Abstract
Aging is a universal biologic process that increases the risk of multiple diseases including cancer. Growing evidence shows that alterations in the genome and epigenome, driven by similar mechanisms, are found in both aged cells and cancer cells. In this review, we detail the genetic and epigenetic changes associated with normal aging and the mechanisms responsible for these changes. By highlighting genetic and epigenetic alterations in the context of tumorigenesis, cancer progression, and the aging tumor microenvironment, we examine the possible impacts of the normal aging process on malignant transformation. Finally, we examine the implications of age-related genetic and epigenetic alterations in both tumors and patients for the treatment of cancer.
Keywords: Aging, epigenetics, genetics, cancer, tumor microenvironment
Aging is a multifactorial, time-dependent biologic process that increases the susceptibility to multiple diseases including cancer [1]. Advancing age is one of the most studied risk factors for the development of most cancers, and it is projected that the total cancer incidence in the United States will increase by 30% from 2020 to 2040, at which point patients 65 years or older will account for ~69% of all new cancer diagnoses [2]. As the global population ages, understanding the processes that drive normal aging and how they affect the development of cancer becomes critically important from both biologic and epidemiologic viewpoints. Aged cells and cancer cells seem fundamentally different since aged cells typically exhibit decreased rates of proliferation and energy production and cancer cells are typified by increased rates of proliferation and energy consumption. However, molecular changes found in aging cells are also drivers of tumorigenesis [3,4]. Importantly, many cancer-causing mutations can be found accumulating with age in normal cells and stem cells, and further, studies have shown that some of these are tissue specific [5,6]. A mathematical calculation of cancer risk based on lifetime accumulation of genetic damage in stem cells, in combination with extrinsic (smoking, UV damage) and intrinsic (heritable genetic changes) factors has been proposed [7]. Additionally, some genes expressed early in life may be beneficial, but later in life expression of those genes may have a detrimental effect, a phenomenon known as antagonistic pleiotropy, and it has been suggested that senescence is a prime example of antagonistic pleiotropy [8]. The switch from beneficial to tumor-promoting gene expression may depend on the tumor microenvironment (TME). Understanding these age-related changes will provide opportunities to further elucidate the mechanisms through which cancers develop and highlight new therapeutic vulnerabilities for the treatment of cancer.
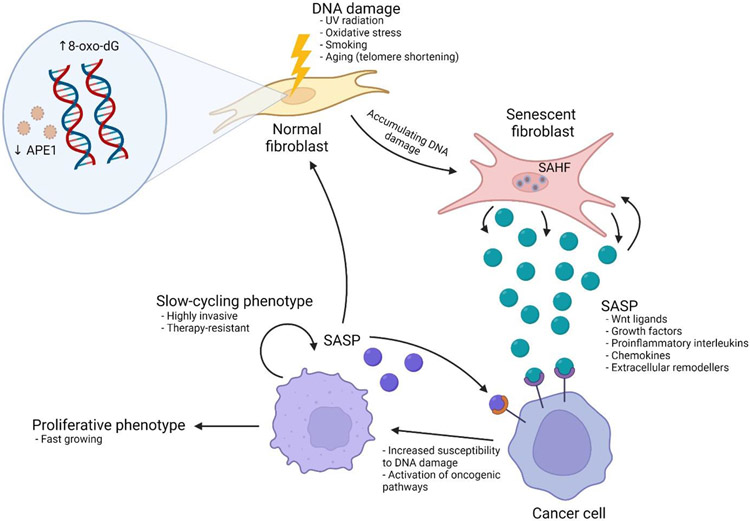
The aging microenvironment as a driver of genetic and epigenetic changes leading to tumorigenesis.
Having recently extensively reviewed age-related changes in the TME that drive tumor progression [9], in this review, we focus largely on the cancer cell-intrinsic genetic and epigenetic changes that are impacted by aging. However, it is critical to point out key microenvironmental factors that drive these changes (Figure 1). In the aging field, the oxidative stress theory of aging refers to the accumulation of damage in response to the accumulation of reactive oxygen and nitrogen species. These may be endogenous, derived from nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, hydrogen peroxidase, and other oxidases, or exogenous, derived from tobacco, alcohol, and other pollutants [10]. Reactive oxygen and nitrogen species can interact with multiple types of molecules from lipids to carbohydrates and proteins, causing damage to these molecules that can result in cellular senescence. Further, reactive oxygen/nitrogen species can interact with and damage DNA, causing genomic instability, as well as transcriptional errors, and this is marked by the accumulation of 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxo-dG), a product of DNA oxidation. Consequently, 8-oxo-dG has been shown to accumulate with age [11]. In order to counteract this damage, fibroblasts secrete molecules such as superoxide dismutase (SOD), catalase, and glutathione peroxidase, but it has been shown that fibroblasts lose expression of these protective molecules over time leading to an accumulation of damage [12,13]. Additionally, the base-excision repair protein, APE-1/Ref1, which specifically deals with DNA damage wrought by oxidative stress is down regulated during aging. In addition to downregulation in fibroblasts, APE-1 in cancer cells can also be downregulated in response to age-related changes in secreted factors from fibroblasts. It has been shown that changes in Wnt signaling in cancer cells exposed to aged fibroblasts result in the loss of activity of the microphthalmia transcription factor (MITF). MITF in turn does not transcribe APE-1, making the cancer cells susceptible to sustaining further damage [14].
Figure 1. Age-related changes in non-malignant cells in the tumor microenvironment alter cancer cell behavior.
Fibroblasts are a principal non-malignant component of the tumor microenvironment. As fibroblasts are exposed to sources of DNA damage including UV radiation, oxidative stress, and the aging process, and this accumulating damage is marked by the presence of 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxo-dG). In addition, the base-excision repair protein APE-1 is downregulated with age, further increasing the susceptibility to DNA damage. These fibroblasts can undergo cellular senescence. Senescent cells are characterized by the presence of senescence-associated heterochromatin foci (SAHF), specialized domains of heterochromatin that contribute to the silencing of proliferation-promoting genes and adopt a senescence-associated secretory phenotype (SASP) which has been shown to promote tumor growth by increasing cancer cell susceptibility to DNA damage and activation of oncogenic signaling. The end result can either be paracrine senescence of cancer cells, though the promotion of a slow-cycling cancer cell phenotype is associated with highly invasive, therapy-resistance cells, or enhancement of cancer cell proliferation.
