SUMMARY
PARPs catalyze ADP-ribosylation—a post-translational modification that plays crucial roles in biological processes, including DNA repair, transcription, immune regulation, and condensate formation. ADP-ribosylation can be added to a wide range of amino acids with varying lengths and chemical structures, making it a complex and diverse modification. Despite this complexity, significant progress has been made in developing chemical biology methods to analyze ADP-ribosylated molecules and their binding proteins on a proteome-wide scale. Additionally, high-throughput assays have been developed to measure the activity of enzymes that add or remove ADP-ribosylation, leading to the development of inhibitors and new avenues for therapy. Real-time monitoring of ADP-ribosylation dynamics can be achieved using genetically encoded reporters, and next-generation detection reagents have improved the precision of immunoassays for specific forms of ADP-ribosylation. Further development and refinement of these tools will continue to advance our understanding of the functions and mechanisms of ADP-ribosylation in health and disease.
eTOC blurb
Dasovich and Leung’s review highlights the latest technical advances in characterizing ADP-ribosylation, such as identifying ADP-ribosylated proteins and sites, utilizing NAD+ analogs to determine PARP substrate specificity, using chemical probes to identify the ADP-ribose binding proteome, and monitoring ADP-ribosylation dynamics both in vitro and in vivo.
BACKGROUND
Post-translational modifications are ideally suited for rapid responses to the environment by using the chemistry of ubiquitous metabolites, suc h as ATP, Acetyl-CoA, and NAD+, to alter protein function. Without the need to synthesize new molecules, protein modifications can be initiated immediately. An example is ADP-ribosylation, an NAD+-dependent protein modification best known for its role in the DNA damage response. DNA damage-dependent ADP-ribosylation is catalyzed by PARP1, an abundant nuclear protein associated with chromatin.1 Once a DNA strand break occurs, PARP1 rapidly (~1 s) binds the break, activating the enzyme by opening its NAD+-binding pocket.2 PARP1 then uses NAD+ to ADP-ribosylate proteins in the vicinity of the DNA break, which initiates a signaling cascade that repairs DNA. While ADP-ribosylation research has historically focused on PARP1 and DNA repair, humans express 16 additional PARPs of emerging biological importance and therapeutic relevance. ADP-ribosylation also plays critical roles in functions unrelated to DNA repair, such as RNA metabolism, protein turnover, and the immune response. PARP inhibitors have been FDA-approved for treating multiple cancers, and pre-clinical data support their use in immune-related diseases, viral infection, and neurodegeneration. Due to intense medical interest over the last two decades, an expanded toolkit is now available for studying ADP-ribosylation with chemical precision. Here, we review the chemical and regulatory complexity of ADP-ribosylation, then delve into the molecular tools developed to uncover its substrates, binding partners, and dynamics.
The Chemical Complexity of ADP-ribosylation
The genetic fingerprints of ADP-ribosylation are found across all kingdoms of life and viruses, with enzymes ADP-ribosylating proteins, small molecules, DNA, and RNA.3 This review focuses on protein ADP-ribosylation catalyzed by human PARPs, though there are other “writer” enzymes that add this modification, including sirtuins, bacterial toxins, and ecto-enzymes.4 PARPs contain the eponymous catalytic domain responsible for ADP-ribose transfer (Figure 1A). PARP domains are found in all six eukaryotic supergroups and knockout of PARPs 1/2 or PARPs 5a/b causes embryonic lethality in mice, hinting that PARPs have conserved and essential functions in eukaryotes.5–7 In humans, PARPs represent the largest group of NAD+-consuming enzymes and can use almost any nucleophilic amino acid (CDEHKRSTY) as acceptor sites for ADP-ribosylation,8 encompassing diverse functional groups from sulfides and ethers to ketoamines and esters (Figure 1B). Mass spectrometry analyses suggest that PARPs display promiscuity. For example, PARP7 modifies C, D, and E in cells and PARP11 ADP-ribosylates C, D, E, and K in an in vitro reaction (Figure 1B).9–11 However, sites identified from in vitro reactions should be interpreted with caution,11 as it is now apparent that PARPs do not always act alone, and instead form complexes with accessory proteins that regulate PARP activity and specificity in cells (e.g., HPF1, discussed below).
Figure 1. The chemical diversity of human PARP metabolism.
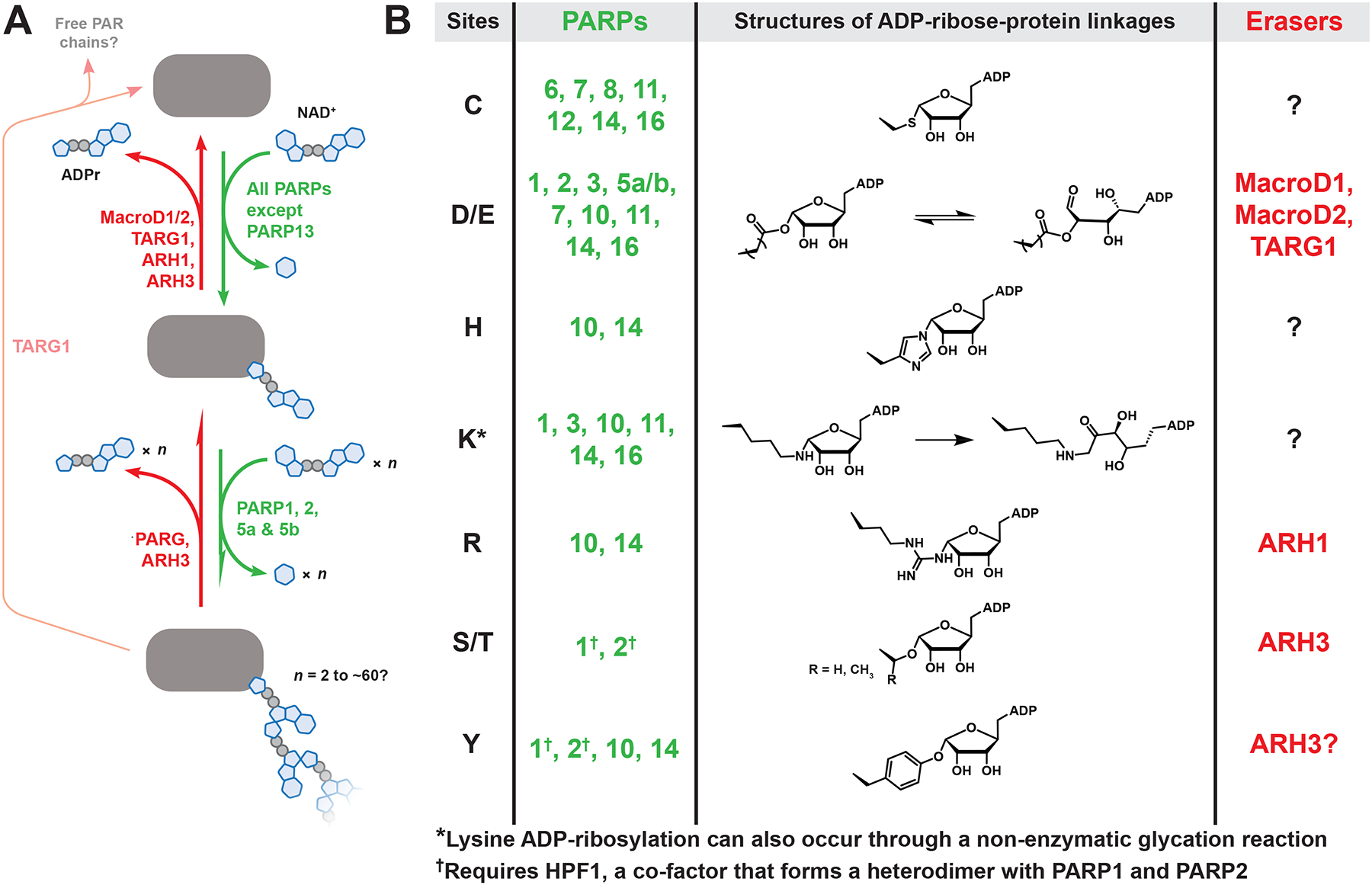
(A) PAR metabolism involves four steps: 1) transfer of a single ADP-ribose from NAD+ to the target protein, resulting in mono-ADP-ribose (MAR). 2) Further ADPr transfers onto MAR, producing linear and branched poly(ADP-ribose) (PAR). 3) PAR degradation, which is primarily carried out by poly(ADP-ribose) glycohydrolase (PARG). Notably, human PARG does not remove the final ADP-ribose attached to the protein, thereby converting PAR to MAR. 4) Cleavage of the ADP-ribose-protein bond by various “eraser” enzymes, each with a preference for specific functional groups. Note: ARH3 can also degrade PAR, although it is less efficient than PARG and unable to remove branch points. (B) The chemical diversity of ADPr-protein bonds synthesized by human PARPs, with known site preferences listed. See Data S1 for related references.
The seventeen human PARPs distinguish themselves with several accessory domains that regulate their functions.12 Notably, most PARPs (except for 5a/b, 6, 8, and 16) contain one or more DNA-, RNA-, and/or ADPr-binding domains, suggesting a direct link between nucleic acid metabolism and PARP-mediated ADP-ribosylation (Figure 2). Many PARPs are transcriptionally regulated by interferon,13 and PARP activity can be altered by NAD+ availability, nucleic acid binding, and accessory factors. For example, regulation of PARP activity can be achieved through NAD+ synthesis by nicotinamide adenylyl transferases (NMNATs 1–3), which are localized to different subcellular locations and control the NAD+ concentration in specific compartments.14 PARP activity can also be tuned by their nucleic acid or protein partners. PARP1 can be activated by nucleic acids (e.g., DNA strand breaks or small nucleolar RNAs), and it can perform catalysis on its own or in complex with either NMNAT1 or histone PARylation factor (HPF1).15–18 Each one of these ternary nucleic acid−PARP1−accessory factor complexes targets different proteins, and in the case of PARP1/HPF1, switching the specificity to Ser.19
Figure 2. An overview of the PARP family, including domains, targets, and phenotypes.
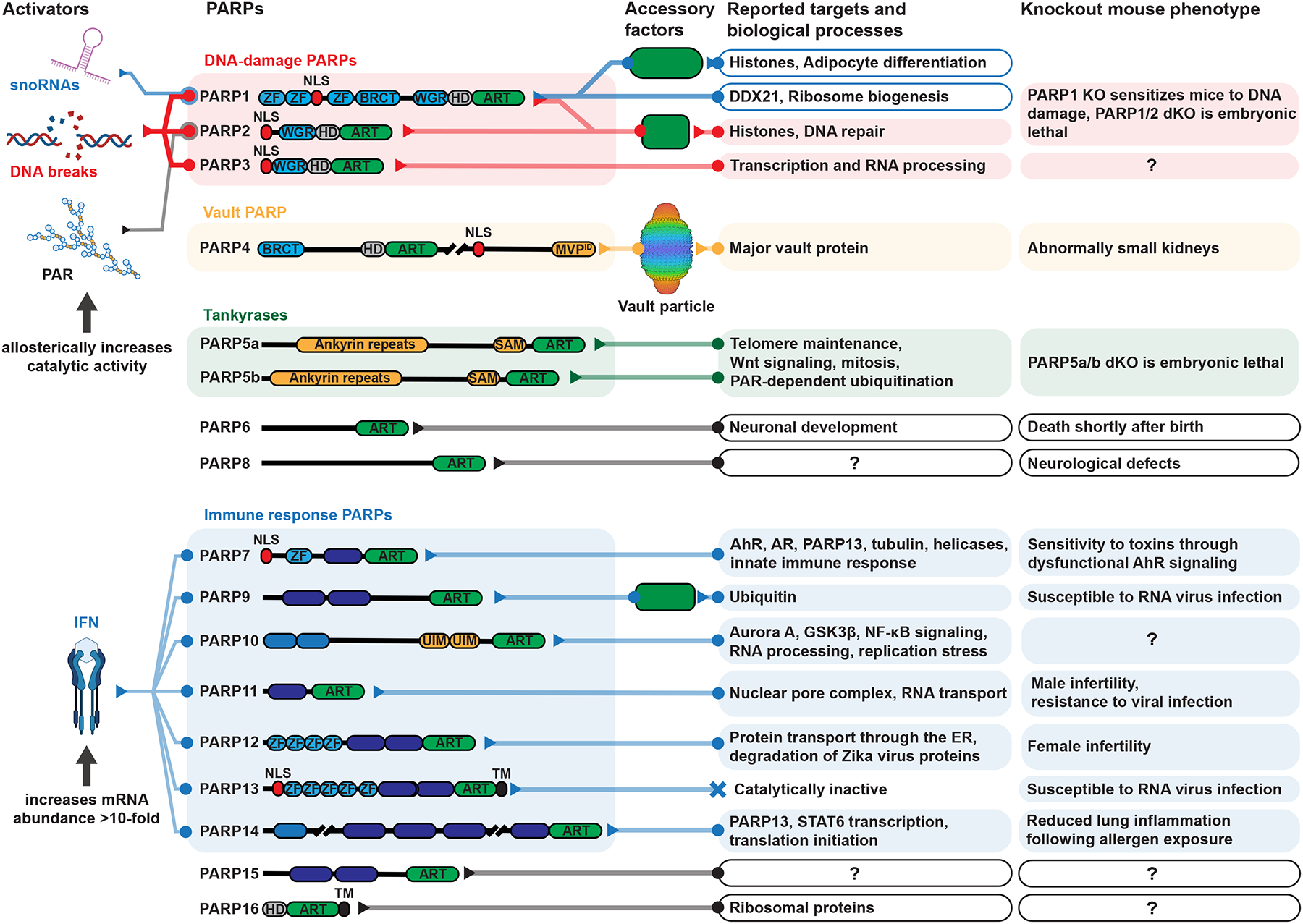
Domain abbreviations: ZF = zinc finger, NLS = nuclear localization signal, BRCT = BRCA1 C-terminus, WGR = the three most conserved amino acids in this DNA-binding domain, HD = helical domain, ART = ADP-ribosyltransferase, MVPID = major vault protein interacting domain, SAM = sterile alpha motif, WWE = the three most conserved amino acids in this PAR-binding domain, Macro = macrodomain, RRM = RNA recognition motif, UIM = ubiquitin interacting motif, TM = transmembrane. See Data S1 for related references.
