Abstract
Mesh-based reconstruction of the cerebral cortex is a fundamental component in brain image analysis. Classical, iterative pipelines for cortical modeling are robust but often time-consuming, mostly due to expensive procedures that involve topology correction and spherical mapping. Recent attempts to address reconstruction with machine learning methods have accelerated some components in these pipelines, but these methods still require slow processing steps to enforce topological constraints that comply with known anatomical structure. In this work, we introduce a novel learning-based strategy, TopoFit, which rapidly fits a topologically-correct surface to the white-matter tissue boundary. We design a joint network, employing image and graph convolutions and an efficient symmetric distance loss, to learn to predict accurate deformations that map a template mesh to subject-specific anatomy. This technique encompasses the work of current mesh correction, fine-tuning, and inflation processes and, as a result, offers a 150× faster solution to cortical surface reconstruction compared to traditional approaches. We demonstrate that TopoFit is 1.8× more accurate than the current state-of-the-art deep-learning strategy, and it is robust to common failure modes, such as white-matter tissue hypointensities.
Keywords: Cortical Surface Reconstruction, Topology, Geometric Deep-Learning
1. Introduction
Many medical image analysis techniques employ mesh representations, as opposed to image-based segmentation maps, for the reconstruction (or fitting) of tissue boundaries around regions of interest. This mesh-based approach is especially relevant in brain MRI processing, where polygonal meshes are often leveraged for reconstructing and studying the cerebral cortex, the outer layer of brain tissue. This layer is characterized by the topology of a 2D sheet, and due to its highly-folded nature, image-space Cartesian distances between points on the cortex can substantially underestimate the true distances along the sheet. Establishing a mesh-based surface representation of the cortex is therefore a fundamental step for downstream and multi-modal analysis of its structure, connectivity, function, and disease pathology.
Classical approaches to cortical reconstruction are robust and well established in neuroimaging pipelines, but tend to involve long runtimes: tens of minutes or even hours to run on just a single image (Fischl, 2012; Gaser and Dahnke, 2016). A substantial portion of this runtime is spent fulfilling the requirement to represent the cortex with a topologically-correct, genus zero manifold, facilitating anatomical plausibility and downstream geometric computations. However, surfaces are generally derived by tessellating volumetric segmentations, providing no guarantee that this condition is met. To address topological errors, cortical modeling tools employ mesh correction strategies that consume a substantial and variable amount of time (Fischl et al., 2001; Ségonne et al., 2007; Yotter et al., 2011).
In recent years, rapid deep-learning solutions have dominated the field of brain MRI analysis for a variety of tasks, but only a small handful of methods tackle cortical surface placement (Cruz et al., 2021; Ma et al., 2021). While these methods offer runtime improvements for specific components of surface-fitting pipelines, they are not complete solutions and require substantial pre- or post-processing to remove topological errors. Thus, cortical surface reconstruction remains a major step in brain MRI analysis without an established rapid, learning-based solution.
In this work, we address this missing link by introducing TopoFit, a novel approach that rapidly fits an accurate and topologically-correct polygonal mesh to the cerebral cortex. By leveraging convolutions in both the image and graph domains, TopoFit learns to estimate deformations from a mesh template, yielding a detailed reconstruction of subject-specific white-matter surfaces in less than 20 seconds on a CPU. The graph network facilitates deformations informed by the geometry and topology of the manifold. We demonstrate considerable robustness to local deviations in image intensity, which often disrupt traditional reconstruction methods, and we establish a 45.8% improvement in accuracy compared to a recent deep-learning approach. Our code is available at https://github.com/ahoopes/topofit, and we also publicly distribute our method within the open-source FreeSurfer package (Fischl, 2012). While TopoFit was developed in the context of neuroimaging, the concepts introduced here can be applied in a wide range of medical imaging contexts.
2. Related Work
Classical Surface Analysis.
Surface-based neuroimaging software, such as FreeSurfer (Fischl, 2012) and CAT12 (Gaser and Dahnke, 2016), construct detailed, per-hemisphere models of cortical gray-matter to facilitate analysis of functional activation, diffusion connectivity, and structural brain morphometry. As illustrated in Figure 1, the traditional surface-modeling pipeline begins by segmenting interior white-matter tissue from a brain MRI image then tessellating the computed segmentation map. After correcting for topological defects, this mesh is fine-tuned using a per-vertex optimization that considers the underlying image intensity gradient while constraining properties of the mesh geometry and topology (Dale et al., 1999). The resulting white-matter surface is subsequently deformed outwards to model the exterior pial tissue boundary. While these steps yield a complete representation of cortical gray-matter at the subject-level, a universal mapping must be computed for further group-level analysis and surface segmentation. This correspondence is achieved in a spherical coordinate system by inflating the white-matter manifold to the geometry of a sphere (Fischl et al., 1999a), which enables a curvature-matching registration with an average atlas (Fischl et al., 1999b; Yeo et al., 2009) and, consequently, establishes a global anatomical mapping across surfaces.