Accumulation of DNA damage downstream of APE-1 can lead to cellular senescence, a cellular phenomenon that is linked to aging [15]. Senescence is a complex phenomenon, with important roles in tumor progression, and would require extensive review in and of itself [16], however, we summarize very briefly its importance here. Senescence is often initiated through a p16 or p53 mediated response to DNA damage, and results in cells going into a growth arrest. During aging, this DNA damage response can be induced by the dysfunction of telomeres, and the accumulation of chromatin marks, both of which are thought to maintain senescence through p53 [16,17]. Senescence is also accompanied by the secretion of various factors, such as IL-6, IL-10, interferons, chemokines, wnt ligands, and other cytokines, collectively known as the Senescence-Associated Secretory Phenotype (SASP) [18]. The SASP has been shown to both suppress tumor growth by inducing a paracrine senescence in tumor cells through an inflammosome-mediated mechanism, but also can paradoxically induce tumor progression [18]. The contributions of the immune microenvironment to senescence are summarized in Box 1. This is due to the fact that not only can senescence of tumor cells make them more resistant to therapy, since many therapies are targeted against rapidly proliferating cells, but also because signaling pathways induced by the SASP can be oncogenic [19].
BOX 1: Age-related epigenetic and genetic changes in the tumor immune microenvironment.
During aging there is a decline in immunity and the development of chronic low-grade inflammation (inflammaging) [96], resulting from the accumulation of both genetic and epigenetic alterations throughout life. The frequency and diversity of both peripheral circulating and tumor-infiltrating CD8+ cytotoxic T cells declines with age [97]. A recent study identified a clonal population of CD8+ T cells associated with inflammaging that have an exhausted-like phenotype, accumulated in tissues, and promote neighboring cells to adopt a senescence associated secretory phenotype through secretion of granzyme K [98], termed immunosenescence. These cells had a specific epigenetic signature that developed in response to an aged host microenvironment in in vivo studies. Multiple studies have highlighted that tumor DNA hypomethylation is associated with increased expression of immune checkpoint molecules or their immune-evasion promoting ligands in malignant cells [99]. Additionally, epigenetic changes in T cell populations may also have impacts on immunotherapy response. For instance, CD8+ memory T-cells, a subset that is exceptionally important for mediating successful tumor clearance, undergo changes in chromatin structure during aging that lead to less accessible DNA in regions known to be critical for response to NF-κB and STAT factors and may result in diminished immune responses [100].
Regulatory T-cells (Tregs) are also affected by aging-related epigenetic changes. Treg dysfunction can contribute to immunosenescence through enhanced suppression of effector T-cell response. Hypomethylation of an enhancer upstream of FoxP3, a major regulator of Treg function, is associated with aging in mice, and aging is associated with increased Treg numbers [101]. It is tempting to assume that with this increase in Treg number, older patients with cancer treated with immunotherapy would have a higher proportion of tumor-infiltrating Tregs in a melanoma model has demonstrated the opposite [102], suggesting that other changes in the tumor microenvironment in older patients may impact Treg trafficking and function. This is further supported by data from a study of patients with head and neck squamous cell carcinoma showing older patients had increased Tregs in the peripheral blood but decreased tumor-infiltrating Tregs [97]. Clinical experience with the use of ICI in older patients has shown ICI to be highly effective in this population [9]. Finally, myeloid-derived suppressor cells (MDSCs), implicated in tumorigenesis, metastasis, and alteration of tumor immune surveillance, increase in frequency with age, possibly due to alterations in epigenetic regulation that favor myeloid differentiation of hematopoietic precursors [103].
It is also important to note that malignant cells can themselves impact normal cells, further propagating a genomically unstable microenvironment. For example, stromal cells near a tumor exist in an environment that may promote the accumulation of DNA alterations. Using single-cell multiomics sequencing, it was discovered that somatic copy number alterations (SCNAs) are present in fibroblasts, endothelial, and immune cells in both the TME and normal tissues of individuals with cancer [20]. Interestingly, the proportion of fibroblasts with SCNAs in close proximity to malignant cells was significantly higher than those residing in adjacent normal tissues. This suggested that cells in the TME may be genetically altered through a mechanism depending on proximity to cancer cells. Fibroblasts with SCNAs may interact with tumor cells to promote cancer progression, and this is an active area of ongoing research. Therefore, the effect on the microenvironment on genetic instability in cancer cells is a complex one - direct damage to initiated tumor cells due to oxidative stress and environmental factors, or through inflammatory pathways such as the SASP from neighboring cells. In turn, the malignant tumor cells can feed back to create further damage to normal cells in their immediate microenvironment. These combined changes, external and internal to the initiated cancer cell, drive increased genomic instability, ultimately leading to tumor progression.
Genomic instability and the accumulation of genetic alterations in aging cells
Normal human cells constantly replicate their genome and divide, increasing the risk of acquiring genetic alterations and DNA damage. DNA damage comes in many forms and includes single-stranded breaks (SSBs), double-stranded breaks (DSBs), alteration of nucleotides, and the formation of bulky DNA adducts [21]. Most of these alterations will not lead to deleterious effects on cellular function and the majority of these alterations will be repaired by the endogenous DNA repair mechanisms found in normal cells. However, some changes to the genome can have far-reaching implications for cells during aging and in multiple diseases including cancer. For instance, unrepaired DNA alterations can impact the function of tumor-suppressor and oncogenes that in turn increase the risk of malignant transformation [22]. Analyses of the genomes from healthy cells have shown that the accumulation of somatic mutations in normal cells is directly correlated with the risk of developing cancer [23]. Many of the same mechanisms known to cause genomic instability in cancer cells have been implicated in the normal aging process [3], highlighting how changes during aging may enhance the risk of developing cancer with advancing age.
Changes in DNA repair pathways during aging
Unrepaired DNA alterations accumulate in an age-dependent fashion, suggesting that DNA damage response and repair pathways become less efficient with age [24]. Extensive evidence exists for age-related changes in multiple DNA damage response pathways. Declines in the activity, fidelity, or expression levels of components of the mismatch repair [25], base excision repair [26], nucleotide excision repair [27], non-homologous end joining (NHEJ) [28], and homologous recombination (HR) [29] repair pathways during aging have been reported in both human and animal models.
In the case of base excision repair, decreased efficiency with age has been linked with a reduction in Sirtuin 6 (SIRT6) levels. SIRT6 is a member of the sirtuin family of nicotinamide adenine dinucleotide+-dependent enzymes that regulate chromatin signaling, metabolism, and aging (Figure 2) [30]. In a study of base excision repair in foreskin fibroblast from donors ranging in age from 20 to 64 years old [31], there was a significant decrease in base excision repair efficiency and a concomitant decrease in SIRT6 expression with age in a PARP1-dependent fashion.