The Structural Complexity of ADP-ribosylation
Adding to this complexity, PARPs 1, 2, 5a, and 5b can synthesize ADP-ribose–ADP-ribose bonds to create poly(ADP-ribose) (PAR). Polymers as large as ~200 units have been reported in cells treated with high concentrations of DNA-damaging agents,20 while a sizable population of PAR oligomers between 2 and 20 units is detected in more biologically relevant conditions.21–23 PAR can also have branch points every 20–30 ADPr units.24 The potential for ADP-ribose modifications on nine different amino acids, combined with the possibility of mono-, oligo-, or poly(ADP-ribose), results in hundreds of possible structures. Therefore, ADP-ribosylation is grouped with other complex post-translational modifications, such as glycosylation and ubiquitination, that have been historically difficult to characterize.25,26
The length of the ADP-ribosylation is tightly regulated by opposing classes of enzymes and cofactors.15,27 PAR is primarily degraded by PAR glycohydrolase (PARG).28 While PARP activity is activated by stress, PARG is constitutively active, keeping PAR concentration low under most circumstances. PARG knockout increases DNA breaks and causes embryonic lethality in mice.29,30 Therefore, the proper dynamics of PARylation—both synthesis and degradation—are critical for DNA repair and cell survival. Interestingly, human PARG does not cleave the ADPr-protein bond. The final mono(ADP-ribose) (MAR) is instead left for several eraser enzymes, each with a preference for specific ADPr-protein linkages (Figure 1B). The eraser specificity reveals a possible regulatory mechanism where the half-life of the modification depends on the conjugated amino acid. For example, MAR on Asp and Glu have a half-life of <1 hour in cells, whereas MAR on Cys persists for several hours.10
The structural diversity of ADP-ribosylation is broadly categorized as MAR or PAR (Figure 1A). It was previously thought that PAR was the most abundant form in cells, but recent studies have suggested that MAR is more common, even after DNA damage.31,32 However, PAR is observed upon PARG knockdown, suggesting some proteins are initially modified with PAR, then PARG rapidly trims the polymer to MAR.31 In situations unrelated to DNA damage (e.g., proteotoxic stress), MARylation may be the most abundant form.33 Consistent with this premise, most PARPs appear to exclusively synthesize MARyation when incubated with NAD+ in vitro.11 Mirroring the stepwise process of PAR removal, a MARylating PARP could transfer the first ADP-ribose, then a second PARylating PARP could add additional ADPr. In this model, a MAR–PAR equilibrium could exist at a single ADP-ribosylation site, regulated by the relative concentrations and rates of PARPs, PARG, MAR-erasing enzymes, and NAD+ (Figure 1A). Accessory factors can also influence the structure of ADPr modifications; PARP1 alone synthesizes PAR, whereas the PARP1/HPF1 complex reduces the ADPr length to primarily MAR.34–38 Notably, unrestrained Ser MARylation induced by ARH3 knockout is viable, yet unrestrained PARylation through additional PARG inhibition is acutely toxic.39 Taken together, these data suggest that MAR and PAR are discrete signaling molecules with profoundly different consequences for cellular function. Detection reagents that differentiate between MAR and PAR have been invented recently.31,40,41 As detection technologies improve, we may uncover the biological significance and mechanism behind the balance between MAR and PAR.
This review will summarize current techniques for teasing apart the molecular details of PARP-related processes, including proteomic methods for identifying ADP-ribosylated proteins, sites, and non-covalent ADPr-protein interactions, as well as the development of detection reagents, small molecule inhibitors, and genetically encoded reporters for monitoring ADP-ribosylation dynamics in living cells.
TECHNIQUES FOR IDENTIFYING ADP-RIBOSYLATED PROTEINS AND SITES
Large-scale proteomic studies of phosphorylation and acetylation have been essential for establishing the function of specific sites and delineating their roles in diseases.42,43 Analyzing ADP-ribosylation to a similar depth has proved challenging due to its chemical heterogeneity and low abundance. However, several methods have recently been developed that overcame these challenges and revealed thousands of ADP-ribosylated proteins and sites.
Key techniques to identify ADP-ribosylation sites
Enrichment
The classic ADP-ribose enrichment method uses boronate agarose to covalently bind cisdiols on ribose at alkaline pH (Figure 3). This method successfully identified thousands of ADP-ribosylated proteins in breast cancer cell lines.44,45 Boronate enrichment has the potential to be highly effective in identifying peptides modified with long PAR, thanks to the increased avidity from the higher number of cis-diols. To further enrich PARylated sites, PARG may be knocked down to boost the amount and length of PAR modifications for analyses.
Figure 3. Key steps in ADP-ribosylomics workflows.
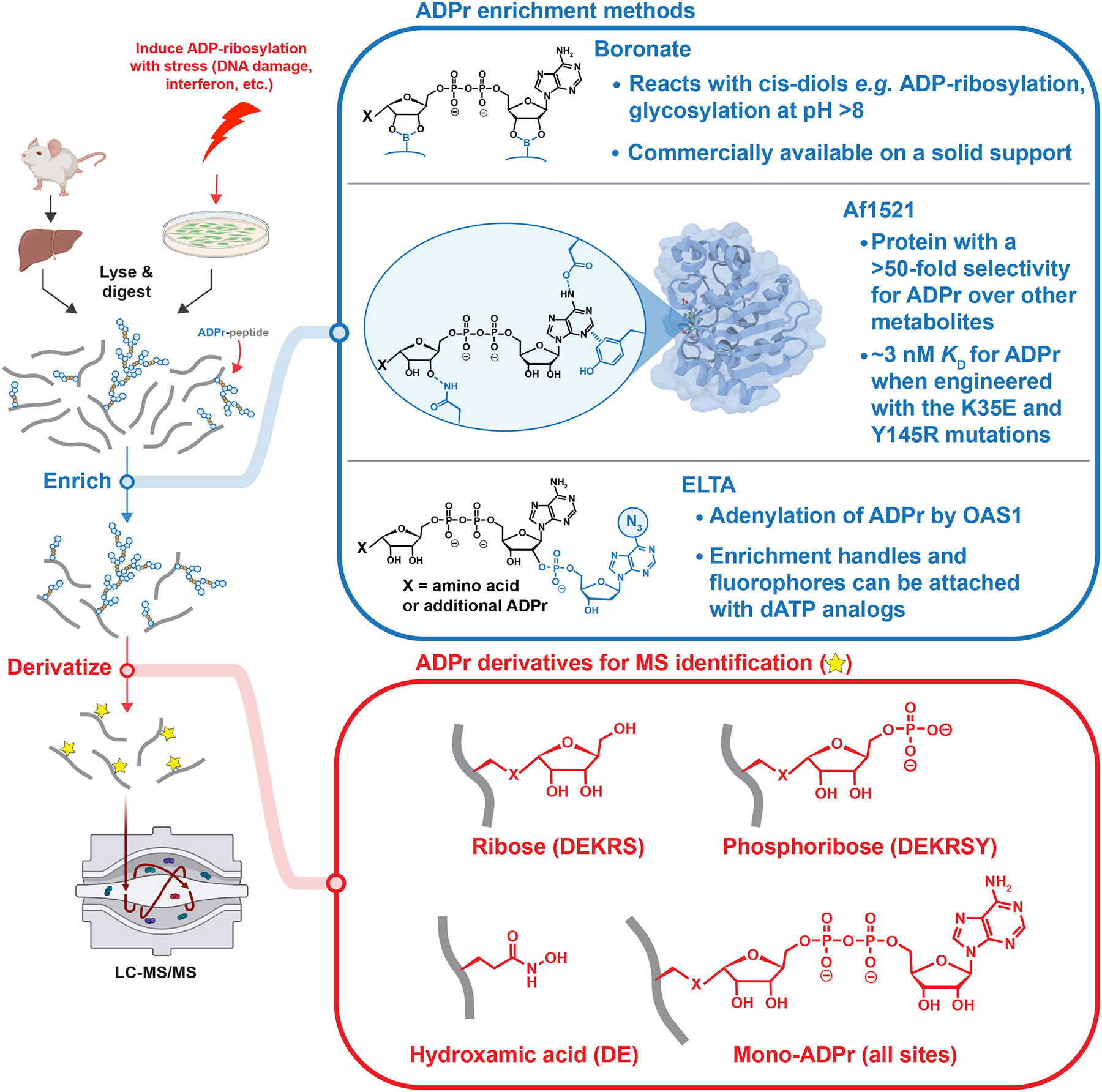
Following protein extraction from cultured cells or tissues and tryptic digestion, ADP-ribosylated peptides are enriched with a variety of methods (blue). Enriched peptides contain heterogeneous ADP-ribose modifications (i.e., MAR and PAR), which are then derivatized to a single, homogenous mass (red) to simplify the interpretation of mass spectra. For each derivative, the amino acids searched in published experiments are in paratheses, though ribose and phosphoribose could be on any amino acid. The structure of Af1521 is from reference 50.
Another enrichment method relies on the archaeal Af1521 macrodomain (Figure 3),46 a protein that binds to MAR and the terminal ADPr of PAR.37 This enrichment is robust, with multiple labs reporting hundreds to thousands of ADP-ribosylated proteins and sites.8,47–50 Af1521 enrichment was recently bolstered by protein engineering to increase the macrodomain–ADPr affinity to low nM.50 However, the Af1521 macrodomain is also an eraser capable of removing MAR at Asp and Glu.51 Therefore, it is important to mitigate the hydrolysis by performing enrichment at 4°C.49 Despite the inherent differences between boronate and macrodomain enrichment, a direct comparison revealed that the majority of the ADP-ribosylated proteins are identified by both techniques.32
The final method discussed here is mediated through enzymatic labeling of terminal ADP-ribose (ELTA; Figure 3). This method relies on oligoadenylate synthase 1, a human protein that polymerizes ATP into 2’–5’ oligoadenylate as part of the immune response.52 OAS1 can efficiently add a variety of ATP analogs to ADP-ribose,53 and using 2’-dATP prevents oligoadenylation to allow attachment of a single analog to ADP-ribosylated molecules.34 To identify ADP-ribosylation sites, ELTA labels ADP-ribosylated peptides with N6-azido-dATP, followed by enrichment on dibenzocyclooctyne-agarose via copper-free click chemistry. With this method, as little as 1 fmol of modified peptide can be detected in cell lysates, and hundreds of ADP-ribosylated proteins were identified. The diversity of these sites and the surrounding sequences can be enhanced by varying the ion-pairing reagents used during solid-phase extraction of tryptic peptides.54
Derivatization
Once ADP-ribosylated peptides are enriched, the next step is to simplify the chemical complexity of ADP-ribosylation for site identification. Variable lengths of PAR on ADP-ribosylated peptides complicate mass spectra interpretation, and their negative charges make them difficult to analyze with the positive ion mode routinely used for proteomics. To circumvent these issues, a chemical or enzymatic reaction is used to derivatize the ensemble of ADPr modifications into a single, defined mass (Figure 3). Hydroxylamine reacts with the Asp/Glu-ADPr ester bond, replacing MAR and PAR modifications with hydroxamic acid.44,45 Hydroxamic acid is a stable, neutrally charged modification that can be sequenced with fragmentation methods such as higherenergy collisional dissociation (HCD). However, hydroxylamine can only identify Asp/Glu-ADPr and may react with unmodified Asp, Glu, Asn, and Gln residues at high concentrations.32 Therefore, sites from hydroxylamine-based proteomics should be supplemented with mutagenesis experiments to ensure that the identified Asp/Glu are bona fide ADP-ribosylation sites.16,17,33 A more general method of derivatizing ADPr is treatment with hydrofluoric acid, which cleaves the phosphate-oxygen bonds, leaving a single ribose at the modified site. While hydrofluoric acid-based derivatization could theoretically identify ADPr at any amino acid, it has only been used to analyze in vitro PARylated PARP1.55
An alternative approach to simplifying ADPr involves enzymatic derivatization. MAR and PAR can be digested with phosphodiesterase to produce phosphoribose.56 While it requires purification before use, a highly potent phosphodiesterase from snake venom is commercially available. On the other hand, human enzymes such as NudT16 and ENPP1, which exhibit lower enzymatic activity, can be produced in significant amounts in laboratories.57–59 Phosphoribose derivatization allows for standard phospho-enrichment, streamlining co-analysis of ADP-ribosylation and phosphorylation sites.60 Many Ser and Tyr sites are co-targeted by these modifications, and their cross-talk on histone proteins has important consequences for gene expression.8,17 However, because phosphorylation is more abundant than ADP-ribosylation, such co-enrichment may reduce sensitivity.
Another enzyme commonly used is PARG, which converts PAR to MAR, resulting in a +541 Da mass tag at all modified sites. Mono(ADP-ribose) modifications are labile during HCD fragmentation, making it difficult to determine the modification site from the mass spectrum.61 Instead, electron transfer dissociation (ETD), which specifically fragments the peptide backbone, provides higher localization probabilities for ADP-ribosylated peptides, albeit with lower sensitivity than HCD. However, hybrid methods such as EThcD and AI-ETD have been developed to improve the sensitivity of ETD-based methods and have confidently localized hundreds of ADP-ribosylation sites in a single experiment.47
Diverse ADP-ribosylation sites revealed by various techniques
Together, these proteomics techniques have extensively characterized ADP-ribosylated proteins in cultured human cells treated with DNA-damaging agents. Under these conditions, the majority of ADP-ribosylation occurs on PARP1 itself, histone proteins, and to a lesser extent on proteins involved in DNA repair, cell cycle control, and RNA metabolism.8 Precisely which sites are modified is, however, less clear. Initial in vitro experiments found Asp and Glu are the acceptor sites on histones and PARP1,62,63 which inspired the development of the boronate/hydroxylamine method.44,45 However, when the PARG/macrodomain method is used and intact ADP-ribose is searched for on all nine amino acids, most ADPr is found on Ser (~90% of sites, 97% of intensity),8 primarily through the activity of the PARP1/HPF1 complex,19,32 while Asp and Glu only constitutes a minor fraction of total ADPr (~20 sites, <0.1% intensity).8
The discrepancy may be explained due to the technical details of the chosen methods. As hydroxylamine-based methods can only detect ADPr on Asp and Glu, they may provide excellent depth and sensitivity for a minor fraction of the total ADP-ribosylome. These studies also often used PARG knockdown cells, which may reveal modifications that would not stably exist in cells with normal PAR turnover.44,45 On the other hand, Asp/Glu sites have an ester, which is more sensitive to hydrolysis than other sites, particularly at basic pH (Figure 1B). ADP-ribosylomics experiments typically involve multiple overnight steps above pH 7 and have employed off-line high-pH fractionation to increase the analysis depth.8 The extent of Asp/Glu-ADPr signal loss due to hydrolysis during these lengthy sample preparation steps has not been assessed. Recently, the synthesis of defined MARylated peptides has been described for Ser, Thr, Tyr, Cys, and Arg.31,64–66 If available for all sites, these chemically defined peptides would be valuable standards for evaluating the stability of different ADPr–peptide linkages during proteomics procedures.
Orthogonal western blot analyses found that Ser ADP-ribosylation is the most common after DNA damage, confirming the proteomics results. However, the same western blots also detected a minor fraction of DNA damage-dependent ADP-ribosylation on Asp/Glu.67 Asp/Glu ADP-ribosylation also has functions unrelated to DNA damage, such as promoting transcription.68 Western blot analyses found most ADP-ribosylation occurred at Asp/Glu in pre-adipocytes, where Asp/Glu-ADPr on histones controls gene expression during differentiation.17 The relative abundance of ADP-ribosylation on other amino acids, such as Lys, Arg, His, and Tyr, can vary depending on the tissue, cell type, cellular compartment, or stress condition (e.g., interferon response).47–49 Bolstered by these proteomics efforts, recent studies have explored the biochemical effects and biological outcomes of site-specific ADP-ribosylation on proteins, highlighting how PARPs regulate essential cellular components, like histones and ribosomes (Table 1).9,16,18,19,32,33,37,68–77 While the biological functions of most ADP-ribosylation sites are still unknown, proteomics studies have revealed a chemical diversity of ADPr-protein linkages, suggesting that ADP-ribosylation plays a role in multiple functional niches, with each linkage type regulated by a unique set of writers and erasers.