Figure 1:
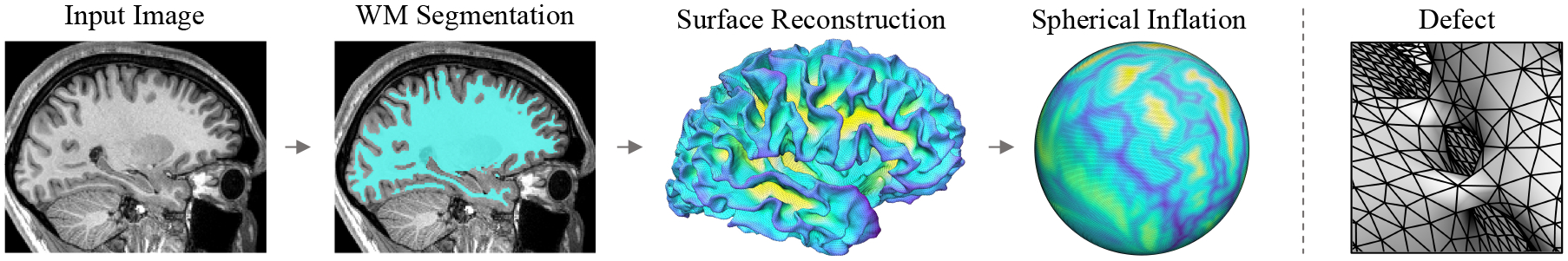
Left: Partial overview of the traditional surface-based cortical processing pipeline, involving white-matter (WM) segmentation, mesh reconstruction, and spherical inflation. Right: Example of a topological defect in which anatomically implausible “bridges” are incorrectly estimated between two gyri.
Topology Correction.
A mesh can only be diffeomorphically inflated and spherically-mapped if it possesses a topology that is homotopic, or continuously deformable, to that of a sphere. Specifically, a topologically-correct surface can be defined as a convex polyhedron with an Euler characteristic of two (genus zero). Since the initial white-matter surface derives from a segmentation with discrete resolution and potential for error, topological defects in the surface connectivity are common. These errors manifest as anatomically-implausible holes in the white-matter (Figure 1) and bridges connecting two banks of a sulcus. Defects must be corrected using time-consuming “manifold-surgery” algorithms that take into account the underlying geometry and image information to appropriately cut or fill regions of the mesh (Fischl et al., 2001; Ségonne et al., 2007; Yotter et al., 2011). In FreeSurfer, topology correction consumes roughly 70% of the total white-matter surface reconstruction time.
Recent research directions explore topology in deep-learning by leveraging differential persistent homology (PH), a technique to compute topological features across a landscape of spatial resolutions (Edelsbrunner et al., 2000). PH has been used to regularize model weights (Gabrielsson et al., 2020) as well as classification decision boundaries (Chen et al., 2019), and it is particularly useful for enforcing topological priors in medical image segmentations (Byrne et al., 2020; Clough et al., 2019). While TopoFit and PH methods both involve constraining the topology of model outputs, PH aims to encourage topological correctness during training, whereas our method avoids the need to correct the topology all together by using a topologically-correct, predefined mesh.
Learning-Based Reconstruction.
Recent deep-learning efforts have concentrated on mesh segmentation and registration (Hao et al., 2020; Lyu et al., 2021; Wu et al., 2019; Zhao et al., 2021) of the cortex, but few focus on the mesh placement itself. The leading method, DeepCSR (Cruz et al., 2021), uses implicit surface representations (Park et al., 2019) to reconstruct both white-matter and pial surfaces, but it requires a time-consuming topology correction. PialNN (Ma et al., 2021) uses explicit surfaces to project the pial boundary, but it still relies on a pre-computed, corrected white-matter mesh. Similar to our method, Voxel2Mesh (Wickramasinghe et al., 2020) addresses general anatomical segmentation by deforming a spherical template to the relevant tissue boundary, guaranteeing topologically-correct outputs, but it is limited to the focus of broad structure segmentation. Our work also builds on concepts from learning-based image (Balakrishnan et al., 2019; de Vos et al., 2019; Mok and Chung, 2020) and point-cloud registration (Hansen et al., 2019; Shen et al., 2021; Wang and Solomon, 2019) tactics that employ networks to align data pairs.
3. Method
Given a 3D brain MR image x, we aim to estimate a high-resolution, topologically-correct 2D manifold y of the white-matter. We employ a function fθ(x, m) = ϕ that computes a deformation field ϕ from a template surface m to the subject-specific anatomy in x. By using a template m with predefined, correct connectivity, the deformed mesh reconstruction is guaranteed to be free of intrinsic topological defects. We design fθ(·,·) as a convolutional network that employs both image-space and graph convolutions, at several scales. We find the optimal network parameters using
| (1) |
where y = m ◦ fθ(x, m), with ◦ representing the deformation procedure, and z is the target surface for an image x in the training set 𝒟. The loss term ℒdist measures surface similarity, and ℒreg, weighted by the hyperparameter λ, encourages geometric regularity of y.
