Abstract
The human cytomegalovirus (HCMV) early UL4 promoter has served as a useful model for studying the activation of early viral gene expression. Previous transient-transfection experiments detected cis-acting elements (the NF-Y site and site 2) upstream of the transcriptional start site (L. Huang and M. F. Stinski, J. Virol. 69:7612–7621, 1995). The roles of two of these sites, the NF-Y site and site 2, in the context of the viral genome were investigated further by comparing mRNA levels from the early UL4 promoter in human foreskin fibroblasts infected by recombinant viruses with either wild-type or mutant cis-acting elements. Steady-state mRNA levels from the UL4 promoter with a mutation in the NF-Y site were comparable to that of wild type. A mutation in an Elk-1 site plus putative IE86 protein binding sites decreased the steady-state mRNA levels compared to the wild type at early times after infection. Electrophoretic mobility shift assays and antibody supershifts detected the binding of cellular transcription factor Elk-1 to site 2 DNA with infected nuclear extracts but not with mock-infected nuclear extracts. The role of cellular transcription factors activated by the mitogen activated protein kinase/extracellular signal-regulated kinase pathway in activating transcription from early viral promoters is discussed.
Human cytomegalovirus, a member of the betaherpesvirus family, is a ubiquitous human pathogen. Primary infection in healthy individuals is usually asymptomatic, but in immunocompromised individuals, human cytomegalovirus (HCMV) infection can cause pneumonitis, hepatitis, retinitis, and gastrointestinal diseases. In utero infections can cause severe congenital defects including mental retardation and hearing loss (4). HCMV replicates in fibroblasts and epithelial, endothelial, smooth muscle, and microglial cells and in macrophages (72, 75). HCMV remains latent in macrophage-granulocyte progenitors and in peripheral blood monocytes (27, 46, 57, 62, 76). Upon differentiation from monocytes to macrophages, lytic gene expression occurs (18, 33, 51, 76).
HCMV contains a linear double-stranded genome of approximately 230 kbp which can code for more than 200 proteins (12). As with other herpesviruses, HCMV genes are expressed in a temporal cascade. The first genes expressed without de novo viral protein synthesis are referred to as immediate-early (IE) genes. The IE proteins are required for subsequent early gene expression. The early genes code for viral DNA replication factors. After viral DNA replication, late gene expression occurs (79).
Two of the IE genes, IE1 and IE2, which are both expressed from the major IE promoter, are transactivators for early viral gene expression as demonstrated by transient-transfection assays (79). The mechanism by which the IE86 protein encoded by the IE2 gene transactivates early viral promoters in the context of the viral genome is still not well understood. Recombinant truncated IE86 fusion proteins produced in bacteria can interact in vitro with basal transcription factors TFIID, TATA-binding protein, TAFII-130, and TFIIB (9, 26, 40, 54), but these interactions have not been demonstrated in vivo. The IE86 protein also interacts in vitro with a variety of other cellular regulatory proteins and some unknown cellular proteins (19, 25, 49, 55, 73, 77, 81). In addition, the IE86 protein interacts in vivo with itself (2, 13) and the viral early UL84 protein (20, 70, 80). Cellular and viral trans-acting proteins and cis-acting regulatory elements are crucial for regulation of early viral gene expression. Many of the HCMV IE86 protein-responsive genes have regulatory elements upstream of the TATA box that contribute to activation in transient-transfection assays. For example, the TRL4, TRL6, UL4, UL112-113, and UL54 promoters have USF/MLTF, AP-1, NF-Y, ATF/CREB, and ATF/Sp1 upstream binding sites, respectively (30, 41–44, 49, 56, 82, 92, 97).
The mitogen-activated protein kinase (MAPKs) signaling pathways are important for cells to respond to various extracellular stimuli such as growth factor stimulation and cellular stress, including heat shock, osmotic shock, UV light, and cytokine stimulation. The cells may respond by cellular differentiation and proliferation, growth inhibition, or apoptosis. These important signaling events are strictly regulated (68, 96). In mammalian cells, three major subfamily of MAPKs—the extracellular signal-regulated kinase 1/2 (ERK1/2), the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), and the p38 kinase have been well studied (68, 96). MAPKs are activated by dual phosphorylation on specific tyrosine and threonine residues upon stimulation. Following activation, the activated MAPKs can be translocated from the cytoplasm to the nucleus, where their transcription factor targets are located (14). The substrates for activated ERK1/2 include transcription factor AP-1, Elk-1, SAP-1a, Ets-1, CREB, c-Myc, Tal, and the signal transducer and activator of transcription (STAT) proteins, etc. (68, 96).
Viruses have evolved numerous ways to take advantage of the host cell machinery to facilitate their own gene expression and replication in host cells. These include the shut-off of host cell protein synthesis and inhibition of cell cycle progression. Evidence from a variety of studies indicates that viruses can stimulate the activation of cellular MAPK pathways and use these pathways to regulate their own viral gene expression. The MAPK/ERK pathway has been demonstrated to be important for viral replication of the bovine papillomavirus (59), simian virus 40 (SV40) (78), and human immunodeficiency virus type 1 (34). In addition, the activation of SAPK/JNK signaling pathway is important for herpes simplex virus type 1 (HSV-1) replication (60, 98).
A recent report by Rodems et al. (69) demonstrated that HCMV infection activates ERK1/2. ERK activity can be detected 15 min after infection and remains elevated for 8 h postinfection (hpi). In addition, the activity of the early viral UL112-113 promoter is reduced in the presence of a MAPK/ERK kinase inhibitor (69). These studies suggested that HCMV-induced activation of the MAPK/ERK pathway might contribute to the activation of early viral gene expression. In addition, HCMV infection can activate the p38 pathway at approximately 8 h after infection and maintain activation through a mechanism of inhibition of dephosphorylation (36, 37). The p38 pathway is also important for HCMV replication because the usage of a p38 inhibitor results in a significant decrease in the viral titer (37). The SAPK/JNK activity is not elevated after HCMV infection (36). The activation of the ERK1/2 and p38 signal transduction pathways by HCMV requires de novo viral protein synthesis (36, 69).
To better delineate the mechanisms of HCMV early viral gene expression and the relationship to the ERK1/2 signal transduction pathway, we have used the viral early UL4 promoter as a model system. Previous transient-transfection assays demonstrated that the NF-Y binding site and the site 2 region upstream of the early viral UL4 promoter (E1) were major upstream regulatory elements. The viral promoter E1 drives the synthesis of a 1.5-kb mRNA, which encodes a 48-kDa virion-associated glycoprotein of unknown function (10, 11, 30). To better understand the regulation of HCMV early promoters in relation to activation of signal transduction pathways, we determined the roles of the upstream cis sites in the context of the viral genome by constructing recombinant viruses with wild-type or mutant upstream regulatory cis sites in an ectopic position of the viral genome. We demonstrate that a recombinant virus with site 2 mutations had a reduced level of transcription at early times after infection compared to a recombinant virus with a wild-type site 2. The NF-Y binding site did not play a significant role in UL4 promoter activity in virus-infected human foreskin fibroblasts (HFFs). An Ets family member, Elk-1 protein, which is a downstream target of the MAPK/ERK pathway, binds to the site 2 region. We propose that the HCMV early UL4 promoter is activated by cellular transcription factors activated by the MAPK/ERK pathway.
MATERIALS AND METHODS
Virus and cell culture.
The maintenance of primary HFFs has been described previously (84). The maintenance and propagation of HCMV Towne strain and all AD169 strain-derived recombinant viruses have been described previously (38, 83).
Enzymes.
Restriction endonucleases were purchased from New England Biolabs Inc. (Beverly, Mass.). T4 DNA ligase, the Klenow fragment of Escherichia coli DNA polymerase I, and calf intestinal alkaline phosphatase were obtained from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Taq or Vent DNA polymerase was purchased from New England Biolabs, Inc., and Fisher (Pittsburgh, Pa.), respectively. RNasin and RNase-free DNase were purchased from Promega (Madison, Wis.). The enzymes were used according to the manufacturers' instructions.
Plasmid constructions.
Plasmids p-220CAT and pΔMSVgpt have been described previously (10, 61). Plasmid p-220CAT has the UL4 (E1) promoter and 220 bp upstream of the transcription start site and the downstream chloramphenicol acetyltransferase (CAT) gene. A 1,225-bp AvrII-SacI DNA fragment (bp 200,578 to 201,803) of HCMV Towne strain containing the US12 and US13 genes and a 1,205-bp HindIII-BamHI DNA fragment (bp 195,838 to 197,043) containing the US6 and US7 genes were cloned into the corresponding sites in p-220CAT to generate plasmid pwt-as. Two fragments were generated by PCR using two sets of primers: primer 5′-GTATCCGGccTGagtggccTCGGCTCTGGTC-3′ with primer 5′-CGCCCCGCCCTGCCACTC-3′ and primer (T7 primer) 5′-TAATACGACTCACTATAGGG-3′ with primer 5′-CCGAggccactCAggCCGGATACGCTACA-3′ (mutant bases are indicated by lowercase letters). Plasmid pwt-as was used as the template. The 2,013-bp SacI-NcoI DNA fragment of pwt-as was replaced with the two PCR subfragments generated as described above—the 1,288-bp SacI-SfiI DNA fragment and the 725-bp SfiI-NcoI DNA fragment—to generate plasmid pdlIE86-as, which contains a mutation in the putative IE86 protein binding site in the site 2 region.