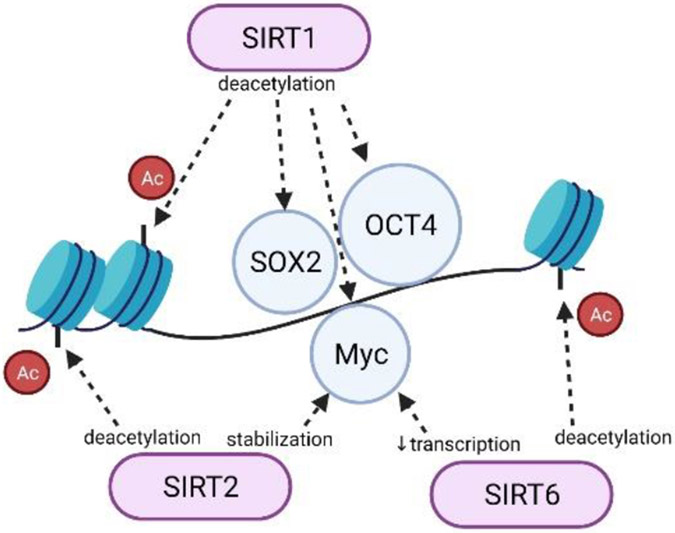
Figure 2. Sirtuins, histone modifications, and the regulation of chromatin and gene transcription.
SIRT1, SIRT2, and SIRT6 are involved in regulating chromatin remodeling. SIRT1 can deacetylate H3K9, H3K56, H1K26, and H4K16. It can also deacetylate the transcription factors SOX2, Myc, and Oct4. SIRT2 deacetylates H3K56, H4K16, and H3K18 and can stabilize Myc through alterations in Myc ubiquitination. SIRT6 deacetylates H3K9 and H3K56 and can repress the transcription of MYC.
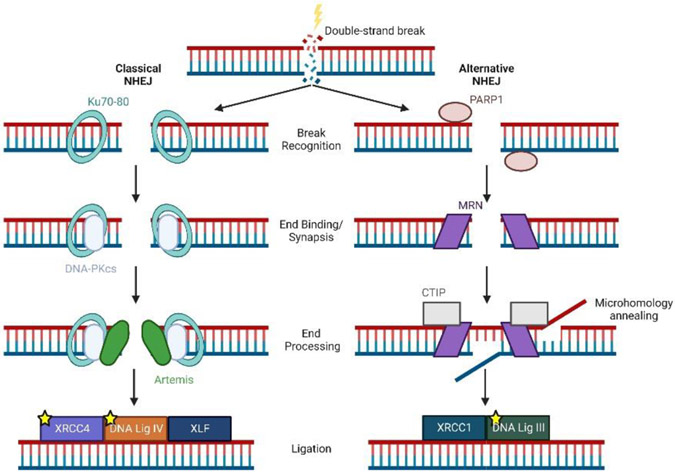
Two major pathways, NHEJ and HR, serve to repair DSBs in human cells, and aged cells have lower DSB repair efficiency [32]. A study examining 50 eyelid fibroblast cell lines isolated from healthy donors ranging in age from 16-75 years old demonstrated an age-related decline in HR efficiency as well as NHEJ efficiency and fidelity in these cells [28]. In the case of NHEJ, mechanistic studies showed that decreased expression of multiple proteins that comprise both the classical and alternative NHEJ pathways (Figure 3) drove this phenotype. Decreased NHEJ and HR efficiency sensitized normal cells from older donors to ionizing radiation, a finding that has implications for older individuals exposed to radiation such as patients receiving cancer radiation therapy. Additionally, the NR4A family of nuclear receptors, which also participate in DNA double stranded break repair, are expressed at lower levels in a number of age-related disease including atherosclerosis and multiple cancers [33].
Figure 3. Classical and alternative nonhomologous end-joining (NHEJ) pathways.
After the formation of a double-strand break, the classical (left) or the alternative (right) nonhomologous end-joining pathways repair DNA damage through break recognition, end binding/synapsis, end processing, and final ligation of DNA. Proteins marked with a yellow star have been shown to have decreased expression during aging.
Aging, the accumulation of genetic alterations, and cancer risk
Somatic genetic alterations that are known drivers of tumorigenesis when found in cancers have been identified in the cells of people without cancer. Some of these alterations have been identified early in life. For instance, the leukemia-associated AML1-ETO gene fusion is present in cord blood in 1% of neonates, however, aging remains the strongest independent predictor for the presence of somatic mutations in normal tissues [34].
Population studies utilizing whole exome sequencing have identified recurrently mutated genes, including TET2, DNMT3A, and other genes known to be involved in the development of hematologic malignancies, in the peripheral blood of healthy, older individuals [35]. Clonal hematopoiesis, where a substantial proportion of mature blood cells arise from a single hematopoietic stem cell, was once thought to be rare, but has now been shown to be ubiquitous. Up to 95% of individuals aged 50-60 years old harbor leukemia-associated mutations found in cells from peripheral blood samples [36]. Not all mutations found in cases of clonal hematopoiesis affect genes known to increase cancer risk. Nonetheless, patients with clonal hematopoiesis have a tenfold increased risk of developing a hematologic malignancy, and even suffer adverse clinical outcomes including increased mortality even in the absence of carrying identifiable leukemia-associated mutations [37].
Somatic DNA alterations in normal tissues from solid organs have been well-described in the context of preneoplastic diseases and tumor-adjacent normal tissues [38]. Normal skin and skin cancers including melanoma are particularly illustrative of the link between DNA damage from environmental exposures and an age-related accumulation genetic alterations as drivers of tumorigenesis. Many mutations that act as drivers in melanomas are linked predominantly to ultraviolet radiation damage, and over time these errors accumulate in DNA after each cell division [39]. Ultradeep sequencing of 74 cancer-associated genes in biopsies of normal, sun-exposed skin showed the presence of somatic mutations in clonal patches [40] identified that 18-32% of normal skin cells harbored positively selected mutations in known melanoma-associated genes including NOTCH1, NOTCH2, FAT1, and TP53. These cells still maintained the functions required in the normal epidermis, showing that aged sun-exposed skin is a patchwork of evolving clones that may one day give rise to cancer.
More recent evidence has linked age as a key determinant of the mutational landscape in melanoma. Melanoma is a disease with many clinical subtypes that are often associated with specific oncogenic mutations. Mutations in BRAF and NRAS are found at high frequencies in cutaneous melanomas, and their presence is typically, though not always, mutually exclusive [41]. BRAF mutations affecting the V600E residue are found more frequently in younger patients [42]. Through the use of mathematical models to analyze genomic data from melanomas from patients across a wide age spectrum, it was shown that NRAS and NF1-driven melanomas exhibit a correlation between increase in UV damage and cell division mutations with age, but BRAF-mutated melanomas were associated only with UV damage [43]. It is conceivable that genetic changes incurred throughout the aging process may dictate the overall mutational landscape of solid tumors and the tumor microenvironment. Another study in head and neck cancers supported this hypothesis by revealing distinct gene networks closely associated with migration and pro-growth signaling are recurrently altered in older patients [44]. Further study in this area is required to determine the generalizability of these findings to other cancer types.
