Summary
Background
SARS-CoV-2 is a single-stranded positive-sense RNA virus. Several negative-sense SARS-CoV-2 RNA species, both full-length genomic and subgenomic, are produced transiently during viral replication. Methodologies for rigorously characterising cell tropism and visualising ongoing viral replication at single-cell resolution in histological sections are needed to assess the virological and pathological phenotypes of future SARS-CoV-2 variants. We aimed to provide a robust methodology for examining the human lung, the major target organ of this RNA virus.
Methods
A prospective cohort study took place at the University Hospitals Leuven in Leuven, Belgium. Lung samples were procured postmortem from 22 patients who died from or with COVID-19. Tissue sections were fluorescently stained with the ultrasensitive single-molecule RNA in situ hybridisation platform of RNAscope combined with immunohistochemistry followed by confocal imaging.
Findings
We visualised perinuclear RNAscope signal for negative-sense SARS-CoV-2 RNA species in ciliated cells of the bronchiolar epithelium of a patient who died with COVID-19 in the hyperacute phase of the infection, and in ciliated cells of a primary culture of human airway epithelium that had been infected experimentally with SARS-CoV-2. In patients who died between 5 and 13 days after diagnosis of the infection, we detected RNAscope signal for positive-sense but not for negative-sense SARS-CoV-2 RNA species in pneumocytes, macrophages, and among debris in the alveoli. SARS-CoV-2 RNA levels decreased after a disease course of 2–3 weeks, concomitant with a histopathological change from exudative to fibroproliferative diffuse alveolar damage. Taken together, our confocal images illustrate the complexities stemming from traditional approaches in the literature to characterise cell tropism and visualise ongoing viral replication solely by the surrogate parameters of nucleocapsid-immunoreactive signal or in situ hybridisation for positive-sense SARS-CoV-2 RNA species.
Interpretation
Confocal imaging of human lung sections stained fluorescently with commercially available RNAscope probes for negative-sense SARS-CoV-2 RNA species enables the visualisation of viral replication at single-cell resolution during the acute phase of the infection in COVID-19. This methodology will be valuable for research on future SARS-CoV-2 variants and other respiratory viruses.
Funding
Max Planck Society, Coronafonds UZ/KU Leuven, European Society for Organ Transplantation
Keywords: Single-stranded RNA virus, RNAscope, Nucleocapsid, Spike
Research in context.
Evidence before this study
Various approaches have been taken to examine SARS-CoV-2 cell tropism and ongoing viral replication in the human lung. Assessing viral replication by detecting subgenomic viral RNA species with qRT-PCR on homogenized tissue samples does not provide spatial information at single-cell resolution and may reflect not only ongoing viral replication but also recent replication. Several studies have reported on the histological visualisation of SARS-CoV-2 infection in the lung with in situ hybridisation (ISH), often with the ultrasensitive single-molecule platform of RNAscope. Immunohistochemistry (IHC) with antibodies against viral nucleocapsid or spike protein has been reported to be less reliable than ISH due to the often-questionable specificity and high background. In the majority of ISH histological studies, single-colour chromogenic brightfield imaging was performed to visualise viral RNA species. A few studies visualised positive-sense viral RNA species or viral protein together with target cell type markers in double-stained sections. Detection of positive-sense SARS-CoV-2 RNA species and viral proteins was reported mostly for ciliated cells, pneumocytes, endothelial cells, and alveolar macrophages. SARS-CoV-2 is a single-stranded positive-sense RNA virus. Several RNAscope probes for negative-sense SARS-CoV-2 RNA species are commercially available and enable the visualisation of ongoing viral replication by the viral RNA-dependent RNA polymerase at single-cell resolution in histological sections. There have been no reports on confocal imaging of lung tissue sections for negative-sense SARS-CoV-2 RNA species together with markers for target cell types.
Added value of this study
We performed four-channel confocal imaging of human lung sections fluorescently stained with RNAscope and IHC for several SARS-CoV-2 RNA species or nucleocapsid protein and various target cell type markers. We visualised perinuclear RNAscope signal representing negative-sense SARS-CoV-2 RNA species in ciliated cells of the bronchiolar epithelium in human lung tissue sections and in ciliated cells of a primary culture of human airway epithelium experimentally infected with SARS-CoV-2. Our findings confirm and extend previous observations that ciliated cells are the major target cell type in the lower respiratory tract, in the sense that they support viral replication.
Implications of all the available evidence
SARS-CoV-2 replication can be visualised at single-cell resolution in histological sections of human lung tissue by virtue of a punctate fluorescent signal residing at a characteristic perinuclear location with RNAscope probes for various negative-sense SARS-CoV-2 RNA species. Confocal imaging of sections stained with a combination of fluorescence RNAscope and IHC is a robust methodology for characterising cell tropism and visualising ongoing viral replication in airways and lung parenchyma. Future SARS-CoV-2 variants of concern may be more virulent owing to changes in cell or tissue tropism, or to differences in spatial or temporal patterns of replication. Our methodology is based on a battery of commercially available probes and antibodies and will be valuable for a rigorous assessment of the virological and pathological phenotypes of future SARS-CoV-2 variants as well as other respiratory viruses, in particular RNA viruses.
Introduction
COVID-19 was characterised a pandemic by the World Health Organisation on 11 March 2020. Since then, several variants of concern have independently emerged from the ancestral SARS-CoV-2 Wuhan-Hu-1 strain. With global virus transmission showing no sign of abatement in the endemic phase, the scenario of the potential emergence of more virulent variants warrants further optimisation of methods to assess accurately the virological and pathological phenotypes in COVID-19 patients. Light-microscopic histological methods have provided insights into the kinetics of the infection in the major target organ of this RNA virus, the human lung, by the visualisation of SARS-CoV-2 RNA species with in situ hybridisation (ISH) and of SARS-CoV-2 proteins with immunohistochemistry (IHC). Samples from COVID-19 patients with a short disease course are more informative in this regard because the SARS-CoV-2 viral load peaks before or around the onset of symptoms.1 Such histological studies are typically conducted on lung samples procured postmortem from patients who died from or with COVID-19. A postmortem cohort study generates a collection of snapshots from several singular disease courses.
Negative-sense full-length genomic and subgenomic RNA species are produced transiently by the viral RNA-dependent RNA polymerase during the life cycle of this single-stranded positive-sense RNA virus.2,3 These negative-sense SARS-CoV-2 RNA species are not incorporated into virions that are released from infected cells such that the presence of negative-sense SARS-CoV-2 RNA species in a given cell reflects viral replication that was ongoing in this cell at the time of tissue fixation. Using postmortem tissue samples of respiratory mucosa and olfactory cleft mucosa of COVID-19 patients who died during the acute phase of the infection, we reported a robust methodology to visualise negative-sense SARS-CoV-2 RNA species at single-cell resolution with a battery of commercially available SARS-CoV-2-N-sense, SARS-CoV-2-S-sense, and SARS-CoV-2-orf1ab-sense RNAscope probes.4,5 We showed that RNAscope puncta representing negative-sense SARS-CoV-2 RNA species reside characteristically at a perinuclear location within a cell, whereas RNAscope puncta representing positive-sense SARS-CoV-2 RNA species and nucleocapsid-immunoreactive signal are diffusely spread throughout the cytoplasm of the cell.4,5 Arguably, the mere presence of positive-sense SARS-CoV-2 RNA species or viral antigen within a cell does not prove that this cell had been exposed to an infectious virus particle, let alone that it was replicating the virus; instead, the cell could have taken up viral RNA or viral protein, intact or fragmented, in any of several ways. Fluorescence staining with ISH and IHC allows for multiplex visualisation of viral RNA or protein together with RNA or protein markers for target cell types. IHC visualises cellular antigens as an immunoreactive signal that often diffusely fills the cells and outlines their contours, thereby facilitating cell type identification. Imaging with confocal microscopy reveals these fluorescent signals within the same plane of the section, thereby minimising the risk of erroneous interpretation of superimposed signals as coexpression or colocalisation, which often occurs in brightfield microscopy and epifluorescence microscopy. With the combined confocal visualisation of RNAscope puncta and IHC signal, a given cell type or a particular cell can be unmistakably identified as a target cell type or infected, respectively.
Taken together, multiplex fluorescence ISH/IHC staining followed by confocal microscopy allows for the rigorous characterisation of cell tropism and the visualisation of ongoing viral replication at single-cell resolution in histological sections. To our knowledge, the combined application of ISH probes for negative-sense SARS-CoV-2 RNA species, multiplex staining, and confocal microscopy has not been reported in the literature on the lungs of COVID-19 patients. Table S1 provides an overview of the relevant literature.
Methods
Patients, study design, and ethics
A cohort of 22 patients who died from or with a qRT-PCR-proven SARS-CoV-2 infection between March 2020 and February 2021 at the University Hospitals Leuven (Leuven, Belgium) were included in the CONTAGIOUS study protocol S63881, which was approved by the Ethics Committee Research, UZ/KU Leuven. The cohort is consecutive in the sense that all patients from whom lung tissue was available were included. These patients are henceforth referred to as "COVID cases". Informed consent was obtained from next of kin. Inclusion of the control cases was under study protocols S52174 and S63978, which were approved by the Ethics Committee Research, UZ/KU Leuven.
Clinical data
The digital health records of the COVID cases and control cases were retrospectively reviewed and analysed to collect relevant information. The collection, processing, and disclosure of personal data, such as patient demographic, health, and medical information, are subject to compliance with Regulation (EU) 2016/679, also referred as the General Data Protection Regulation, and with the Belgian Law on the protection of natural persons regarding the processing of personal data. Therefore, combinations of data deemed to be identificatory to specific persons cannot be disclosed.