For construction of plasmid pwt-xs, a 1,411-bp XbaI-SacI DNA fragment (bp 200,392 to 201,803) of HCMV Towne replaced the AvrII-SacI of pwt-as. The NF-Y binding site and site 2 mutations were introduced into the UL4 promoter by site-directed mutagenesis using PCR and pwt-xs as the template. The primer pairs used to generate the UL4 promoter mutants were as follows: for plasmids pdlNF-Y-xs (NF-Y mutant), primer 5′-GAGGAATTCTCAGGGGATGATATGGGAagatcagcgctcATAAGACAAG-3′ and primer 5′-GCCATACGGAATTCCGGATGAGCA-3′; for plasmid pdlElk-1/IE86-xs (site 2 mutant), primer 5′-CaTATCatGgcctGAATgGcctactagTGGTCaGGGGGATAGTGA-3′ and primer 5′-GCCATACGGAATTCCGGATGAGCA-3′; for the T7 primer and primer, 5′-agtaggCcATTCaggcCatgATAtGCTACATACCT-3′. All primers were purchased from Life Technologies (Grand Island, N.Y.) unless otherwise specified. All mutations were confirmed by automated dideoxynucleotide sequencing (University of Iowa DNA core).
A 2,115-bp BsrGI-BamHI DNA fragment containing the guanine phosphoribosyltransferase gene (gpt) under the control of a minimal SV40 promoter was isolated from pΔMSVgpt. A BamHI linker was added to the blunt end of BsrGI. The 2.1-kb DNA fragment was subcloned into the BamHI-HpaI-digested plasmid pwt-as. The resulting plasmid was designated pHBgpt-220CAT AS.
HCMV recombination and plaque purification.
Recombinant virus RVwt-as and RVdlIE86-as were generated by the blue-white isolation method as described previously (45). Plasmids pwt-as and pdlIE86-as were linearized with XhoI and cotransfected into HFFs with infectious DNA from recombinant virus RV7150 (a gift from Thomas R. Jones, American Cyanamid Company, Pearl River, N.Y.) using the calcium phosphate precipitation method of Graham and Van der Eb (22). Approximately 3 days after 100% cytopathic effect (CPE), the viral supernatant was harvested and stored as viral stock at −70°C. Recombinant viruses were plaque purified on HFF monolayers grown under medium containing 0.5% agarose. Individual plaques were picked and transferred to HFFs in 24-well culture dishes at approximately 14 days after infection. The cells were overlaid with medium containing 0.5% agarose and 75 μg of X-Glu (5-bromo-4-chloro-3-indolyl-β-d-glucuronide; Sigma) per ml. White plaques were picked. CAT assays, dot blot analyses and Southern blot analyses were performed as described previously (10, 21, 50, 61).
Recombinant virus RVUL4CATgpt was isolated using a combination of the blue-white isolation method and the gpt positive selection method (23). Subconfluent HFFs were cotransfected with infectious RV7150 viral DNA and pHBgpt-220CAT AS. After 100% CPE, the viruses were harvested. Enrichment of gpt containing virus was performed by infecting HFFs with the virus stock in medium containing 40 μg of mycophenolic acid (Sigma) per ml and 200 μg of xanthine (Sigma) per ml. After three rounds of enrichment for gpt-containing virus, the viruses were used to infect HFFs grown in 60-mm-diameter plates. Recombinant viruses were isolated by the blue-white isolation method and identified further by dot blot hybridization as described above.
Recombinant viruses RVwt-xs, RVdlNF-Y-xs, and RVdlElk-1/IE86-xs were generated by the method of Greaves and Mocarski (24). HFFs were cotransfected with infectious RVUL4CATgpt viral DNA and plasmid shuttle vectors pwt-xs, pdlNF-Y-xs, or pdlElk-1/IE86-xs. After 100% CPE, the viruses were harvested and diluted 1:10 or 1:20. Lesch-Nyhan cells GM02291 (Coriell Cell Repository, Camden, N.J.) in 100-mm-diameter plates in medium containing 6-thioguanine (50 μg/ml; Sigma) were infected. Virus plaques were picked and transferred to HFFs grown in 24-well dishes. Cell-associated viral DNA was screened by dot blot hybridization and cell lysates were screened by CAT assay as described (21, 50, 61). Positive plaques were subjected to two additional rounds of plaque purifications.
Southern blot analysis.
Viral supernatant was collected and centrifugation was used to pellet virus particles as described (83). The viral DNA was isolated from the viral pellet as described previously (61). Then, the viral DNAs were digested with restriction endonuclease HindIII, SfiI, or HaeII and subjected to electrophoresis in 0.6 or 3% agarose gels. The DNA was transferred to maximum strength NYTRAN (Schleicher & Schuell, Keene, N.H.), and Southern blot analysis was performed as described (5, 61). All probes used in dot blot as well as Southern blot analysis were prepared by randomly labeling gel-purified DNA fragments using [32P]dCTP (Amersham, Arlington Heights, Ill.) and the multiprime DNA labeling system (Amersham). Unincorporated nucleotides were removed by pushing the probe through a Nuctrap purification column (Stratagene, La Jolla, Calif.). The 32P-HindIIIX probe was prepared using the 5,019-bp HindIII-HindIII DNA fragment of pMSDT DG (87). The 32P-XA probe was prepared by labeling the 186-bp XbaI-AvrII DNA fragment from plasmid pwt-xs.
Northern blot analysis.
Cytoplasmic RNAs from mock-infected or HCMV-infected HFFs were isolated as described previously (10, 29). Eight micrograms of cytoplasmic RNA was subjected to electrophoresis in a 1% agarose gel containing 2.2 M formaldehyde and transferred to maximum strength NYTRAN (Schleicher & Schuell). Northern blot analysis was performed as described previously (61). UL4- and CAT-specific DNA probes were derived from the 230-bp AvaII-DraI DNA fragment of pEgp48 (10) and the 305-bp NcoI-BspEI DNA fragment of pHB-220CAT AS, respectively. All probes were labeled with the multiprime DNA labeling system (Amersham). The same blot was serially stripped and rehybridized with different probes.
RNase protection assays.
Antisense actin and IE1 riboprobes have been described previously (53, 88). For the CAT antisense probe, a 384-bp EcoRI-EcoRI fragment from plasmid pwt-as, which spans 116 bp upstream and 268 bp downstream from the transcription start site of the UL4-CAT promoter, was cloned in vector pBluescript-II KS(+) (Stratagene). The riboprobes were synthesized using [32P]UTP (Amersham) as described previously (47).
Cytoplasmic RNA was harvested from four 100-mm-diameter plates of HFFs either mock infected or infected with HCMV recombinant viruses at a multiplicity of infection (MOI) of 5 PFU/cell at various times after infection. Twenty micrograms of RNA was hybridized to 32P-labeled antisense CAT, IE1, or actin probes at room temperature (RT) overnight before digestion with RNase T1 (100 U) as described previously (53). The protected RNA fragments were subjected to electrophoresis in denaturing 6% polyacrylamide gels followed by autoradiography on Hyperfilm MP (Amersham). Signals were quantitated by an electronic Autoradiographic Instant Imager (Packard Instant Imager, Meridan, Conn.).
EMSA.
DNA fragments used in electrophoretic mobility shift assay (EMSA) were generated by using primer 5′-GCAGGTATcTAGaGTATCCG-3′ and primer 5′-CCTCACgcTagCCCGGACCAGAGCCG-3′ (31) for PCR amplification of pwt-as for the wild type and pdlIE86-as for dlIE86. After digestion with NheI and XbaI, the PCR product was fractionated in 3% LMP agarose gel (Eastman Kodak Company, Rochester, N.Y.) and further purified with the MERmaid kit (Bio 101, Vista, Calif.). For all other probes or competitor DNAs (TNSM6, dlElk-1/IE86, or M3), synthetic oligonucleotides were purchased from Life Technologies. The sequence for the TNSM6 probe was 5′-CCTTTATAAAGGCCGGAAACGCTGAAAGGG-3′ (forward) and 5′-CCCTTTCAGCGTTTCCGGCCTTTATAAAG-3′ (reverse). The probes and nonradioactive competitor DNAs were denatured at 95°C and annealed gradually by cooling down to RT. Double-stranded DNA was purified as described above. The concentrations of the DNA fragments were estimated by ethidium dot assay (71) and spectrophotometry. Probes were prepared by 3′ end labeling using the Klenow fragment of the E. coli DNA polymerase I and [32P]dCTP or [32P]dGTP (Amersham). Unincorporated deoxynucleoside triphosphates were removed by a Nuctrap column (Stratagene).