Epigenetic alterations during aging
In addition to DNA alterations, epigenetic changes that do not alter an organism’s DNA sequence are important contributors to the normal aging process [45]. As organisms age, there is a general loss of histone proteins, widespread chromatin remodeling, changes in histone modifications, and a change in the DNA methylation pattern throughout the genome [46]. These epigenetic alterations lead to distinct transcriptional changes associated with an aged phenotype. Here we will highlight a subset of the known age-related epigenetic changes.
Histone and heterochromatin loss during aging
DNA in human cells is wrapped around histone proteins to form nucleosomes. Each nucleosome is comprised of DNA wound around eight core histone proteins (H2A, H2B, H3, and H4, and nucleosomes) can then be organized into more complex three-dimensional structures known as chromatin [47]. The number of histones as well as changes that occur in chromatin structure affect the accessibility of DNA and are key elements in the regulation of gene expression and genome stability during DNA replication [48] (Box 2). Heterochromatin, a tightly packed form of DNA and histone protein complexes, is a key structural feature of eukaryotic chromosomes important for regulation of DNA repair, chromosome segregation during cellular division, and the restriction of lineage-specific gene expression [49].
BOX 2: Histone variants.
In addition to the loss or post-translational modification of core histones with aging, core histones are often replaced with variant histones in aging organisms [71]. Histone variants differ in primary sequence compared to their canonical counterparts and can have unique effects on the regulation of gene expression. For instance, the H3 histone family has seven variants in humans [104]. In contrast to the canonical H3.1 and H3.2, the H3.3 variant is incorporated in the genome in a replication-independent manner and is the major form of H3 found in the chromatin of senescent human cells [105]. H3.3 has been shown to accumulate during aging in mouse somatic tissues to the point of nearly completely replacing the core H3 histone by the time the mice were 18 months old [106]. Furthermore, H3.3 levels increase in human brain tissue over the first decade of life, though levels remained stable across individuals who were 14-72 years old [107]. Functional effects of H3.3 accumulation during aging was demonstrated in C. elegans where H3.3 positively regulated the lifespan of long-lived through activation of diverse lifespanextending signaling pathways, and lack of H3.3 results in a much shorter lifespan [108], suggesting that its accumulation may be a key feature of successful normal aging.
One proposed model to explain the link between epigenetics and aging is the heterochromatin loss model. Multiple studies have supported the hypothesis that changes in histone expression and occupancy alter the structure of chromatin away from the tightly packed heterochromatin form in aged cells [50]. A study of replicative aging employing the use of micrococcal nuclease-DNA sequencing (MNase-seq) in budding yeast cells showed that nucleosome occupancy decreased by approximately 50% across the entire genome, leading to a rearrangement of nucleosome binding, widespread transcriptional activation, and genomic instability [51]. In fact, nucleosome occupancy is altered in aged mouse cells even in the absence of appreciable changes in canonical histone expression levels. Through the use of chromatin immunoprecipitation sequencing (CHIP-seq), it was demonstrated that aged mouse tissues from 29-month-old mice show a change in the H3 occupancy despite having similar overall H3 expression levels as tissue from 3-month-old mice. The changes in histone occupancy were associated with an increase in chromatin accessibility at pro-inflammatory genes [52], highlighting that age-related changes in chromatin organization lead to changes in cellular functioning that recapitulate aged phenotypes.
Changes in histone modification with aging
Post-translational modification (methylation, phosphorylation, ubiquitination, and acetylation, among others) of the N-terminal tails of core histones are important activators and repressors of transcription [53]. The balance of either activating or repressive modifications of histones has been shown to vary with age and is linked with lifespan [45]. The link between changes in global histone acetylation and aging is among the best understood. The addition of acetyl groups to lysine residues of histones by histone acetyltransferases (HATs) decreases the interaction between DNA and histones and activates transcription. Conversely, histone deacetylases (HDACs) act to remove acetyl groups from histones and repress transcription [54].
Histone H3 can be acetylated at lysine 9 (H3K9ac) and is then associated with active chromatin and aged cells [55]. In addition to its role in regulating base excision repair during aging, SIRT6 is a H3K9 deacetylase that has been shown to impact cellular longevity and survival through epigenetic mechanisms including a decrease in H3K9ac [56]. SIRT6 deficiency decreases lifespan [57], and SIRT6 overexpression leads to increased lifespan in mice [58]. SIRT6 also has deacetylase-independent functions that affect the epigenome. Depletion of SIRT6 results in altered telomere structure similar to those found in cells from patients with Werner syndrome, a premature aging syndrome [59]. SIRT6 can also promote heterochromatin silencing at retrotransposable elements. Specifically, SIRT6 can bind and post-translationally modify regulatory elements for Long Interspersed Element-1 (LINE-1) leading to suppressed transposition [60]. However, in both replicatively senescent human dermal fibroblasts and in 24-month-old mice, SIRT6 is depleted from LINE-1 regulatory sequences resulting in heterochromatin activation and transposition compared to young controls, events that increase the risk of genomic instability.
Chromatin remodeling
Chromatin undergoes extensive remodeling to support critical cellular processes including DNA replication, DNA damage repair, and transcription. These changes in chromatin structure are regulated by ATP-dependent chromatin remodeling complexes [61]. Despite the fact that aging and premature aging syndromes are characterized by changes in chromatin structure, the role chromatin remodeling complexes play in aging remains incompletely understood. Chromatin remodeling complexes implicated in the aging process include Mi2 (CHD-3/CHD-4). Depletion of the C. elegans Mi2 homolog, LET-418/Mi2, results in enhanced lifespan and stress resistance that was at least partially dependent on the longevity-associated DAF-16/FOXO transcription factor [62]. DAF-16/FOXO is also dependent on co-localization with chromatin remodeling SWI/SNF subfamily members at DAF-16/FOXO target promoters to activate genes associated with stress resistance and longevity [63].
Another developing field of aging research concerns changes in 3D chromatin structure and how it changes during aging. Using Hi-C, it has been shown that chromatin in human cells is spatially segregated into two compartments and further partitioned into topologically associated domains [64]. One of the first examinations of this structure in the context of aging came from a study of late passage cells from patients with the premature aging disease Hutching-Gilford progeria syndrome. These cells exhibited a global loss of chromosome compartmentalization and disassociation of heterochromatin from the nuclear lamina [65].