Table 1 |.
Functional characterization of ADP-ribosylation sites identified with proteomics
| NELF-E |
E121
E151 E152 E171 E172 E374 |
The negative elongation factor complex (NELF) inhibits transcription elongation by RNA polymerase II. PARP1 ADP-ribosylated NELF-E, which disrupted the complex and increased transcription at select promoters. | In vitro ADP-ribosylation assay, EMSA, ChlP-seq | 68 |
| DDX21 |
E13
D15 E43 E67 E196 |
DDX21 is a nucleolar RNA helicase that localizes to rRNA loci and promotes rRNA transcription. PARP1 ADP-ribosylated DDX21, which disrupted DDX21-RNA interactions and prevented DDX21 from localizing to rRNA loci. This reduced rRNA levels, protein translation, and cell growth. | In vitro ADP-ribosylation assay, IP, RIP-qPCR, EMSA, ChIP-qPCR, RNA Bioanalyzer, puromycilation assay, cell proliferation assay | 16 |
|
Histone
H2B |
E35 | PARP1 ADP-ribosylated Histone H2B at E35 in preadipocytes, which prevented differentiation to adipocytes by blocking AMPK-mediated phosphorylation of H2BS36. | In vitro ADP-ribosylation and phosphorylation assay, IP, RNA-seq, ChlP-qPCR, Oil Red O staining | 18 |
|
RPL24
RPS6 |
E4
E35 |
RPL24 & RPS6 are ribosome components at the interface of the 60S and 40S subunits. PARP16 ADP-ribosylated RPL24 & RPS6, which prevented ribosome assembly and reduced translation. This promoted cell survival by limiting the toxicity associated with excessive protein synthesis. | IP, IF, puromycilation assay, ribosome profiling, cell proliferation assay | 33 |
| Tubulin |
D69
E71 E79 |
PARP7 ADP-ribosylated α-tubulin, which contributed to microtubule depolymerization. | IP, IF | 9 |
| STAT1α |
E393
E394 D721 |
STAT1α is a key transcriptional regulator of pro-inflammatory gene expression mediated by interferon γ. PARP1 ADP-ribosylated the DNA-binding and transactivation domains of STAT1α, both of which increased interferon-regulated gene expression by promoting STAT1α interactions with proteins and DNA. STAT1α ADP-ribosylation drove pro-inflammatory phenotypes in macrophages, increasing their ability to phagocytose bacteria. | IP, ChIP-seq, RT-qPCR, RNA-seq, phagocytosis assay, seahorse assay, oligonucleotide-binding assay | 69 |
|
U2AF35
U2AF65 |
E162
E425 |
U2 snRNP auxiliary factor (U2AF) is an essential splicing factor that recognizes splice sites and initiates ribosome assembly. PARP1 ADP-ribosylated U2AF, which altered U2AF-RNA interactions and regulated RNA splicing events in stem cells. | In vitro ADP-ribosylation assay, IP, EMSA, splicing assay | 70 |
|
Histone
H1 |
S150
S188 |
The PARP1/HPF1 complex ADP-ribosylated histone Hi during the DNA damage response. ADP-ribosylation did not directly affect Hi-DNA interactions but did decrease chromatin compaction. | In vitro ADP-ribosylation assay, IP, sedimentation assay, Native PAGE | 32, 71 |
|
Histone
H2B |
S6 | The PARP1/HPF1 complex ADP-ribosylated histone H2B during the DNA damage response. H2B ADP-ribosylation reduced chromatin compaction and primed H2B for recognition and remodeling by the ADP-binding chromatin remodeler ALC1. | Sedimentation assay, fluorescence polarization, IP, nucleosome remodeling assay | 37, 72 |
|
Histone
H3 |
S10 | The PARP1/HPF1 complex ADP-ribosylated histone H3 during the DNA damage response. H3 ADP-ribosylation reduced chromatin compaction and primed H3 for recognition and remodeling by the ADPr-binding chromatin remodeler ALC1. ADP-ribosylation of H3Si0 also blocked acetylation or methylation of H3K9. Conversely, acetylation or methylation of H3K9, phosphorylation of H3T11, and acetylation of H3K14 block H3S10ADPr. |
In vitro ADP-ribosylation assay, western blotting, IP, sedimentation assay, fluorescence polarization, nucleosome remodeling assay, in vitro acetylation and methylation assays | 19, 37, 72–74 |
| PARP1 |
S499
S507 S519 |
The PARP1/HPF1 complex auto-ADP-ribosylated PARP1 during the DNA damage response. ADP-ribosylation of PARP1 at these three serines promoted the dissociation of PARP1 from DNA breaks and decreasds the toxicity of PARP1 inhibitors by reducing the amount of PARP1 trapped on DNA. | Western blotting, live-cell microscopy, cell survival assay | 32, 75 |
| AR |
C125
C131 C284 C290 C327 C406 C519 C596 C602 C620 C670 |
PARP7 ADP-ribosylated the androgen receptor (AR), which promoted interactions with the ADPr-binding PARP9/DTX3L complex and modulated androgen-dependent gene expression. | Western blotting, AR-dependent luciferase reporter assay | 76 |
| HPX | R218 | Hemopexin (HPX) is a heme-binding protein that transports heme to the liver for recycling and iron recovery. HPX is ADP-ribosylated by the ARTC1 ecto-ADP-ribosyltransferase. In vitro, HPX ADP-ribosylation by ARTC1 inhibited HPX-heme binding. | In vitro ADP-ribosylation assay, heme-binding assay | 77 |
Harnessing NAD+ analogs to identify PARP substrate specificity
Which PARP enzymes are responsible for these diverse site modifications? PARPs have diverse accessory domains, unique site preferences, and appear to target specific subsets of proteins (Figure 2). While proteome arrays can be used to identify in vitro substrates of individual PARPs,78,79 several labs have used NAD+ analogs to study PARP substrate specificity in cellular contexts (Figure 4).80,81 Early work found that PARPs tolerate modifications at the nicotinamide and adenine rings of NAD+.82,83 Jiang and colleagues extended this work by demonstrating that PARPs can accept N6-alkyne-NAD+, which is then linked through click chemistry to various moieties, including fluorescent dyes and enrichment handles to detect and purify ADP-ribosylated proteins.84,85 However, N6-alkyne-NAD+ alone does not differentiate which targets are modified by specific PARPs because multiple PARPs can use this analog.
Figure 4. NAD+ analogs used to study ADP-ribosylation.
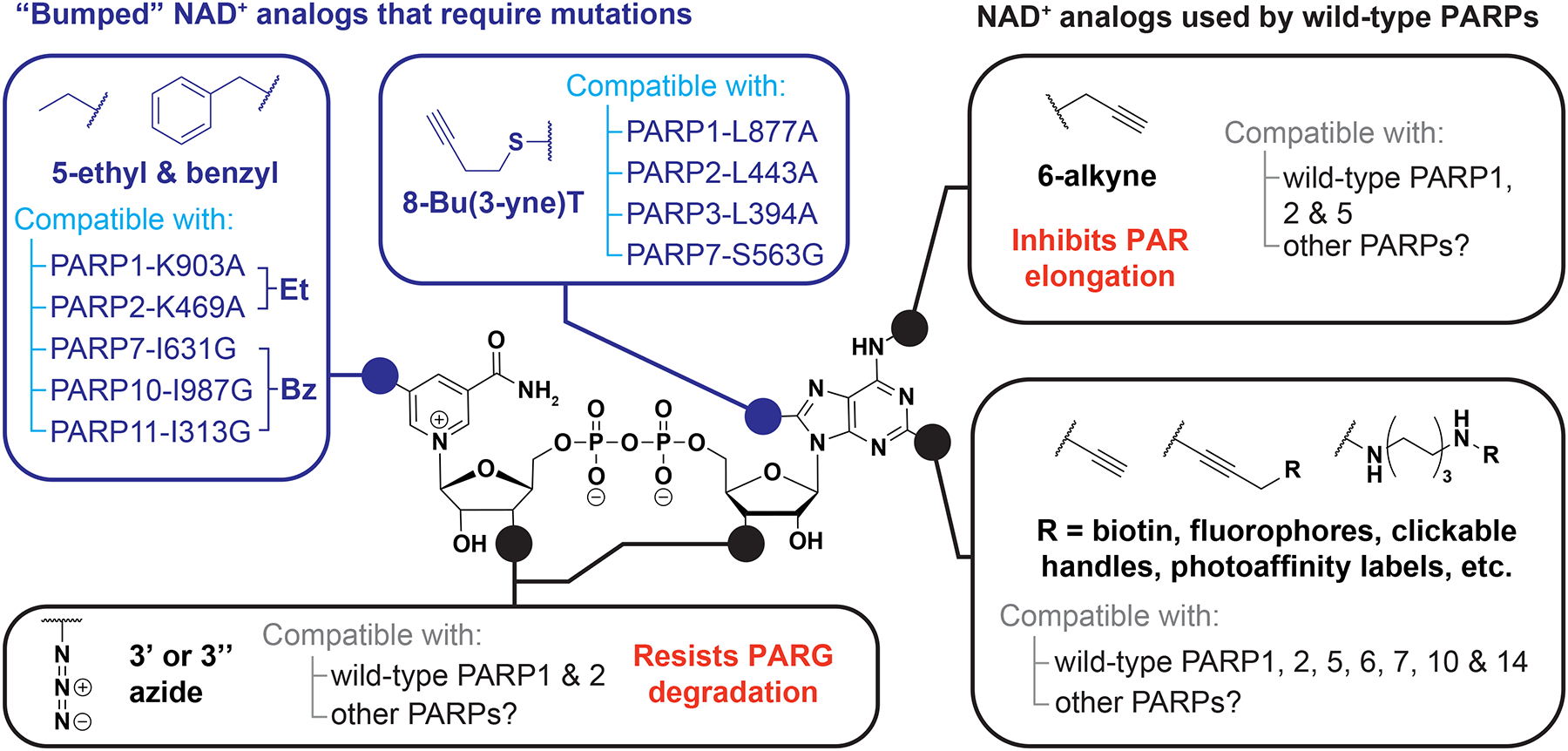
Analogs with substitutions to the N6, C2, and 3’-OH positions can be used with wild-type PARP proteins, whereas substitutions to the C5 and C8 positions require mutations in the NAD+-binding pocket to confer analog sensitivity.
Inspired by “analog-sensitive” approaches used to identify targets of other proteinmodifying enzymes,86 Carter-O’Connell and colleagues applied similar technology to investigate modifications catalyzed by specific PARPs.87 By mutating a lysine within the active site, PARP1 and PARP2 can accept NAD+ analogs with an ethyl group at the C5 position of the nicotinamide ring (Figure 4), while the wild-type enzymes cannot modify proteins with this analog. By combining this approach with the N6-alkyne modification, they obtained the first proteomics data set of PARP1 and PARP2 targets from human cell lysates.87 The Cohen lab went on to characterize the targets of PARPs 7, 10, 11, and 14.10,88,89 This revealed that these six PARPs have distinct target preferences and occupy specific cellular niches. Notably, they created a chimera of the PARP11 accessory domains fused to the PARP10 catalytic domain. Their PARP10–PARP11 chimera modified a subset of PARP10 and PARP11 targets, indicating both the accessory and catalytic domains regulate target preferences.88
N6-alkyne-NAD+, however, inhibits PAR elongation,85,87 which is a significant issue for PARylating PARPs 1, 2, 5a, and 5b.11 Gibson and colleagues solved this problem by exploring the C8 position of the adenine ring. Interestingly, amine substitutions at C8 result in poor reactivity, but sulfide substitutions are well tolerated by PARP1 once an active-site leucine is mutated to alanine (Figure 4).68,85 They applied their C8 analog to intact nuclei to identify PARP1, PARP2, and PARP3 targets and modification site preferences with proteomics, as well as which regions of the genome are ADP-ribosylated by PARP1 by coupling enrichment of ADP-ribosylated histones with DNA sequencing. This method detected strong correlations between PARP1 ADP-ribosylation and actively transcribed promoters, where PARP1 releases paused RNA polymerase II through ADP-ribosylation of negative elongation factor E.68
Rational analog design is possible, now that the structure of the wild-type PARP1 catalytic domain bound to a non-hydrolyzable NAD+ has been solved.90 The structure agrees well with previous findings: the C5 nicotinamide and C8 adenine positions are buried deep within the domain, and therefore mutation of residues near these positions produces analog-sensitive PARPs. On the other hand, the C2, N3, and C4 positions on adenine and the ribose hydroxyls are exposed to solvent, suggesting that modifications at these positions will be tolerated by wild-type PARP1. For example, C2-functionalized NAD+ analogs are broadly compatible, as they have shown activity with all PARPs tested thus far (Figure 4).10,91–94 Zhang and colleagues replaced the 3’-hydroxyl with an azide to create NAD+ analogs with high PARylation activity.95,96 The 3’-azide-NAD+ specifically showed activity with PARP1 and little activity with PARP2 or PARP5a.96 Yet, 3’-azide-NAD+ modified proteins in lysates when PARP1 was knocked out,96 suggesting this analog can also be used by other ADP-ribosylation enzymes in cells.
NAD+ analogs have traditionally been difficult to use in living cells due to their inability to cross the cell membrane. However, the Marx and Zhang labs have developed methods for delivering NAD+ analogs into living cells, including partially permeabilizing the membrane with detergent or using the cationic lipid DOTAP as a transfection reagent.91,94,95 Through combination with analog-sensitive technologies, NAD+ analog delivery is an exciting development for studying the substrates and functions of individual PARPs in living cells, especially in contexts such as the immune response, where ADP-ribosylation levels are low and multiple PARPs are involved.
SYNTHETIC ADP-RIBOSYLATED PROBES TO IDENTIFY THE ADP-RIBOSE BINDING PROTEOME
Besides altering the activities of covalently modified substrates, ADP-ribosylation can also affect the function of unmodified proteins through non-covalent interactions. PAR serves as a scaffold for macromolecular complexes and biomolecular condensates.97 For example, PARP1 ADP-ribosylates proteins surrounding DNA breaks, which can then recruit DNA repair proteins through their PAR-binding domains.98 Some E3 ubiquitin ligases, such as RNF146, activate their catalytic activity upon binding to PARylated proteins, resulting in PAR-dependent ubiquitination.99,100 Emerging data indicate ADPr-binding domains govern the specificity and timing of PARP signaling events (Figure 5). For instance, macrodomains bind ADPr, but significant sequence divergence within the family leads to unique binding preferences and functions (e.g., the PARG macrodomain degrades PAR chains, but has no affinity for MAR, while the PARP14 macrodomains have no catalytic activity and only bind to MAR).101–104 The fact that the PARP14 macrodomains are specific for MAR suggests the sequence context surrounding the MARylation site may also be important for macrodomain recognition.40,41,102 The WWE domain in RNF146 binds to the internal repeating unit of PAR,105 while the tandem PARP13 WWE domain prefers to “cap” the PAR chain by binding to the end.106 The PBZ domains within APLF specifically bind to PAR, with a preference for branch points.107,108 Altogether, these examples suggest that the length and branching of PAR modifications control which proteins are recruited at which times, introducing the possibility of a “PAR code” where the structure of the ADPr modification impacts downstream signaling.97 Therefore, tools are much needed to evaluate the extent and specificity of ADPr–protein interactions on a proteome-wide scale.