EMSA with nuclear extracts was performed essentially as described (31) with minor modifications. Two micrograms of nuclear extract was preincubated with nonradioactive competitor DNA at 10- or 50-fold molar excess relative to the probe in the presence of 1 μg of sheared salmon sperm DNA and buffer I (6.25 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.01% Nonidet P-40, and 9% glycerol) at RT for 15 min. Then, 176 fmol of radioactive probe was added to the reaction mixture and incubated at RT for 15 min. The DNA-protein complexes were separated from free probe by electrophoresis in a 5% nondenaturing polyacrylamide gel in 0.5× TAE (20 mM Tris-acetate, pH 7.2, containing 1.0 mM EDTA) at 4°C. Gels were dried and exposed to Hyperfilm MP (Amersham).
In antibody supershift experiments, 2 μg of either mock- or HCMV-infected nuclear extract was incubated with 1 μg of sheared salmon sperm DNA and buffer I in the presence or absence of nonradioactive DNA at RT for 15 min. One or 5 μl of anti-immunoglobulin G (IgG) control antibody or anti-Elk polyclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) was added to the reaction mixture at RT for 30 min before 176 fmol of probe (∼50,000 cpm) was added. The reaction mixture was allowed to incubate at RT for 15 min. Electrophoresis was performed as described above. The total amount of protein in each reaction mixture was balanced by adding bovine serum albumin.
RESULTS
Recombinant viruses with UL4 promoter mutations.
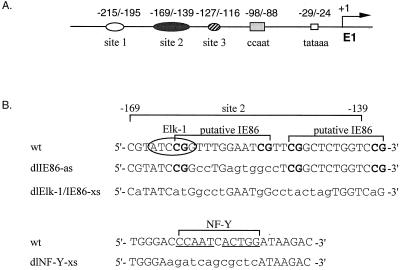
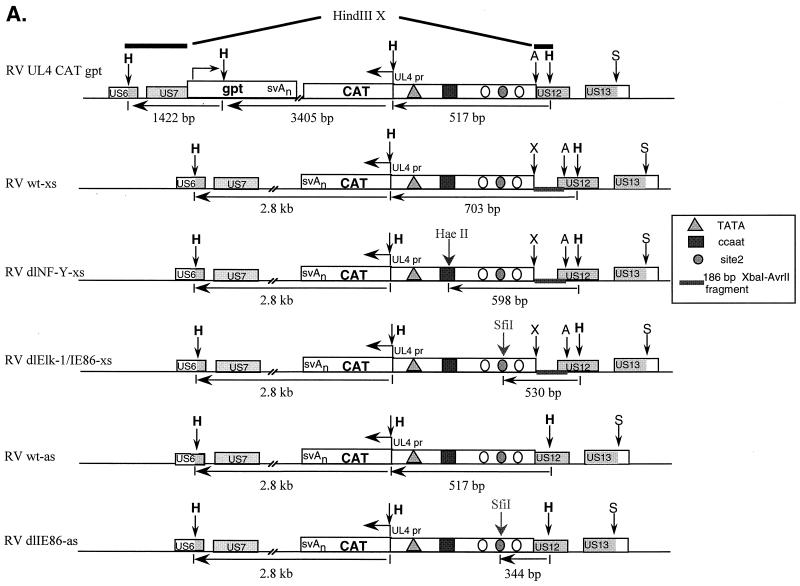
Previous deletion mutation analysis indicated that elements upstream of the HCMV UL4 promoter were important for promoter activation. DNase I footprinting analysis demonstrated three cellular protein binding sites, designated sites 1, 2, and 3 (31). In addition, the viral IE86 protein expressed as a truncated form in E. coli can bind in vitro to site 2 DNA (31). Deletion mutation analysis and transient-transfection assays indicated that site 2 and the NF-Y binding site were critical for promoter activity (30, 31) (Fig. 1A). We performed site-directed mutagenesis of site 2 and the NF-Y site as indicated in Fig. 1B. Putative IE86 protein binding sites, CGN9CG, between −163 and −151, and −148 and −136, relative to the transcription start site, were also targeted for mutagenesis. The potential role of these sites was determined in the context of the viral genome using a series of recombinant viruses containing the UL4 promoter driving transcription of the CAT gene (UL4-CAT) (Fig. 2A). Although the UL4 gene of HCMV is reported to be nonessential (66, 86), we replaced the UL4 gene with the CAT gene and made upstream promoter mutations in an ectopic position of the viral genome because the marker gene coding for β-glucuronidase was cloned in this region of the viral genome (38) and facilitated recombinant virus isolation. We took advantage of the blue-white plaque selection method (45) as well as the gpt selection method (23) to isolate recombinant viruses. The UL4 promoter-CAT constructs were inserted by homologous recombination between the US8 and US11 region of the viral genome, which has been shown to be dispensable for viral replication in tissue culture (38, 39).
FIG. 1.
Sequences upstream of the early HCMV UL4 promoter. (A) Schematic representation of the early viral UL4 promoter (E1). The TATA box and the imperfect dyad NF-Y binding site (CCAAT box) are designated by the open and shaded box, respectively. Cellular protein binding sites 1, 2, and 3 are labeled. The numbers above the symbols are positions relative to the transcription start site (arrow). (B) DNA sequences for the wild-type (wt) and mutated site 2 and CCAAT box (NF-Y site) regions. Wild-type site 2, the putative IE86 protein binding sites, and the NF-Y site are indicated by brackets. A core Elk-1 binding site is circled. The CGs, which might be essential for IE86 protein binding, are in boldface type. All mutations in the DNA sequences are indicated by lowercase letters.
FIG. 2.
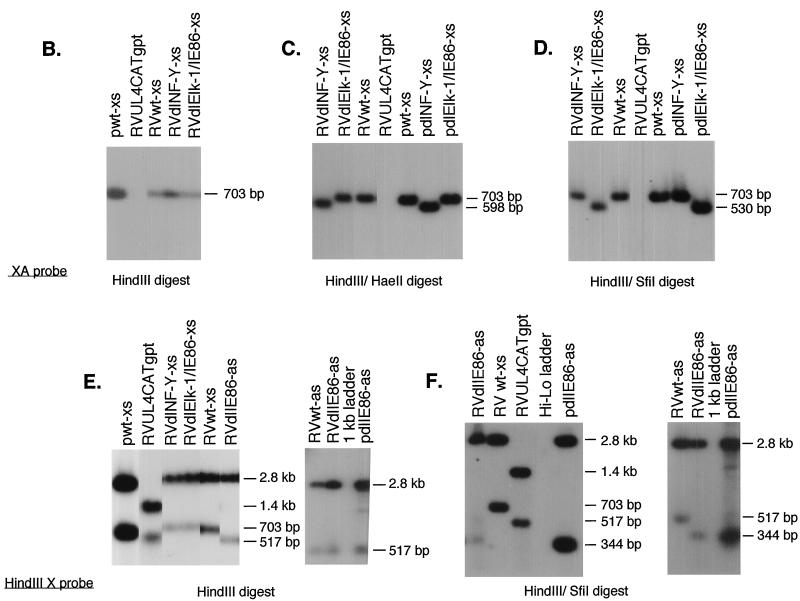
Structural analysis of recombinant viruses RVwt-xs, RVdlNF-Y-xs, RVdlElk-1/IE86-xs, RVwt-as, and RVdlIE86-as. (A) Maps of RVUL4CATgpt, RVwt-xs, RVdlNF-Y-xs, RVdlElk-1/IE86-xs, RVwt-as, and RVdlIE86-as. The sizes of the DNA fragments resulting from HindIII, HaeII, or SfiI restriction endonuclease digestion are indicated in base pairs. The genes involved in homologous recombination in shuttle vectors are shown by shaded boxes. A, S, H, and X stand for the restriction endonuclease sites AvrII, SacI, HindIII, and XbaI, respectively. (B to F) Individual autoradiograms of Southern blots to identify the recombinant viruses using either 32P-labeled XA probe (B to D) or HindIII X probe (E and F). Lanes containing viral DNA fragments from different recombinant viruses were spliced together from the same gel. Shuttle vectors pwt-xs, pdlNF-Y-xs, pdlElk-1/IE86-xs, and pdlIE86-as were used as positive controls.