3.1. Architecture
We design fθ as a network that jointly leverages convolutional operations on the image grid and on the graph of the template mesh (Figure 2). Specifically, a first network , with input image x, employs image-space convolutions to extract useful features on the image grid. A second network employs blocks of graph convolutions that operate on the features identified by , interpolated at intermediate locations of the template mesh m, and output a series of mesh deformations ϕ.
Figure 2:
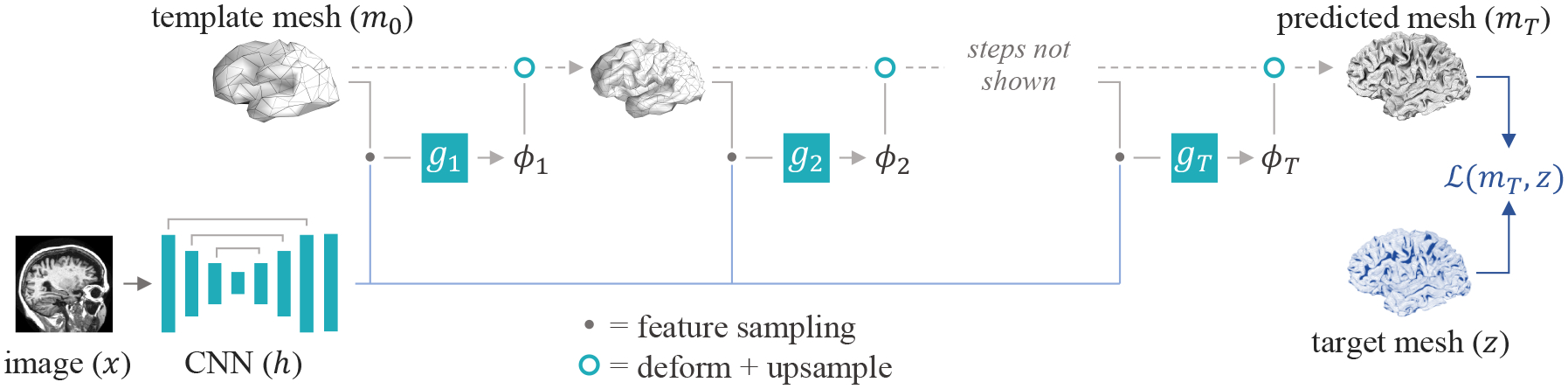
TopoFit architecture: a series of graph-convolutional (GCN) blocks iteratively warp a template surface m0 to fit the anatomy of an input image x. Each block gt predicts a deformation ϕt by sampling spatial image features from a volumetric U-Net h at the positions of mt−1. The template mesh is up-sampled between each deformation step.
To preserve geometric regularity, we adopt a scale-space strategy for the GCN . We employ a low-resolution template mesh m0 = m derived from a high-resolution icosphere mesh, corresponding to the average geometry across a set of surfaces. At each deformation step t = {1‥T }, a GCN block gt takes as input the feature activations sampled at the spatial coordinates of mt−1, and predicts the corresponding deformation ϕt. We then form mt = u(mt−1◦ϕt), where u(·) is a mesh upsampling operation based on the connectivity of the icosphere, such that y = mT. We illustrate this framework in Figure 2.
3.2. Loss Function
We minimize a symmetric surface distance between the estimated mesh y and the ground truth z. The symmetric Chamfer distance (Fan et al., 2017) is frequently used in existing methods, but is often computationally impractical for two high-resolution manifolds (with more than 150k points each). Instead, we take advantage of topological consistency across all training surfaces, which are aligned to a spherical cortical template and ensure that a given vertex represents the same anatomical region for all surfaces. This enables us to precompute a neighborhood 𝒩v of k closest vertices for each vertex v ∈ 𝒱 and efficiently minimize the localized symmetric distance between the predicted and target surface:
| (2) |
where is the Cartesian coordinate of i-th vertex of mesh j.
We also employ geometric regularization that minimizes self-intersecting faces to encourage an anatomically-accurate, smooth manifold. Specifically, we use a hinge-spring term that maximizes the angle between all neighboring triangles a, b in the set of mesh edges E:
| (3) |
where is the unit normal of the i-th triangle in y. This metric encourages smoothness without shrinking the mesh towards the center of mass, a side-effect present when using a vertex-based spring term (Fischl et al., 1999a).
3.3. Implementation
We implement a U-Net-like convolutional network h (Ronneberger et al., 2015), comprised of four down-sampling and up-sampling convolutional layers with skip connections, followed by one more convolutional layer. Each layer involves 64 channels and LeakyReLU activations.