Figure 5. Protein domains that bind to ADP-ribosylated molecules.
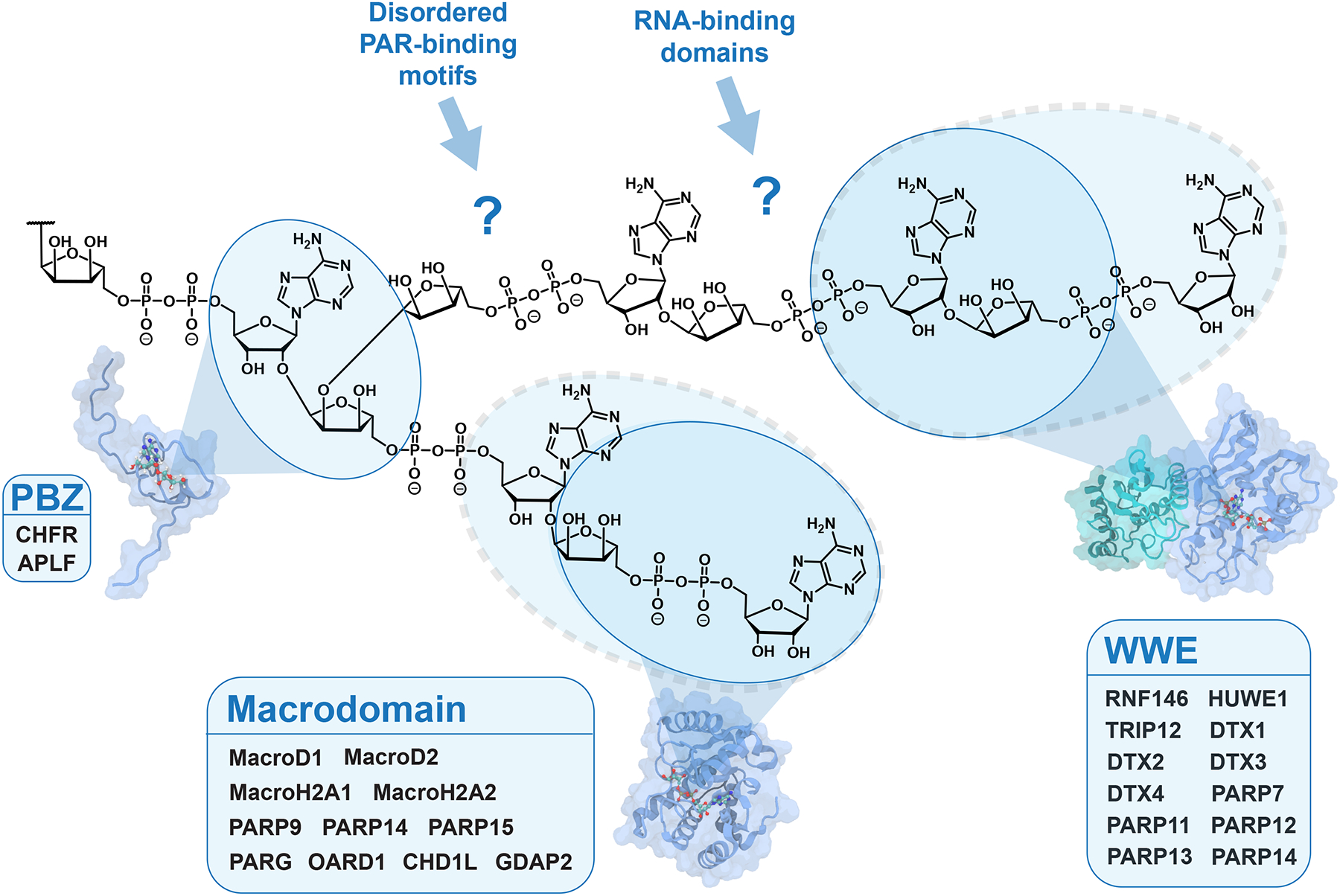
The PAR-binding zinc finger (PBZ), WWE domain, and macrodomain are well-defined folds that recognize specific forms of ADP-ribosylation. The blue circles indicate the binding preferences for each domain based on structural data, with dashed circles indicating variations in binding preference within the family. For instance, most macrodomains bind ADP-ribose, MARylated proteins, and/or the terminal ADPr of PAR, except for CHD1L (ALC1), which specifically binds PAR.109 Several RNA-binding domains and intrinsically disordered PAR-binding motifs are reported to bind PAR, but how these domains engage PAR is unclear due to a lack of structural data. The structures of Af1521, RNF146 RING-WWE, and APLF PBZ are from references 50, 99, and 107, respectively.
Chemical probes reveal the MAR- and PAR-binding proteomes
Pioneering efforts identified PAR-binding proteins by overlaying radioactive PAR on protein membranes transferred from gels of cell extracts and used peptide arrays to define clusters of basic and hydrophobic amino acids as a PAR-binding motif.110,111 Later proteomics studies identified proteins within PARylated complexes from cells with anti-PAR antibodies.112,113 However, these pulled-down proteins may not necessarily be direct binders or “readers”, as they could be covalently PARylated, PAR-binding, or pulled down through indirect protein-protein interactions. Recently, a range of synthetic MAR and PAR probes have been developed and used to directly identify ADPr readers with proteomics (Figure 6).114
Figure 6. Techniques for identifying MAR and PAR readers with chemically and enzymatically synthesized probes.
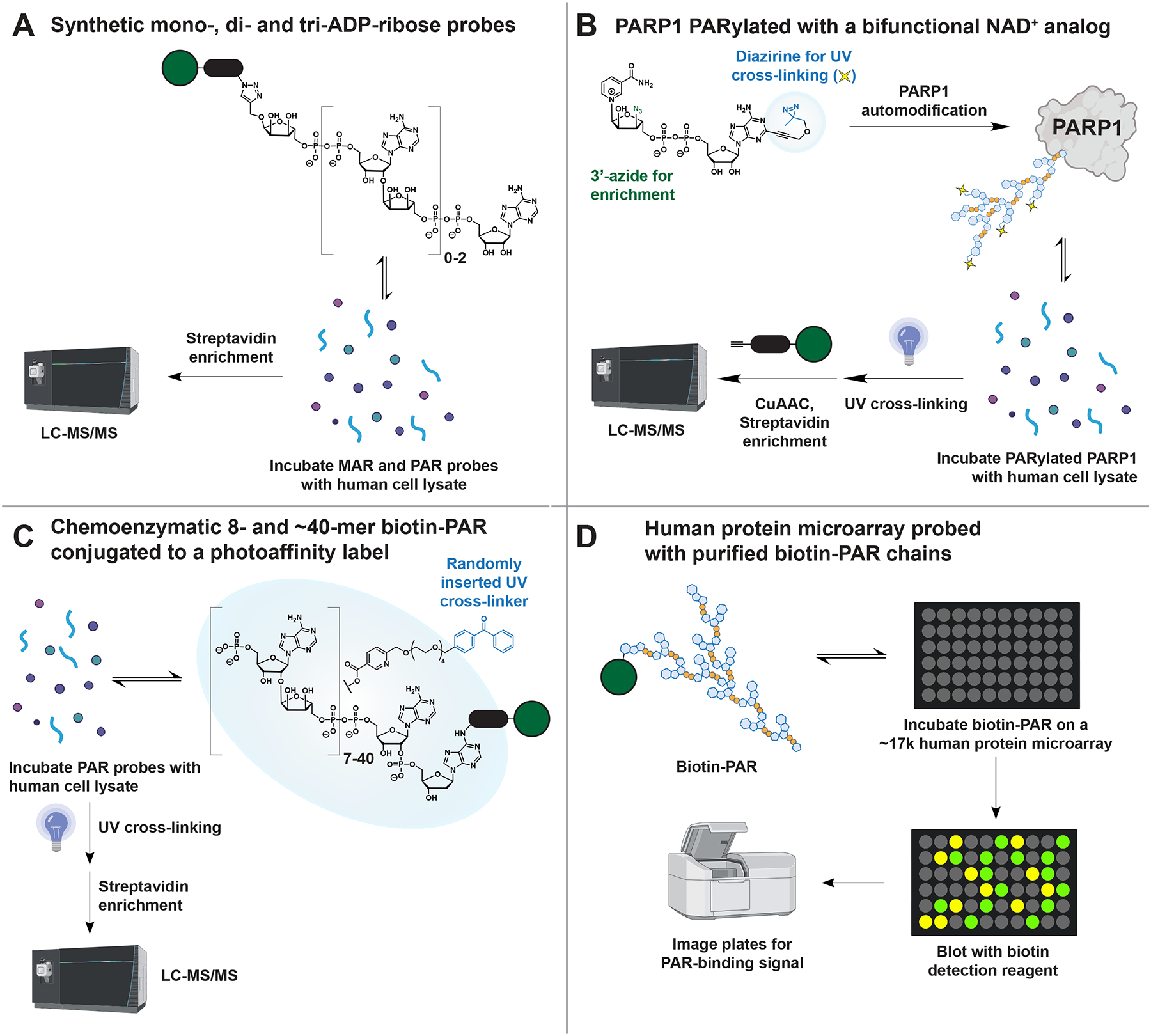
(A) Kliza et al.,100 (B) Lam et al.,93 (C) Dasovich et al.,119 and (D) Kang et al.,120 have developed complementary methods that together have identified 1,918 MAR- and PAR-binding candidate proteins.
One challenge in developing these probes has been the difficulty of synthesizing ADP-ribosylated molecules. Commercial PAR is costly and contains a mixture of lengths (~10–300+), making it impractical for use in large-scale proteomics. However, recent advances have made it possible to synthesize milligrams of single-length PAR oligomers using chemical or enzymatic methods. The core building block for chemical synthesis contains a protected phosphoramidite and an α(2→1) adenosine-ribose bond, which can be linked through solid-phase P(III)-P(V) couplings and terminated with an adenosine phosphoramidite.115,116 Using this synthetic strategy, Kliza and colleagues used biotinylated mono-, di-, and tri-ADP-ribose to identify their respective binders from human cell lysates with mass spectrometry (Figure 6A).100 The tri-ADP-ribose probe identified many proteins involved in DNA strand break repair, while the mono-ADP-ribose probe identified proteins involved in metabolism and transcription initiation. These data suggest unique functions for MAR and PAR, highlighting how structurally defined probes improve our understanding of ADPr signaling.
The low yield of pyrophosphate coupling (~70%, compared to >99% for DNA/RNA synthesis) has, however, limited the longest chemically synthesized PAR to a pentamer.116 To obtain longer lengths of PAR, several labs have used in vitro reactions with NAD+ and purified PARP1 or PARP5a.21,34,117 Enzymatically synthesized PAR can then be functionalized with enrichment handles and photoaffinity labels through organic synthesis. Lam and colleagues synthesized a bifunctional NAD+ analog with an azide for click chemistry-based enrichment and a diazirine for efficient photo-cross-linking (Figure 6B).93 PARP1 PARylated with this analog is then added to human cell lysates and irradiated to covalently capture PAR readers for identification. One advantage of this method is that the synthesized PAR remains protein-conjugated, which is more biologically relevant than purified PAR that has been detached from the protein. However, a disadvantage is that the synthesized PAR has varying lengths and branch points, which provides no insight into the structural specificity of interactions identified with this approach.
Alternatively, PAR can be synthesized as a mixed chain polymer using PARPs and NAD+ and detached from proteins with hydroxylamine or hydroxide. The PAR mixture is fractionated into defined lengths using anion exchange HPLC.34,117 While this method is inherently inefficient for synthesizing a single length of PAR, the range of polymer lengths produced in the enzymatic reaction can be adjusted by adding HPF1, histones, or PARP1 fragments.34,117 This enzymatic approach remains the only method for obtaining PAR longer than pentamer, with lengths up to 63-mer reported.118 Our group has used the enzymatic method to synthesize 8- and ~40-mer PAR for identifying length-specific PAR binding proteomes (Figure 6C).119 These polymers were converted into probes for protein capture by ELTA with biotin-dATP and chemical acylation with a photo-cross-linker. The probes were incubated with human cell lysate, cross-linked to capture PAR readers, enriched, and analyzed with mass spectrometry. Many proteins were identified with both lengths of the probe, but some readers were only captured by the 8-mer oligomer or the 40-mer polymer. The 8-mer readers were involved in central metabolism (e.g., glycolysis), while the 40-mer readers were often associated with RNA metabolism and DNA repair. These data, along with those from Kliza and colleagues,100 support a hypothesis where the length of PAR regulates the timing and specificity of PARP signaling pathways.
Other proteome-based screens complement existing approaches
While cell lysates are a useful source of endogenous human proteins, they provide poor sampling of proteins that have low solubility or abundance in the cell types typically used for lysate preparation. To address this issue, Kang and colleagues used a microarray-based method that individually expresses and purifies ~17,000 human proteins from yeast and spots them onto chips for analysis.120 The chip is then overlaid with biotin-PAR and detected with fluorescent streptavidin (Figure 6D). This method has the advantage of directly observing each interaction, and it offers a better sampling of proteins with low protein copy numbers that may be difficult to detect with mass spectrometry-based proteomics. Indeed, this array-based method uncovered several PAR-protein interaction motifs not found in other screens, including the cysteine-rich motifs CNXC and CPXC.
Although these probes have made considerable progress by identifying thousands of ADPr-binding candidates, the probes in use mostly lack the native peptide context. Even for the study by Lam and colleagues, the use of in vitro PARylated PARP1 may not recapitulate biologically relevant PARP1 modification states. To address these issues, future research may tap into new reagents such as peptides MARylated at specific residues31,64–66 or proteins with site-specific MAR and PAR modifications.37,71,72 If used in pull-downs in cell extracts, these recombinant ADP-ribosylated substrates may reveal new readers that require both ADPr and the surrounding protein for recognition.
METHODS FOR MONITORING ADP-RIBOSYLATION DYNAMICS IN VITRO AND IN VIVO
Covalent substrates and their non-covalent interactions with unmodified proteins form an ADPr-centric network that regulates various cellular processes. This network can be fine-tuned by the form of ADP-ribosylation (MAR or PAR). To better understand the function of this dynamic network, researchers are developing tools including antibodies and genetically encoded reporters to detect specific forms of ADPr. Additionally, high-throughput assays have led to the discovery of small molecule inhibitors that can be used to study enzymes that control ADPr dynamics and assess their potential as therapeutic targets.
Reagents detect specific forms of ADP-ribosylation
Historically, ADP-ribosylation was studied using bulk detection methods, such as radioactively labeled precursors (e.g., ATP, NMN, NAD+), which were used to discover PARylation in isolated nuclei.121 The sensitivity of PAR detection was further improved by boronate enrichment and chloroacetaldehyde-mediated conversion to fluorescent poly(ethenoADP-ribose), which established a link between PAR synthesis and DNA repair.122 Monoclonal and polyclonal antibodies against PAR, such as 10H, were developed for immunoassays (western blotting, immunofluorescence, ELISA, etc.).123 However, these methods tend to bias towards PARylation. The radioactive and etheno-ADP-ribose-based methods have a linear relationship between signal and PAR length, and the 10H antibody prefers PAR. As a result, the first five decades of PARP research largely excluded mono- and oligo(ADP-ribose) from their analyses.