Tissue sampling
For 15 COVID cases, lung samples were harvested bedside via a right anterolateral thoracotomy in the fourth or fifth intercostal space. A cubic sample of 3 × 3 × 3 cm was taken from the right lower lobe and divided in a large piece that was transferred in 4% paraformaldehyde (PFA) for overnight fixation and in six small pieces that were snap-frozen. For 7 COVID cases, lung samples from all five lobes were procured during conventional autopsy. For the control cases, whole lungs were removed during the course of multi-organ donation and were transported to the laboratory where they were inflated to total lung capacity and frozen in the fumes of liquid nitrogen. After storage at −80 °C, the frozen lungs were sliced with a band saw in two cm axial discs and cylinders with a diameter of two cm were sampled. These cylinders were split in half and one half was transferred to 4% PFA for overnight fixation. After PFA fixation, lung samples were processed into formalin-fixed paraffin-embedded (FFPE) blocks, which were sectioned with a semi-automated Leica microtome at 5 μm, collected on BOND Plus slides (Leica Biosystems, Cat#S21.2113. A) and heated at 56 °C overnight. Slides were stored at 4 °C in dry conditions until staining. For COVID case 21, lung samples were transferred into containers with 10% neutral buffered formalin (Sigma–Aldrich, Cat#HT5011) for >72 h. Samples were treated for cryoprotection by immersing serially in 15%, 25%, and 30% sucrose (Sigma–Aldrich, Cat#S0389-1KG) in 1 x PBS over a period of 6–8 days and then embedded in Tissue-Tek O.C.T. compound (Sakura, Cat#4583) on dry ice. Cryosections of 6–8 μm thickness were cut on a Leica CM3050 S cryostat and collected on SuperFrost Plus Gold slides (Thermo Fisher Scientific/Menzel Gläser, Cat#K5800AMNZ72). Slides were air-dried at room temperature and stored at −80 °C.
Primary cultures of human airway epithelium
Four SmallAir inserts (Epithelix, Cat#SA082201) containing ∼500,000 bronchiolar airway epithelial cells from a 48-year-old male without history of smoking and without comorbidities were inoculated with ∼30,000 TCID50/insert of strain BetaCoV/Belgium GHB-03021/2020 passage 4, resulting in a multiplicity of infection of ∼0.06. The apical side of the cultures was inoculated with virus for 2 h followed by washing of the inserts with PBS. Four days after inoculation, cultures were fixed in 4% PFA for 30 min and processed into FFPE blocks. Serving as non-infected control, a culture was established in the prepandemic era from primary bronchial epithelial cells of a 58-year-old male whose lungs were deemed unsuitable for lung transplantation due to pneumonia; methods were as described6 except that cultures were expanded in Pneumacult Ex-Plus medium (StemCell Technologies, Cat#05040) and that freezing was in 10% DMSO/FBS.
RNAscope platform for RNA in situ hybridisation
Sections of lung samples and cultures were stained with the RNAscope manual assay using the Multiplex Fluorescent Detection Kit v2 (Advanced Cell Diagnostics, Cat#323110) according to manufacturer protocols and as we described previously.4,5,7 Briefly, slides with FFPE sections were baked at 60 °C for 1 h, then deparaffinised in Xylene (Leica Biosystems, Cat#3803665EG). Slides with cryosections were baked at 55° overnight. For FFPE sections, slides were pretreated with hydrogen peroxide at room temperature for 15 min, followed by permeabilization in target retrieval reagent (Advanced Cell Diagnostics, Cat#322000) for 15 min in a steamer, and digestion with Protease Plus (Advanced Cell Diagnostics, Cat#322330) at 40 °C for 20 min. For cryosections, slides were pretreated with hydrogen peroxide at room temperature for 15 min, followed by permeabilisation in target retrieval reagent for 3 min in a steamer, and digestion with Protease III (Advanced Cell Diagnostics, Cat#322337) at 40 °C for 15 min. A combination of probes for target RNA detection was hybridized at 40 °C for 2 h. Probes in the C4 channel were developed with the RNAscope 4-Plex Ancillary Kit (Advanced Cell Diagnostics, Cat#323120). Signal amplification was followed by development of appropriate HRP channels with dyes Opal 520 (Akoya Biosciences, Cat#FP1487001KT), Opal 570 (Akoya Biosciences, Cat#FP1488001KT), and Opal 690 (Akoya Biosciences, Cat#FP1497001KT). DAPI (Thermo Fisher Scientific, Cat#D1306) served as nuclear stain. Slides were mounted in Mount Solid antifade (abberior, Cat#MM-2011-2X15ML). The commercially available panel of RNAscope probes was for: SARS-CoV-2-N (nucleocapsid, giving rise to puncta hereafter abbreviated as N puncta), SARS-CoV-2-S (spike; S puncta), SARS-CoV-2-M (membrane; M puncta), SARS-CoV-2-orf1ab (open reading frames 1a and 1b; orf1ab puncta), SARS-CoV-2-N-sense (N-sense puncta), SARS-CoV-2-S-sense (S-sense puncta), and SARS-CoV-2-orf1ab-sense (orf1ab-sense puncta). Catalogue numbers of RNAscope probes can be found in the Key Resources Table of Khan et al.4 and Khan et al.5 RNAscope probes hybridise with a single RNA molecule, which is visualised as a fluorescent or chromogenic dot or punctum (plural "puncta") as a result of signal amplification.8 In an initial study, two sections from at least one lung tissue sample of each case were stained with an RNAscope probe for positive-sense SARS-CoV-2-N. The RNAscope signal was scored in a semiquantified manner as ‘+++’ for diffuse, ‘++’ for focal, ‘+’ for rare and ‘0’ for the absence of N puncta. A nearby section was stained with H&E for conventional histopathology. In the next phase, the five COVID cases with diffuse presence of N puncta, the two control cases, and sections from the infected and non-infected cultures were subjected to further stainings and imaging.
Immunohistochemistry
For co-detection of RNA and protein, IHC was performed after the final step of HRP blocker application in the RNAscope Multiplex Fluorescent Detection protocol. Slides were blocked in 10% donkey serum (Sigma–Aldrich, Cat#S30-100ML) in 0.1% Triton/PBS at room temperature for 1 h. The following primary antibodies were diluted in 2% donkey serum in 0.1% Triton/PBS and incubated at 4 °C overnight: Nucleocapsid (Sino Biological, Cat# 40143-R001) at 1:100, Cytokeratin 5/6 (Novus Biologicals, Cat#NBP2-77439) at 1:300, Cytokeratin 8 (R&D Systems, Cat#MAB3165-SP) at 1:200, Cytokeratin 8 (Novus Biologicals, Cat#NBP2-67468) at 1:100, MUC5AC (Thermo Fisher Scientific, Cat#MA5-12178) at 1:300, CD31 (Abcam, Cat#ab134168) at 1:100, Surfactant protein C (abbexa, Cat# abx027534-80) at 1:250, CD68 (BioLegend, Cat# 916104) at 1:300, CD68 (Sigma–Aldrich, Cat#SAB5500070) at 1:100, and club cell secretory protein (CC16)/Uteroglobin (ThermoFisher; Cat# PA5-95864) at 1:300. Slides were then washed in 0.1% Triton/PBS 3 x 5 min each followed by incubation with appropriate secondary antibodies at 1:500 in 2% donkey serum in 0.1% Triton/PBS for 1 h at room temperature. Secondary antibodies were Alexa Fluor Plus 647 donkey anti-rabbit (Thermo Fisher Scientific, Cat#A32795), Alexa Fluor Plus 647 donkey anti-mouse (Thermo Fisher Scientific, Cat#A32787), Alexa Fluor Plus 555 donkey anti-rabbit (Thermo Fisher Scientific, Cat#A32794), and Alexa Fluor Plus 555 donkey anti-mouse (Thermo Fisher Scientific, Cat#A32773). Slides were washed in 0.1% Triton/PBS 3 x 5 min each followed by nuclear staining with DAPI (Thermo Fisher Scientific, Cat#D1306). Slides were mounted in Mount Solid antifade (abberior, Cat#MM-2011-2X15ML).
Imaging
Confocal images were taken under a Zeiss LSM 800 confocal microscope with the Zeiss ZEN 2.6 system. From each case, at least two FFPE or cryosections labeled with the RNAscope SARS-CoV-2-N probe were imaged. One section, adjacent to the sections for RNAscope and IHC, was stained using a fully automated H&E platform (Dako CoverStainer, Agilent). The H&E-stained sections were imaged using a PANNORAMIC MIDI II scanner (3DHistech) in brightfield mode.
Tissue qRT-PCR
Viral RNA from frozen lung tissue was extracted using the QIAgen RNeasy mini kit (QIAgen, Cat#74106) according to the protocol Purification of Total RNA from Animal Tissues. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed on a QuantStudio 7 Flex Real Time PCR system (Applied Biosystems, ThermoFisher). A reaction mix was made using 5 μl TaqMan Fast Virus 1-step Master Mix (Applied Biosystems, Cat #4444434), 1.5 μl primer/probe mix from the 2019-nCoV CDC EUA kit (IDT, Cat#10006606) and supplemented with 8.5 μl RNase free water until a total of 15 μl was obtained. Thermal cycling conditions were 5 min at 50 °C, 2 min at 95 °C, followed by 45 cycles of 3 s at 95 °C and 30 s at 60 °C. Analysis was with QuantStudio Real-Time PCR Software (Applied Biosystems, ThermoFisher).
Serology
For the COVID cases, blood samples taken prior to death were stored at −20 °C. In the serum sample obtained closest to the time of death, anti-nucleocapsid (anti-N) IgG antibodies were measured on the Abbott Architect platform with the Chemiluminescence Microparticle Immuno-assay (CMIA; Cat#6R86). A signal/cut-off ratio of ≥1.4 was interpreted as positive and a ratio of <1.4 as negative.
Role of funders
Funders were not involved in study design, data collection, data analysis, data interpretation, and manuscript writing.
Results
Cohort
We procured postmortem lung samples from a cohort of 22 hospitalised patients who died from or with a diagnosed SARS-CoV-2 infection in the era prior to the emergence of variants of concern and prior to the rollout of vaccines. The cohort comprises predominantly male patients (82%) with ages ranging between 47 and 90 years [median 74; interquartile range 67–82]. Patients died within 2–51 days after PCR diagnosis (median 22; interquartile range 14–34). Samples were obtained in 15 cases bedside and in 7 cases during conventional autopsy. The lung samples of the bedside cases were obtained with a postmortem interval ranging between 15 and 115 min (median 35; interquartile range 23–50). Table S2 provides information about demographics, comorbidities, disease course, treatment, and tissue sampling of the COVID cases. Table S3 provides similar information about the control cases.