In our experiments we use T = 7 GCN blocks gt, each with a similar U-Net-like architecture employing edge-convolutions (Wang et al., 2019). Given the input activations of h sampled at vertices mt, each block gt comprises at most three down-sampling and up-sampling LeakyReLU-activated layers, each with 64 channels, and a final, linearly activated layer with three channels that estimates ϕt. Resolution levels of the template surface are defined by the orders of icosphere tessellation, which facilitates up- and down-sampling of graph features using a universal adjacency map. In our experiments, the template m0 resolution corresponds to icosphere order 1 (42 vertices), and the final target mesh mT corresponds to order 7 (163,842 vertices). In this configuration, the gt U-Nets at t = {1, 2} cannot down-sample beyond icosphere order 1 and therefore contain only one and two resolution levels, respectively. We use vertex neighborhood size k = 100, which maximizes surface placement accuracy as determined by a grid hyperparameter search.
During training, we sample an image and corresponding target surface (x, z) from 𝒟 at each mini-batch and augment x with random added Gaussian noise (max σ = 0.1). We use the Adam optimizer (Kingma and Ba, 2014) with an initial learning rate of 10−4, which is reduced by a factor of two for every 10,000 iterations that do not exhibit a decrease in validation loss. We use the TensorFlow Graphics (Abadi et al., 2016; Valentin et al., 2019), Keras (Chollet et al., 2015), and Neurite (Dalca et al., 2018) packages, conducting all experiments using Intel Xeon Silver 4214R CPUs and Nvidia RTX 8000 GPUs.
4. Experiments
We evaluate the ability of TopoFit to accurately and rapidly reconstruct white-matter surfaces. We compare to the recent state-of-the-art deep-learning method and test robustness to common sources of error for conventional vertex placement techniques. Lastly, we perform an analysis of model hyperparameters and predicted mesh regularity.
Data: We employ a set of 1,232 subjects with T1-weighted MRI scans gathered from the publicly-available OASIS (Marcus et al., 2007), IXI1, MCIC (Gollub et al., 2013), and Buckner40 (Fischl et al., 2002) cohorts. We split these data into train, validate, and test subsets of sizes 788, 40, and 404, respectively, and additionally hold out the Buckner40 subjects entirely until final evaluation. For each scan, we use FreeSurfer v7.2 to derive a bias-corrected input image, an affine alignment to the Talairach atlas space, and white-matter surfaces for both hemispheres of the brain. For all training surfaces, we remesh to the universal icosphere topology, mapped by their computed spherical alignments with the FreeSurfer cortical atlas. We conform per-hemisphere images to 1mm isotropic voxel sizes with intensities normalized between [0, 1] and crop them to a 96 × 144 × 208 region.
Evaluation: We train two models, one for each hemisphere, and evaluate their performance for the entire set of test subjects, using manually-corrected FreeSurfer surfaces as a “silver-standard” reference set. We further compare TopoFit to DeepCSR, the primary established learning-based approach to white-matter reconstruction. We train and evaluate DeepCSR following online instructions and source code, adapting the training scheme to account for our data augmentation protocol and learning-rate decay schedule to yield the best DeepCSR results.
Metrics: We measure reconstruction accuracy using the mean, 99th-percentile (P99), and max (Hausdorff) symmetric distances between predicted and reference surfaces. Since surface modeling near the medial wall is noisy and unused in cortical analysis, we only include vertices for cortical regions marked by FreeSurfer. We also report the Dice overlap (Dice, 1945) between the interior of the filled surfaces, using 0.75 mm voxel sizes. Finally, we quantify surface regularity by the percent of self-intersecting faces in the mesh.
4.1. Reconstruction Accuracy
In the first experiment, we evaluate the ability of our proposed method to fit a topologically-correct surface to the white-matter boundary. Table 1 shows that TopoFit computes highly accurate surfaces and substantially outperforms DeepCSR, which tends to collapse into or broadly extend the white-matter in the presence of tissue abnormalities or intensity variation (Figure 3). The performance of TopoFit is consistent across each subject set (Table A1), generalizing well to the unseen Buckner40 cohort and on MCIC subjects. To fit white-matter surfaces to both hemispheres of the brain, TopoFit requires 17.3 ± 0.2 total seconds on a CPU and 1.2 ± 0.1 seconds on a GPU. This is in stark contrast to FreeSurfer, which requires 1, 086.6 ± 852.0 CPU seconds for the same process. Furthermore, since our method leverages the pre-defined topology of an icosphere, it can avoid the additional 1, 518.3 ± 750.6 seconds required for spherical mapping, resulting in a roughly 150× total speed-up over FreeSurfer. We find similar runtime improvements compared to DeepCSR, as shown in Table 1.