Engineering more specific ADPr detection reagents became possible following the discovery of protein domains that bind defined ADPr structures (Figure 5). Detection reagents based on fusing WWE or macrodomains to antibody moieties are now available for use in immunoassays.40 Anti-ADPr antibodies were also generated with phage display of MARylated peptides.31 This approach created antibodies that are MAR-specific, bind both MAR and PAR, and for the first time, antibodies that recognize MARylation at specific sites. The increased specificity of these detection reagents improves the precision of experiments and, more crucially, their commercial availability makes ADP-ribosylation research accessible to the broader scientific community.
High-throughput assays monitor the activity of ADPr writer and erasers
While antibody-based methods are useful for basic biological research, they may not be suitable for drug discovery, which requires quantitative, high-throughput detection of ADPr activity. High-throughput enzymatic assays typically convert the enzymatic activity of interest into an optical signal (e.g., fluorescence and luminescence) that can be easily adapted to automated drug screening platforms in 384-well formats.
Strategies for converting PARP activity to light take advantage of NAD+ analogs (Figure 4). In general, recombinant PARPs are expressed and immobilized on plate. Biotin-NAD+ is added to initiate the PARP reaction, which is then converted to a fluorescent signal using detection reagents like europium-labeled streptavidin (Figure 7A).124,125 However, the low auto-modification activity of some PARP family members can be an issue, which is addressed through forced proximity or an active-site displacement assay.126,127 Potent and selective inhibitors for PARP1/2, 7, 10, 11, and 14 are now available, which are important tools for understanding the functions of individual PARPs.128 These inhibitors are also the foundation for new therapeutics, with PARP1/2 inhibitors achieving FDA approval and a PARP7 inhibitor currently in phase I clinical trials for cancer.129,130
Figure 7. Methods for visualizing ADPr in vitro and in vivo.
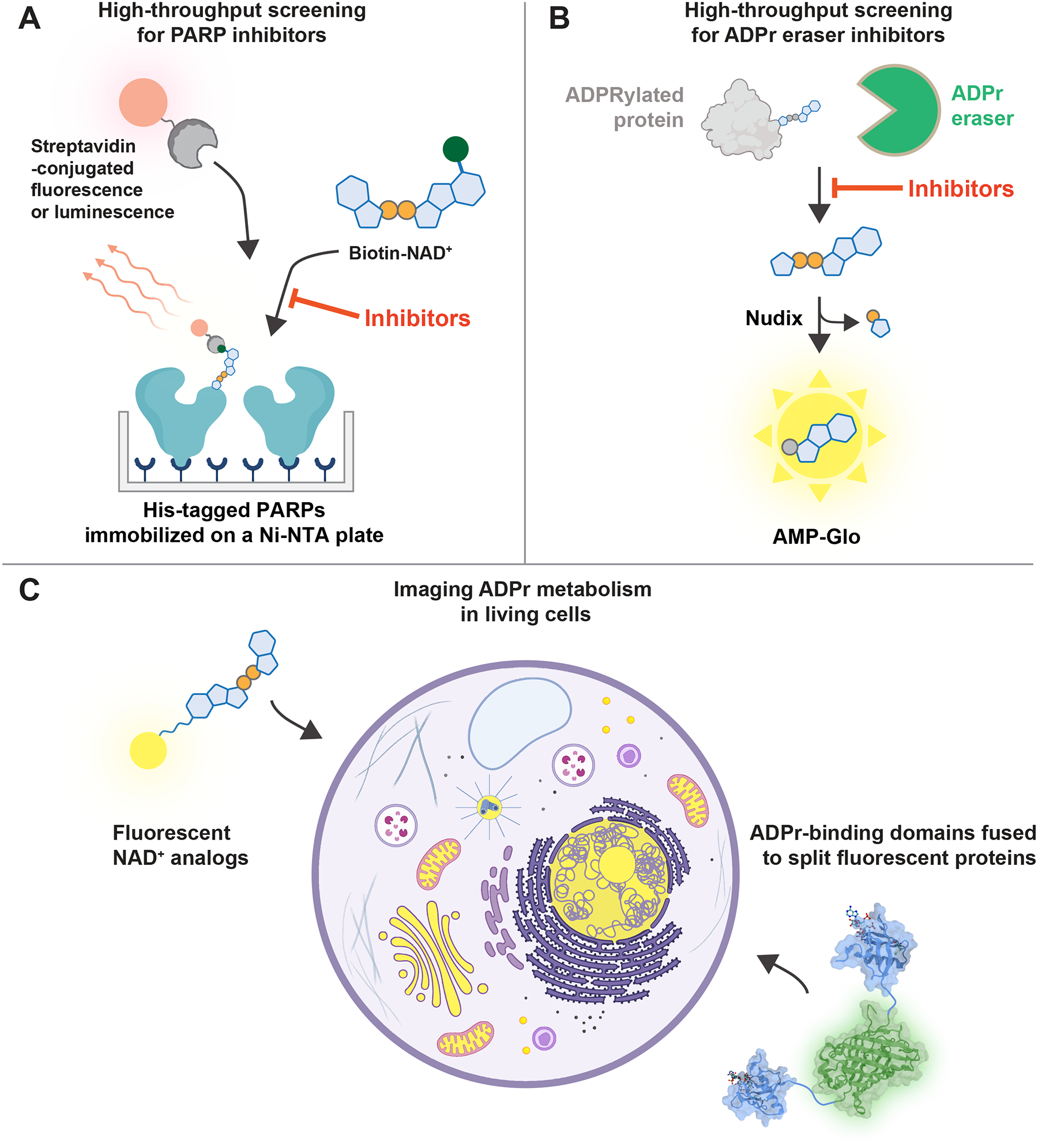
(A) A high-throughput ADP-ribosylation assay identifies PARP inhibitors by measuring the number of biotin-ADPr modifications. (B) A high-throughput ADP-ribosylhydrolase assay identifies inhibitors by converting hydrolyzed ADP-ribose into luminescence. (C) Small molecule and genetically encoded methods for imaging ADP-ribosylation dynamics in living cells. The structures of RNF146 WWE and split GFP are from references 105 and 137, respectively.
While PARP inhibition has been studied for over 40 years,129 the inhibition of ADP-ribosylhydrolases, enzymes that remove ADPr, is less studied.131 There is now significant interest in discovering small-molecule inhibitors of ADPr eraser enzymes as their roles in the progression of cancer and viral infections have recently been recognized. Multiple high-throughput assays for ADP-ribosylhydrolase activity are now available. The conversion of ADPr removal into light can be achieved through multiple routes: the enzymatic cleavage of ADP-ribose into phosphoribose and AMP, which then converts AMP to luminescence with the luciferase-based AMP-Glo (Figure 7B), chemical conversion of α-NAD+ into a fluorescent compound, or the use of a bespoke ADP-ribose analog that releases a fluorophore upon hydrolysis.64,132–136
The most well-developed inhibitor program for ADP-ribosylhydrolases targets human PARG, and a cell-permeable compound with nanomolar potency is now commercially available.138 This inhibitor has been used to study the function of PARG in living cells and effectively kills breast and prostate cancer cells.139,140 Additionally, viral macrodomains, which are conserved in some RNA virus families (e.g., Coronaviridae) and play a critical role in viral replication and pathogenesis, are another druggable ADPr eraser of therapeutic interest.141–143 Viruses expressing catalytically-deficient macrodomains are less lethal in mouse models, suggesting inhibitors of viral macrodomains could reduce the severity of infection.141,142,144 Several inhibitors of the SARS-CoV-2 macrodomain have recently been reported and are being evaluated as a new strategy for antiviral therapy.134,145,146
Visualization of ADPr dynamics in cells
ADPr is a reversible, dynamic modification. For example, PARP1 is recruited to DNA breaks within seconds, and PAR synthesis begins immediately, reaches a maximum level within minutes before degrading within 1 hour.147 The temporal changes of PAR length can be followed by isolating the polymers from other nucleic acids and separating them through electrophoresis or chromatography. In the past, the length of intact PAR chains was visualized by feeding cells radiolabeled adenine,20,148 but this approach may induce unintended radio-damage and trigger PARylation. Instead, isolated PAR can be stained with nucleic acid dyes or labeled with fluorescent or radioactive moieties with ELTA.34 However, ADP-ribosylation can be difficult with traditional assays, as they often involve cell lysis or fixation, and may not reveal the full repertoire of ADPr structures. To directly visualize ADPr modifications in cells, fluorescent NAD+ analogs can be delivered through mild permeabilization or transfection (Figure 4 and 7C).92,94,95 However, this method may also detect other NAD+-dependent processes unrelated to PARP activity.
Genetically encoded reporters, such as fluorescent proteins fused to ADPr reader domains (Figure 5 and 7C), enable the real-time monitoring ADP-ribosylation dynamics in live cells.149–151 PAR-specific biosensors have been developed using split-protein systems, where fluorescence only occurs when the two halves of the split fluorophore complement each other to form a fully folded structure on the same PAR molecule. Split PAR sensors were validated in a proof-of-principle experiment using cell lysate,152 and later confirmed the rapid metabolism of PARylation following DNA damage in live cells.153 Notably, a PAR sensor can also be used to detect specific PARylated proteins by fusing one half of the sensor to the gene of interest and the other half to a PAR-binding domain. The fluorescence produced by this technique acts as a “memory” of the PARylation event, even after the PAR has been degraded, due to the longer half-life of split fluorescent protein dimerization compared to the PARylation event (~10 minutes).153
Challa, Ryu, and colleagues recently improved the design of split-GFP PAR sensors by systematically analyzing different combinations of ADPr-binding domains.154 They also developed a split nanoluciferase (NanoLuc) reagent with improved signal-to-noise ratio for use in tissues and animals. These sensors were used to study the PARylation dynamics in cellular differentiation and in live animals following gamma-irradiation. While the dynamic range of these sensors is relatively modest (~3−5-fold), their direct detection of PAR in vivo simplifies experimental design and reduces the risk of artifacts.154 Together, the high spatial and temporal resolution of these sensors complements the proteomic techniques used to identify ADP-ribosylation substrates and binders, providing a more comprehensive, temporal view of PAR signaling networks in living systems.
REMAINING CHALLENGES AND FUTURE DIRECTIONS
Over the past several years, techniques for studying ADP-ribosylation have advanced rapidly and delved into the targets, binding partners, and metabolism of a once enigmatic protein modification (Table 2). Proteomics advances have illuminated a surprisingly complex view of ADP-ribosylation with thousands of protein targets and nine possible modification sites that vary depending on cell type, cellular compartment, and environmental cues. However, current techniques for identifying ADP-ribosylation sites derivatize the heterogeneous mixture of ADPr modifications into defined mass tags (Figure 3). As a result, details of the interplay between MAR and PAR and their regulation in cells remain largely uncharacterized. While techniques have been developed to preserve both the length and identity of repeating units in other complex post-translational modifications, traditional mass spectrometry is not set up for analyzing the negative charge of PAR chains due to the use of positive ion mode, which is optimized for basic tryptic peptides. One possible solution is to explore the negative ion mode, which has been successful in sequencing negatively charged glycans and glycopeptides.155
Table 2 |.
Overview of techniques used to study ADP-ribosylation
| Boronate + hydroxylamine | Enrich ADP-ribosylated peptides from cell lysates with boronate, then derivatize with hydroxylamine. | May improve the depth of ADPr detection at aspartate and glutamate. Identifies sites from living cells. |
Only detects ADPr at aspartate and glutamate, and may be biased towards the detection of PARylation. Novel derivation methods may allow identification of ADPr sites other than those at the acidic residues. |
| PARG + Af1521 | Trim ADP-ribosylated peptides to mono-ADPr with PARG, then enrich with Af1521. | Detects ADPr on all sites. Identifies sites from living cells and tissues. |
Thousands of sites have been mapped, but only a handful have been functionally characterized. Most of the experiments involving ADPr site mapping have been done in cancer cell lines under genotoxic stress, indicating the need for further research in cells exposed to other stresses and specific mouse tissues. |
| ELTA-MS | Functionalize ADP-ribosylated peptides by labeling them with a clickable handle using oligoadenylate synthase 1. Pull-down labeled peptides and derivatize them with phosphodiesterase. | Detects ADPr on all sites. Identifies sites from living cells. |
ELTA may label other adenosine-containing molecules in cells, thereby requiring the derivation for ADPr site identification. Novel derivatization methods that preserve the structure may allow for the site-specific characterization of different forms of ADP-ribosylation. |
| Analog-sensitive PARPs | Combine a “bumped” NAD+ analog with a mutated PARP that has a “hole” in the binding pocket. ADP-ribosylated proteins are then enriched with an affinity tag on the analog. | Identifies proteins targeted by a specific PARP. | The membrane impermeability of NAD+ analogs has limited experiments to cell lysates and intact nuclei. Recently reported NAD+ transfection reagents may enable to use of bumped NAD+ analogs in living cells. |
| Synthetic ADPr probes | Prepare ADP-ribosylated molecules with defined structures, then use them as baits to capture ADPr-binding proteins. | Can measure whether binding proteins prefer specific ADPr structures. | The membrane impermeability of ADP-ribosylated molecules has limited experiments to cell lysates. |
| Photo-cross-linking NAD+ analogs | Synthesize photo-cross-linking NAD+, use it to modify PARPs in vitro, then use it as bait to capture binding proteins. | The ADPr probe is protein conjugated. | Future experiments should focus on identifying the ADPr binding sites for mutagenesis to better understand their physiological roles in cells. |
| ADPr biosensors | Genetically encode ADPr-binding domains fused to fluorescent proteins and express in cells or tissues of interest. | Measures the highly dynamic, reversible nature of ADPr in living cells and animals. | While biosensors with PAR-binding domains have been largely successful, biosensors with MAR-binding domains have had limited success in cells. This may be due to off-target binding to free ADP-ribose and NAD+. The use of next-generation MAR detection reagents that specifically bind protein MARylation, without affinity for free ADP-ribose, may allow for the imaging of cellular MAR dynamics. |
With the identification of nearly 2,000 ADPr reader candidates, we are now in a position to make significant progress in understanding the non-covalent interactions between proteins and ADP-ribosylation. By determining the binding sites of these readers and studying the effects of binding-deficient mutant proteins in live cells and tissues,16,156 we will gain valuable insights into the biological roles of ADPr-protein interactions. Additionally, transfecting a photo-cross-linking NAD+ analog into a cell line expressing an analog-sensitive PARP can provide further information about the coordination of both covalent and non-covalent ADP-ribosylation events by specific PARPs. Mapping ADPr-binding sites to pathological mutations may uncover the physiological significance of these binding interfaces and potential drug-targeting opportunities.
Another revelation is the importance of protein MARylation. MAR-specific detection reagents are now available,40,41 including antibodies that recognize specific MARylation sites within histone and PARP1.31 Further development of MAR-specific reagents and their use in imaging will complement proteomics data by measuring the cellular dynamics of MARylation. Emergent data also revealed that the chain length of PAR is linked to pathological states (e.g., slower PAR degradation in cancerous tissue),147,157 highlighting the need to develop tools for monitoring changes in both the site and length of ADP-ribosylation as clinical biomarkers. These tools could have a significant impact on patient selection and treatment, particularly in light of ongoing efforts to pursue drugs targeting the synthesis and degradation of ADP-ribosylation.