The genome structures of the recombinant viruses were analyzed by HindIII restriction endonuclease digestion of viral DNAs followed by Southern blot hybridization using a 32P-labeled XA probe. The predicted sizes of viral DNA fragments are indicated in Fig. 2A. The resulting RVwt-xs, RVdlNF-Y-xs, and RVdlElk-1/IE86-xs differ from the parental RVUL4CATgpt by the presence of the XbaI-AvrII DNA fragment and the absence of the gpt gene (Fig. 2B and data not shown). Site-specific mutations generated in the UL4 promoter region were confirmed by DNA sequencing in the shuttle vector and by double restriction endonuclease digestion of the recombinant viral DNAs as illustrated in Fig. 2C and D. The genome structures of RVwt-xs, RVdlNF-Y-xs, and RVdlElk-1/IE86-xs were also confirmed by using the HindIII X probe, which spans part of the US6 and US12 in the recombinant viruses (Fig. 2E and F). RVwt-as and RVdlE86-as were confirmed by Southern blot analysis as illustrated in Fig. 2E and F. All of these recombinant viruses had growth kinetics similar to their parental RV7150 and to the wild-type construct (data not shown).
UL4 promoter activity in both the ectopic and natural positions.
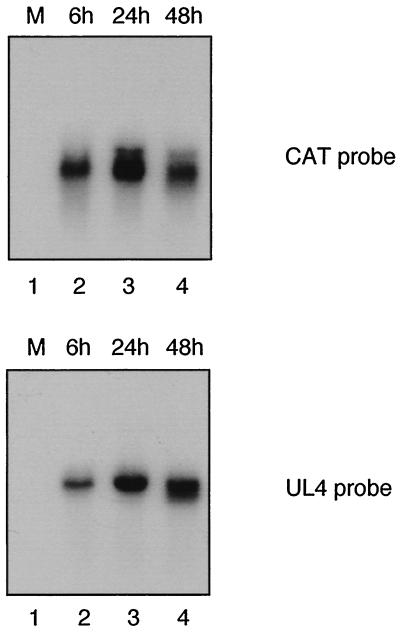
To determine whether the expression pattern of the ectopic UL4-CAT promoter correlates with that of UL4 promoter in the unique long component of the viral genome, the UL4 and CAT transcripts were assayed in permissive HFFs infected with RVwt-xs at 5 PFU/cell. Cytoplasmic RNAs were isolated at 6, 24, and 48 h.p.i. and subjected to Northern blot analysis using 32P-labeled DNA probes. Both CAT and UL4 RNAs were detected at 6 h and increased at 24 and 48 h after infection as shown in Fig. 3. The highest amount of steady-state viral mRNA was detected at 24 hpi. The steady-state level of transcript from the UL4-CAT promoter in the ectopic position was qualitatively similar to that of the transcript from the UL4 promoter in the unique long component of the HCMV genome. An equal amount of RNA was loaded onto each lane and confirmed by ethidium bromide staining of the 28S and 18S rRNA present in each lane (data not shown). We conclude that transcription from the ectopic UL4-CAT promoter in the recombinant viruses was qualitatively similar to that from the UL4 promoter in the natural position of the viral genome.
FIG. 3.
Northern blot analysis of UL4-CAT or UL4 RNA expression in HFFs infected with RVwt-xs. HFFs were either mock infected or infected with RVwt-xs (5 PFU/cell). Cytoplasmic RNA was harvested at various times after infection and analyzed by Northern blot hybridization with either CAT or UL4 probes as described in Materials and Methods. Lanes: 1, mock-infected RNA; 2 to 4, RNA harvested at 6, 24, and 48 h after infection, respectively.
Effect of the NF-Y mutation.
To determine whether the NF-Y binding site was important for activation of the UL4 promoter in the context of the viral genome, we analyzed the steady-state level of RNA transcribed from the UL4-CAT promoter in HFFs infected with either a wild-type (RVwt-xs) or mutant (RVdlNF-Y-xs) at 5 PFU/cell. Cytoplasmic RNAs were isolated from infected cells under treatment with phosphonoacetic acid (200 μg/ml) for 48 h or without treatment for 6 and 24 h. An RNase protection assay of HFFs infected with RVwt-xs or RVdlNF-Y-xs at early times after infection (6, 24, or 48 h.p.i. plus PAA) is shown in Fig. 4. The expression from the UL4-CAT promoter displays a typical pattern of an early viral promoter with the highest level of steady-state RNA at 24 hpi in both the RVwt-xs and RVdlNF-Y-xs infected HFFs. The internal control for multiplicity of viral infection, the protected IE1 RNA, had significantly higher levels at 6 hpi than at 24 and 48 hpi as expected. The protected CAT RNA level from RVwt-xs infected cells was similar to that of RVdlNF-Y-xs which contains a mutant NF-Y site (Fig. 4A, compare lane 6, 8, and 10 with lanes 7, 9, and 11 respectively). The levels of protected CAT RNA after normalization to the internal IE1 RNA or to actin RNA exhibited little to no difference as shown in Fig. 4B. Other independently plaque-purified RVdlNF-Y-xs virus isolates behaved similarly. We conclude that mutation of the NF-Y binding site, which is a positive element in transient-transfection assays, does not play a critical role in UL4 promoter activation in the context of the viral genome.
FIG. 4.
Steady-state RNA levels transcribed from the UL4-CAT promoter with either a wild-type or mutant NF-Y binding site at early times after infection. Cytoplasmic RNA was isolated at the indicated time points after infection with RVwt-xs or RVdlNF-Y-xs and then subjected to RNase protection assays as described in Materials and Methods. (A) Autoradiogram of RNase protection assay. Lanes: 1, 32P-labeled DNA standard molecular weight markers; 2 to 4, 32P-labeled CAT, actin, and IE1 riboprobes not treated with RNase, respectively; 5, mock infected; 6, RVwt-xs at 6 hpi; 7, RVdlNF-Y-xs at 6 hpi; 8, RVwt-xs at 24 hpi; 9, RVdlNF-Y-xs at 24 hpi; 10, RVwt-xs at 48 hpi with PAA treatment; 11, RVdlNF-Y-xs at 48 hpi with phosphonoacetic acid (PAA) treatment. The sizes of the protected RNAs are indicated (in nucleotides [nt]). Lanes 6 and 7, both the CAT and IE1 probes were added to the reaction mixture. Lanes 5 and 8 to 11, both the CAT and actin probes were added to the reaction mixture. (B) Image acquisition analysis. The CAT RNA signals from RVwt-xs and RVdlNF-Y-xs was normalized to protected IE1 (6 hpi) or actin (24 hpi, 48 hpi + PAA) RNA.
Effect of site 2 mutations.
The function of site 2 in the context of the viral genome was not known. Site 2 has two putative IE86 protein binding sites (CGN9CG) between −163 and −151 and −148 and −136, relative to the transcription start site (Fig. 1). However, these sites are not the consensus CGN10CG, which is critical for efficient IE86 protein binding (93). Recombinant virus RVdlIE86-as contains a wild-type Elk-1 site and site-specific mutations in the distal putative IE86 protein binding site. Since a previous report demonstrated that mutation of both the CG dinucleotides of the consensus CGN10CG sequence are necessary for complete abolishment of the binding of IE86 protein in vitro (93), we mutated all four CGs and some of the N9 sequence between the CGs in RVdlElk-1/IE86-xs. We tested by EMSA as described previously (31) for in vitro binding of purified r-maltose-IE86 fusion protein to DNA probes containing either dlIE86 or dlElk-1/IE86 and detected no binding to the mutated DNA (data not shown).