Table 1:
White-matter surface placement accuracy and GPU runtime for both hemispheres.
| Mean Dist. (mm) | P99 Dist. (mm) | Hausdorff (mm) | Dice (%) | Runtime (s) | |
|---|---|---|---|---|---|
| DeepCSR | 0.24 ± 0.09 | 1.56 ± 1.13 | 7.53 ± 2.01 | 96.0 ± 1.2 | 818.2 ± 134.9 |
| TopoFit | 0.13 ± 0.07 | 0.78 ± 0.35 | 4.35 ± 1.41 | 97.6 ± 0.6 | 1.2 ± 0.1 |
Figure 3:
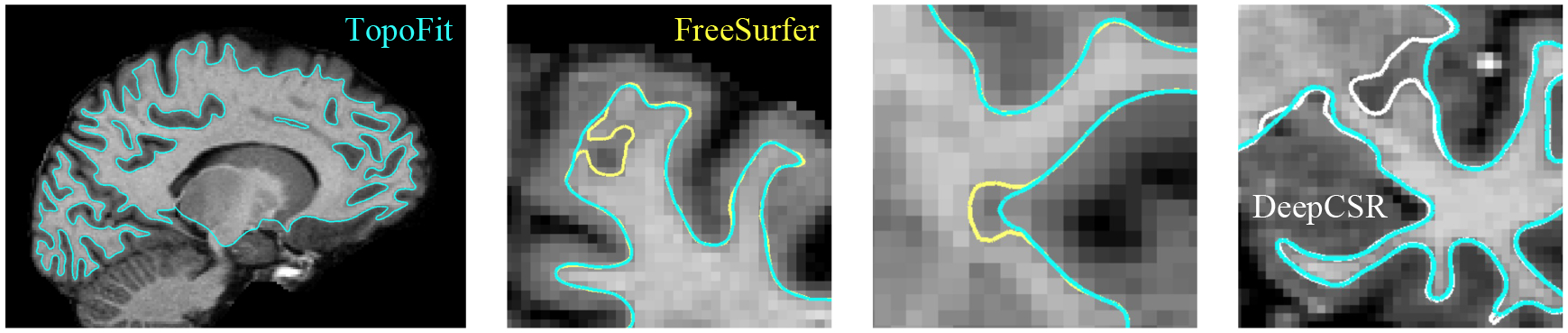
Representative reconstruction examples for TopoFit (blue), FreeSurfer (yellow), and DeepCSR (white). FreeSurfer and DeepCSR tend to get stuck in local minima around unexpected white-matter tissue abnormalities.
4.2. Robustness
A disadvantage of iterative mesh optimization techniques is their tendency to get stuck in local minima near regions of unexpected intensity distribution, requiring end-users to manually inspect each surface and correct errors. We test whether TopoFit is robust to these failure modes by evaluating on 20 randomly-selected test surfaces that required manual edits during the preprocessing stage. In this experiment, we determine accuracy by using a human rater to indicate whether our method makes the same mistake as FreeSurfer and by measuring the mean symmetric distance to the fixed reference surface within a 4 mm radius of the error region. We find that TopoFit produces correct white-matter reconstructions in 90% (18/20) of these regions, resulting in a negligible 0.18 ± 0.04 mm local deviation from the corrected reference. As highlighted in Figure 3, we find that FreeSurfer often fails to correctly fit surfaces in regions where white-matter lesions neighbor the cortex, whereas TopoFit is able to account for the global spatial context of the image.
4.3. Mesh Regularity and Hyperparameters
In addition to surface placement accuracy, maximizing mesh regularity is important for reproducing realistically smooth manifolds and ensuring that mesh connectivity corresponds to underlying anatomy. We perform a hyperparameter grid search to determine the optimal ℒreg smoothing weight λ*. Figure 4 shows that the optimal λ* = 1.0, used in our models, enforces a smooth mesh and minimizes the amount of self-intersecting faces (only 0.04%) without sacrificing reconstruction accuracy. Using λ = 0 results in predicted surfaces in which 5.1% of faces are self-intersecting. We also investigate how scale-space architecture choices impact mesh regularity by training models that use a different icosphere order R for the template mesh at the first GCN step m0. We find that a scale-space approach helps predict regular and accurate meshes and hypothesize that the decrease in performance as R increases is the result of far-reaching initial deformations that tangle nearby vertices.