One outstanding question remaining in the field is how PAR—a homopolymer—achieves specificity with its binding partners.97,158 The length of PAR seems to be a crucial regulator during the initiation and elongation steps of building an ADPr-centric network. However, the three-dimensional structure of this homopolymer has remained elusive, with previous efforts only able to resolve dimeric ADP-ribose.101 Recent developments in synthesizing length-defined PAR and labeling both termini have enabled the use of biophysical techniques that precisely measure PAR-protein interactions with single-molecule precision.34,159–161 Fully synthetic PAR synthesis allows for complete control over its structure, including the ability to add defined branch points and alter chemical moieties.162 Creating chemical probes to identify branch readers and developing them into biosensors will enable us to explore the significance of this unique PAR structure. By synthesizing PAR analogs, we can also determine the importance of specific components, such as the phosphate group, base, and sugar, in binding interactions.83,93,95
Lastly, exciting recent work has revealed that DNA and RNA are ADP-ribosylated in mammalian cells.163,164 MARylated proteins can also be modified with ubiquitin through a noncanonical O-linkage between the C-terminal glycine of ubiquitin and the 3’-hydroxyl of ADPr.165 These examples illustrate the exceptional versatility and high-energy nature of NAD+, which facilitates chemical reactions with diverse biomolecules that challenge our conventional definitions of post-translational modifications. The biological significance of non-canonical ADP-ribosylation in human cells is just beginning to emerge, but this new direction supports further development of detection reagents that are specific for DNA-ADPr, RNA-ADPr, or ubiquitin-ADPr. Coupling these reagents with proteomics, sequencing, and live-cell imaging will drive cutting-edge basic science efforts in the field and may reveal novel applications for ADPr in biotechnology and medicine.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Marc Greenberg and Sua Myong for critical reading of the initial draft of the manuscript. Several figures used artwork from Biorender. The ADP-ribosylation work in the Leung Lab has been supported by Catalyst Award, Discovery Award, and COVID-19 PreClinical Research Discovery Fund from the Johns Hopkins University as well as NIH grants T32GM080189, R01GM104135, and RF1AG071326.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
Anthony K. L. Leung holds a patent related to the ELTA technology used for labeling ADP-ribosylated molecules.
REFERENCES
- 1.Krishnakumar R, & Kraus WL (2010). The PARP Side of the Nucleus: Molecular Actions, Physiological Outcomes, and Clinical Targets. Mol. Cell, 39, 8–24. 10.1016/j.molcel.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pascal JM (2018). The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst.) 71, 177–182. 10.1016/j.dnarep.2018.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuller M & Ahel I (2022). Beyond protein modification: the rise of non-canonical ADP-ribosylation. Biochem. J 479, 463–477. 10.1042/BCJ20210280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lüscher B, Bütepage M, Eckei L, Krieg S, Verheugd P, & Shilton BH (2018). ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease. Chem. Rev 118, 1092–1136. 10.1021/acs.chemrev.7b00122 [DOI] [PubMed] [Google Scholar]
- 5.Perina D, Mikoč A, Ahel J, Ćetković H, Žaja R, & Ahel I (2014). Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair (Amst.), 23, 4–16. 10.1016/j.dnarep.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ménissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Amé JC, Dierich A, LeMeur M, Sabatier L, Chambon P, & de Murcia G (2003). Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263. 10.1093/emboj/cdg206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S, & Hodes RJ (2008). Tankyrase 1 and Tankyrase 2 Are Essential but Redundant for Mouse Embryonic Development. PLoS One, 3, e2639. 10.1371/journal.pone.0002639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendriks IA, Larsen SC, & Nielsen ML (2019). An Advanced Strategy for Comprehensive Profiling of ADP-ribosylation Sites Using Mass Spectrometry-based Proteomics. Mol. Cell. Proteomics, 18, 1010–1026. 10.1074/mcp.TIR119.001315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palavalli Parsons LH, Challa S, Gibson BA, Nandu T, Stokes MS, Huang D, Lea JS, & Lee Kraus W (2021). Identification of PARP-7 substrates reveals a role for MARylation in microtubule control in Ovarian cancer cells. eLife, 10, 60481. 10.7554/eLife.60481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez KM, Buch-Larsen SC, Kirby IT, Siordia IR, Hutin D, Rasmussen M, Grant DM, David LL, Matthews J, Nielsen ML, & Cohen MS (2021). Chemical genetics and proteome-wide site mapping reveal cysteine MARylation by PARP-7 on immune-relevant protein targets. eLife, 10, e60480. 10.7554/eLife.60480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, & Chang P (2014). Family-wide analysis of poly(ADP-ribose) polymerase activity. Nature Communications 5, 4426. 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lüscher B, Ahel I, Altmeyer M, Ashworth A, Bai P, Chang P, Cohen M, Corda D, Dantzer F, Daugherty MD, et al. (2021). ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 289, 7399–7410. 10.1111/febs.16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanderson DJ, & Cohen MS (2020). Mechanisms governing PARP expression, localization, and activity in cells. Crit. Rev. Biochem. Mol. Biol 55, 541–554. 10.1080/10409238.2020.1818686 [DOI] [PubMed] [Google Scholar]
- 14.Cambronne XA, & Kraus WL (2020). Location, Location, Location: Compartmentalization of NAD+ Synthesis and Functions in Mammalian Cells. Trends Biochem. Sci 45, 858–873. 10.1016/j.tibs.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs-Seymour I, Fontana P, Rack JGM, & Ahel I (2016). HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity. Mol. Cell, 62, 432–442. 10.1016/j.molcel.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DS, Camacho C. v., Nagari A, Malladi VS, Challa S, & Kraus WL (2019). Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol. Cell, 75, 1270–1285.e14. 10.1016/j.molcel.2019.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang D, Camacho C. v., Setlem R, Ryu KW, Parameswaran B, Gupta RK, & Kraus WL (2020). Functional Interplay between Histone H2B ADP-Ribosylation and Phosphorylation Controls Adipogenesis. Mol. Cell, 79, 934–949.e14. 10.1016/j.molcel.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D, Kim DS, & Kraus WL (2020). Specific Binding of snoRNAs to PARP-1 Promotes NAD+-Dependent Catalytic Activation. Biochemistry, 59, 1559–1564. 10.1021/acs.biochem.0c00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, & Matic I (2017). Serine ADP-Ribosylation Depends on HPF1. Mol. Cell, 65, 932–940.e6. 10.1016/j.molcel.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Gonzalez R & Jacobson MK (1987). Characterization of Polymers of Adenosine Diphosphate Ribose Generated in Vitro and in Vivo. Biochemistry, 26, 3218–3224. 10.1021/bi00385a042 [DOI] [PubMed] [Google Scholar]
- 21.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, & Jacobson MK (1993). High Resolution Fractionation and Characterization of ADP-Ribose Polymers. Anal. Biochem 208, 26–34. 10.1006/abio.1993.1004 [DOI] [PubMed] [Google Scholar]
- 22.Kleczkowska HE, Malanga M, Szumiel I, & Althaus FR (2002). Poly ADP-ribosylation in two L5178Y murine lymphoma sublines differentially sensitive to DNA-damaging agents. Int. J. Radiat. Biol 78, 527–534. 10.1080/095530002317577349 [DOI] [PubMed] [Google Scholar]
- 23.Malanga M, & Althaus FR (1994). Poly(ADP-ribose) molecules formed during DNA repair in vivo. J. Biol. Chem 269, 17691–17696. 10.1016/S0021-9258(17)32496-1 [DOI] [PubMed] [Google Scholar]
- 24.Miwa M, Saikawa N, Yamaizumi Z, Nishimura S, & Sugimura T (1979). Structure of poly(adenosine diphosphate ribose): identification of 2’-[1”-ribosyl-2”-(or 3”-)(1”’-ribosyl)]adenosine-5’,5”,5”’-tris(phosphate) as a branch linkage. Proc. Natl. Acad. Sci. USA 76, 595–599. 10.1073/pnas.76.2.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, Costello CE, Cravatt BF, Fenselau C, Garcia BA, et al. (2018). How many human proteoforms are there? Nat. Chem. Biol 14, 206–214. 10.1038/nchembio.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seydel C (2022). Diving deeper into the proteome. Nat. Methods 19, 1036–1040. 10.1038/s41592-022-01599-9 [DOI] [PubMed] [Google Scholar]
- 27.Kozlowski M, Corujo D, Hothorn M, Guberovic I, Mandemaker IK, Blessing C, Sporn J, Gutierrez-Triana A, Smith R, Portmann T, et al. (2018). MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 19, e44445. 10.15252/embr.201744445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Sullivan J, Tedim Ferreira M, Gagné JP, Sharma AK, Hendzel MJ, Masson JY, & Poirier GG (2019). Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun 10, 1182. 10.1038/s41467-019-08859-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stöger T, Poirier GG, Dawson VL, & Dawson TM (2004). Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 101, 17699–17704. 10.1073/pnas.040618210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirai H, Poetsch AR, Gunji A, Maeda D, Fujimori H, Fujihara H, Yoshida T, Ogino H, & Masutani M (2013). PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell Death Dis. 4, e656. 10.1038/cddis.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonfiglio JJ, Leidecker O, Dauben H, Longarini EJ, Colby T, San Segundo-Acosta P, Perez KA, & Matic I (2020). An HPF1/PARP1-Based Chemical Biology Strategy for Exploring ADP-Ribosylation. Cell, 183, 1086–1102.e23. 10.1016/j.cell.2020.09.055 [DOI] [PubMed] [Google Scholar]
- 32.Hendriks IA, Buch-Larsen SC, Prokhorova E, Elsborg JD, Rebak AKLFS, Zhu K, Ahel D, Lukas C, Ahel I, & Nielsen ML (2021). The regulatory landscape of the human HPF1- and ARH3-dependent ADP-ribosylome. Nat. Commun 12, 5893. 10.1038/s41467-021-26172-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Challa S, Khulpateea BR, Nandu T, Camacho C. v., Ryu KW, Chen H, Peng Y, Lea JS, & Kraus WL (2021). Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell, 184, 4531–4546.e26. 10.1016/j.cell.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ando Y, Elkayam E, McPherson RL, Dasovich M, Cheng S-J, Voorneveld J, Filippov D. v, Ong S-E, Joshua-Tor L, & Leung AKL (2019). ELTA: Enzymatic Labeling of Terminal ADP-Ribose. Mol. Cell, 73, 845–856.e5. 10.1016/j.molcel.2018.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langelier M-F, Billur R, Sverzhinsky A, Black BE, & Pascal JM (2021). HPF1 dynamically controls the PARP1/2 balance between initiating and elongating ADP-ribose modifications. Nat. Commun 12, 6675. 10.1038/s41467-021-27043-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudolph J, Roberts G, Muthurajan UM, & Luger K (2021). HPF1 and nucleosomes mediate a dramatic switch in activity of PARP1 from polymerase to hydrolase. eLife, 10, e65773. 10.7554/eLife.65773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohapatra J, Tashiro K, Beckner RL, Sierra J, Kilgore JA, Williams NS, & Liszczak G (2021). Serine ADP-ribosylation marks nucleosomes for ALC1-dependent chromatin remodeling. eLife, 10, e71502. 10.7554/eLife.71502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suskiewicz MJ, Zobel F, Ogden TEH, Fontana P, Ariza A, Yang JC, Zhu K, Bracken L, Hawthorne WJ, Ahel D, et al. (2020). HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602. 10.1038/s41586-020-2013-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prokhorova E, Agnew T, Wondisford AR, Tellier M, Kaminski N, Beijer D, Holder J, Groslambert J, Suskiewicz MJ, Zhu K, et al. (2021). Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease. Mol. Cell, 81, 2640–2655.e8. 10.1016/j.molcel.2021.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibson BA, Conrad LB, Huang D, & Kraus WL (2017). Generation and Characterization of Recombinant Antibody-like ADP-Ribose Binding Proteins. Biochemistry, 56, 6305–6316. 10.1021/acs.biochem.7b00670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weixler L, Ikenga NJ, Voorneveld J, Aydin G, Mhr Bolte T, Momoh J, Bütepage M, Golzmann A, Lüscher B, Filippov D. v, et al. (2023). Protein and RNA ADP-ribosylation detection is influenced by sample preparation and reagents used. Life Sci. Alliance, 6, e202201455. 10.26508/lsa.202201455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chunaram C, Chanchal K, Florian G, Nielsen LM, Michael R, Walther CT, Jesper VO, & Matthias M (2009). Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science, 325, 834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 43.Ochoa D, Jarnuczak AF, Viéitez C, Gehre M, Soucheray M, Mateus A, Kleefeldt AA, Hill A, Garcia-Alonso L, Stein F, et al. (2020). The functional landscape of the human phosphoproteome. Nat. Biotechnol 38, 365–373. 10.1038/s41587-019-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Wang J, Ding M, & Yu Y (2013). Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods, 10, 981–984. 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
- 45.Zhen Y, Zhang Y, & Yu Y (2017). A Cell-Line-Specific Atlas of PARP-Mediated Protein Asp/Glu-ADP-Ribosylation in Breast Cancer. Cell Rep. 21, 2326–2337. 10.1016/j.celrep.2017.10.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, & Ladurner AG (2005). The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920. 10.1038/sj.emboj.7600664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buch-Larsen SC, Hendriks IA, Lodge JM, Rykær M, Furtwängler B, Shishkova E, Westphall MS, Coon JJ, & Nielsen ML (2020). Mapping Physiological ADP-Ribosylation Using Activated Ion Electron Transfer Dissociation. Cell Rep. 32, 108176. 10.1016/j.celrep.2020.108176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higashi H, Maejima T, Lee LH, Yamazaki Y, Hottiger MO, Singh SA, & Aikawa M (2019). A Study into the ADP-Ribosylome of IFN-I-Stimulated THP-1 Human Macrophage-like Cells Identifies ARTD8/PARP14 and ARTD9/PARP9 ADP-Ribosylation. J. Proteome Res 18, 1607–1622. 10.1021/acs.jproteome.8b00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martello R, Leutert M, Jungmichel S, Bilan V, Larsen SC, Young C, Hottiger MO, & Nielsen ML (2016). Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun 7, 12917. 10.1038/ncomms12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nowak K, Rosenthal F, Karlberg T, Bütepage M, Thorsell AG, Dreier B, Grossmann J, Sobek J, Imhof R, Lüscher B, et al. (2020). Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins. Nat. Commun 11, 5199. 10.1038/s41467-020-18981-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, & Ladurner AG (2013). A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol 20, 508–514. 10.1038/nsmb.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donovan J, Dufner M, & Korennykh A (2013). Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Natl. Acad. Sci. USA, 110, 1652–1657. 10.1073/pnas.1218528110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cayley PJ, & Kerr IM (1982). Synthesis, Characterisation and Biological Significance of (2′−5′)Oligoadenylate Derivatives of NAD+, ADP-Ribose and Adenosine(5′)Tetraphospho(5′)Adenosine. Eur. J. Biochem 122, 601–608. 10.1111/j.1432-1033.1982.tb06481.x [DOI] [PubMed] [Google Scholar]