Changes in DNA methylation during aging
Methylation of DNA is a widely studied epigenetic modification that has been well-characterized in the context of aging. DNA is methylated through the covalent linkage of a methyl group to the fifth position of the cytosine resulting in the generation of 5-methylcytosine (5mC) [66]. The presence of 5mC can interfere with site-specific binding of transcription factors which in certain contexts can lead to a repressive, constitutive heterochromatin state. Additional effects of DNA methylation on genome function, including at non-promoter/enhancer regions, are not necessarily associated with transcriptional repression [67], and their link to cancer and aging is a topic of continued research.
DNA methylation plays a key role in development where it can silence expression of unneeded genes in a tissue-specific manner [68]. During aging in mammals, there is a general decrease in DNA methylation across the genome, leading to activation of normally silenced genes and other DNA sequences including transposable elements [69]. In studies of identical twins, the patterns of DNA methylation loss diverge between genetically identical individuals over time, likely due to environmental factors or random errors in the transmission of DNA methylation during genome replication [70]. However, there are also specific regions of the genome that undergo changes in methylation patterns during aging that are reproducible across individuals. This loss of CpG methylation at transposable elements increases the risk of retrotransposition events, providing a link between age-related changes in the epigenome and alteration of the genome itself [69,71].
A decrease in the expression levels of the DNA methyltransferase DNMT1, which helps maintain genomic methylation stability, during aging may in part explain the progressive decrease in global DNA methylation with age [72]. Dnmt1 haploinsufficient mice exhibit age-dependent decreases in bone mineral density and impaired learning and memory function without suffering a significant increase in mortality, suggesting that DNMT1 activity is involved in maintaining age-related physiologic health [72]. In addition to hypomethylation, aging has also been associated with site-specific hypermethylation at genes associated with development of age-related diseases [73,74].
These and numerous other studies have rapidly advanced our knowledge of the mechanisms that are responsible for the multitude of changes in the epigenome that occur with aging. However, a major outstanding question in the field is whether or not these changes directly impact longevity-associated genes and cause aging, or if the epigenetic changes are a consequence of genetic alterations of those same longevity-associated genes.
Epigenetic alterations in aging and their impact on tumorigenesis
The link between cancer and epigenetic dysregulation has been thoroughly explored and remains an important topic of research. Changes in epigenetic regulation of genes has been shown to increase expression of oncogenes, silence tumor suppressors, and drive tumorigenesis [75]. Many of the same epigenetic alterations observed in studies of aging organisms have been implicated in the development and progression of cancer [76,77], helping illustrate a possible connection between age-dependent epigenetic changes and increased cancer risk in older individuals.
The comparison of changes in DNA methylation patterns during aging with those found in cancer cells further supports the interconnectivity between an aging epigenome and cancer (Figure 4). Just as hypomethylation is a major feature of aging cells, hypomethylation was one of the earliest described epigenetic changes in human cancers that has been reported across multiple tumor types [78]. In addition, site-specific hypermethylation in cancer is also implicated in tumorigenesis through the silencing of tumor suppressor genes. DNA methylation patterns in non-neoplastic gastric epithelia have an age-dependent increase in methylation of the promoters in multiple tumor suppressor genes including LOX, p16/CDKN2A, RUNX3, and TIG1, suggesting that a subpopulation of normal cells that have already undergone changes in methylation may be more likely to undergo oncogenic transformation [79].
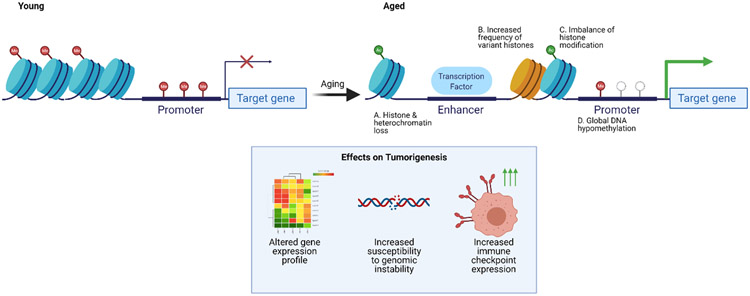
Figure 4. Age-related epigenetic changes and their effects on tumorigenesis.
(A-D) Aging is characterized by multiple alterations in the epigenome including (A) loss of histones and heterochromatin, (B) an increased frequency of non-canonical, variant histones, (C) a change in the balance of activating and repressing histone modifications, and (D) global DNA hypomethylation among others. These changes have been implicated in tumor growth and progression through the alteration of gene expression, genomic instability, and an increase in immune checkpoint molecule expression in cancer cells (inset box).
Perhaps the most-well known epigenetic link between aging and cancer is site-specific hypermethylation of developmental genes. This hypermethylation is characteristic of aging tissues and occurs predominantly at Polycomb-target sites that are associated with so-called “bivalent” chromatin – histones that are dually marked with both active and inactive modifications [80]. This same pattern of hypermethylation at bivalent chromatin domain promoters is also a key feature of multiple cancers, with a proposed model that aberrant gene silencing events that disrupt normal cellular homeostasis are stabilized as heritable gene silencing events in evolving abnormal clones that promote tumorigenesis [81].
A recent study, however, has shown that DNA methylation changes observed in aged cells and cancer cells may differ more than previously thought. This study found that aged cells showed a propensity toward global DNA hypermethylation and cancer cells showed strong bidirectional methylation changes compared to normal tissues, though a limitation of the study was a lack of ability to assay methylation at repetitive DNA regions [82]. Even when considering only differentially hypomethylated DNA sequences, there were surprising differences between aged and cancer cells when it came to the chromatin signature at areas of hypomethylation; genomic regions marked with the activating posttranslational histone modification H3K4me1 in aged cells, while in cancer, loss of DNA methylation was primarily associated with the repressive H3K9me3 modification. Furthermore, DNA hypomethylation in cancer was associated with heterochromatin regions while chromatin marks of DNA hypomethylation in aging were associated with enhancer regions, further complicating the link between DNA hypomethylation and tumorigenesis.
Impact of age-related epigenetic alterations for the treatment of older patients with cancer
While the majority of cancer diagnoses are made in patients over 65 years old, older patients are vastly underrepresented in clinical trials [83], there is a lack of robust clinical knowledge regarding toxicity risks in the average older patient with multiple co-morbidities [84], and evidence suggests older patients have inferior disease-specific outcomes even when controlling for variables such as stage and patient performance status [85]. Finding important intersections between normal aging and the genetic and epigenetic alterations found in cancers may provide avenues for improvement in the care of older patients with cancer.