Levels of viral RNA signal
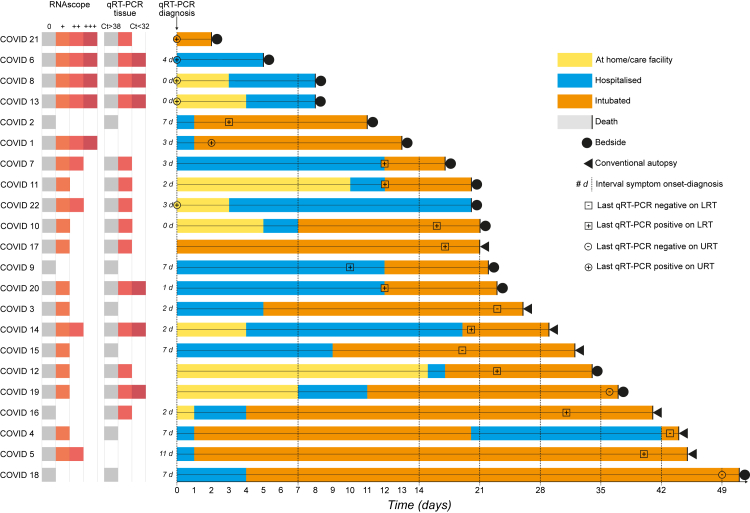
We correlated the disease course of the COVID cases with abundance of RNAscope signal and qRT-PCR amplification of SARS-CoV-2 RNA in lung tissue. Fig. 1 shows an overview in the form of a swimmer plot. Table S4 lists the Ct values of the qRT-PCR assays and semiquantified scores of the abundance of RNAscope N puncta. Both assays consistently detected viral RNA up to a diagnosis-to-death interval of 21 days (COVID 17), with one exception: no viral RNA was detected in COVID 2, who died with a diagnosis-to-death interval of 11 days and 18 days after the presumed onset of symptoms, henceforth referred to as presumptive duration of illness. Viral RNA was detected with RNAscope for a diagnosis-to-death interval as long as 45 days (COVID 5) and with qRT-PCR for a diagnosis-to-death interval as long as 41 days (COVID 16). The level of viral RNA in lung tissue decreased with longer diagnosis-to-death intervals and neither assay detected viral RNA in COVID 18, the case with the longest diagnosis-to-death interval (51 days) and a presumptive duration of illness of 58 days. No viral RNA was detected in the control cases, as expected.
Fig. 1.
Swimmer plot of the COVID cases and results of assays detecting SARS-CoV-2 RNA in lung tissue. The disease course of each COVID case is shown on a horizontal bar. The reference point 0 is the time of diagnosis of the SARS-CoV-2 infection with qRT-PCR. Whenever feasible, the interval in days between the presumed onset of symptoms and the diagnosis is indicated as "x d". The time point of the last qRT-PCR test, the result, and the sampling site (LRT, lower respiratory tract; URT, upper respiratory tract) are indicated with the respective symbols. Positive-sense nucleocapsid RNA was visualised fluorescently with the RNAscope platform of ultrasensitive single-molecule RNA in situ hybridisation and was scored in a semiquantified way as ‘+++’ for diffuse, ‘++’ for focal, ‘+’ for rare and ‘0’ for absence of fluorescent N puncta. Tissue qRT-PCR was performed with the CDC set of N1 and N2 primers. A cycling threshold (Ct) value of N1 and N2 >38 was considered negative. No frozen samples were available for qRT-PCR in COVID 1 and 5.
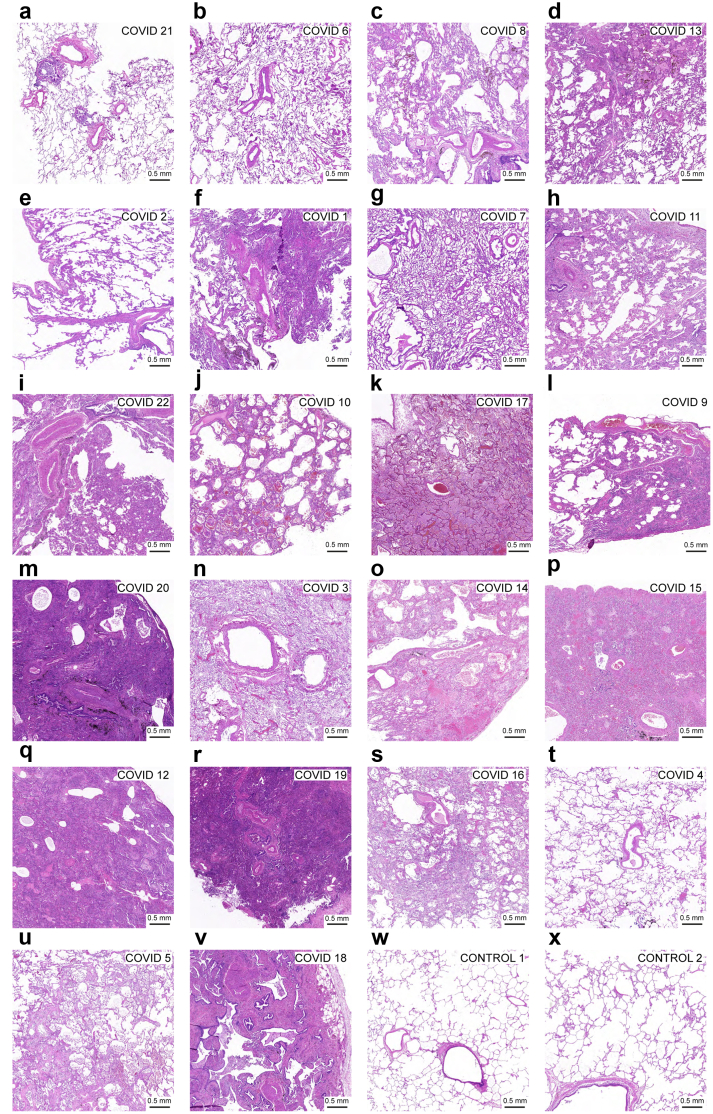
Histopathology
H&E-stained sections of lung samples of the COVID cases were analysed histopathologically (Fig. 2 and Table S5). We identified a shift in the pattern of lung injury during disease progression. Up to a diagnosis-to-death interval of 17 days (COVID 7), the injury was of an acute exudative to early proliferative pattern reminiscent of diffuse alveolar damage and located mainly in the bronchioles and centrilobular area of the parenchyma. The injury then shifted towards a fibroproliferative pattern affecting the mediolobular and peripheral lobular parenchyma.
Fig. 2.
Brightfield images of H&E-stained sections of the 22 COVID cases and 2 control cases. (a–v) Brightfield images of the COVID cases are ordered from shortest to longest diagnosis-to-death interval. Observations are summarised in Table S5. (w and x) Brightfield images from control cases. At the earliest stage of the infection, histopathological injury is found in the bronchioles and the centrilobular portion of the alveoli. Oedema, neutrophil infiltrate, and a mixed neutrophil and mononuclear infiltrate are predominant in the acute exudative phase of diffuse alveolar damage. At a later stage, the histopathological changes migrate to the peripheral alveolar areas. Mononuclear infiltration and fibrosis become predominant and reflect the fibroproliferative phase of diffuse alveolar damage. In the majority of cases, the disease course correlated well with the histopathological injury and no pre-existing histopathological lung abnormalities were found. (a-d and f) In COVID 21, 6, 8, 13 and 1, cases with diffuse RNAscope N puncta, histopathological injury is mild and situated in the early exudative phase, with a complete absence of pathological findings in COVID 6. Some cases deviated from the described diffuse alveolar damage pattern because of preexisting comorbidities with pulmonary manifestation. (c) The centrilobular to panlobular distribution of chronic histopathological changes in COVID 8 can be explained by congestive vasculopathy that existed prior to COVID-19. (e) In COVID 2, only signs of pulmonary hypertension were found, without signs of viral pneumonia and viral RNA; positive anti-N IgG serology confirmed the diagnosis of COVID-19. (t) With the exception of limited oedema, no abnormalities were found in COVID 4; with a long diagnosis-to-death interval of 44 days, the biopsied area of the lung might well have healed at the time of death. (k, n, and u) In COVID cases 17, 3 and 5, a superinfection with signs of (broncho)pneumonia was detected.
Cell type markers
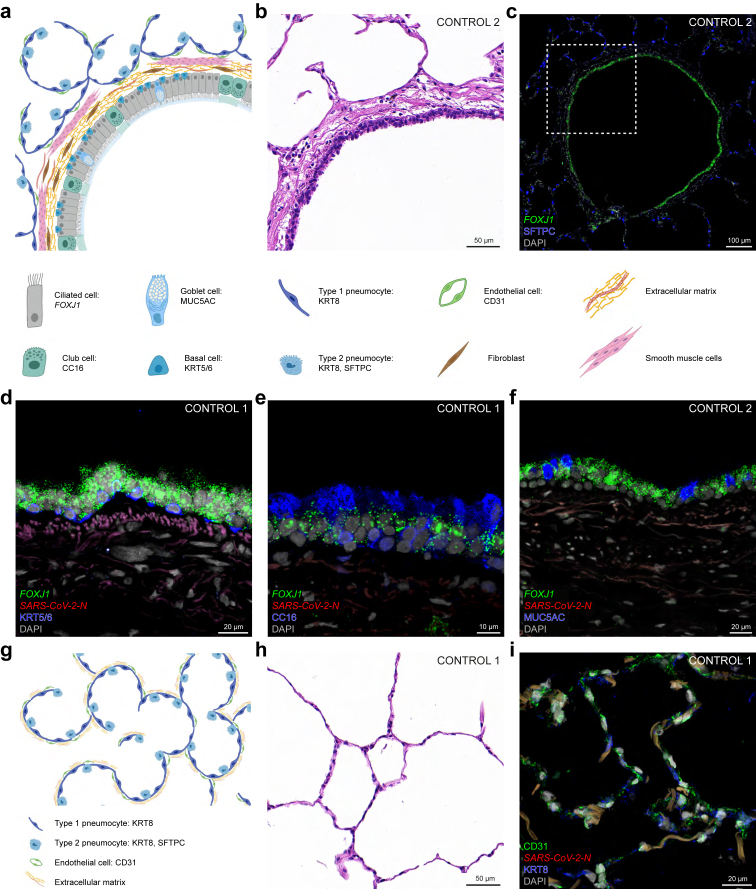
Lung samples procured in the prepandemic era were used as non-infected control cases to verify the specificity of our battery of commercially available RNAscope probes and antibodies in sections of bronchioles and alveoli (Fig. 3). We identified the relevant cell types by a combination of their expression of markers for RNA (names in italics) and antigens/proteins (names in roman), their morphology, and their position within the tissue. The wall of the bronchioles consists of a pseudostratified epithelium lining the airway lumen and a basal lamina with extracellular matrix produced by fibroblasts and smooth muscle cells (Fig. 3a and b). Ciliated cells are the dominant cell type in the bronchiolar epithelium and harbour RNAscope puncta for FOXJ1, a transcription factor required for ciliogenesis (Fig. 3c–f). Basal cells, which serve as progenitor cells for ciliated cells and other cell types in the epithelium, abut the basal lamina and contain KRT5/6-immunoreactive signal (Fig. 3d). Club cells secrete antimicrobial club cell secretory protein (CC16) and contain CC16-immunoreactive signal (Fig. 3e). Goblet cells secrete mucin 5AC and contain MUC5AC-immunoreactive signal (Fig. 3f). This glycoprotein is a component of the mucus that covers the epithelium and is swept to the throat by the ciliated cells, such that MUC5AC-immunoreactive signal is also visible extracellularly. The lung parenchyma consists of alveoli, air-filled sacks organised in a honeycomb pattern (Fig. 3g and h). The alveolar walls comprise type 1 and 2 pneumocytes, both containing KRT8-immunoreactive signal (Fig. 3i). These walls are supported by an extracellular matrix consisting of collagen and elastin fibres, which give rise to autofluorescent signal. Type 2 pneumocytes secrete surfactant protein C (SFTPC), which reduces surface tension and prevents alveolar collapse (Fig. 3c). Endothelial cells lining the lumen of alveolar capillaries can be identified with CD31-immunoreactive signal (Fig. 3i).