Figure 4:
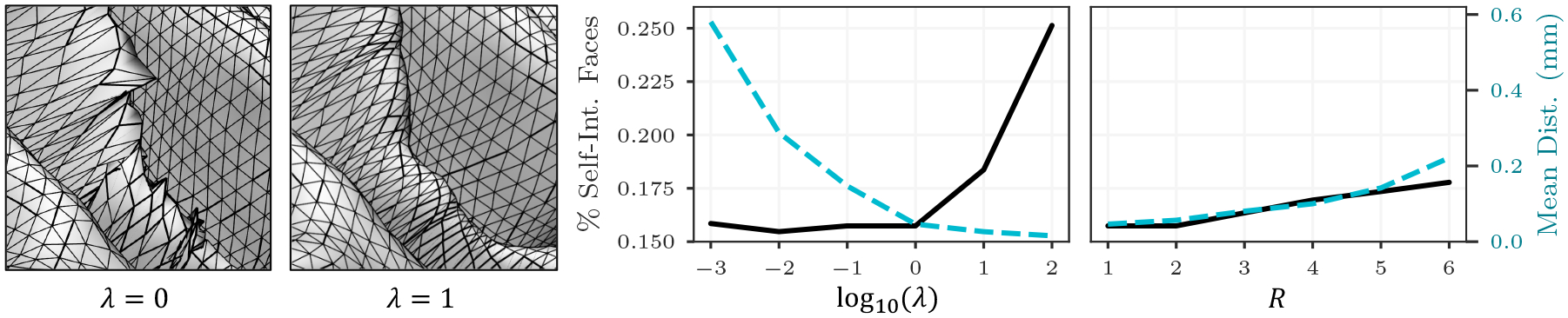
Model results given different hyperparameter values. The optimal regularization weight λ and template resolution level R aim to maximize surface placement accuracy while constraining the number of self-intersecting faces in the mesh.
5. Conclusion
We introduce TopoFit, a learning-based solution for reconstructing cerebral white-matter folds with a topologically-correct manifold. In a matter of seconds on a CPU, TopoFit can accomplish the tasks of traditional reconstruction pipelines that often require 40 minutes or more, by deforming a template mesh with intrinsic mapping to a sphere. This template surface is fit to the subject with high accuracy, even in regions that commonly lead to errors in FreeSurfer. We plan to extend our framework to include a pial surface reconstruction, and by further exploring the integration of learning-based cortical segmentation and spherical registration methods with TopoFit, we also plan to create an end-to-end model that encompasses the entirety of the cortical processing workflow. Lastly, we aim to extend this strategy to reconstruct the tissue boundaries of other, subcortical brain structures.
Acknowledgments
Support for this research was provided by the National Institute for Biomedical Imaging and Bioengineering (P41 EB015896, 1R01 EB023281, R01 EB006758, R21 EB018907, R01 EB019956, P41 EB030006), the National Institute on Aging (1R56 AG064027, 1R01 AG064027, 5R01 AG008122, R01 AG016495, 1R01 AG070988), the BRAIN Initiative Cell Census Network (U01 MH117023), the National Institute of Mental Health (R01 MH123195, R01 MH121885, 1RF1 MH123195), the National Institute for Neurological Disorders and Stroke (R01 NS0525851, R21 NS072652, R01 NS070963, R01 NS083534, 5U01 NS086625, 5U24 NS10059103, R01 NS105820), the NIH Blueprint for Neuroscience Research (5U01 MH093765), and Shared Instrumentation Grants 1S10 RR023401, 1S10 RR019307, and 1S10 RR023043.
Appendix A.
Table A1:
Dataset-specific comparison of total white-matter reconstruction accuracy for both hemispheres of the brain.
| Buckner40 (40 test subjects) | ||||
|---|---|---|---|---|
| Mean Dist. (mm) | P99 Dist. (mm) | Hausdorff (mm) | Dice (%) | |
| DeepCSR | 0.25 ± 0.17 | 1.60 ± 1.27 | 7.64 ± 2.12 | 95.95 ± 2.04 |
| TopoFit | 0.11 ± 0.02 | 0.73 ± 0.37 | 4.34 ± 1.45 | 97.69 ± 0.51 |
| IXI (158 test subjects) | ||||
| Mean Dist. (mm) | P99 Dist. (mm) | Hausdorff (mm) | Dice (%) | |
| DeepCSR | 0.26 ± 0.10 | 1.37 ± 0.74 | 7.47 ± 1.80 | 95.90 ± 1.38 |
| TopoFit | 0.15 ± 0.09 | 0.81 ± 0.37 | 4.41 ± 1.55 | 97.37 ± 1.25 |
| OASIS (130 test subjects) | ||||
| Mean Dist. (mm) | P99 Dist. (mm) | Hausdorff (mm) | Dice (%) | |
| DeepCSR | 0.22 ± 0.07 | 1.71 ± 1.43 | 7.60 ± 2.21 | 96.29 ± 0.87 |
| TopoFit | 0.11 ± 0.02 | 0.68 ± 0.27 | 4.31 ± 1.36 | 97.81 ± 0.41 |
| MCIC (76 test subjects) | ||||
| Mean Dist. (mm) | P99 Dist. (mm) | Hausdorff (mm) | Dice (%) | |
| DeepCSR | 0.25 ± 0.06 | 1.55 ± 0.75 | 7.45 ± 1.82 | 95.76 ± 0.86 |
| TopoFit | 0.15 ± 0.05 | 0.92 ± 0.35 | 4.25 ± 0.98 | 97.30 ± 0.73 |
Footnotes
Acquired from http://brain-development.org/ixi-dataset.
