. 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreano E., Piccini G., Licastro D., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espenhain L., Funk T., Overvad M., et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.50.2101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing L., Xu X., Xu W., et al. A five-helix-based SARS-CoV-2 fusion inhibitor targeting heptad repeat 2 domain against SARS-CoV-2 and its variants of concern. Viruses. 2022;14:597. doi: 10.3390/v14030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H., Li Y., Liu P., et al. Structural basis of HCoV-19 fusion core and an effective inhibition peptide against virus entry. Emerg. Microb. Infect. 2020;9:1238–1241. doi: 10.1080/22221751.2020.1770631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zenarruzabeitia O., Astarloa-Pando G., Terrén I., et al. T cell Activation, highly armed cytotoxic cells and a shift in, onocytes CD300 receptors expression is characteristic of patients with severe COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.655934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georg P., Astaburuaga-García R., Bonaguro L., et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell. 2021;185:493–512. doi: 10.1016/j.cell.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cargnello M., Roux P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao M., Liu Y., Yuan J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 28.Ji S., Li Z., Song W., et al. Bioactive constituents of Glycyrrhiza uralensis (Licorice): Discovery of the effective components of a traditional herbal medicine. J. Nat. Prod. 2016;79:281–292. doi: 10.1021/acs.jnatprod.5b00877. [DOI] [PubMed] [Google Scholar]
- 29.Yi Y., Li J., Lai X., et al. Natural triterpenoids from licorice potently inhibit SARS-CoV-2 infection. J. Adv. Res. 2022;36:201–210. doi: 10.1016/j.jare.2021.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi Y., Zhang M., Xue H., et al. Schaftoside inhibits 3CLpro and PLpro of SARSCoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19. Acta Pharm. Sin. B. 2022;12:4154–4164. doi: 10.1016/j.apsb.2022.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenwick C., Turelli P., Ni D., et al. Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys. Nat. Microbiol. 2022;7:1376–1389. doi: 10.1038/s41564-022-01198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 33.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 34.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation energy formula into a functional of the electron density. Phys. Rev. B. 1998;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 35.Miehlich B., Savin A., Stoll H., et al. Results obtained with the correlation-energy density functionals of becke and lee, Yang and parr. Chem. Phys. Lett. 1989;157:200–206. [Google Scholar]
- 36.McLean A.D., Chandler G.S. Contracted Gaussian-basis sets for molecular calculations. 1. 2nd row atoms, Z=11-18. J. Chem. Phys. 1980;72:5639–5648. [Google Scholar]
- 37.Frisch M.J., Trucks G.W., Schlegel H.B., et al. 2009. https://www.gaussian.com (Gaussian, Inc., Wallingford CT). [Google Scholar]
- 38.Morris G.M., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rappe A.K., Casewit C.J., Colwell K.S., et al. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992;114:10024–10035. [Google Scholar]
- 40.Chan J.F.W., Kok K., Zhu Z., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J.B., Zhao S., Teng T., et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cinatl J., Morgenstern B., Bauer G., et al. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ardito F., Giuliani M., Perrone D., et al. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review) Int. J. Mol. Med. 2017;40:271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giri R., Bhardwaj T., Shegane M., et al. Understanding COVID-19 via comparative analysis of dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses. Cell. Mol. Life Sci. 2021;78:1655–1688. doi: 10.1007/s00018-020-03603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai Z., Li P., Wen J., et al. Inhibitory effects and mechanisms of the anti-covid-19 traditional Chinese prescription, Keguan-1, on acute lung injury. J. Ethnopharmcol. 2022;285 doi: 10.1016/j.jep.2021.114838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuadrado A., Nebreda A.R. Mechanisms and functions of p38 MAPK signaling. Biochem. J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 47.Huang G.H., Shi L.Z.C., Chi H.B. Regulation of JNK and p38 MAPK in the immune system: Signal integration, propagation and termination. Cytokine. 2009;48:161–169. doi: 10.1016/j.cyto.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veerdonk F.L., Giamarellos-Bourboulis E., Pickkers P., et al. A guide to immunotherapy for COVID-19. Nat. Med. 2022;28:39–50. doi: 10.1038/s41591-021-01643-9. [DOI] [PubMed] [Google Scholar]
- 49.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao D., Xu W., Zhang X., et al. Understanding the phase separation characteristics of nucleocapsid protein provides a new therapeutic opportunity against SARS-CoV-2. Protein Cell. 2021;12:734–740. doi: 10.1007/s13238-021-00832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.