Fig. 3.
Bronchiolar and alveolar architecture and histology of lung samples of control cases. Schematic illustrations (a,g; created with BioRender.com), brightfield images of H&E stained sections (b,h), and confocal images of sections (c-f,i) of control 1 and 2 stained with multiplex fluorescence RNAscope and IHC and DAPI as nuclear stain. (a) Schematic illustration of a transverse section through a bronchiole. (b–f) Images of sections through a bronchiole. (b) Brightfield H&E image of the bronchiolar wall. (c) FOXJ1 is a marker for ciliated cells in the bronchiolar epithelium and SFTPC is a marker for type 2 pneumocytes in the alveoli surrounding a bronchiole. The dashed square in (c) corresponds to the brightfield image of the same bronchiolar wall in a nearby section (b). In the bronchiolar epithelium, KRT5/6 is a marker for basal cells (d), CC16 for club cells (e) and MUC5AC for goblet cells (f). As expected, there are no N puncta (d–f). (g) Schematic illustration of a section through alveoli of the lung parenchyma. (h,i) Images of sections through alveoli. (h) Brightfield H&E image of alveoli. (i) KRT8 is a marker for type 1 and type 2 pneumocytes lining the alveoli and CD31 is a marker for endothelial cells lining the lumen of capillaries coursing through the alveolar walls; the weak orange-brown signal in the alveolar walls represents autofluorescence; no N puncta are detected. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
Informative cases
We selected COVID cases 21, 6, 8, 13, and 1, with increasing duration of diagnosis-to-death intervals, as the most informative cases for our purposes based on the +++ score of the abundance of SARS-CoV-2-N puncta. COVID 8 and COVID 13 have the same diagnosis-to-death interval of 8 days, and we present only images of COVID 8. The salient data of these five cases are summarised in Table 1.
Table 1.
Summary of the five informative COVID cases.
| Case ID | 1 | 6 | 8 | 13 | 21 |
|---|---|---|---|---|---|
| Age | 71 | 89 | 90 | 87 | 47 |
| Immuno-compromised | Rituximab for diffuse large B-cell lymphoma | Steroids for autoimmune disease | No | No | Steroids for brain oedema |
|
Days intubated |
12 | 0 | 0 | 0 | 6 |
| Presumptive duration of illness (days) | 16 | 9 | 8 | 8 | Not reported |
| Diagnosis-to-death interval (days) | 13 | 5 | 8 | 8 | 2 |
| Cause of death | Respiratory failure | Respiratory failure | Respiratory failure | Respiratory failure | Brain tumour |
| Result of last SARS-CoV-2 qRT-PCR | Positive | Positive | Positive | Positive | Positive |
| Postmortem tissue sampling | Bedside | Bedside | Bedside | Bedside | Bedside |
| Degree of histopathological injury | Exudative to proliferative phase in bronchioles and centrilobular parenchyma | Absent | Early exudative phase in bronchioles and centrilobular parenchyma with superimposed injury in proliferative phase in all regions of parenchyma caused by congestive vasculopathy | Early exudative phase in bronchioles and centrilobular parenchyma | Early exudative phase in bronchioles |
| Result of SARS-CoV-2 qRT-PCR on lung tissue | Not reported | Positive | Positive | Positive | Positive |
| Abundance RNAscope N puncta | +++ | +++ | +++ | +++ | +++ |
| Histological and cellular location of SARS-CoV-2 RNAscope signal | Positive-sense RNA among sloughed-off cell debris and in macrophages in alveolar lumen | Positive-sense RNA in pneumocytes of the alveolar walls and in macrophages in the alveolar lumen | Positive-sense RNA among sloughed-off cell debris and in macrophages in alveolar lumen | Positive-sense RNA among sloughed-off cell debris and in macrophages in alveolar lumen | Positive-sense RNA and negative-sense RNA in ciliated cells of the bronchiolar epithelium and macrophages in the airway lumen |
Hyperacute phase of the infection
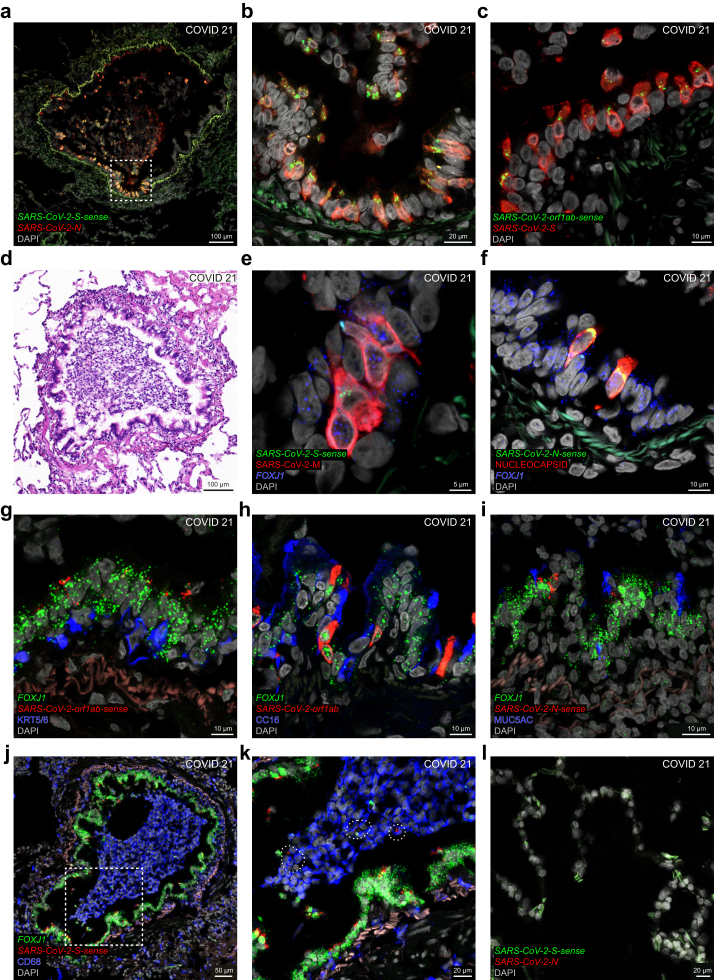
We detected abundant RNAscope signal for several SARS-CoV-2 RNA species in the bronchioles of COVID 21, who had a diagnosis-to-death interval of only two days. This patient tested positive on a nasopharyngeal swab taken during screening on an intensive care unit, where he had been intubated because of brain oedema that had developed soon after a stereotactic biopsy of a brain tumour. He died two days after diagnosis despite treatment with high-dosage methylprednisolone. The phase of the infection at the time of death is hyperacute, based on the following four observations: the patient tested negative by qRT-PCR on nasopharyngeal swabs taken 35 days, 13 days, and 8 days before the time of death; daily chest X-rays in the final days did not reveal signs of pulmonary infiltrates; serology for anti-N IgG the day before the time of death was negative; and a serum sample taken a few hours prior to the time of death had a qRT-PCR Ct-value of 33.6, possibly reflecting viremia. COVID 21 is an example of a patient who died with, rather than from, COVID-19. In view of these clinical circumstances, it was not possible to determine the onset of COVID-19 symptoms; therefore, we cannot provide a presumptive duration of illness.
The distribution of infected cells in the bronchiolar epithelium of COVID 21 is patchy (Fig. 4), as we also found to be the case for respiratory mucosa samples that were procured endoscopically postmortem from the nasal cavity of this patient, who was called COVID #60 in Khan et al.4 and Khan et al.5 The dense packing of N puncta and S puncta reflect high levels of intracellular viral RNA, and the perinuclear S-sense puncta and orf1ab-sense puncta, which represent the corresponding negative-sense viral RNA species, reflect ongoing viral replication in a large subset of cells (Fig. 4a–c). A brightfield image reveals injury of the bronchiolar mucosa suggestive of cytopathic changes and a clump of leukocytic debris within the lumen (Fig. 4d). Ciliated cells are the main target cell type in the bronchiolar epithelium (Fig. 4e–k). Densely packed M puncta cluster with perinuclear S-sense puncta in cells harbouring FOX1J puncta (Fig. 4e). Nucleocapsid-immunoreactive cells harbour FOXJ1 puncta and perinuclear N-sense puncta (Fig. 4f). Clusters of orf1ab-sense puncta, orf1ab puncta, and N-sense puncta occur in cells harbouring FOXJ1 puncta but not in cells containing KRT5/6-immunoreactive signal (Fig. 4g), CC16-immunoreactive signal (Fig. 4h), or MUC5AC-immunoreactive signal (Fig. 4i). The clump of cells in the bronchiolar lumen comprises numerous CD68-immunoreactive macrophages (Fig. 4j). Viewed at higher magnification, a few of these macrophages appear to harbour S-sense puncta but these puncta are not located perinuclearly and may reflect phagocytosed cell debris rather than viral replication within the macrophages (Fig. 4k). We did not observe N puncta or S-sense puncta in the alveoli (Fig. 4l).
Fig. 4.