- 54.McPherson RL, Ong S-E, & Leung AKL, (2019). Ion-Pairing with Triethylammonium Acetate Improves Solid-Phase Extraction of ADP-Ribosylated Peptides. J. Proteome Res 19, 984–990. 10.1021/acs.jproteome.9b00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gagné JP, Langelier MF, Pascal JM, & Poirier GG (2018). Hydrofluoric Acid-Based Derivatization Strategy to Profile PARP-1 ADP-Ribosylation by LC-MS/MS. J. Proteome Res 17, 2542–2551. 10.1021/acs.jproteome.8b00146 [DOI] [PubMed] [Google Scholar]
- 56.Daniels CM, Ong SE, & Leung AKL (2014). Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J. Proteome Res 13, 3510–3522. 10.1021/pr401032q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daniels CM, Thirawatananond P, Ong SE, Gabelli SB, & Leung AKL (2015). Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci. Rep 5, 18271. 10.1038/srep18271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palazzo L, Thomas B, Jemth AS, Colby T, Leidecker O, Feijs KLH, Zaja R, Loseva O, Vert JC, Matic I, Helleday T, & Ahel I (2015). Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J 468, 293–301. 10.1042/BJ20141554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palazzo L, Daniels CM, Nettleship JE, Rahman N, McPherson RL, Ong SE, Kato K, Nureki O, Leung AKL, & Ahel I (2016). ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 283, 3371–3388. 10.1111/febs.13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daniels CM, Kaplan PR, Bishof I, Bradfield C, Tucholski T, Nuccio AG, Manes NP, Katz S, Fraser IDC, & Nita-Lazar A (2020). Dynamic ADP-Ribosylome, Phosphoproteome, and Interactome in LPS-Activated Macrophages. J. Proteome Res 19, 3716–3731. 10.1021/acs.jproteome.0c00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, Palazzo L, Stockum A, Ahel I, & Matic I (2016). Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol 12, 998–1000. 10.1038/nchembio.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogata N, Ueda K, Kagamiyama H, & Hayaishi O (1980). ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem 255, 7616–7620. 10.1016/S0021-9258(19)43873-8 [DOI] [PubMed] [Google Scholar]
- 63.Tao Z, Gao P, & Liu HW (2009). Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: Analysis and implications. J. Am. Chem. Soc 131, 14258–14260. 10.1021/ja906135d [DOI] [PubMed] [Google Scholar]
- 64.Voorneveld J, Rack JGM, Ahel I, Overkleeft HS, van der Marel GA, & Filippov DV (2018). Synthetic α- And β-Ser-ADP-ribosylated Peptides Reveal α-Ser-ADPr as the Native Epimer. Org. Lett 20, 4140–4143. 10.1021/acs.orglett.8b01742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voorneveld J, Rack JGM, van Gijlswijk L, Meeuwenoord NJ, Liu Q, Overkleeft HS, van der Marel GA, Ahel I, & Filippov DV (2021). Molecular Tools for the Study of ADP-Ribosylation: A Unified and Versatile Method to Synthesise Native MonoADP-Ribosylated Peptides. Chemistry, 27, 10621–10627. 10.1002/chem.202100337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voorneveld J, Kloet MS, Wijngaarden S, Kim RQ, Moutsiopoulou A, Verdegaal M, Misra M, Đikić I, van der Marel GA, Overkleeft HS, et al. (2022). Arginine ADP-Ribosylation: Chemical Synthesis of Post-Translationally Modified Ubiquitin Proteins. J. Am. Chem. Soc 144, 20582–20589. 10.1021/jacs.2c06249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, & Ahel I (2018). Serine is the major residue for ADP-ribosylation upon DNA damage. eLife, 7.e34334. 10.7554/eLife.34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, & Kraus WL (2016). Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science, 353, 45–50. 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupte R, Nandu T, & Kraus WL (2021). Nuclear ADP-ribosylation drives IFNγdependent STAT1α enhancer formation in macrophages. Nat. Commun 12, 3931. 10.1038/s41467-021-24225-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones A, & Kraus WL (2022). Multiomics analysis of the NAD+–PARP1 axis reveals a role for site-specific ADP-ribosylation in splicing in embryonic stem cells. Genes Dev. 36, 601–617. 10.1101/gad.349335.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tashiro K, Mohapatra J, Brautigam CA, & Liszczak G (2022). A Protein Semisynthesis-Based Strategy to Investigate the Functional Impact of Linker Histone Serine ADP-Ribosylation. ACS Chem. Biol 17, 810–815. 10.1021/acschembio.2c00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hananya N, Daley SK, Bagert JD, & Muir TW (2021). Synthesis of ADP-Ribosylated Histones Reveals Site-Specific Impacts on Chromatin Structure and Function. Journal of the American Chemical Society, 143, 10847–10852. 10.1021/jacs.1c05429 [DOI] [PubMed] [Google Scholar]
- 73.Bartlett E, Bonfiglio JJ, Prokhorova E, Colby T, Zobel F, Ahel I, & Matic I (2018). Interplay of Histone Marks with Serine ADP-Ribosylation. Cell Rep. 24, 3488–3502.e5. 10.1016/j.celrep.2018.08.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liszczak G, Diehl KL, Dann GP, & Muir TW (2018). Acetylation blocks DNA damage–induced chromatin ADP-ribosylation. Nat. Chem. Biol 14, 837–840. 10.1038/s41589-018-0097-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prokhorova E, Zobel F, Smith R, Zentout S, Gibbs-Seymour I, Schützenhofer K, Peters A, Groslambert J, Zorzini V, Agnew T, et al. (2021). Serine-linked PARP1 auto-modification controls PARP inhibitor response. Nat. Commun 12, 4055. 10.1038/s41467-021-24361-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang CS, Jividen K, Kamata T, Dworak N, Oostdyk L, Remlein B, Pourfarjam Y, Kim IK, Du KP, Abbas T, Sherman NE, Wotton D, & Paschal BM (2021). Androgen signaling uses a writer and a reader of ADP-ribosylation to regulate protein complex assembly. Nat. Commun 12, 2705. 10.1038/s41467-021-23055-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leutert M, Menzel S, Braren R, Rissiek B, Hopp AK, Nowak K, Bisceglie L, Gehrig P, Li H, Zolkiewska A, et al. (2018). Proteomic Characterization of the Heart and Skeletal Muscle Reveals Widespread Arginine ADP-Ribosylation by the ARTC1 Ectoenzyme. Cell Rep. 24, 1916–1929.e5. 10.1016/j.celrep.2018.07.048 [DOI] [PubMed] [Google Scholar]
- 78.Feijs KL, Kleine H, Braczynski A, Forst AH, Herzog N, Verheugd P, Linzen U, Kremmer E, & Lüscher B (2013). ARTD10 substrate identification on protein microarrays: Regulation of GSK3β by mono-ADP-ribosylation. Cell Commun. Signal 11, 5. 10.1186/1478-811x-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Troiani S, Lupi R, Perego R, Re Depaolini S, Thieffine S, Bosotti R, & Rusconi L (2011). Identification of candidate substrates for poly(ADP-ribose) polymerase-2 (PARP2) in the absence of DNA damage using high-density protein microarrays. FEBS J. 278, 3676–3687. 10.1111/j.1742-4658.2011.08286.x [DOI] [PubMed] [Google Scholar]
- 80.Depaix A, & Kowalska J (2019). NAD Analogs in Aid of Chemical Biology and Medicinal Chemistry. Molecules 24, 4187. 10.3390/molecules24224187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodriguez KM, & Cohen MS (2021). Chemical genetic methodologies for identifying protein substrates of PARPs. Trends Biochem Sci. 47, 390–402. 10.1016/j.tibs.2021.07.002 [DOI] [PubMed] [Google Scholar]
- 82.Bauer PI, Hakam A, & Kun E (1986). Mechanisms of poly(ADP-ribose) polymerase catalysis; mono-ADP-ribosylation of poly(ADP-ribose) polymerase at nanomolar concentrations of NAD. FEBS Lett. 195, 331–338. 10.1016/0014-5793(86)80188-0 [DOI] [PubMed] [Google Scholar]
- 83.Oei SL, Griesenbeck J, Buchlow G, Jorcke D, Mayer-Kuckuk P, Wons T, & Ziegler M (1996). NAD+ analogs substituted in the purine base as substrates for poly(ADP-ribosyl) transferase. FEBS Lett. 397, 17–21. 10.1016/s0014-5793(96)01137-4 [DOI] [PubMed] [Google Scholar]
- 84.Meldal M, & Tomøe CW (2008). Cu-catalyzed azide-alkyne cycloaddition. Chemical Reviews, 108, 2952–3015. 10.1021/cr0783479 [DOI] [PubMed] [Google Scholar]
- 85.Jiang H, Kim JH, Frizzell KM, Kraus WL, & Lin H (2010). Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J. Am. Chem. Soc 132, 9363–9372. 10.1021/ja101588r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Islam K (2018). The Bump-and-Hole Tactic: Expanding the Scope of Chemical Genetics. Cell Chemical Biology, 25, 1171–1184. 10.1016/j.chembiol.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carter-O’Connell I, Jin H, Morgan RK, David LL, & Cohen MS, (2014). Engineering the Substrate Specificity of ADP-Ribosyltransferases for Identifying Direct Protein Targets. J. Am. Chem. Soc 136, 5201–5204. 10.1021/ja412897a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carter-O’Connell I, Jin H, Morgan RK, Zaja R, David LL, Ahel I, & Cohen MS (2016). Identifying Family-Member-Specific Targets of Mono-ARTDs by Using a Chemical Genetics Approach. Cell Rep. 14, 621–631. 10.1016/j.celrep.2015.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carter-O’Connell I, Vermehren-Schmaedick A, Jin H, Morgan RK, David LL, & Cohen MS (2018). Combining Chemical Genetics with Proximity-Dependent Labeling Reveals Cellular Targets of Poly(ADP-ribose) Polymerase 14 (PARP14). ACS Chem. Biol 13, 2841–2848. 10.1021/acschembio.8b00567 [DOI] [PubMed] [Google Scholar]
- 90.Langelier MF, Zandarashvili L, Aguiar PM, Black BE, & Pascal JM (2018). NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun 9, 844. 10.1038/s41467-018-03234-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wallrodt S, Buntz A, Wang Y, Zumbusch A, & Marx A (2016). Bioorthogonally Functionalized NAD+ Analogues for In-Cell Visualization of Poly(ADP-Ribose) Formation. Angew. Chem. Int. Ed. Engl 55, 7660–7664. 10.1002/anie.201600464 [DOI] [PubMed] [Google Scholar]
- 92.Wallrodt S, Simpson EL, & Marx A (2017). Investigation of the action of poly(ADP-ribose)-synthesising enzymes on NAD+ analogues. Beilstein J. Org. Chem 13, 495–501. 10.3762/bjoc.13.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lam AT, Zhang X-N, Courouble VV, Strutzenberg TS, Pei H, Stiles BL, Louie SG, Griffin PR, & Zhang Y (2021). A Bifunctional NAD+ for Profiling Poly-ADP-Ribosylation-Dependent Interacting Proteins. ACS Chem. Biol 16, 389–396. 10.1021/acschembio.0c00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lehner M, Rieth S, Höllmüller E, Spliesgar D, Mertes B, Stengel F, & Marx A (2022). Profiling of the ADP-Ribosylome in Living Cells. Angew. Chem. Int. Ed. Engl 61, e202200977. 10.1002/anie.202200977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang X-N, Cheng Q, Chen J, Lam AT, Lu Y, Dai Z, Pei H, Evdokimov NM, Louie SG, & Zhang Y (2019). A ribose-functionalized NAD+ with unexpected high activity and selectivity for protein poly-ADP-ribosylation. Nat. Commun 10, 4196. 10.1038/s41467-019-12215-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang XN, Lam AT, Cheng Q, Courouble V. v., Strutzenberg TS, Li J, Wang Y, Pei H, Stiles BL, Louie SG, Griffin PR, & Zhang Y (2022). Discovery of an NAD+ analogue with enhanced specificity for PARP1. Chem. Sci 13, 1982–1991. 10.1039/d1sc06256e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leung AKL (2020). Poly(ADP-ribose): A Dynamic Trigger for Biomolecular Condensate Formation. Trends Cell Biol. 30, 370–383. 10.1016/j.tcb.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chaudhuri AR, & Nussenzweig A (2017). The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol 18, 610–621. 10.1038/nrm.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, & Xu W (2014). Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature, 517, 223–226. 10.1038/nature13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kliza KW, Liu Q, Roosenboom LWM, Jansen PWTC, Filippov DV, & Vermeulen M (2021). Reading ADP-ribosylation signaling using chemical biology and interaction proteomics. Mol. Cell, 81, 4552–4567.e8. 10.1016/j.molcel.2021.08.037 [DOI] [PubMed] [Google Scholar]
- 101.Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, Lafite P, Ahel M, Mitchison TJ, Ahel I et al. (2013). Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat. Commun 4, 2164. 10.1038/ncomms3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forst AH, Karlberg T, Herzog N, Thorsell AG, Gross A, Feijs KLH, Verheugd P, Kursula P, Nijmeijer B, Kremmer E, et al. (2013). Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains. Structure, 21, 462–475. 10.1016/j.str.2012.12.019 [DOI] [PubMed] [Google Scholar]
- 103.Rosenthal F, Feijs KLH, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, et al. (2013). Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol 20, 502–507. 10.1038/nsmb.2521 [DOI] [PubMed] [Google Scholar]
- 104.Rack JGM, Perina D, & Ahel I (2016). Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu. Rev. Biochem 85, 431–454. 10.1146/annurev-biochem-060815-014935 [DOI] [PubMed] [Google Scholar]
- 105.Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, & Xu W (2012). Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240. 10.1101/gad.182618.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuttiyatveetil JRA, Soufari H, Dasovich M, Uribe IR, Mirhasan M, Cheng SJ, Leung AKL, & Pascal JM (2022). Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose). Cell Rep. 41, 111529. 10.1016/j.celrep.2022.111529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, & Neuhaus D (2010). Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat. Struct. Mol. Biol 17, 241–243. 10.1038/nsmb.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Q, Kassab MA, Dantzer F, & Yu X (2018). PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun 9, 3233. 10.1038/s41467-018-05588-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Singh HR, Nardozza AP, Möller IR, Knobloch G, Kistemaker HAV, Hassler M, Harrer N, Blessing C, Eustermann S, Kotthoff C, et al. (2017). A Poly-ADP-Ribose Trigger Releases the Auto-Inhibition of a Chromatin Remodeling Oncogene. Mol. Cell, 68, 860–871.e7. 10.1016/j.molcel.2017.11.019 [DOI] [PubMed] [Google Scholar]