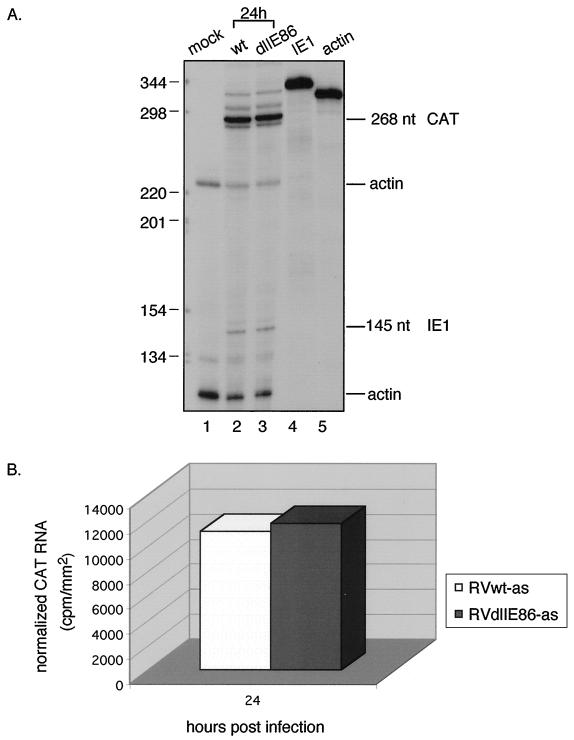
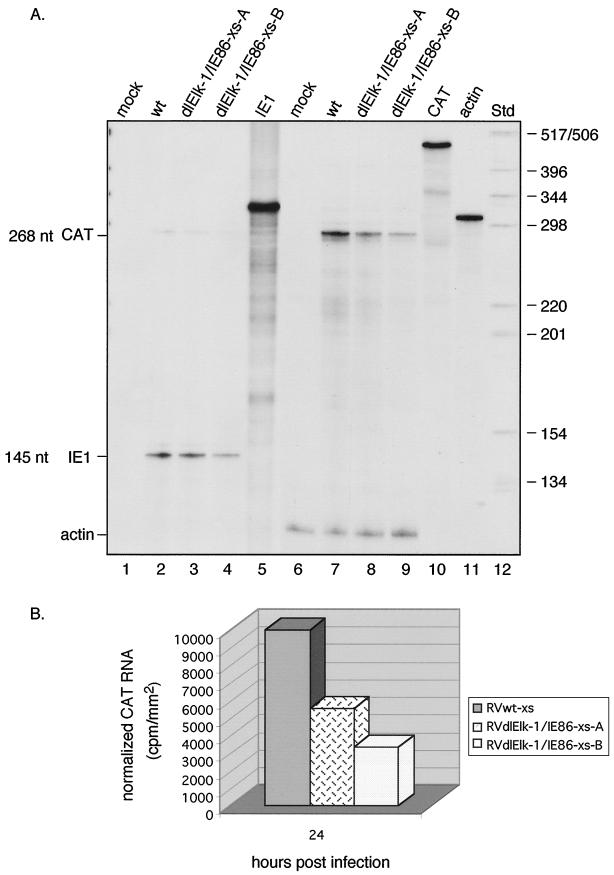
At 24 hpi, the steady-state level of CAT RNA was equal between values for RVwt-as and RVdlIE86-as (Fig. 5). Therefore, mutations in the region of the putative distal IE86 protein binding site had no effect. RVdlElk-1/IE86-xs contains mutations in both the distal and proximal putative IE86 protein binding sites as well as the Elk-1 site (Fig. 1B). HFFs were infected with the recombinant viruses at approximately 5 PFU/cell and cytoplasmic RNAs were analyzed by RNase protection assays as described in the Materials and Methods. The steady-state level of CAT RNA was higher with RVwt-xs than RVdlElk-1/IE86-xs-A at 24 hpi. (Fig. 6A, lane 7 and 8). To confirm this result, another recombinant RVdlElk-1/IE86-xs-B virus was derived from a separate transfection and plaque purified. At 24 h pi, both RVdlElk-1/IE86-xs-A and -B viruses exhibited a reduction in steady-state CAT RNA relative to wild type (Fig. 6A, lanes 7 to 9, and 6B). The same MOI was established by detection of the IE1 RNA signal from RVwt-xs and RVdlElk-1/IE86-xs-A, but with RVdlElk-1/IE86-xs-B, the IE1 RNA was lower (Fig. 6A, lanes 2 to 4). This explains why there was more of a reduction in the CAT RNA with RVdlElk-1/IE86-xs-B. The more extensive site 2 mutation, which disrupts both the putative IE86 protein binding sites and the Elk-1 site, reduced the level of steady-state mRNA from the UL4-CAT promoter in the context of the unique short component of the viral genome. We conclude that site 2 in the context of the viral genome plays a regulatory role in the activity of the UL4 promoter.
FIG. 5.
Steady-state RNA levels transcribed from the UL4-CAT promoter with either wild-type or a site 2 mutation at early times after infection. HFFs were infected with 5 PFU of RVwt-as or RVdlIE86-as per cell. Cytoplasmic RNA was analyzed as described in the legend to Fig. 4. (A) Autoradiogram of RNase protection assay. Lanes: 1, mock infected; 2, RVwt-as at 24 hpi; 3, RVdlIE86-as at 24 hpi; 4 and 5, 32P-labeled IE1 and actin riboprobes not treated with RNase, respectively. The sizes of the protected RNAs are indicated (in nucleotides [nt]). Lanes 1 to 3, the 32P-labeled CAT, IE1, and actin riboprobes were added to the reaction mixture. (B) Image acquisition analysis of the CAT signal normalized to actin.
FIG. 6.
Steady-state RNA levels transcribed from the UL4-CAT promoter with either a wild-type site 2 or a more extensive mutation of the site 2 region at early times after infection. HFFs were infected with 5 PFU of RVwt-xs, RVdlElk-1/IE86-xs-A, or RVdlElk-1/IE86-xs-B per cell. (A) Autoradiogram of RPA. Lanes: 1, mock infected; 2, RVwt-xs at 24 hpi; 3, RVdlElk-1/IE86-xs-A at 24 hpi; 4, RVdlElk-1/IE86-xs-B at 24 hpi; 5, 32P-labeled IE1 riboprobe not treated with RNase; 6, mock infected; 7, RVwt-xs at 24 hpi; 8, RVdlElk-1/IE86-xs-A at 24 hpi; 9, RVdlElk-1/IE86-xs-B at 24 hpi; 10 and 11, 32P-labeled CAT and actin riboprobes not treated with RNase, respectively; 12, 32P-labeled DNA standard molecular weight markers. Lanes 1 to 4, only the 32P-labeled IE1 riboprobe was added to the reaction mixture; lanes 6 to 9, both the 32P-labeled CAT and actin riboprobes were added to the reaction mixture. Lanes 1 to 5 were from a longer exposure to show the IE1 signal. The sizes of the protected RNAs are indicated (in nucleotides [nt]). RVdlElk-1/IE86-xs-A and RVdlElk-1/IE86-xs-B were isolated from two independent transfections. (B) Image acquisition analysis of the CAT signal normalized to actin. This figure represents the results from one of at least three experiments performed.
Cellular nuclear protein(s) binding to site 2.
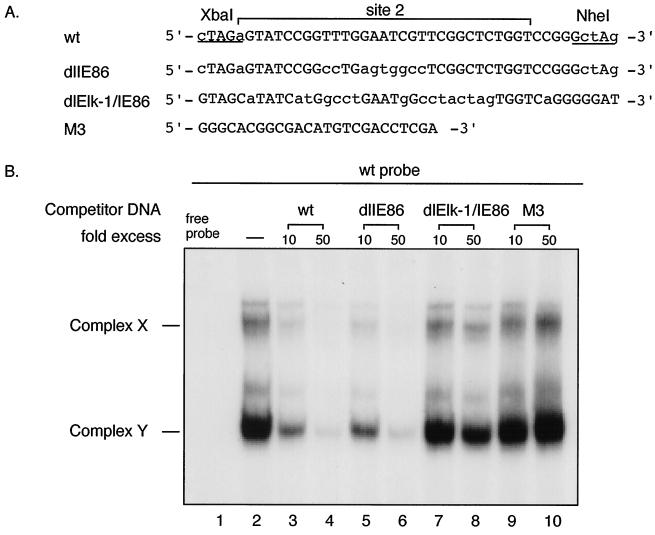
Previous DNase I footprint assays demonstrated that cellular proteins from both HeLa cell and HFF nuclear extracts bound site 2 DNA sequence (31). EMSA detected two DNA-protein complexes, designated complex X and Y. The identity of the cellular protein(s) was not known. We performed EMSA and competition assays with wild-type site 2 or mutant site 2 (dlIE86 or dlElk-1/IE86) DNA sequences using HeLa or HFF nuclear extracts. Nuclear extract was incubated with or without 10- or 50-fold molar excess of each of nonradiolabeled competitor DNAs (Fig. 7A) prior to the addition of the radioactive wild-type site 2 probe. Without the addition of the competitor DNA, EMSA detected two DNA-protein complexes, X and Y, as expected (Fig. 7B, lane 2). In competition assays, the nonradioactive wild-type and dlIE86 DNA fragments competed efficiently for the formation of the complexes X and Y (Fig. 7B, lanes 3 and 4 and lanes 5 and 6, respectively). In contrast, dlElk-1/IE86 DNA fragments did not compete (Fig. 7B, lanes 7 and 8). In addition, the heterologous control DNA M3 did not compete (Fig. 7B, lanes 9 and 10). These assays were confirmed by EMSA using radiolabeled wild type, dlIE86 or dlElk-1/IE86 probes with HeLa cell or HFF nuclear extract (data not shown). In summary, cellular protein binding to site 2 was disrupted in dlElk-1/IE86 DNA and intact in dlIE86 DNA. This binding difference might be responsible for the difference in UL4-CAT promoter expression between RVdlElk-1/IE86-xs and wild type or RVdlIE86-as.
FIG. 7.
EMSA and competition assay with wild-type (wt) and mutant site 2 DNA using HeLa cell nuclear extract. (A) Sequences of the wt and mutant (dlIE86 and dlElk-1/IE86) DNA probes. Probes and competitor DNAs were generated as described in Materials and Methods. (B) Autoradiogram of EMSA and competition assay using HeLa cell nuclear extract. Lanes: 1, free wt site 2 probe alone; 2, wt site 2 probe plus HeLa cell nuclear extract; 3, 5, 7, and 9, wt site 2 probe plus HeLa cell nuclear extract in the presence of a 10-fold molar excess of nonradioactive wt, dlIE86, dlElk-1/IE86, and M3 control DNA fragments, respectively; 4, 6, 8, and 10, wt probe plus HeLa cell nuclear extract in the presence of a 50-fold molar excess of nonradioactive wt, dlIE86, dlElk-1/IE86, and M3 control DNA fragments, respectively. The specific complexes X and Y are indicated. Free probe is at the bottom of the gel, which is not shown.