Hyperacute SARS-CoV-2 infection of the bronchiolar epithelium in a COVID-19 patient. Confocal and brightfield images of sections through lung tissue of COVID 21, who died two days after diagnosis of infection. The extracellular matrix in the bronchiolar wall emits a weak green or red autofluorescent signal. (a) N puncta reveal a patchy pattern of cells in the bronchiolar epithelium, and the dense packing of N puncta reflects high levels of intracellular viral RNA. The dashed square in (a) is magnified in (b). Most cells harbouring N puncta also harbour perinuclear S-sense puncta, reflecting ongoing viral replication. (c) S puncta are densely packed in many cells, and some of these harbour perinuclear orf1ab-sense puncta. (d) The bronchiole in (a) is shown in a brightfield image of a nearby H&E-stained section. There is marked atypia without necrosis in the bronchiolar epithelium and the surrounding alveoli are not affected. (e) M puncta are densely packed in ciliated cells harbouring FOXJ1 puncta, and some of these harbour perinuclear S-sense puncta. (f) Nucleocapsid-immunoreactive signal diffusely fills two ciliated cells harbouring FOXJ1 puncta and perinuclear N-sense puncta. (g) Basal cells containing KRT5/6-immunoreactive signal do not harbour orf1ab-sense puncta; by contrast, orf1ab-sense puncta cluster with FOXJ1 puncta in ciliated cells. (h) Club cells contain CC16-immunoreactive signal and do not harbour orf1ab puncta; by contrast, orf1ab puncta cluster with FOXJ1 puncta in ciliated cells. (i) MUC5AC-immunoreactive signal labels secreted mucus lining the mucosa; MUC5AC-immunoreactive goblet cells do not harbour N-sense puncta; a few cells harbouring FOXJ1 puncta also harbour N-sense puncta. (j) A clump of CD68-immunoreactive macrophages fills the bronchiolar lumen. The dashed square in (j) is magnified in (k). A few CD68-immunoreactive macrophages harbour S-sense puncta in areas indicated with a stippled line. (l) N puncta and S-sense puncta are not detected in the alveoli. DAPI served as nuclear stain. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
Primary culture of human airway epithelium
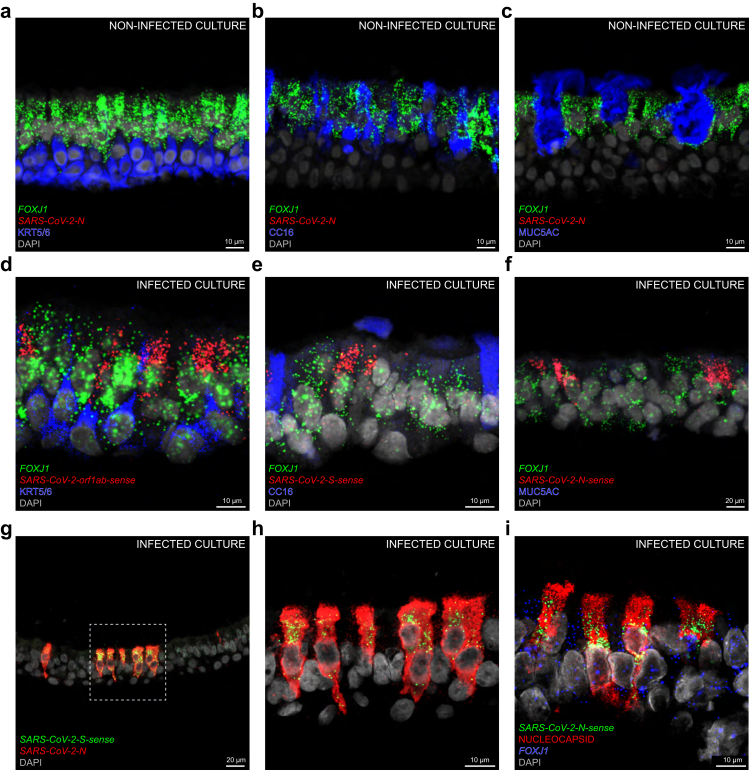
Confocal images of a primary culture of human airway epithelium in an air-liquid interface setup that was experimentally infected with SARS-CoV-2 four days earlier strikingly resemble those of the hyperacute phase of the infection in the bronchiolar epithelium of COVID 21 (Fig. 5). A non-infected culture served as negative control for the SARS-CoV-2-N probe (Fig. 5a–c). In a culture infected with BetaCoV/Belgium/GHB-03021/202, a SARS-CoV-2 Wuhan-Hu-1 strain, the clustering of perinuclear orf1ab-sense puncta, S-sense puncta, and N-sense puncta with FOXJ1 puncta confirms that ciliated cells are the major target cell type (Fig. 5d–f,i). We did not observe viral puncta in KRT5/6-immunoreactive basal cells (Fig. 5d) or in CC16-immunoreactive club cells (Fig. 5e). MUC5AC-immunoreactive goblet cells were not present in this culture (Fig. 5f), consistent with its origin from distal bronchioles, which contain a lower fraction of goblet cells. The pattern of infection is patchy across the culture, with dense packing of N puncta throughout the cytoplasm and a characteristic perinuclear location of S-sense puncta (Fig. 5g and h). Perinuclear N-sense puncta cluster with FOXJ1 puncta in cells diffusely filled with nucleocapsid-immunoreactive signal (Fig. 5i).
Fig. 5.
Experimental SARS-CoV-2 infection of a primary culture of human airway epithelium. (a–c) Confocal images of sections through a non-infected culture in an air-liquid interface setup; FOXJ1 puncta mark ciliated cells; N puncta are absent. KRT5/6-immunoreactive signal marks basal cells (a), CC16-immunoreactive signal marks club cells (b) and MUC5AC-immunoreactive signal marks goblet cells (c). (d–i) Confocal images of sections through a culture experimentally infected with an aliquot of a SARS-CoV-2 stock four days earlier. Ongoing viral replication is reflected by perinuclear orf1ab-sense puncta (d), S-sense puncta (e), and N-sense puncta (f) clustering with FOXJ1 puncta in ciliated cells. KRT5/6-immunoreactive basal cells do not harbour orf1ab-sense puncta (d) and CC16-immunoreactive club cells do not harbour S-sense puncta (e). (f) MUC5AC-immunoreactive goblet cells are absent in the infected culture. (g) N puncta are densely packed in a subset of cells. The dashed square in (g) is magnified in (h); perinuclear S-sense puncta cluster with N puncta. (i) In another section, four cells are diffusely filled with nucleocapsid-immunoreactive signal and harbour perinuclear N-sense puncta clustering with FOXJ1 puncta. DAPI served as nuclear stain. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
SARS-CoV-2-N puncta in alveoli
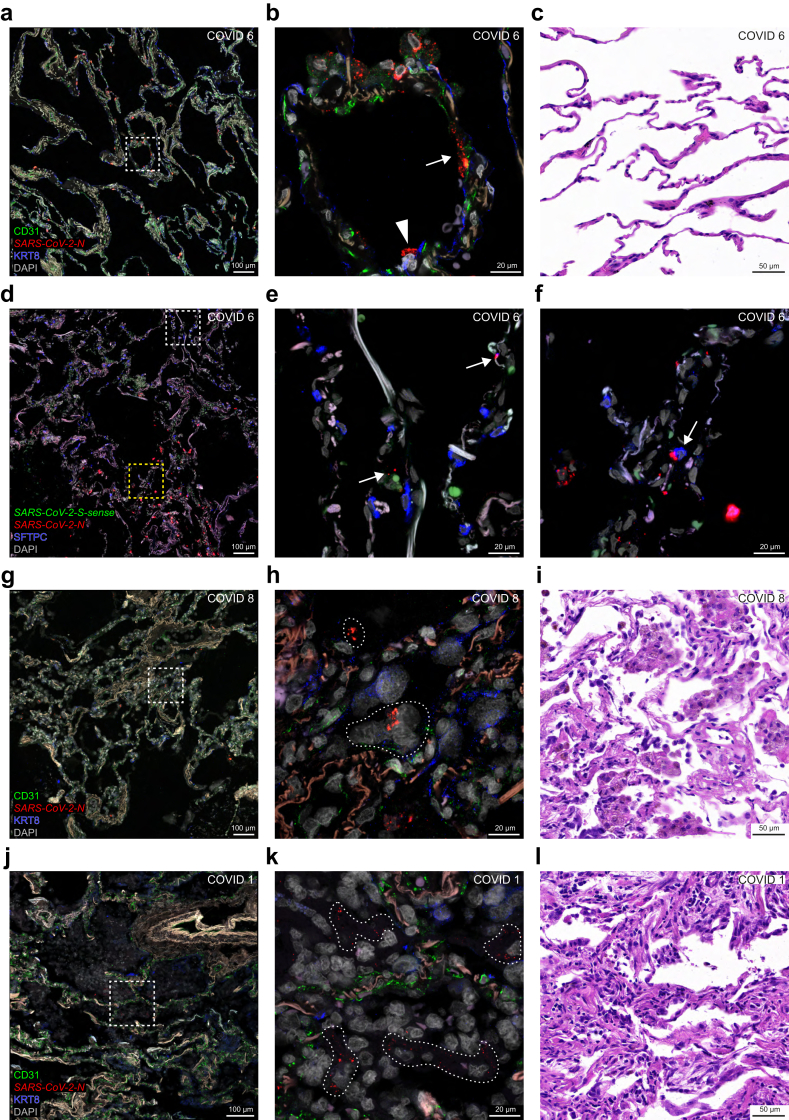
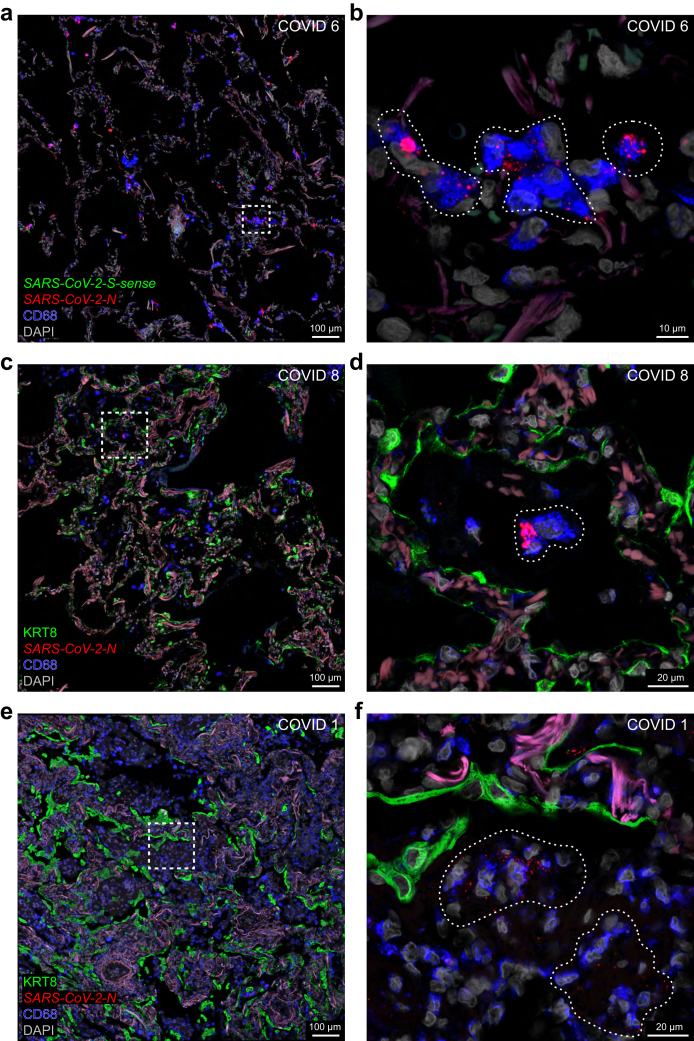
The most acute case in which we detected SARS-CoV-2 RNA species at the level of alveoli is COVID 6, who had a diagnosis-to-death interval of 5 days and a presumptive duration of illness of 9 days (Fig. 6a–f). This case was the deceased patient with acute COVID-19 described as the control case in Ceulemans et al.9 N puncta are spread diffusely across the alveolar walls (Fig. 6a). Viewed at higher magnification, N puncta occur in KRT8-immunoreactive pneumocytes (Fig. 6b). No overt pathological changes were found in a brightfield H&E-stained image of a nearby section (Fig. 6c). The absence of S-sense puncta suggests an absence of ongoing viral replication (Fig. 6d–f). N puncta occur in SFTPC-immunoreactive type 2 pneumocytes cells (Fig. 6f).