Aging, DNA methylation changes, and cancer risk have also been examined through DNA methylation clocks. These are mathematical models derived from epigenetic DNA methylation markers that correlate strongly with chronologic age. For example, individuals with “age acceleration,” meaning their DNA methylation clock profile was similar to that of someone older by chronologic age, had an increased risk of developing pancreatic cancer [86]. Clocklike signatures in cancer may shape the overall mutational landscape of a tumor and thus affect a patient’s ability to be treated with certain targeted therapies such as tyrosine kinase inhibitors. A recent analysis of the impact of clock-like mutational signatures in both melanoma and non-small cell lung cancer has highlighted another therapeutic implication of aging-related mutational signatures: response to immune checkpoint inhibitors (ICI). ICI, a class of immunotherapy that utilizes monoclonal antibodies that block immune checkpoints on CD8+ cytotoxic T-cells, has rapidly been adopted in standard of care treatment regimens for multiple cancers [87]. Analysis of whole-exome sequencing data from melanoma and non-small cell lung cancer tumors obtained from patients undergoing treatment with ICI identified the presence of an age-related clock-like mutational signature, characterized by enrichment of C→T mutations at NpCpG trinucleotides [88]. This was associated with lower tumor mutational burden, reduction in cytotoxic T cell tumor infiltration, increased regulatory T cells, and worse prognosis after treatment with ICI. While this data may help subtyping patients based on genomic data and guide therapy choices, further work is required to determine if an age-related mutational signature is the cause these unfavorable biomarkers and clinical outcomes after ICI treatment.
Given the importance of epigenetic alterations in cancer, drugs targeting epigenetic modifying pathways and proteins have been developed and successfully used in the clinic [89]. Notable examples include the DNA methyltransferase inhibitors (DNMTi) and histone deacetylase inhibitors (HDACi). A more recently appreciated intersection between the epigenetics of aging and cancer therapy is the effect of epigenetic alterations in the cells of the immune system and response to immunotherapy (Box 1).
Concluding remarks
Aging is an extremely complex biologic process. Here, we have focused on a subset of these changes with a focus on genetic and epigenetic alterations associated with age. Research over the past decades has shown that there is an age-dependent accumulation of genetic and epigenetic alterations in the genome. It has become increasingly apparent that the control of multiple longevity pathways and determinants of cellular function are regulated by these age-related genetic and epigenetic changes. In addition, a better understanding of these age-related changes has further strengthened the link between aging and cancer risk as many of the same alterations found in normal cells during aging are thought to be drivers of tumorigenesis in cancer cells.
Recent work has given us unique insights into the implications of age-related genomic and epigenomic alterations found in aged cells for the use of common cancer treatments including immunotherapies. Despite detailed understanding of many of the specific mechanisms and that cause these alterations, there is still a wide range of phenotypic variability among the older population. Continued genetic, epigenetic, and functional studies in both normal and cancer cells performed in different age groups are needed to not only add to our understanding of normal aging, but help refine and improve cancer treatment strategies for older patients (Outstanding Questions Box). Overall, age cannot be ignored as an important variable when designing pre-clinical experiments, or clinical trials.
Outstanding Questions.
Despite the increased understanding of the shared hallmarks between aging and cancer, direct links that will ultimately affect how cancer is treated remain active areas of investigation.
Do epigenetic in longevity-associated changes in aged cells directly cause aged phenotypes, or are these epigenetic changes driven by the accumulation of genetic alterations?
How do age-related changes in non-malignant cells in the tumor microenvironment, such as fibroblasts, alter the behavior of cancer cells and other components of the TME?
Can aging-related genetic and epigenetic changes be targeted or reversed to decrease the incidence of cancer in humans?
How can clinical trial participation by older patients with cancer be increased to enable better study of the link between aging, cancer, and cancer therapy?
Highlights.
Multiple changes in genome stability and epigenetic regulation are shared between both biologic aging and cancer development, providing further evidence behind the mechanisms of the age-related increase in cancer incidence.
Age-related changes in the tumor microenvironment have been identified as key contributors to cancer progression and response to targeted and immunotherapies. Older patients are underrepresented in clinical trials, possibly limiting generalizability when considering the demographics of study populations.
Further study of aging and cancer will improve highlight new targets for cancer therapy and improve patient outcomes.
Acknowledgements:
Figures were created using BioRender.com. DJZ is supported by 5T32CA009071-39. EMJ is supported by P01CA247886-01A1 and R01CA197296-06. ATW is supported by R01CA207935, P01114046, and R01CA232256.
Glossary:
- Antagonistic Pleiotropy
Refers to genes that confer fitness early in life, only to reduce fitness later in life, causing an aging phenotype
- Tumor microenvironment
The collection of non-malignant cells (fibroblasts, immune cells, etc.), molecules, and blood vessels that comprise a tumor in addition to cancer cells
- Endogenous sources of DNA damage
DNA damaging agents present inside of cells including reactive oxygen species and free radicals
- Exogenous sources of DNA damage
Factors such as exposure to ionizing radiation, ultraviolet radiation, and chemicals including cigarette smoke that have potential to damage DNA
- Senescence
A stable cell cycle arrest that can be triggered by various stimuli as well as developmental signals. It is a dynamic process in which cells remain viable, but have alterations in metabolic activity and gene expression [90].
- Inflammasome
Multiprotein complex involving innate immune system receptors and sensors that regulate activity of inflammatory cysteine-dependent aspartate-directed proteases (caspases) after detection of pathogenic microorganisms or other danger signals in cells [91].
- BRAF
A proto-oncogene mutated at high frequency in multiple cancers such including melanoma. The gene encodes the serine/threonine-protein kinase B-Raf which is involved in directing cell growth [92].
- NRAS
A proto-oncogene frequently mutated in melanoma and other cancers that encodes the NRAS enzyme which is part of a family of G-like regulatory proteins involved in the normal control of cell growth through its GTPase activity [93].
- Sirtuins
a family of protein deacetylases and ADP-ribosyltransferases involved in multiple cellular processes including metabolic and epigenetic regulation. Sirtuin family members have been implicated in regulating the mammalian lifespan, however the role of sirtuin family members in longevity is still disputed [94].
- HI-C
A method to comprehensively detect chromatin interactions in the mammalian nucleus in which chromatin is crosslinked, digested, and re-ligated so that only DNA fragments that are covalently linked together form ligation products. This technique provides information on where in the genomic sequence chromatin interactions occur, but also where in the 3D organization of the genome they reside [95].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests:
DJZ declares no competing interests.
EMJ is a paid consultant for Adaptive Biotech, CSTONE, Achilles, DragonFly, Candel Therapeutics, NextCure Biotech, and Genocea. She receives funding from Lustgarten Foundation and Bristol Myer Squibb. She is the Chief Medical Advisor for Lustgarten and SAB advisor to the Parker Institute for Cancer Immunotherapy (PICI) and for the C3 Cancer Institute. She is a founding member of Abmeta.