Cellular protein(s) in complex Y.
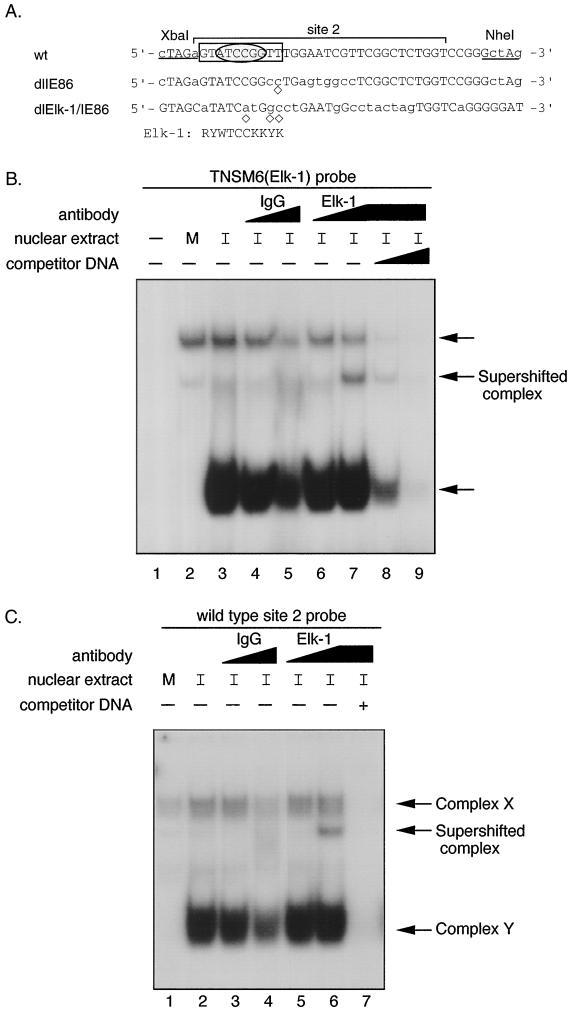
To identify candidate cellular site 2 binding proteins, a transcription factor database was searched. By using the TESS program, the site 2 sequence was predicted to bind cellular transcription factors Elk-1 or GATA-1. The consensus sequence for Elk-1 DNA binding contains the core sequence 5′-GGA-3′ as shown in Fig. 8A. There is a second 5′-GGA-3′ trinucleotide motif present in the wild-type site 2 region, but the flanking sequence does not fit the predicted Elk-1 consensus binding site, and therefore, this second site was not investigated further. Since recent reports indicated that the MAPK/ERK pathway was activated by HCMV infection (69) and Elk-1 is a target for phosphorylation at the end of this pathway, we determined whether Elk-1 could be one of the cellular proteins bound to the site 2 DNA sequence. A probe (TNSM6) containing a known Elk-1 site (48) was used in EMSA. Nuclear extracts from mock-infected or HCMV Towne-infected HFFs (24 hpi) were incubated with or without 10- or 50-fold molar excess of nonradioactive wild-type site 2 DNA prior to the addition of the rabbit polyclonal antibody against Elk-1. After incubation with TNSM6 probe, protein-DNA complexes similar to X and Y were detected, and anti-Elk-1 antibody, but not control IgG, supershifted a portion of the faster-migrating complex (Fig. 8B, lane 7). The formation of the new complex disappeared gradually as the amount of nonradioactive competitive wild-type site 2 DNA increased (Fig. 8B, lane 8 and 9). When the site 2 wild-type DNA was used as a probe, only the anti-Elk-1 antibody caused a supershift (Fig. 8C, lane 6). The new complex can be competed away by wild-type nonradioactive site 2 DNA fragments (Fig. 8C, lane 7). These data indicated that Elk-1 could specifically bind to the site 2 element. The amount of supershift with the Elk-1 polyclonal antibody was not complete, suggesting that the antibody is not very efficient in recognizing the Elk-1 antigen under the conditions used or complex Y is also formed by another member of the Ets family. EMSA also revealed a striking result when mock-infected versus HCMV-infected HFF nuclear extracts were used. The formation of the fast-migrating DNA-protein complex Y was greatly induced by HCMV infection (Fig. 8B, compare lanes 2 and 3, or Fig. 8C, compare lanes 1 and 2).
FIG. 8.
Transcription factor Elk-1 in DNA-protein complex Y. (A) Comparison of the DNA sequence of dlIE86 and dlElk-1/IE86 with wild-type (wt) site 2 DNA. Site 2 (−169 to −139) is designated by a bracket. The mutated nucleotides are shown in lowercase letters. The computer-predicted Elk-1 consensus binding site is shown and also boxed in the wt probe. Abbreviations: R = A or G; Y = C or T; W = A or T; K = G or T. ◊, nucleotide which does not fit the Elk-1 consensus sequence. The sequence in an oval reflects the core consensus for Elk-1/SAP-1 binding. (B) EMSA and competition assay with TNSM6 (Elk-1) probe. Nuclear extract from either mock-infected or HCMV-infected (24 hpi) HFFs was incubated with or without 10- or 50-fold molar excess of nonradioactive wild-type site 2 DNA at RT for 15 min before either control IgG or anti-Elk-1 IgG antibody was added. The IgGs were at the same protein concentrations. The reaction mixture was incubated at RT for 30 min, and then probe was added. The complexes were fractionated as described in Materials and Methods. Lanes: 1, TNSM6 probe alone; 2, TNSM6 probe plus mock-infected HFF nuclear extract; 3, TNSM6 probe plus HCMV-infected HFF nuclear extract; 4 and 5, same as lane 3, plus 1 μl (lane 4) or 5 μl (lane 5) of control IgG; 6 and 7, same as lane 3, plus 1 μl (lane 6) or 5 μl (lane 7) of anti-Elk-1 (IgG) polyclonal antibody; 8 and 9, same as 7, plus a 10- or 50-fold molar excess of nonradioactive wild-type site 2 DNA, respectively. M, mock infection; I, 24 h HCMV infection. (C) EMSA and competition with wild-type probe and IgG or anti-Elk-1 (IgG) polyclonal antibody as described for panel B, except lane 7 contains a 50-fold molar excess of nonradioactive wild-type site 2 DNA. Complexes X and Y are indicated by arrows.
DISCUSSION
Previous transient-transfection experiments demonstrated two cis-regulatory sites for the transcriptional regulation of the HCMV early UL4 promoter, the NF-Y binding site, and the site 2 region. Since NF-Y can interact with histone acetyltransferases (HATs) (15), the binding of NF-Y between −98 to −88 relative to the transcription start site could recruit HATs to the HCMV UL4 promoter and be responsible for the activation of this early viral promoter. Alternatively, the IE86 protein encoded by the IE2 gene could bind to a putative IE86 protein binding sites and function by interacting directly or indirectly with HAT. We determined the role of the NF-Y binding and a putative distal IE86 protein binding site, and to our surprise, recombinant viruses with mutations had promoter activity similar to the wild-type UL4-CAT promoter. Therefore, the NF-Y site did not play a role in this early viral promoter activation in the context of the unique short component of the viral genome.
The site 2 region can specifically bind to cellular protein(s) (31). We have identified one of the cellular proteins as Elk-1. Recombinant virus with mutations in both of the putative IE86 protein binding sites and the Elk-1 protein binding site in the site 2 region had a significant reduction in UL4-CAT promoter activity. So far, we cannot absolutely rule out the possibility that the putative IE86 protein binding site might play a role in the activation of the UL4-CAT promoter, but we consider this mechanism unlikely. Transcription factor binding site database search analysis (http://mpap1.trc.rwcp.or.jp/research/db/TFSEARCHJ.html) did not reveal any repressor protein binding sites in the sequence of dlElk-1/IE86 or any specific activator binding sites in the putative IE86 protein binding sites of the wild-type sequence.
Supershift experiments demonstrated that cellular transcription factor Elk-1 binds to site 2 and is in a DNA-protein complex designated complex Y. The formation of DNA-protein complex Y was greatly induced after HCMV infection. Elk-1 is one of the members of the Ets family of transcription factors, which can bind to DNA motifs containing a core 5′-GGA-3′ trinucleotide (16, 74). We cannot rule out the possibility of other DNA binding proteins. Several candidate proteins, such as cellular transcription factors Sp1, SRF, and SAP-1 were not detected in complex Y by EMSA, by supershift assay, or by competition assays (data not shown).