Fig. 6.
SARS-CoV-2-N puncta at the level of the alveoli. Confocal and brightfield images of sections through alveoli of COVID 6 (top two rows), COVID 8 (third row), and COVID 1 (bottom row). The extracellular matrix in the alveolar walls emits a weak beige to reddish-brown autofluorescent signal. DAPI served as nuclear stain. (a) In COVID 6, N puncta occur in the alveolar walls but not in the alveolar spaces, consistent with an acute phase of the infection. The dashed square in (a) is magnified in (b): N puncta occur in a cell containing KRT8-immunoreactive signal (arrowhead) and either within or close to a cell containing CD31-immunoreactive signal (arrow). (c) The brightfield image of an area in an H&E-stained nearby section reveals no overt histopathological injury. (d) SFTPC-immunoreactive signal is observed mostly in the upper half of the image; clusters of N puncta occur in the alveolar walls in the lower half of the image; S-sense puncta are absent. The upper, white dashed square in (d) is magnified in (e): SFTPC-immunoreactive signal labels type 2 pneumocytes in the alveolar walls; the few N puncta (arrows) reflect a low level of viral RNA. The lower, yellow dashed square in (d) is magnified in (f): an SFTPC-immunoreactive cell (arrow) harbours an aggregate of N puncta; there are no S-sense puncta. The weak green autofluorescent signal in (e) and (f) originates from erythrocytes. (g,h,j,k) In COVID 8 and COVID 1, N puncta are scattered within the alveolar spaces, consistent with a subacute phase of the infection. The dashed squares in (g) and (j) are magnified in (h) and (k) respectively: the alveolar walls can still be identified via CD31-immunoreactive endothelial cells and autofluorescent signal from the extracellular matrix; KRT8-immunoreactive signal originating from sloughed-off pneumocytes weakly appears in the alveoli; N puncta are present in areas indicated with a stippled line. (i,l) Brightfield images of H&E-stained sections of COVID 8 and COVID 1 shows early fibroproliferative diffuse alveolar damage. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
COVID case 8 had a diagnosis-to-death interval of 8 days and a presumptive duration of illness of also 8 days. N puncta occur among sloughed-off cell debris residing within the alveolar spaces (Fig. 6g and h). The H&E-stained image shows alveolar exudates with mononuclear infiltrates associated with siderophages (Fig. 6i). The chronic interstitial inflammation was the result of congestive vasculopathy due to chronic left ventricular failure that existed prior to the SARS-CoV-2 infection.
The longest survivor among the five informative cases in the cohort is COVID 1, with a diagnosis-to-death interval of 13 days and a presumptive duration of illness of 16 days. The delay in viral clearance may be related to the immunocompromised state resulting from the treatment with anti-CD20 monoclonal antibodies for diffuse large B cell lymphoma. N puncta occur diffusely in the alveolar spaces among cell debris (Fig. 6j and k). The H&E-stained image shows progression towards a mononuclear infiltrate and fibroproliferative diffuse alveolar damage (Fig. 6l).
Nucleocapsid-immunoreactive signal
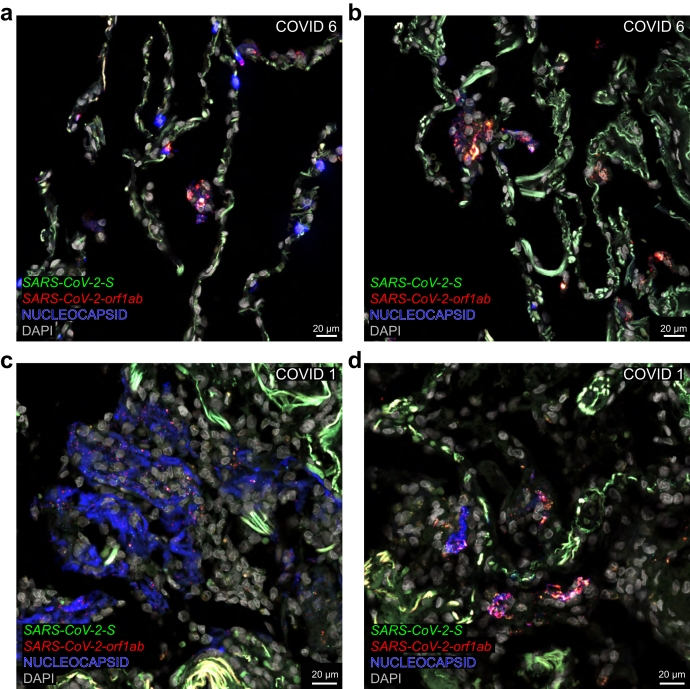
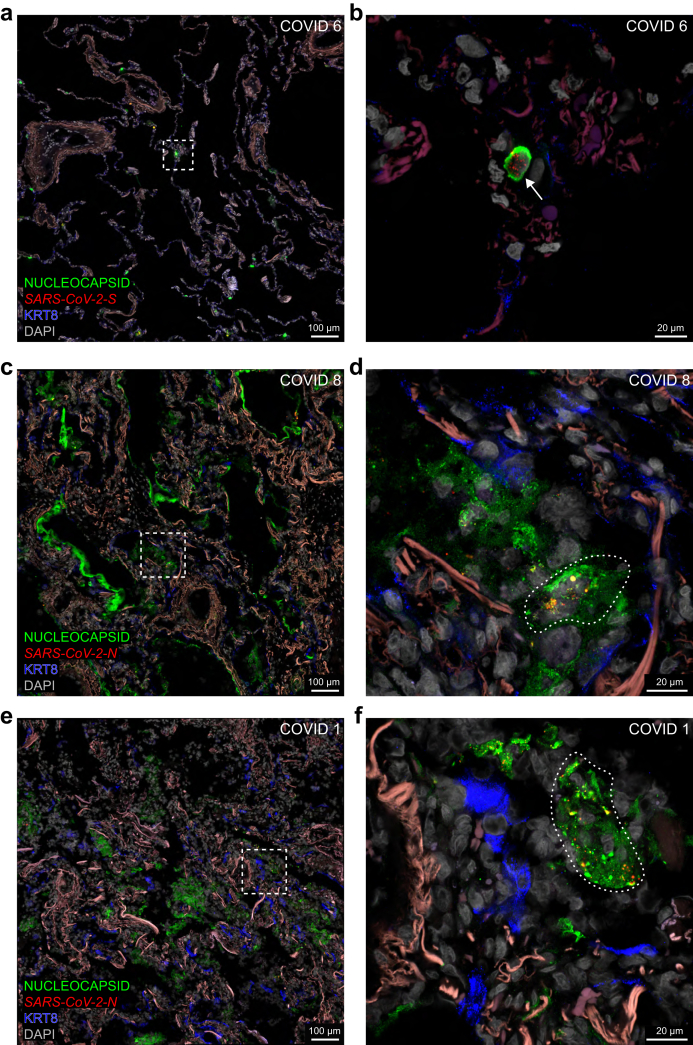
Traditional approaches in the literature to characterise cell tropism and visualise ongoing viral replication rely, often exclusively, on the surrogate parameter of immunoreactive signal for nucleocapsid or spike (Table S1). In COVID 6, nucleocapsid-immunoreactive signal is confined to the alveolar walls where it colocalises with S puncta and orf1ab puncta (Fig. 7a and b) and an individual cell filled with nucleocapsid-immunoreactive signal harbours S puncta (Fig. 8a and b). In COVID 8, nucleocapsid-immunoreactive signal prominently labels debris lining alveolar spaces (Fig. 8c) and N puncta occur within nucleocapsid-immunoreactive areas (Fig. 8d). COVID 1 exhibits an extensive alveolar presence of nucleocapsid-immunoreactive signal colocalising with S puncta and orf1ab puncta (Fig. 7c and d) or N puncta (Fig. 8e and f). Taken together, despite the prominent nucleocapsid-immunoreactive signal in these cases, sense puncta are absent, indicating an absence of ongoing viral replication.
Fig. 7.