ATW is a member of the board of directors of ReGAIN Therapeutics and is on the scientific advisory board of Phoremost Technologies.
References
- 1.Partridge L et al. (2018) Facing up to the global challenges of ageing. Nature 561, 45–56 [DOI] [PubMed] [Google Scholar]
- 2.Garner WB et al. (2019) Predicting future cancer incidence by age and gender. J. Clin. Oncol 37, 1559–1559 [Google Scholar]
- 3.Aunan JR et al. (2017) The Biology of Aging and Cancer: A Brief Overview of Shared and Divergent Molecular Hallmarks. Aging Dis. 8, 628–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risques RA and Kennedy SR (2018) Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet. 14, e1007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie M et al. (2014) Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med 20, 1472–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokzijl F et al. (2016) Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomasetti C et al. (2017) Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giaimo S and d’Adda di Fagagna F (2012) Is cellular senescence an example of antagonistic pleiotropy? Aging Cell 11, 378–383 [DOI] [PubMed] [Google Scholar]
- 9.Fane M and Weeraratna AT How the ageing microenvironment influences tumour progression. Nature Reviews Cancer. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liguori I et al. (2018) Oxidative stress, aging, and diseases. Clin. Interv. Aging 13, 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie B et al. (2013) Age-dependent accumulation of 8-oxoguanine in the DNA and RNA in various rat tissues. Oxid. Med. Cell. Longev 2013, 303181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ighodaro OM and Akinloye OA (2018) First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alexandria Journal of Medicine 54, 287–293 [Google Scholar]
- 13.Fujiwara T et al. (2019) Age-associated intracellular superoxide dismutase deficiency potentiates dermal fibroblast dysfunction during wound healing. Exp. Dermatol 28, 485–492 [DOI] [PubMed] [Google Scholar]
- 14.Kaur A et al. (2016) sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 532, 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Li M et al. (2018) APE1 deficiency promotes cellular senescence and premature aging features. Nucleic Acids Res. 46, 5664–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salama R et al. (2014) Cellular senescence and its effector programs. Genes Dev. 28, 99–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodier F et al. (2011) DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci 124, 68–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acosta JC et al. (2013) A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol 15, 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webster MR et al. (2020) Paradoxical role for wild-type p53 in driving therapy resistance in melanoma. Mol. Cell 77, 633–644.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y et al. (2020) Single-Cell Multiomics Sequencing Reveals Prevalent Genomic Alterations in Tumor Stromal Cells of Human Colorectal Cancer. Cancer Cell DOI: 10.1016/j.ccell.2020.09.015 [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee N and Walker GC (2017) Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen 58, 235–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu AK (2018) DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milholland B et al. (2015) Age-related somatic mutations in the cancer genome. Oncotarget 6, 24627–24635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijg J and Suh Y (2013) Genome instability and aging. Annu. Rev. Physiol 75, 645–668 [DOI] [PubMed] [Google Scholar]
- 25.Neri S et al. (2005) Mismatch repair system and aging: microsatellite instability in peripheral blood cells from differently aged participants. J. Gerontol. A Biol. Sci. Med. Sci 60, 285–292 [DOI] [PubMed] [Google Scholar]
- 26.Lagunas-Rangel FA and Bermúdez-Cruz RM (2019) The Role of DNA Repair in Cellular Aging Process. In DNA Repair (Mognato M, ed), IntechOpen [Google Scholar]
- 27.Deng X-D et al. (2017) The age-related expression decline of ERCC1 and XPF for forensic age estimation: A preliminary study. J. Forensic Leg. Med 49, 15–19 [DOI] [PubMed] [Google Scholar]
- 28.Li Z et al. (2016) Impaired DNA double-strand break repair contributes to the age-associated rise of genomic instability in humans. Cell Death Differ. 23, 1765–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sukup-Jackson MR et al. (2014) Rosa26-GFP direct repeat (RaDR-GFP) mice reveal tissue- and age-dependence of homologous recombination in mammals in vivo. PLoS Genet. 10, e1004299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan R.l. et al. (2018) A Review of the Recent Advances Made withSIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z et al. (2015) SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 14, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorbunova V and Seluanov A (2016) DNA double strand break repair, aging and the chromatin connection. Mutat. Res 788, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin K et al. (2017) NR4A2 promotes DNA double-strand break repair upon exposure to UVR. Mol. Cancer Res 15, 1184–1196 [DOI] [PubMed] [Google Scholar]
- 34.Mori H et al. (2002) Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl. Acad. Sci. U. S. A 99, 8242–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genovese G et al. (2014) Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med 371, 2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young AL et al. (2016) Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun 7, 12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook EK et al. (2020) Clonal hematopoiesis and inflammation: Partners in leukemogenesis and comorbidity. Exp. Hematol 83, 85–94 [DOI] [PubMed] [Google Scholar]
- 38.Oh J-H and Sung CO (2020) Comprehensive characteristics of somatic mutations in the normal tissues of patients with cancer and existence of somatic mutant clones linked to cancer development. J. Med. Genet DOI: 10.1136/jmedgenet-2020-106905 [DOI] [PubMed] [Google Scholar]
- 39.Sample A and He Y-Y (2018) Mechanisms and prevention of UV-induced melanoma. Photodermatol. Photoimmunol. Photomed 34, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martincorena I et al. (2015) Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raaijmakers MIG et al. (2016) Co-existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget 7, 77163–77174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Network (2015) Genomic Classification of Cutaneous Melanoma. Cell 161, 1681–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lotz M et al. (2020) Molecular subtype, biological sex and age shape melanoma tumour evolution. Br. J. Dermatol DOI: 10.1111/bjd.19128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meucci S et al. (2016) Mutational load and mutational patterns in relation to age in head and neck cancer. Oncotarget 7, 69188–69199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen P et al. (2016) Epigenetic Mechanisms of Longevity and Aging. Cell 166, 822–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W et al. (2020) The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol 21, 137–150 [DOI] [PubMed] [Google Scholar]
- 47.Li B et al. (2007) The role of chromatin during transcription. Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 48.Lai WKM and Pugh BF (2017) Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol 18, 548–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allshire RC and Madhani HD (2018) Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol 19, 229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J-H et al. (2020) Heterochromatin: an epigenetic point of view in aging. Exp. Mol. Med 52, 1466–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Z et al. (2014) Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 28, 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y et al. (2020) Remodeling of the H3 nucleosomal landscape during mouse aging. Transl. med. aging 4, 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrence M et al. (2016) Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 32, 42–56 [DOI] [PubMed] [Google Scholar]