Previous studies have demonstrated that Elk-1 can be activated by the p38 pathway (35, 65, 90, 95). In HCMV-infected cells including HFFs and human embryonic lung fibroblasts, both the ERK1/2 and p38 MAPK pathways were activated early after infection (36, 69). Involvement of the SAPK/JNK pathway in Elk-1 activation is unlikely, since SAPK/JNK is not activated up to 48 hpi with HCMV (36). In the absence of prestimulation of the infected cells, the ERK1/2 activation can be detected at 15 min postinfection and maintained elevated through 8 hpi. The p38 activity can be detected at 8 hpi and maintained throughout late times after infection (36, 69). The involvement of kinases in the phosphorylation and activation of Elk-1 protein is under further investigation. In addition, a recombinant virus with just the Elk-1 site mutated is being isolated.
The second possible mechanism of UL4 promoter activation could be the IE86 protein that transactivates early viral promoters by interacting with the basal transcription complex at the TATA box. The IE86 protein has been shown to be phosphorylated in vitro by ERK2, a member of MAPKs (28). An adenovirus vector preexpressing the IE86 protein alone cannot efficiently activate the UL4 promoter as determined by gene array assay (E. A. Murphy, G. C. Bullock, W. A. Bresnahan, D. N. Streblow, J. A. Nelson, T. E. Shenk, and M. F. Stinski, submitted for publication). This suggests that another unidentified viral or cellular protein(s) or a combination of the IE86 protein with other unidentified viral or cellular proteins is necessary for activation of this early viral promoter.
Studies of other viruses have demonstrated regulation of viral gene expression by the cellular MAPK/ERK pathway. For example, Friend spleen focus-forming virus, coxsackievirus B3, human hepatitis B virus, SV40, adenovirus, and human immunodeficiency virus type 1 induce an activation of the MAPK/ERK pathway after infection (6, 32, 52, 63, 78, 94). In addition to the HCMV IE86 protein, adenovirus E1A protein has been found to be phosphorylated by the activated ERKs (28, 94). A recent report indicated that the early HCMV promoter, UL112-113, which drives the synthesis of a 2.2-kb RNA, is a MAPK/ERK-responsive promoter (69). The regulatory site identified is a CREB/ATF site, which is a target of several signaling pathways, including the cyclic AMP-dependent protein kinase A, calcium or calmodulin kinases, the ERK, SAPK/JNK and p38 pathways (68, 85, 90, 91).
Different viruses can induce activation of one or more of the MAPKs. For example, the simian immunodeficiency virus can induce activation of all three major MAPKs upon binding to cell surface receptor (64). Among herpesviruses, cells infected with HSV-1 can have both the p38 and SAPK/JNK pathways activated; however, the activation of ERK was not detected (60, 98). The ERK, SAPK/JNK, and p38 MAPK pathways were all activated following infection with Epstein-Barr virus (1, 17).
For HSV-1, the mechanisms for activation of the SAPK/JNK pathway have been narrowed down to the point that early and late viral proteins are not necessary for this activation (60). More interestingly, recent reports by Adamson et al. (1) demonstrated that the Epstein-Barr virus IE protein BZLF1(Z) or BRLF1(R) expressed by adenovirus vectors can activate the SAPK/JNK and the p38 signaling pathways in HeLa cells. Some investigators suggest that the latent membrane protein 1 might be responsible for the activation of ERKs in rodent fibroblasts (68). Which viral IE proteins of HCMV are responsible for activation of either ERK or p38 signal transduction pathways is not known.
So far, studies have focused on transcriptional regulation of the UL4 promoter and translational regulation of glycoprotein gp48 encoded by the UL4 gene (3, 7, 8, 30, 31, 58), but the putative function of this viral glycoprotein has not been determined yet. Although the HCMV UL4 gene is not essential for viral growth in tissue culture, whether or not this viral glycoprotein is dispensable for infection and pathogenesis in a human host is not known yet.
In summary, these results indicate that the UL4 promoter of HCMV contains an Elk-1 protein-binding site. The activation of the HCMV UL4 promoter is affected by the activation of an Ets family of transcription factors, such as Elk-1, and the MAPK/ERK pathway may play a role in HCMV early viral promoter activation. The activation of the MAPK/ERK pathway by these viruses may contribute to the regulation of the host cell cycle and viral pathogenesis (89).
ACKNOWLEDGMENTS
We thank members of the laboratory for helpful discussion and Richard Roller for critical reading of the manuscript.
This work was supported by grant AI-13562 from the National Institutes of Health.
REFERENCES
- 1.Adamson A L, Darr D, Holley-Guthrie E, Johnson R A, Mauser A, Swenson J, Kenney S. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J Virol. 2000;74:1224–1233. doi: 10.1128/jvi.74.3.1224-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J-H, Chiou C-J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. doi: 10.1016/s0378-1119(98)00056-0. [DOI] [PubMed] [Google Scholar]
- 3.Alderete J P, Jarrahian S, Geballe A P. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J Virol. 1999;73:8330–8337. doi: 10.1128/jvi.73.10.8330-8337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, et al., editors. Raven Press. 1990. pp. 1981–2010. Ltd., New York, N.Y. [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York: John Wiley & Sons; 1989. pp. 2.9.1–2.9.5. [Google Scholar]
- 6.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J, Geballe A P. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA. 1998;4:181–188. [PMC free article] [PubMed] [Google Scholar]
- 9.Caswell R, Hagemeier C, Chiou C-J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE2) protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via region of IE2 required for transcription regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 10.Chang C-P, Malone C L, Stinski M F. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J Virol. 1989;63:281–290. doi: 10.1128/jvi.63.1.281-290.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C-P, Vesole D H, Nelson J A, Oldstone M B A, Stinski M F. Identification and expression of a human cytomegalovirus early glycoprotein. J Virol. 1989;63:3330–3337. doi: 10.1128/jvi.63.8.3330-3337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chee M S, Bankier S, Beck S, Bohni R, Brown C R, Horsnell T, Hutchisno III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Chiou C J, Zong J, Waheed I, Hayward G S. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–361. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- 15.Currie R A. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 16.Dhulipal P D K. Ets oncogene family. Indian J Exp Biol. 1997;35:315–322. [PubMed] [Google Scholar]
- 17.Fenton M, Sinclair A J. Divergent requirements for the MAPK/ERK signal transduction pathway during initial virus infection of quiescent primary B cells and disruption of Epstein-Barr virus latency by phorbol esters. J Virol. 1999;73:8913–8916. doi: 10.1128/jvi.73.10.8913-8916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fish K N, Depto A S, Moses A V, Britt W, Nelson J A. Growth kinetics of human cytomegalovirus are altered in monocyte-derived macrophages. J Virol. 1995;69:3737–3743. doi: 10.1128/jvi.69.6.3737-3743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furnari B A, Poma E, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol. 1993;67:4981–4991. doi: 10.1128/jvi.67.8.4981-4991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J Virol. 1997;71:7048–7060. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorman M C, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham F L, van der Eb A J. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 23.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the E. coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 24.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harel N Y, Alwine J C. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J Virol. 1998;72:5481–5492. doi: 10.1128/jvi.72.7.5481-5492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, Malone C L, Stinski M F. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J Virol. 1994;68:2108–2117. doi: 10.1128/jvi.68.4.2108-2117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Stinski M F. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J Virol. 1995;69:7612–7621. doi: 10.1128/jvi.69.12.7612-7621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber M, Watson K A, Selinka H C, Carthy C M, Klingel K, Mcmanus B M, Kandolf R. Cleavage of RasGAP and phosphorylation of mitogen-activated protein kinase in the course of coxsackievirus B3 replication. J Virol. 1999;73:3587–3594. doi: 10.1128/jvi.73.5.3587-3594.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibanez C E, Schrier R, Ghazal P, Wiley C, Nelson J A. Human cytomegalovirus productively infects primary differentiated macrophages. J Virol. 1991;65:6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacque J M, Mann A, Enslen H, Sharuva N, Brichacek B, Davis R J, Stevenson M. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 1998;17:2607–2618. doi: 10.1093/emboj/17.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janknecht R, Hunter T. Convergence of MAP kinase pathways on the ternary complex factor Sap-1a. EMBO J. 1997;16:1620–1627. doi: 10.1093/emboj/16.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson R A, Huong S-M, Huang E-S. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J Virol. 2000;74:1158–1167. doi: 10.1128/jvi.74.3.1158-1167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson R A, Huong S-M, Huang E-S. Inhibitory effect of 4-(4-fluorophenyl)-2-(4-hydroxyphenyl)-5-(4-pyridyl)1H-imidazole on HCMV DNA replication and permissive infection. Antivir Res. 1999;41:101–111. doi: 10.1016/s0166-3542(99)00002-9. [DOI] [PubMed] [Google Scholar]