Covisualising SARS-CoV-2 RNA species and nucleocapsid-immunoreactive signal in alveoli. Confocal images of sections through alveolar parenchyma of COVID 6 and COVID 1. The extracellular matrix in the alveolar walls emits a bright and diffuse green autofluorescent signal. Nucleocapsid-immunoreactive signal is in blue, where it is in green in the related Fig. 8. (a,b) Confocal images of sections of COVID 6. (a) Nucleocapsid-immunoreactive signal is confined to the alveolar walls; a group of nucleocapsid-immunoreactive cells at the center of the image harbours S puncta and orf1ab puncta. (b) A group of nucleocapsid-immunoreactive cells in another region of the same section harbours S puncta and orf1ab puncta. (c,d) Confocal images of sections of COVID 1. (c) Large groups of nucleocapsid-immunoreactive cells and debris fill the alveolar spaces; distinct S puncta and orf1ab puncta are scattered within the nucleocapsid-immunoreactive signal. (d) Groups of cells and debris in the alveolar space contain nucleocapsid-immunoreactive signal and harbour clusters of S puncta and orf1ab puncta. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
Fig. 8.
Covisualising SARS-CoV-2 RNA species and nucleocapsid-immunoreactive signal in alveoli. Confocal images of sections through alveolar parenchyma of COVID 6, 8, and 1. The dashed squares in the images on the left are the magnified areas on the right. The extracellular matrix in the alveolar walls emits a weak reddish-brown autofluorescent signal. Nucleocapsid-immunoreactive signal is in green, whereas it is in blue in the related Fig. 7. (a,b) Confocal images of sections of COVID 6. (a) Nucleocapsid-immunoreactive signal occurs in the alveolar walls. (b) A single cell is diffusely filled with nucleocapsid-immunoreactive signal and harbours S puncta (arrow); a weak KRT8-immunoreactive signal labels pneumocytes in the alveolar walls. (c,d) Confocal images of sections of COVID 8. (c) Nucleocapsid-immunoreactive signal prominently stains debris lining and filling the alveoli. (d) An area with nucleocapsid-immunoreactive signal (indicated with a stippled line) harbours N puncta; a weak KRT8-immunoreactive signal labels pneumocytes in the alveolar walls. (e,f) Confocal images of sections of COVID 1. (e) Nucleocapsid-immunoreactive signal prominently stains large groups of cells and debris in the alveolar spaces. (f) An area with nucleocapsid-immunoreactive signal (indicated with a stippled line) harbours N puncta; the KRT8-immunoreactive signal labels pneumocytes in the alveolar walls. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
Macrophages
These leukocytes differentiate from monocytes and reside within alveolar spaces and bronchioles where they phagocytose viral particles and sloughed-off cell debris during the course of a SARS-CoV-2 infection. Macrophages identified with CD68-immunoreactive signal constitute a predominant cell type harbouring N puncta species (Fig. 9). Groups of alveolar macrophages harbour aggregates and clusters of N puncta in COVID 6 but do not appear to replicate the virus, as assessed by the absence of S-sense puncta (Fig. 9a and b). The groups of alveolar macrophages are slightly larger in COVID 8 and harbour aggregates and clusters of N puncta (Fig. 9c and d). The CD68-immunoreactive signal in COVID 1 fills a large portion of the alveolar spaces (Fig. 9e) and large groups of macrophages harbour distinct N puncta (Fig. 9f). Taken together, despite the presence of N puncta in macrophages in these cases, sense puncta are absent, indicating an absence of viral replication in these cells.
Fig. 9.
Macrophages in alveolar walls and alveolar spaces. Confocal images of sections through alveolar parenchyma of COVID 6, 8, and 1. The dashed squares in the images on the left are magnified on the right. The extracellular matrix in the alveolar walls emits a weak fuchsia autofluorescent signal. DAPI served as nuclear stain. (a–d) In COVID 6 and COVID 8, groups of CD68-immunoreactive macrophages are observed in close relation to the alveolar walls. (a,c) The CD68-immunoreactive signal is prominent but does not completely fill the alveolar spaces. (b,d) Macrophages harbour clusters and aggregates of N puncta in areas indicated with a stippled line. (b) S-sense puncta are not observed and reflect the absence of viral replication in macrophages. (e,f) In COVID 1, alveolar spaces are filled with CD68-immunoreactive macrophages. (e) KRT8-immunoreactive signal labels pneumocytes that line the alveolar walls. (f) Macrophages in the alveolar spaces harbour N puncta in areas indicated with a stippled line. The names of RNAscope probes are in italics and the names of antigens/proteins that are revealed as immunoreactive signal by antibodies are in roman.
Discussion
We subjected postmortem lung tissue samples of a cohort of 22 hospitalised patients who died from or with a SARS-CoV-2 infection to complementary analyses by confocal microscopy, tissue qRT-PCR, and conventional H&E histopathology. We visualised various SARS-CoV-2 RNA species, nucleocapsid protein, and markers for relevant cell types in sections fluorescently stained with multiplex RNAscope and IHC and imaged under a confocal microscope.
The abundance of RNAscope N puncta markedly declined 2–3 weeks after diagnosis and puncta were detected for as long as 6 weeks. We previously reported the presence of N puncta and S puncta in lung tissue of a donor 105 days after the onset of COVID-19.9 Others also consistently visualised SARS-CoV-2 RNA with ISH within the first 2–4 weeks of the disease course.10, 11, 12, 13, 14, 15 Orthogonal detection of viral RNA with qRT-PCR was largely in agreement with the ISH results. High levels of viral RNA are detected with qRT-PCR until the second or third week of disease and lower levels remain detectable for as long as two months after the onset of symptoms.10,11,14,16, 17, 18, 19, 20 As the nucleocapsid open reading frame is the most abundantly transcribed2 and translated21 part of the SARS-CoV-2 genome, assaying nucleocapsid RNA in lung tissue with ISH or qRT-PCR is the most sensitive method. We also used RNAscope probes for orf1ab, S, and M to support our findings with the N probe. In our cohort, SARS-CoV-2 RNA was best detected in cases with a short diagnosis-to-death interval and without histopathological injury or with features of acute exudative diffuse alveolar damage. Progression towards fibroproliferative diffuse alveolar damage occurs after 2–3 weeks as the infection and viral RNA is being cleared. Our findings are generally consistent with reports of other cohorts of COVID-19 cases (Table S1) and confirm and extend previous observations that the host response not only clears the virus but can also injure the lung parenchyma.14,18,22,23
A highlight of our study is catching the virus in the act of infecting the distal bronchiolar epithelium in the hyperacute phase of the infection in COVID 21 and in a primary culture of human airway epithelium that was experimentally inoculated four days earlier. Perinuclear puncta for various negative-sense SARS-CoV-2 RNA species provide robust evidence at single-cell resolution for ongoing viral replication in ciliated cells but not in basal cells, club cells, and goblet cells. We have previously reported similar observations of perinuclear puncta in ciliated cells of the nasal respiratory epithelium and in sustentacular cells of the olfactory epithelium in postmortem tissue samples of COVID-19 cases.4,5 The perinuclear location is consistent with the replication and transcription of the SARS-CoV-2 genome within perinuclear double-membrane vesicles.3,24 RNAscope probes for negative-sense spike RNA were applied in several studies and visualisation of intracellular puncta was reported in some of these studies, but without correlation with the disease course (Table S1).19,20,22,25,26 Visualisation of positive-sense intracellular RNA species10,11,13,14,19,25, 26, 27, 28, 29 or nucleocapsid/spike-immunoreactive signal10,12, 13, 14, 15,23,26,30, 31, 32, 33 has frequently been proposed to identify target cell types of SARS-CoV-2 in the lung (Table S1) but does not provide robust evidence about cell tropism and ongoing viral replication.23,31,34
In this regard, our confocal images address the complexities stemming from traditional approaches in the literature to characterise cell tropism and visualise ongoing viral replication by the surrogate indirect parameters of nucleocapsid-immunoreactive signal or ISH for positive-sense viral RNA species. During viral replication in a host cell, newly-synthesised nucleocapsid protein molecules package a single positive-sense full-length genomic RNA molecule into a single SARS-CoV-2 virion.3 Nucleocapsid-immunoreactive signal (or, for that matter, spike-immunoreactive signal) is often and inappropriately equated with "SARS-CoV-2 infected",33 in part because of the well-established routine of performing IHC and brightfield or epifluorescence microscopy as opposed to fluorescence ISH and confocal imaging in the clinical setting. But nucleocapsid-immunoreactive signal reflects a variety of origins of the antigen including but not limited to intracellular protein within cells that are productively replicating the virus; a structural component of virions, intracellular or extracellular; intracellular protein or protein fragments within cells that have phagocytosed infected cellular debris; or extracellular protein or protein fragments trapped or encapsulated within debris. Costaining with viral ISH probes helps clarify the interpretation of the nucleocapsid-immunoreactive signal. The ISH detection of positive-sense viral RNA species supports the presence of molecular material of viral origin but arguably, claims and interpretations about cell tropism and ongoing viral replication require evidence of the presence of negative-sense viral RNA species. Our collection of confocal images showcases a variety of constellations and configurations of nucleocapsid-immunoreactive signal and positive-sense RNA puncta among sloughed-off cell debris and in CD68-immunoreactive macrophages within the alveolar walls and spaces.
We have previously speculated that this debris in the lung may shield SARS-CoV-2 RNA species from further degradation, thereby explaining the so-called viral "persistence" in lung tissue, which however may not reflect a persistence of infectivity.9,35 This issue is becoming increasingly apparent in the clinical practice of lung transplantation with regard to the assessment of the suitability of potential donor lungs from individuals with a SARS-CoV-2 infection in the past - which is nowadays an element in the clinical history of most individuals around the globe.36 We surmise that persistent positive-sense viral RNA molecules (intact or fragmented) may underlie the persistent weakly-positive qRT-PCR results on bronchoalveolar lavage fluid screening samples from candidate donors. In the absence of a longitudinal series of qRT-PCR results over a period of a few days, it is not possible to distinguish conclusively between a persistent viral RNA signal vs. a recently contracted infection. Such ambiguous results may hamper the decision-making process about accepting these lungs for transplantation because of the lingering concern of iatrogenically infecting the recipient, who is profoundly immunosuppressed at the time of transplantation.37,38 It has been speculated that viral persistence may be a driver of long COVID symptoms.39
We argue that convincing evidence for cell tropism and ongoing viral replication requires the implementation of a robust methodology based on multi-channel confocal microscopy to co-detect positive-sense viral RNA species, negative-sense viral RNA species, and nucleocapsid protein together with target cell type markers at single-cell resolution within the same tissue section. Brightfield imaging of single-colour chromogenic IHC or ISH has been used most frequently in the COVID-19 lung literature (Table S1). In a few studies, multiplex IHC or ISH was followed by brightfield imaging of chromogenic signal29,32 or epifluorescence imaging with a scanner10,16,19,27 or a confocal microscope.18 One study performed multiplex IHC and RNAscope staining and imaged with three-channel confocal microscopy.25
Limitations of the study
The patients got infected with SARS-CoV-2 during the first two waves of the pandemic in Belgium starting in March 2020 and October 2020 prior to the emergence of variants of concern. Our findings may be different for the various variants that have emerged since then and that may emerge in the future. We analysed one lung sample per patient but recognise that SARS-CoV-2 infection in the lung is characterised by temporal and spatial heterogeneity.20,31 We took a virocentric approach and visualised viral RNA and protein and target cell type markers but did not stain for ACE2, TMPRSS2, or other host factors that are necessary for viral cell entry.24 Our controls did not suffer from acute respiratory distress syndrome, or disease due to overt infection with other pathogens. Given the specificity of the reagents used, and given our experience with visualizing SARS-CoV-2 infection in the respiratory mucosa, olfactory mucosa, olfactory bulb, and frontal lobe of the brain, we are confident in the specificity of the RNAscope puncta and nucleocapsid-immunoreactive signal.4,5
Conclusion
Confocal imaging of sections stained with RNAscope sense probes hybridising to negative-sense SARS-CoV-2 RNA species and with relevant cell type markers is the pillar of a robust methodology for characterising cell tropism and visualising ongoing replication at single-cell resolution in histological sections of lung tissue samples. This methodology can be applied to future SARS-CoV-2 variants as well as to other respiratory viruses, in particular to RNA viruses.