- 54.Marmorstein R and Zhou M-M (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol 6, a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawakami K et al. (2009) Age-related difference of site-specific histone modifications in rat liver. Biogerontology 10, 415–421 [DOI] [PubMed] [Google Scholar]
- 56.Kawahara TLA et al. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W et al. (2018) SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560, 661–665 [DOI] [PubMed] [Google Scholar]
- 58.Kanfi Y et al. (2012) The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 [DOI] [PubMed] [Google Scholar]
- 59.Michishita E et al. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Meter M et al. (2014) SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun 5, 5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clapier CR and Cairns BR (2009) The biology of chromatin remodeling complexes. Annu. Rev. Biochem 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 62.De Vaux V et al. (2013) The Caenorhabditis elegans LET-418/Mi2 plays a conserved role in lifespan regulation. Aging Cell 12, 1012–1020 [DOI] [PubMed] [Google Scholar]
- 63.Riedel CG et al. (2013) DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol 15, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun L et al. (2018) Chromatin architectural changes during cellular senescence and aging. Genes (Basel) 9, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCord RP et al. (2013) Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 23, 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dantas Machado AC et al. (2015) Evolving insights on how cytosine methylation affects protein-DNA binding. Brief. Funct. Genomics 14, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenberg MVC and Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol 20, 590–607 [DOI] [PubMed] [Google Scholar]
- 68.Jung M and Pfeifer GP (2015) Aging and DNA methylation. BMC Biol. 13, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahmood W et al. (2020) Aging-associated distinctive DNA methylation changes of LINE-1 retrotransposons in pure cell-free DNA from human blood. Sci. Rep. 10, 22127. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Wang Y et al. (2019) Comprehensive longitudinal study of epigenetic mutations in aging. Clin. Epigenetics 11, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pal S and Tyler JK (2016) Epigenetics and aging. Sci Adv 2, e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu L et al. (2011) Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clin. Epigenetics 2, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weidner CI and Wagner W (2014) The epigenetic tracks of aging. Biol. Chem 395, 1307–1314 [DOI] [PubMed] [Google Scholar]
- 74.Ciccarone F et al. (2018) DNA methylation dynamics in aging: how far are we from understanding the mechanisms? Mech. Ageing Dev 174, 3–17 [DOI] [PubMed] [Google Scholar]
- 75.Cheng Y et al. (2019) Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 4, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Locke WJ et al. (2019) DNA methylation cancer biomarkers: Translation to the clinic. Front. Genet 10, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Z and Shilatifard A (2019) Epigenetic modifications of histones in cancer. Genome Biol. 20, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Witte T et al. (2014) Pan-cancer patterns of DNA methylation. Genome Med. 6, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.So K et al. (2006) Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci. 97, 1155–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rakyan VK et al. (2010) Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20, 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baylin SB and Jones PA (2016) Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pérez RF et al. (2018) Distinct chromatin signatures of DNA hypomethylation in aging and cancer. Aging Cell 17, e12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sedrak MS et al. (2021) Older adult participation in cancer clinical trials: A systematic review of barriers and interventions. CA Cancer J. Clin 71, 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hurria A et al. (2015) Improving the Evidence Base for Treating Older Adults With Cancer: American Society of Clinical Oncology Statement. J. Clin. Oncol 33, 3826–3833 [DOI] [PubMed] [Google Scholar]
- 85.Wang H et al. (2020) Survival of pancreatic cancer patients is negatively correlated with age at diagnosis: a population-based retrospective study. Sci. Rep 10, 7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung M et al. (2021) DNA methylation ageing clocks and pancreatic cancer risk: pooled analysis of three prospective nested case-control studies. Epigenetics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waldman AD et al. (2020) A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol DOI: 10.1038/s41577-020-0306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chong W et al. (2021) Association of clock-like mutational signature with immune checkpoint inhibitor outcome in patients with melanoma and NSCLC. Mol. Ther. Nucleic Acids 23, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ahuja N et al. (2016) Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu. Rev. Med 67, 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumari R and Jat P (2021) Mechanisms of cellular senescence: Cell cycle arrest and senescence associated secretory phenotype. Front. Cell Dev. Biol 9, 645593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Broz P and Dixit VM (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol 16, 407–420 [DOI] [PubMed] [Google Scholar]
- 92.Zaman A et al. (2019) Targeting oncogenic BRAF: Past, present, and future. Cancers (Basel) 11, 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muñoz-Couselo E et al. (2017) NRAS-mutant melanoma: current challenges and future prospect. Onco. Targets. Ther 10, 3941–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dang W (2014) The controversial world of sirtuins. Drug Discov. Today Technol 12, e9–e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Belton J-M et al. (2012) Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aiello A et al. (2019) Immunosenescence and its hallmarks: Howto oppose aging strategically? A review of potential options for therapeutic intervention. Front. Immunol 10, 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jeske SS et al. (2020) Age-related changes in T lymphocytes of patients with head and neck squamous cell carcinoma. Immun. Ageing 17, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mogilenko DA et al. (2020) Comprehensive Profiling of an Aging Immune System Reveals Clonal GZMK+ CD8+ T Cells as Conserved Hallmark of Inflammaging. Immunity DOI: 10.1016/j.immuni.2020.11.005 [DOI] [PubMed] [Google Scholar]
- 99.Villanueva L et al. (2020) The contribution of epigenetics to cancer immunotherapy. Trends Immunol. 41,676–691 [DOI] [PubMed] [Google Scholar]
- 100.Ucar D et al. (2017) The chromatin accessibility signature of human immune aging stems from CD8+ T cells. J. Exp. Med 214, 3123–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garg SK et al. (2014) Aging is associated with increased regulatory T-cell function. Aging Cell 13, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kugel CH 3rd et al. (2018) Age Correlates with Response to Anti-PD1, Reflecting Age-Related Differences in Intratumoral Effector and Regulatory T-Cell Populations. Clin. Cancer Res 24, 5347–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rauh MJ et al. (2019) Myeloid-derived suppressor cells in aged humans. In Handbook of Immunosenescence pp. 733–744, Springer International Publishing [Google Scholar]
- 104.Hake SB et al. (2006) Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem 281, 559–568 [DOI] [PubMed] [Google Scholar]
- 105.Bano D et al. (2017) The histone variant H3.3 claims its place in the crowded scene of epigenetics. Aging (Albany NY) 9, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tvardovskiy A et al. (2017) Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 45, 9272–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maze I et al. (2015) Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Piazzesi A et al. (2016) Replication-independent histone variant H3.3 controls animal lifespan through the regulation of pro-longevity transcriptional programs. Cell Rep. 17, 987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]