- 110.Pleschke JM, Kleczkowska HE, Strohm M, & Althaus FR (2000). Poly(ADP-ribose) Binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem 275, 40974–40980. 10.1074/jbc.m006520200 [DOI] [PubMed] [Google Scholar]
- 111.Gagné JP, Hunter JM, Labrecque B, Chabot B, & Poirier GG (2003). A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J 371, 331–340. 10.1042/bj20021675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gagné JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, & Poirier GG (2008). Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36, 6959–6976. 10.1093/nar/gkn771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gagné JP, Pic É, Isabelle M, Krietsch J, Éthier C, Paquet É, Kelly I, Boutin M, Moon KM, Foster LJ, et al. (2012). Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 40, 7788–7805. 10.1093/nar/gks486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cohen MS (2021). Catching mono- and poly-ADP-ribose readers with synthetic ADP-ribose baits. Mol. Cell, 81, 4351–4353. 10.1016/j.molcel.2021.10.016 [DOI] [PubMed] [Google Scholar]
- 115.Kistemaker H. A. v., Lameijer LN, Meeuwenoord NJ, Overkleeft HS, van der Marel GA, & Filippov DV (2015). Synthesis of Well-Defined Adenosine Diphosphate Ribose Oligomers. Angew. Chem. Int. Ed. Engl 127, 4997–5000. 10.1002/anie.201412283 [DOI] [PubMed] [Google Scholar]
- 116.Liu Q, Knobloch G, Voorneveld J, Meeuwenoord NJ, Overkleeft HS, van der Marel GA, Ladurner AG, & Filippov DV (2021). Chemical synthesis of linear ADP-ribose oligomers up to pentamer and their binding to the oncogenic helicase ALC1. Chem. Sci 12, 12468–12475. 10.1039/d1sc02340c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tan ES, Krukenberg KA, & Mitchison TJ (2012). Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers. Anal. Biochem 428, 126–136. 10.1016/j.ab.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fahrer J, Kranaster R, Altmeyer M, Marx A, & Bürkle A (2007). Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 35, e143. 10.1093/nar/gkm944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dasovich M, Beckett MQ, Bailey S, Ong S-E, Greenberg MM, & Leung AKL, (2021a). Identifying Poly(ADP-ribose)-Binding Proteins with Photoaffinity-Based Proteomics. J. Am. Chem. Soc 143, 3037–3042. 10.1021/jacs.0c12246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kang BG, Kang S-U, Kim JJ, Kwon J-S, Gagné J-P, Lee SY, Kim S, Sangwon KL, Ha S, Jeong JS, et al. (2022). Proteome-wide microarray-based screening of PAR-binding proteins. BioRxiv, 2022.06.06.494829. 10.1101/2022.06.06.494829 [DOI] [Google Scholar]
- 121.Chambon P, Weill JD, Doly J, Strosser MT, & Mandel P (1966). On the formation of a novel adenylic compound by enzymatic extracts of liver nuclei. Biochem. Biophys. Res. Communs 25, 638–643. 10.1016/0006-291X(66)90502-X [DOI] [Google Scholar]
- 122.Juarez-Salinas H, Sims JL, & Jacobson MK (1979). Poly(ADP-ribose) levels in carcinogen-treated cells. Nature, 282, 740–741. 10.1038/282740a0 [DOI] [PubMed] [Google Scholar]
- 123.Kawamitsu H, Hoshino HO, Okada H, Miwa M, Sugimura T, & Momoi H (1984). Monoclonal Antibodies to Poly(adenosine diphosphate ribose) Recognize Different Structures. Biochemistry, 23, 3771–3777. 10.1021/bi00311a032 [DOI] [PubMed] [Google Scholar]
- 124.Kirby IT, Sanderson DJ, & Cohen MS (2021). PASTA: PARP activity screening and inhibitor testing assay. STAR Protoc. 2, 100344. 10.1016/j.xpro.2021.100344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wigle TJ, Church WD, Majer CR, Swinger KK, Aybar D, Schenkel LB, Vasbinder MM, Brendes A, Beck C, Prahm M, et al. (2020). Forced Self-Modification Assays as a Strategy to Screen MonoPARP Enzymes. SLAS Discov. 25, 241–252. 10.1177/2472555219883623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wigle TJ, Blackwell DJ, Schenkel LB, Ren Y, Church WD, Desai HJ, Swinger KK, Santospago AG, Majer CR, Lu AZ, Niepel M, Perl NR, Vasbinder MM, Keilhack H, & Kuntz KW (2020). In Vitro and Cellular Probes to Study PARP Enzyme Target Engagement. Cell Chem. Biol 27, 877–887.e14. 10.1016/j.chembiol.2020.06.009 [DOI] [PubMed] [Google Scholar]
- 127.Galera-Prat A, Alaviuhkola J, Alanen HI, & Lehtiö L (2022). Protein engineering approach to enhance activity assays of mono-ADP-ribosyltransferases through proximity. Protein Eng. Des. Sel 35, gzac006. 10.1093/protein/gzac006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nizi MG, Maksimainen MM, Lehtiö L, & Tabarrini O (2022). Medicinal Chemistry Perspective on Targeting Mono-ADP-Ribosylating PARPs with Small Molecules. J. Med. Chem 65, 7532–7560. 10.1021/acs.jmedchem.2c00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Curtin NJ, & Szabo C (2020). Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat. Rev. Drug Discov 19, 711–736. 10.1038/s41573-020-0076-6 [DOI] [PubMed] [Google Scholar]
- 130.Gozgit JM, Vasbinder MM, Abo RP, Kunii K, Kuplast-Barr KG, Gui B, Lu AZ, Molina JR, Minissale E, Swinger KK, et al. (2021). PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity. Cancer Cell, 39, 1214–1226.e10. 10.1016/j.ccell.2021.06.018 [DOI] [PubMed] [Google Scholar]
- 131.Glumoff T, Sowa ST, & Lehtiö L (2022). Assay technologies facilitating drug discovery for ADP-ribosyl writers, readers and erasers. BioEssays, 44, 2100240. 10.1002/bies.202100240 [DOI] [PubMed] [Google Scholar]
- 132.Drown BS, Shirai T, Rack JGM, Ahel I, & Hergenrother PJ (2018). Monitoring Poly(ADP-ribosyl)glycohydrolase Activity with a Continuous Fluorescent Substrate. Cell Chem. Biol 25, 1562–1570.e19. 10.1016/j.chembiol.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kasson S, Dharmapriya N, & Kim IK (2021). Selective monitoring of the protein-free ADP-ribose released by ADP-ribosylation reversal enzymes. PLoS One, 16, e0254022. 10.1371/journal.pone.0254022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dasovich M, Zhuo J, Goodman JA, Thomas A, McPherson RL, Jayabalan AK, Busa VF, Cheng SJ, Murphy BA, Redinger KR, et al. (2022). High-Throughput Activity Assay for Screening Inhibitors of the SARS-CoV-2 Mac1 Macrodomain. ACS Chem. Biol 17, 17–23. 10.1021/acschembio.1c00721 [DOI] [PubMed] [Google Scholar]
- 135.Wazir S, Maksimainen MM, Alanen HI, Galera-Prat A, & Lehtiö L (2021). Activity-Based Screening Assay for Mono-ADP-Ribosylhydrolases. SLAS Discov. 26, 67–76. 10.1177/2472555220928911 [DOI] [PubMed] [Google Scholar]
- 136.Rack JGM, & Ahel I (2023). A Simple Method to Study ADP-Ribosylation Reversal: From Function to Drug Discovery. Methods Mol. Biol 2609, 111–132. 10.1007/978-1-0716-2891-1_8 [DOI] [PubMed] [Google Scholar]
- 137.Lin C-Y, Romei MG, Oltrogge LM, Mathews II, & Boxer SG, (2019). Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins. J. Am. Chem. Soc 141, 15250–15265. 10.1021/jacs.9b07152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.James DI, Smith KM, Jordan AM, Fairweather EE, Griffiths LA, Hamilton NS, Hitchin JR, Hutton CP, Jones S, Kelly P, et al. (2016). First-in-class chemical probes against poly(ADP-ribose) glycohydrolase (PARG) inhibit DNA repair with differential pharmacology to olaparib. ACS Chem. Biol 11, 3179–3190. 10.1021/acschembio.6b00609 [DOI] [PubMed] [Google Scholar]
- 139.Houl JH, Ye Z, Brosey CA, Balapiti-Modarage LPF, Namjoshi S, Bacolla A, Laverty D, Walker BL, Pourfarjam Y, Warden LS, et al. (2019). Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat. Commun 10, 5654. 10.1038/s41467-019-13508-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Waszkowycz B, Smith KM, McGonagle AE, Jordan AM, Acton B, Fairweather EE, Griffiths LA, Hamilton NM, Hamilton NS, Hitchin JR, et al. (2018). Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides. J. Med. Chem 61, 10767–10792. 10.1021/acs.jmedchem.8b01407 [DOI] [PubMed] [Google Scholar]
- 141.Fehr AR, Channappanavar R, Jankevicius G, Fett C, Zhao J, Athmer J, Meyerholz DK, Ahel I, & Perlman S (2016). The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio, 7, e01721–16. 10.1128/mbio.01721-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McPherson RL, Abraham R, Sreekumar E, Ong SE, Cheng SJ, Baxter VK, Kistemaker HAV, Filippov DV, Griffin DE, & Leung AKL (2017). ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. USA, 114, 1666–1671. 10.1073/pnas.1621485114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leung AKL, Griffin DE, Bosch J, Fehr AR, & Feinstone WH (2022). The Conserved Macrodomain Is a Potential Therapeutic Target for Coronaviruses and Alphaviruses. Pathogens, 11, 94. 10.3390/pathogens11010094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Abraham R, Hauer D, McPherson RL, Utt A, Kirby IT, Cohen MS, Merits A, Leung AKL, & Griffin DE (2018). ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. USA, 115, E10457–E10466. 10.1073/pnas.1812130115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roy A, Alhammad YM, McDonald P, Johnson DK, Zhuo J, Wazir S, Ferraris D, Lehtiö L, Leung AKL, & Fehr AR (2022). Discovery of compounds that inhibit SARS-CoV-2 Mac1-ADP-ribose binding by high-throughput screening. Antiviral Res. 203, 105344. 10.1016/j.antiviral.2022.105344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gahbauer S, Correy GJ, Schuller M, Ferla MP, Doruk YU, Rachman M, Wu T, Diolaiti M, Wang S, Neitz RJ, et al. (2023). Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2. Proc. Nal. Acad. Sci. USA, 120, e2212931120. 10.1073/pnas.2212931120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martello R, Mangerich A, Sass S, Dedon PC, & Bürkle A (2013). Quantification of cellular poly(ADP-ribosyl)ation by stable isotope dilution mass spectrometry reveals tissue- and drug-dependent stress response dynamics. ACS Chem. Biol 8, 1567–1575. 10.1021/cb400170b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aboul-Ela N, Jacobson EL, & Jacobson MK (1988). Labeling methods for the study of poly- and mono(ADP-ribose) metabolism in cultured cells. Anal. Biochem 174, 239–250. 10.1016/0003-2697(88)90541-6 [DOI] [PubMed] [Google Scholar]
- 149.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EHK, Scheffzek K, et al. (2009). A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol 16, 923–929. 10.1038/nsmb.1664 [DOI] [PubMed] [Google Scholar]
- 150.Aguilera-Gomez A, van Oorschot MM, Veenendaal T, & Rabouille C (2016). In vivo vizualisation of mono-ADP-ribosylation by dPARP16 upon amino-acid starvation. eLife, 5, e21475. 10.7554/elife.21475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Koczor CA, Saville KM, Andrews JF, Clark J, Fang Q, Li J, Al-Rahahleh RQ, Ibrahim M, McClellan S, Makarov M. v., et al. (2021). Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD+/SIRT6 axis. Cell Rep. 37, 109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Furman JL, Mok PW, Shen S, Stains CI, & Ghosh I (2010). A turn-on split-luciferase sensor for the direct detection of poly(ADP-ribose) as a marker for DNA repair and cell death. Chem. Commun 47, 397–399. 10.1039/c0cc02229b [DOI] [PubMed] [Google Scholar]
- 153.Krastev DB, Pettitt SJ, Campbell J, Song F, Tanos BE, Stoynov SS, Ashworth A, & Lord CJ (2018). Coupling bimolecular PARylation biosensors with genetic screens to identify PARylation targets. Nat. Commun 9, 2016. 10.1038/s41467-018-04466-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Challa S, Ryu KW, Whitaker AL, Abshier JC, Camacho CV, & Kraus WL (2022). Development and characterization of new tools for detecting poly(ADP-ribose) in vitro and in vivo. eLife, 11, e72464. 10.7554/elife.72464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kuo C-W, & Khoo K-H (2020). Strategic Applications of Negative-Mode LC-MS/MS Analyses to Expedite Confident Mass Spectrometry-Based Identification of Multiple Glycosylated Peptides. Anal. Chem 92, 7612–7620. 10.1021/acs.analchem.0c00236 [DOI] [PubMed] [Google Scholar]
- 156.Fu H, Liu R, Jia Z, Li R, Zhu F, Zhu W, Shao Y, Jin Y, Xue Y, Huang J, et al. (2022). Poly(ADP-ribosylation) of P-TEFb by PARP1 disrupts phase separation to inhibit global transcription after DNA damage. Nat. Cell Biol 24, 513–525. 10.1038/s41556-022-00872-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, & Poirier GG (2008). PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem 283, 1197–1208. 10.1074/jbc.m706734200 [DOI] [PubMed] [Google Scholar]
- 158.Reber JM, & Mangerich A (2021). Why structure and chain length matter: on the biological significance underlying the structural heterogeneity of poly(ADP-ribose). Nucleic Acid Res. 49, 8432–8448. 10.1093/nar/gkab618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Krüger A, Stier A, Fischbach A, Bürkle A, Hauser K, & Mangerich A (2019). Interactions of p53 with poly(ADP-ribose) and DNA induce distinct changes in protein structure as revealed by ATR-FTIR spectroscopy. Nucleic Acids Res. 47, 4843–4858. 10.1093/nar/gkz175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Abraham R, McPherson RL, Dasovich M, Badiee M, Leung AKL, & Griffin DE (2020). Both ADP-ribosyl-binding and hydrolase activities of the alphavirus nsp3 macrodomain affect neurovirulence in mice. mBio, 11, e03253–19. 10.1128/mbio.03253-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rhine K, Dasovich M, Yoniles J, Badiee M, Skanchy S, Ganser LR, Ge Y, Fare CM, Shorter J, Leung AKL, & Myong S (2022). Poly(ADP-ribose) drives condensation of FUS via a transient interaction. Mol. Cell, 82, 969–985.e11. 10.1016/j.molcel.2022.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Q, Kistemaker HAV, Overkleeft HS, van der Marel GA, & Filippov DV (2017). Synthesis of ribosyl-ribosyl-adenosine-5’,5”,5”‘(triphosphate)-the naturally occurring branched fragment of poly(ADP ribose). Chemical Commun. 53, 10255–10258. 10.1039/c7cc05755e [DOI] [PubMed] [Google Scholar]
- 163.Musheev MU, Schomacher L, Basu A, Han D, Krebs L, Scholz C & Niehrs C (2022). Mammalian N1-adenosine PARylation is a reversible DNA modification. Nat. Commun 13, 6138. 10.1038/s41467-022-33731-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Weixler L, Feijs KLH, & Zaja R (2022). ADP-ribosylation of RNA in mammalian cells is mediated by TRPT1 and multiple PARPs. Nucleic Acids Res. 50, 9426–9441. 10.1093/nar/gkac711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhu K, Suskiewicz MJ, Hloušek-Kasun A, Meudal H, Mikoč A, Aucagne V, Ahel D, & Ahel I (2022). DELTEX E3 ligases ubiquitylate ADP-ribosyl modification on protein substrates. Sci. Adv 8, 4253. 10.1126/sciadv.add4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