References
- Abadi Martín, Agarwal Ashish, Barham Paul, Brevdo Eugene, Chen Zhifeng, Citro Craig, Corrado Greg S, Davis Andy, Dean Jeffrey, Devin Matthieu, et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467, 2016.
- Balakrishnan Guha, Zhao Amy, Sabuncu Mert R, Guttag John, and Dalca Adrian V. Voxelmorph: a learning framework for deformable medical image registration. IEEE Transactions on Medical Imaging, 38(8):1788–1800, 2019. [DOI] [PubMed] [Google Scholar]
- Byrne Nick, Clough James R, Montana Giovanni, and King Andrew P. A persistent homology-based topological loss function for multi-class cnn segmentation of cardiac mri. In International Workshop on Statistical Atlases and Computational Models of the Heart, pages 3–13. Springer, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Chao, Ni Xiuyan, Bai Qinxun, and Wang Yusu. A topological regularizer for classifiers via persistent homology. In The 22nd International Conference on Artificial Intelligence and Statistics, pages 2573–2582. Proceedings of Machine Learning Research, 2019. [Google Scholar]
- Chollet Francois et al. Keras, 2015. URL https://github.com/fchollet/keras.
- Clough James R, Byrne Nicholas, Oksuz Ilkay, Zimmer Veronika A, Schnabel Julia A, and King Andrew P. A topological loss function for deep-learning based image segmentation using persistent homology. arXiv preprint arXiv:1910.01877, 2019. [DOI] [PMC free article] [PubMed]
- Cruz Rodrigo Santa, Lebrat Leo, Bourgeat Pierrick, Fookes Clinton, Fripp Jurgen, and Salvado Olivier. Deepcsr: A 3d deep learning approach for cortical surface reconstruction. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pages 806–815, 2021. [Google Scholar]
- Dalca Adrian V, Guttag John, and Sabuncu Mert R. Anatomical priors in convolutional networks for unsupervised biomedical segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 9290–9299, 2018. [Google Scholar]
- Dale Anders M, Fischl Bruce, and Sereno Martin I. Cortical surface-based analysis: I. segmentation and surface reconstruction. Neuroimage, 9(2):179–194, 1999. [DOI] [PubMed] [Google Scholar]
- de Vos Bob D, Berendsen Floris F, Viergever Max A, Sokooti Hessam, Staring Marius, and Išgum Ivana. A deep learning framework for unsupervised affine and deformable image registration. Medical Image Analysis, 52:128–143, 2019. [DOI] [PubMed] [Google Scholar]
- Dice Lee R. Measures of the amount of ecologic association between species. Ecology, 26 (3):297–302, 1945. [Google Scholar]
- Edelsbrunner Herbert, Letscher David, and Zomorodian Afra. Topological persistence and simplification. In Proceedings 41st Annual Symposium on Foundations of Computer Science, pages 454–463. IEEE, 2000. [Google Scholar]
- Fan Haoqiang, Su Hao, and Guibas Leonidas J. A point set generation network for 3d object reconstruction from a single image. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 605–613, 2017. [Google Scholar]
- Fischl Bruce. Freesurfer. Neuroimage, 62(2):774–781, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl Bruce, Sereno Martin I, and Dale Anders M. Cortical surface-based analysis: Ii: inflation, flattening, and a surface-based coordinate system. Neuroimage, 9(2):195–207, 1999a. [DOI] [PubMed] [Google Scholar]
- Fischl Bruce, Sereno Martin I, Tootell Roger BH, and Dale Anders M. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapping, 8(4):272–284, 1999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl Bruce, Liu Arthur, and Dale Anders M. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging, 20(1):70–80, 2001. [DOI] [PubMed] [Google Scholar]
- Fischl Bruce, Salat David H, Busa Evelina, Albert Marilyn, Dieterich Megan, Haselgrove Christian, Van Der Kouwe Andre, Killiany Ron, Kennedy David, Klaveness Shuna, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron, 33(3):341–355, 2002. [DOI] [PubMed] [Google Scholar]
- Gabrielsson Rickard Brüel, Nelson Bradley J, Dwaraknath Anjan, and Skraba Primoz. A topology layer for machine learning. In International Conference on Artificial Intelligence and Statistics, pages 1553–1563. Proceedings of Machine Learning Research, 2020. [Google Scholar]
- Gaser Christian and Dahnke Robert. Cat - a computational anatomy toolbox for the analysis of structural mri data. Human Brain Mapping, 2016:336–348, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub Randy L, Shoemaker Jody M, King Margaret D, White Tonya, Ehrlich Stefan, Sponheim Scott R, Clark Vincent P, Turner Jessica A, Mueller Bryon A, Magnotta Vince, et al. The mcic collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics, 11(3):367–388, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen Lasse, Dittmer Doris, and Heinrich Mattias P. Learning deformable point set registration with regularized dynamic graph cnns for large lung motion in copd patients. In International Workshop on Graph Learning in Medical Imaging, pages 53–61. Springer, 2019. [Google Scholar]