Contributors
Conceptualisation by JVS, SF, EWa, JW, LJC, and PM. Human tissue sample collection by SF, GDH, EWa, and JW. In vitro experiments by VG, EVa, KV, LA, and BV. PCR assays by EWo. Histological staining and imaging by JVS, MK, SC, and PM. Histopathological review by EVe. Clinical data acquisition by JVS, CV, and SF. Literature search by JVS, MK, and PM. Verification of data by JVS, MK, and PM. Writing original draft by JVS, MK, LJC, and PM. Review & editing draft by VG, SF, EVa, KV, CDF, JK, DVR, RV, BV, and JW. Writing final draft by PM. All authors read and approved the final version of the manuscript.
Data sharing statement
Data supporting our findings will be made available by the corresponding author upon reasonable request.
Declaration of interests
SF received support for travel from Pfizer and Gilead. RV received consulting fees from AstraZeneca. JW received consulting and speaker's fees and support for travel from Pfizer, Gilead and MSD.
Acknowledgments
JVS was supported by a study scholarship of the European Society for Organ Transplantation. VG was supported by a fellowship of Research Foundation Flanders (11L9822N). SF was supported by a fellowship of Research Foundation Flanders (11M6922N). RV was supported by a fellowship and grant of Research Foundation Flanders (G060322N, 12G8715N). JW was supported by a grant of Coronafonds UZ Leuven/KU Leuven. LJC was supported by a Medtronic named chair, a postdoctoral grant of University Hospitals Leuven (KOOR) and a grant of Research Foundation Flanders (G090922N). PM acknowledges the generous financial support of the Max Planck Society. We thank Edisa Turkovic, Ling Li, and Seun-Jun Yoo for their help with processing tissue sections for RNAscope and immunohistochemistry; Mandy Bloemen (KU Leuven, Leuven, Belgium) for performing the qRT-PCR assays; Cato Jacobs (University Hospitals Leuven, Leuven, Belgium) for the collection of clinical data; and Celine Aelbrecht and Kathleen Van den Eynde (KU Leuven, Leuven, Belgium) for help with the preparation of tissue sections.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104608.
Appendix A. Supplementary data
References
- 1.Killingley B., Mann A.J., Kalinova M., et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 2.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan M., Yoo S.-J., Clijsters M., et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184:5932–5949.e15. doi: 10.1016/j.cell.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan M., Clijsters M., Choi S.-J., et al. Anatomical barriers against SARS-CoV-2 neuroinvasion at vulnerable interfaces visualized in deceased COVID-19 patients. Neuron. 2022;110:3919–3935.e6. doi: 10.1016/j.neuron.2022.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathyssen C., Serré J., Sacreas A., et al. Vitamin D modulates the response of bronchial epithelial cells exposed to cigarette smoke extract. Nutrients. 2019;11:2138. doi: 10.3390/nu11092138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feys S., Gonçalves S.M., Khan M., et al. Lung epithelial and myeloid innate immunity in influenza-associated or COVID-19-associated pulmonary aspergillosis: an observational study. Lancet Respir Med. 2022;10:1147–1159. doi: 10.1016/S2213-2600(22)00259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F., Flanagan J., Su N., et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceulemans L.J., Khan M., Yoo S.-J., et al. Persistence of SARS-CoV-2 RNA in lung tissue after mild COVID-19. Lancet Respir Med. 2021;9:e78–e79. doi: 10.1016/S2213-2600(21)00240-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berezowska S., Lefort K., Ioannidou K., et al. Postmortem cardiopulmonary pathology in patients with COVID-19 infection: single-center report of 12 autopsies from Lausanne, Switzerland. Diagnostics. 2021;11:1357. doi: 10.3390/diagnostics11081357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatnagar J., Gary J., Reagan-Steiner S., et al. Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 replication and tropism in the lungs, airways, and vascular endothelium of patients with fatal Coronavirus Disease 2019: an autopsy case series. J Infect Dis. 2021;223:752–764. doi: 10.1093/infdis/jiab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bussani R., Schneider E., Zentilin L., et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. eBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caniego-Casas T., Martinez-Garcia L., Alonso-Riano M., et al. RNA SARS-CoV-2 persistence in the lung of severe COVID-19 patients: a case series of autopsies. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.824967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai N., Neyaz A., Szabolcs A., et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun. 2020;11:6319. doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borczuk A.C., Salvatore S.P., Seshan S.V., et al. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol. 2020;33:2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorward D.A., Russell C.D., Um I.H., et al. Tissue-specific immunopathology in fatal COVID-19. Am J Respir Crit Care Med. 2021;203:192–201. doi: 10.1164/rccm.202008-3265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remmelink M., De Mendonça R., D'Haene N., et al. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Critical Care. 2020;24:495. doi: 10.1186/s13054-020-03218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Cleemput J., Van Snippenberg W., Lambrechts L., et al. Organ-specific genome diversity of replication-competent SARS-CoV-2. Nat Commun. 2021;12:6612. doi: 10.1038/s41467-021-26884-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong D.W.L., Klinkhammer B.M., Djudjaj S., et al. Multisystemic cellular tropism of SARS-CoV-2 in autopsies of COVID-19 patients. Cells. 2021;10:1900. doi: 10.3390/cells10081900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Jamal S.M., Pujadas E., Ramos I., et al. Tissue-based SARS-CoV-2 detection in fatal COVID-19 infections: sustained direct viral-induced damage is not necessary to drive disease progression. Hum Pathol. 2021;114:110–119. doi: 10.1016/j.humpath.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkel Y., Mizrahi O., Nachshon A., et al. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 22.Hirschbühl K., Dintner S., Beer M., et al. Viral mapping in COVID-19 deceased in the Augsburg autopsy series of the first wave: a multiorgan and multimethodological approach. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schurink B., Roos E., Radonic T., et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baggen J., Vanstreels E., Jansen S., Daelemans D. Cellular host factors for SARS-CoV-2 infection. Nat Microbiol. 2021;6:1219–1932. doi: 10.1038/s41564-021-00958-0. [DOI] [PubMed] [Google Scholar]
- 25.Hou Y.J., Okuda K., Edwards C.E., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massoth L.R., Desai N., Szabolcs A., et al. Comparison of RNA in situ hybridization and immunohistochemistry techniques for the detection and localization of SARS-CoV-2 in human tissues. Am J Surg Pathol. 2021;45:14–24. doi: 10.1097/PAS.0000000000001563. [DOI] [PubMed] [Google Scholar]
- 27.Schuler B.A., Habermann A.C., Plosa E.J., et al. Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. J Clin Invest. 2021;131 doi: 10.1172/JCI140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zito Marino F., De Cristofaro T., Varriale M., et al. Variable levels of spike and ORF1ab RNA in post-mortem lung samples of SARS-CoV-2-positive subjects: comparison between ISH and RT-PCR. Virchows Arch. 2022;480:597–607. doi: 10.1007/s00428-021-03262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwab C., Domke L.M., Rose F., Hausser I., Schirmacher P., Longerich T. Cell tropism and viral clearance during SARS-CoV-2 lung infection. Pathol Res Pract. 2022;236 doi: 10.1016/j.prp.2022.154000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barisione E., Grillo F., Ball L., et al. Fibrotic progression and radiologic correlation in matched lung samples from COVID-19 post-mortems. Virchows Arch. 2021;478:471–485. doi: 10.1007/s00428-020-02934-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauter J.L., Baine M.K., Butnor K.J., et al. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology. 2020;77:915–925. doi: 10.1111/his.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer I.M., Padera R.F., Solomon I.H., et al. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol. 2020;33:2104–2114. doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bussani R., Zentilin L., Correa R., et al. Persistent SARS-CoV-2 infection in patients seemingly recovered from COVID-19. J Pathol. 2023;259:254–263. doi: 10.1002/path.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Agnillo F., Walters K.-A., Xiao Y., et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abj7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceulemans L.J., Van Slambrouck J., De Leyn P., et al. Successful double-lung transplantation from a donor previously infected with SARS-CoV-2. Lancet Respir Med. 2021;9:315–318. doi: 10.1016/S2213-2600(20)30524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Slambrouck J., Van Raemdonck D., Wauters J., Vos R., Mombaerts P., Ceulemans L.J. Lung donation and SARS-CoV-2 transmission: missed detection versus missed opportunity? Immun Inflamm Dis. 2022;10:e603. doi: 10.1002/iid3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Querrey M., Kurihara C., Manerikar A., et al. Lung donation following SARS-CoV-2 infection. Am J Transplant. 2021;21:4073–4078. doi: 10.1111/ajt.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peled Y., Kittleson M.M. Transplanting COVID-19 positive donors: expanding our experience to widen the donor pool. J Heart Lung Transplant. 2022;41:1382–1384. doi: 10.1016/j.healun.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis H.E., McCorkell L., Vogel J.M., Topol E.J. Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol. 2023;21:133–146. doi: 10.1038/s41579-022-00846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.