- 38.Jones T R, Muzithras V P, Gluzman Y. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J Virol. 1991;65:5860–5872. doi: 10.1128/jvi.65.11.5860-5872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones T R, Muzithras V R. A cluster of dispensable genes within the human cytomegalovirus genome short component: ISR1, US1 through US5, and the US6 family. J Virol. 1992;66:2541–2546. doi: 10.1128/jvi.66.4.2541-2546.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerry J A, Priddy M A, Jervey T Y, Kohler C P, Staley T L, Vanson C D, Jones T R, Iskenderian A C, Anders D G, Stenberg R M. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996;70:373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klucher K M, Rabert D K, Spector D H. Sequences in the human cytomegalovirus 2.7-kilobase RNA promoter which mediate its regulation as an early gene. J Virol. 1989;63:5334–5343. doi: 10.1128/jvi.63.12.5334-5343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klucher K M, Spector D H. The human cytomegalovirus 2.7-kilobase RNA promoter contains a functional binding site for the adenovirus major late transcription factor. J Virol. 1990;64:4189–4198. doi: 10.1128/jvi.64.9.4189-4198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohler C P, Kerry J A, Carter M, Muzithras V P, Jones T R, Stenberg R M. Use of recombinant virus to assess human cytomegalovirus early and late promoters in the context of the viral genome. J Virol. 1994;68:6589–6597. doi: 10.1128/jvi.68.10.6589-6597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krieg P A, Melton D A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–414. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 48.Kujoth G C, Robinson D F, Fahl W E. Binding of ETS family members is important for the function of the c-sis/platelet-derived growth factor-B TATA neighboring sequence in 12-O-tetradecanoylphorbol-13-acetate-treated K562 cells. Cell Growth Differ. 1998;9:523–534. [PubMed] [Google Scholar]
- 49.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lashmit P E, Stinski M F, Murphy E A, Bullock G C. A cis repression sequence adjacent to the transcription start site of the human cytomegalovirus US3 gene is required to down regulate gene expression at early and late times after infection. J Virol. 1998;72:9575–9584. doi: 10.1128/jvi.72.12.9575-9584.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lathey J L, Spector S A. Unrestricted replication of human cytomegalovirus in hydrocortisone-treated macrophages. J Virol. 1991;65:6371–6375. doi: 10.1128/jvi.65.11.6371-6375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C J, Ueda Y, Shi B, Borodyansky L, Huang L, Li Y Z, Pardee A B. Tat protein induces self-perpetuating permissivity for productive HIV-1 infection. Proc Natl Acad Sci USA. 1997;94:8116–8120. doi: 10.1073/pnas.94.15.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lukac D M, Harel N Y, Tanese N, Alwine J C. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luu P, Flores O. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J Virol. 1997;71:6683–6691. doi: 10.1128/jvi.71.9.6683-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maciejewski J P, Bruening E E, Donahue R E, Mocarski E S, Young N S, St. Jeor S C. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood. 1992;80:170–178. [PubMed] [Google Scholar]
- 58.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin P, Vass W C, Schiller J T, Lowry D R, Velu T J. The bovine papillomavirus E5 transforming protein can stimulate the transforming activity of EGF and CSF receptors. Cell. 1989;59:21–32. doi: 10.1016/0092-8674(89)90866-0. [DOI] [PubMed] [Google Scholar]
- 60.McLean T I, Bachenheimer S L. Activation of c-Jun N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J Virol. 1999;73:8415–8426. doi: 10.1128/jvi.73.10.8415-8426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meier J L, Stinski M F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minton E J, Tysoe C, Sinclair J H, Sissons J G. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muszynski K W, Ohashi T, Hanson C, Ruscetti S K. Both the polycythemia- and anemia-inducing strains of Friend spleen focus-forming virus induce constitutive activation of the Raf-1/mitogen-activated protein kinase signal transduction pathway. J Virol. 1998;72:919–925. doi: 10.1128/jvi.72.2.919-925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popik W, Pitha P M. Early activation of mitogen-activated protein kinase kinase, extracellular signal-regulated kinase, p38 mitogen-activated protein kinase, and c-Jun N-terminal kinase in response to binding of simian immunodeficiency virus to Jurkat T cells expressing CCR5 receptor. Virology. 1998;252:210–217. doi: 10.1006/viro.1998.9466. [DOI] [PubMed] [Google Scholar]
- 65.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 1996;15:6552–6562. [PMC free article] [PubMed] [Google Scholar]
- 66.Ripalti A, Mocarski E S. The products of human cytomegalovirus genes UL1-UL7, including gp48, are dispensable for growth in cell culture. In: Landini M P, editor. Progress in cytomegalovirus research: proceedings of the Third International Cytomegalovirus Workshop. Amsterdam, The Netherlands: Elsevier Science Publishers; 1991. pp. 57–62. [Google Scholar]
- 67.Roberts M L, Cooper N R. Activation of a Ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology. 1998;240:93–99. doi: 10.1006/viro.1997.8901. [DOI] [PubMed] [Google Scholar]
- 68.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 69.Rodems S M, Spector D H. Extracellular signal-regulated kinase activity is sustained early during human cytomegalovirus infection. J Virol. 1998;72:9173–9180. doi: 10.1128/jvi.72.11.9173-9180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samaniego L A, Tevethia M J, Spector D J. The human cytomegalovirus 86-kilodalton immediate-early 2 protein: synthesis as a precursor polypeptide and interaction with a 75-kilodalton protein of probable viral origin. J Virol. 1994;68:720–729. doi: 10.1128/jvi.68.2.720-729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 72.Schmidbauer M, Budka H, Ulrich W, Ambros P. Cytomegalovirus (CMV) disease of the brain in AIDS and connatal infection: a comparative study by histology, immunocytochemistry, and in situ DNA hybridization. Acta Neuropathol Berlin. 1989;79:286–293. doi: 10.1007/BF00294663. [DOI] [PubMed] [Google Scholar]
- 73.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86 kDa protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shore P, Sharrocks A D. The ETS-domain transcription factors Elk-1 and SAP-1 exhibit differential DNA binding specificities. Nucleic Acids Res. 1995;23:4698–4706. doi: 10.1093/nar/23.22.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sinzger C, Plachter B, Grefte A, Houwe The T, Jahn G. Tissue macrophages are infected by human cytomegalovirus. J Infect Dis. 1996;173:240–245. doi: 10.1093/infdis/173.1.240. [DOI] [PubMed] [Google Scholar]
- 76.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 77.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the MAP kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 79.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 80.Spector D J, Tevethia M J. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J Virol. 1994;68:7549–7553. doi: 10.1128/jvi.68.11.7549-7553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Speir E, Modali R, Huang E, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 82.Staprans S I, Rabert D K, Spector D H. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J Virol. 1988;62:3463–3473. doi: 10.1128/jvi.62.9.3463-3473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stinski M F. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J Virol. 1976;19:594–609. doi: 10.1128/jvi.19.2.594-609.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stinski M F. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J Virol. 1978;26:686–701. doi: 10.1128/jvi.26.3.686-701.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 86.Takekoshi M, Maeda-Takekoshi F, Ihara S, Sakuma S, Watanabe Y. Site-specific stable insertion into the human cytomegalovirus genome of a foreign gene under control of the SV40 promoter. Gene. 1991;101:209–213. doi: 10.1016/0378-1119(91)90413-6. [DOI] [PubMed] [Google Scholar]
- 87.Thomsen D R, Stinski M F. Cloning of the human cytomegalovirus genome as endonuclease XbaI fragments. Gene. 1981;16:207–216. doi: 10.1016/0378-1119(81)90077-9. [DOI] [PubMed] [Google Scholar]
- 88.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate early gene (US3) by a silencer/enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tibbles L A, Woodgett J R. The stress-activated protein kinase pathways. Cell Mol Life Sci. 1999;55:1230–1254. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 91.Vanhoutte P, Barnier J V, Guibert B, Pages C, Besson M J, Hipskind R A, Caboche J. Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinase-dependent pathway in brain slices. Mol Cell Biol. 1999;19:136–146. doi: 10.1128/mcb.19.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wade E J, Klucher K M, Spector D H. An AP-1 binding site is the predominant cis-acting regulatory element in the 1.2-kilobase early RNA promoter of human cytomegalovirus. J Virol. 1992;66:2407–2417. doi: 10.1128/jvi.66.4.2407-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waheed I, Chiou C, Ahn J, Hayward G S. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology. 1998;252:235–257. doi: 10.1006/viro.1998.9448. [DOI] [PubMed] [Google Scholar]
- 94.Whalen S G, Marcellus R C, Whalen A, Ahn N G, Ricciardi R P, Branton P E. Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J Virol. 1997;71:3545–3553. doi: 10.1128/jvi.71.5.3545-3553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Whitmarsh A J, Yang S-H, Su M S S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Widmann C, Gibson S, Jarpe M B, Johnson G L. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 97.Wu J, O'Neill J, Barbosa M S. Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells. J Virol. 1998;72:236–244. doi: 10.1128/jvi.72.1.236-244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zachos G, Clements B, Conner J. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J Biol Chem. 1999;274:5097–5103. doi: 10.1074/jbc.274.8.5097. [DOI] [PubMed] [Google Scholar]