- Hao Lingyan, Bao Shunxing, Tang Yucheng, Gao Riqiang, Parvathaneni Prasanna, Miller Jacob A, Voorhies Willa, Yao Jewelia, Bunge Silvia A, Weiner Kevin S, et al. Automatic labeling of cortical sulci using spherical convolutional neural networks in a developmental cohort. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pages 412–415. IEEE, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma Diederik P and Ba Jimmy. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- Lyu Ilwoo, Bao Shuxing, Hao Lingyan, Yao Jewelia, Miller Jacob A, Voorhies Willa, Taylor Warren D, Bunge Silvia A, Weiner Kevin S, and Landman Bennett A. Labeling lateral prefrontal sulci using spherical data augmentation and context-aware training. Neuroimage, 229:117758, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Qiang, Robinson Emma C, Kainz Bernhard, Rueckert Daniel, and Alansary Amir. Pialnn: A fast deep learning framework for cortical pial surface reconstruction. In International Workshop on Machine Learning in Clinical Neuroimaging, pages 73–81. Springer, 2021. [Google Scholar]
- Marcus Daniel S, Wang Tracy H, Parker Jamie, Csernansky John G, Morris John C, and Buckner Randy L. Open access series of imaging studies (oasis): cross-sectional mri data in young, middle aged, nondemented, and demented older adults. Journal of Cognitive Neuroscience, 19(9):1498–1507, 2007. [DOI] [PubMed] [Google Scholar]
- Mok Tony CW and Chung Albert CS. Large deformation diffeomorphic image registration with laplacian pyramid networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 211–221, 2020. [Google Scholar]
- Park Jeong Joon, Florence Peter, Straub Julian, Newcombe Richard, and Lovegrove Steven. Deepsdf: Learning continuous signed distance functions for shape representation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 165–174, 2019. [Google Scholar]
- Ronneberger Olaf, Fischer Philipp, and Brox Thomas. U-net: Convolutional networks for biomedical image segmentation. In MICCAI: Medical Image Computing and Computer Assisted Interventions, pages 234–241. Springer, 2015. [Google Scholar]
- Ségonne Florent, Pacheco Jenni, and Fischl Bruce. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE transactions on medical imaging, 26(4):518–529, 2007. [DOI] [PubMed] [Google Scholar]
- Shen Zhengyang, Feydy Jean, Liu Peirong, Curiale Ariel Hernán, Estepar Ruben San Jose, Estepar Raul San Jose, and Niethammer Marc. Accurate point cloud registration with robust optimal transport. In Thirty-Fifth Conference on Neural Information Processing Systems, 2021. [Google Scholar]
- Valentin Julien, Keskin Cem, Pidlypenskyi Pavel, Makadia Ameesh, Sud Avneesh, and Bouaziz Sofien. Tensorflow graphics: Computer graphics meets deep learning. 2019.
- Wang Yue and Solomon Justin M. Deep closest point: Learning representations for point cloud registration. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 3523–3532, 2019. [Google Scholar]
- Wang Yue, Sun Yongbin, Liu Ziwei, Sarma Sanjay E, Bronstein Michael M, and Solomon Justin M. Dynamic graph cnn for learning on point clouds. ACM Transactions On Graphics, 38(5):1–12, 2019. [Google Scholar]
- Wickramasinghe Udaranga, Remelli Edoardo, Knott Graham, and Fua Pascal. Voxel2mesh: 3d mesh model generation from volumetric data. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 299–308. Springer, 2020. [Google Scholar]
- Wu Zhengwang, Zhao Fenqiang, Xia Jing, Wang Li, Lin Weili, Gilmore John H, Li Gang, and Shen Dinggang. Intrinsic patch-based cortical anatomical parcellation using graph convolutional neural network on surface manifold. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 492–500. Springer, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT Thomas, Sabuncu Mert R, Vercauteren Tom, Ayache Nicholas, Fischl Bruce, and Golland Polina. Spherical demons: fast diffeomorphic landmark-free surface registration. IEEE transactions on medical imaging, 29(3):650–668, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotter Rachel Aine, Dahnke Robert, Thompson Paul M, and Gaser Christian. Topological correction of brain surface meshes using spherical harmonics. Human Brain Mapping, 32 (7):1109–1124, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Fenqiang, Wu Zhengwang, Wang Fan, Lin Weili, Xia Shunren, Shen Dinggang, Wang Li, and Li Gang. S3reg: Superfast spherical surface registration based on deep learning. IEEE Transactions on Medical Imaging, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]


